Abstract
Nicotinamide phosphoribosyltransferase (NAMPT) and nicotinate phosphoribosyltransferase domain containing 1 (NAPRT1) are the main human NAD salvage enzymes. NAD regulates energy metabolism and cell signaling, and the enzymes that control NAD availability are linked to pathologies such as cancer and neurodegeneration. Here, we have screened normal and tumor samples from different tissues and populations of origin for mutations in human NAMPT and NAPRT1, and evaluated their potential pathogenicity. We have identified several novel polymorphisms and showed that NAPRT1 has a greater genetic diversity than NAMPT, where any alteration can have a greater functional impact. Some variants presented different frequencies between normal and tumor samples that were most likely related to their population of origin. The novel mutations described that affect protein structure or expression levels can be functionally relevant and should be considered in a disease context. Particularly, mutations that decrease NAPRT1 expression can predict the usefulness of Nicotinic Acid in tumor treatments with NAMPT inhibitors.
Nicotinamide phosphoribosyltransferase (NAMPT) and nicotinate phosphoribosyltransferase domain containing 1 (NAPRT1) are major enzymes in the cellular metabolism. Their substrates, nicotinamide (Nam) and nicotinic acid (NA), respectively, are important precursors in Nicotinamide Adenine Dinucleotide (NAD) biosynthesis1. Several pathways contribute to the replenishment of the NAD pool and, in mammalian cells, Nam is the predominant precursor2,3 while NA is more effective in increasing NAD levels in some tissues4. Given that NAD participates as a coenzyme in oxidation-reduction reactions, but is also a substrate of NAD-consuming enzymes that are involved in gene expression regulation, DNA repair or cell death, it is expected that NAD availability directly influence pathological conditions5,6. Increasing evidence points to a role of NAD salvage enzymes in cancer and neurodegeneration5,6,7,8.
Originally, NAMPT was identified as pre-B-cell colony enhancing factor 1 (PBEF1)9 and as visfatin10, and its role in NAD biosynthesis was recognized later11. NAMPT functions in immunity, metabolism, and stress responses in physiology and pathophysiology12. As energy generation and NAD–dependent signaling are crucial for cell proliferation and cancer progression, NAMPT inhibitors have emerged as promising antitumor drugs5,13,14.
NAPRT1 increases intracellular NAD levels and prevents oxidative stress4. Since NA increases NAD levels via NAPRT1 action, the oral administration of NA was suggested to ameliorate NAD depletion conditions, namely, as cytoprotective agent in cancer treatments with NAMPT inhibitors13,14,15.
Genotype-phenotype associations in a disease context raise interest in single nucleotide polymorphisms (SNPs) detection. SNPs in human NAMPT associated with pathological conditions such as glucose and lipid metabolism alterations16,17, acute lung injury18, coronary artery disease19 and type 2 diabetes20, are located in non-coding regions. Polymorphisms in NAMPT promoter are related to plasma insulin levels21 and increased cholesterol22. For NAPRT1 the only association with disease, namely, attention deficit hyperactivity disorder23, is the synonymous SNP rs2290416 (G428, exon 10), responsible for differences in protein expression24.
Motivated by the importance of these enzymes in metabolism and homeostasis, and their involvement in several human diseases, we present the results of large-scale analyses that involved more than 200 samples from both normal and tumor origin in order to characterize potentially relevant mutations. The impact of novel variants at the structural and/or functional levels is discussed.
Results
NAMPT and NAPRT1 gene polymorphisms
Despite all the available literature on NAMPT polymorphisms, public databases show that NAMPT is less diverse than NAPRT1. Yet, little is known about the impact of NAPRT1 polymorphisms. We analyzed 96 DNA samples from a control population, focusing in mutation hotspots of the NAMPT and NAPRT1 genes (Supplementary Table S1). At the beginning of this study, the regions analyzed were chosen based on the information retrieved from public databases and the literature. For NAMPT, we found four intronic variants with minor allele frequency (MAF) lower than 0.05 (Table 1). One frequent SNP was detected in exon 7 (rs2302559, g.21735A>G), corresponding to Ser301. The frequencies observed in our samples were consistent with results from the 1000 Genomes Project (Table 1).
Table 1. Allele frequencies of human NAMPT genetic variants. Four intronic and one silent variants were found in human NAMPT from healthy donors (n = 96). The frequencies from 1000 Genomes were retrieved from the Ensembl Genome Browser.
| Ref ID | Variant type | Exon or intron | Nucleotide change | Amino acid change | Frequencies n = 96 | 1000 Genomes |
|---|---|---|---|---|---|---|
| rs41430346 | Non-coding | intron 3 | g.12484G>C | - | 0.979/0.021 | 0.971/0.029 |
| rs375379216 | Non-coding | intron 3 | g.12498C>T | - | 0.995/0.005 | - |
| rs112487390 | Non-coding | intron 4 | g.12774C>G | - | 0.989/0.011 | 0.958/0.042 |
| rs2302559 | Silent | exon 7 | g.21735A>G | Ser301Ser | 0.375/0.625 | 0.335/0.665 |
| rs144888107 | Non-coding | intron 7 | g.21850C>G | - | 0.979/0.021 | 0.994/0.006 |
The above mentioned 96 samples and additional 53 normal tissue samples, consisting in blood, stomach and colon from different ethnic groups, were analyzed for mutations in NAPRT1 (Table 2 and Figure 1). Among the five silent substitutions detected, one at Val142 codon (g.676C>G) remains unreported. The synonymous variant g.1803C>T at Leu305 (rs872935) is the most frequent and found in nearly 63% of the alleles, consistent with 1000 Genomes data for the Caucasian population. Two missense variants (rs200364051, p.Val106Met and rs35975875, p.Arg332Cys) were detected with frequencies of 0.5% and 0.3%, respectively, and four non-coding variants were also identified. From the eleven NAPRT1 variants found, five had MAF<0.05, including two missense, two silent and one non-coding, in accordance to 1000 Genomes data (Table 2). Sequencing the entire NAPRT1 gene allowed the discovery of five additional non-coding variants, plus one novel deletion in intron 8 (g.2542_2544delCCC) (Table 2).
Table 2. Allele frequencies of human NAPRT1 genetic variants. Five intronic, five silent and two missense variants were found in normal (n = 149) and tumor (n = 80) samples, including two novel polymorphic sites. Five additional intronic variants were found in the complete sequence of five samples (n.d., frequencies not determined), plus a previously undescribed deletion. The frequencies from 1000 Genomes were retrieved from the Ensembl Genome Browser.
| Ref ID | Variant type | Exon or intron | Nucleotide change | Amino acid change | Normal n = 149 | Tumor n = 80 | 1000 Genomes |
|---|---|---|---|---|---|---|---|
| rs2015562 | Non-coding | intron 1 | g.257C>A | - | 0.872/0.128 | 0.631/0.369 | 0.761/0.239 |
| rs896953 | Non-coding | intron 1 | g.325T>C | - | 0.316/0.684 | 0.300/0.700 | 0.299/0.701 |
| rs896954 | Silent | exon 2 | g.468C>T | Ala98Ala | 0.851/0.149 | 0.881/0.119 | 0.846/0.154 |
| rs200364051 | Missense | exon 2 | g.490G>A | Val106Met | 0.995/0.005 | - | n.a. |
| rs145565666 | Silent | exon 2 | g.516C>A | Leu114Leu | 0.997/0.003 | - | 0.998/0.002 |
| rs2305496 | Non-coding | intron 2 | g.565G>T | - | 0.875/0.125 | 0.675/0.325 | 0.779/0.221 |
| - | Silent | exon 3 | g.676C>G | Val142Val | 0.915/0.085 | - | - |
| rs12678314 | Non-coding | intron 3 | g.906T>C | - | n.d. | n.d. | 0.272/0.728 |
| rs744650 | Silent | exon 7 | g.1784C>T | Pro298Pro | 0.990/0.010 | - | 0.982/0.018 |
| rs872935 | Silent | exon 7 | g.1803C>T | Leu305Leu | 0.375/0.625 | 0.738/0.263 | 0.551/0.449 |
| rs35975875 | Missense | exon 7 | g.1884C>T | Arg332Cys | 0.997/0.003 | - | 0.997/0.003 |
| rs114291348 | Non-coding | intron 7 | g.2009G>A | - | 0.969/0.031 | 0.987/0.013 | 0.972/0.028 |
| - | Non-coding | intron 7 | g.2013A>G | - | - | 0.994/0.006 | - |
| rs896955 | Non-coding | intron 7 | g.2144G>A | - | n.d. | n.d. | 0.763/0.237 |
| - | Non-coding | intron 8 | g.2542_2544 delCCC | - | n.d. | n.d. | - |
| rs2290417 | Non-coding | intron 11 | g.3245C>G | - | n.d. | n.d. | 0.315/0.685 |
| rs77951814 | Non-coding | intron 12 | g.3362G>C | - | n.d. | n.d. | 0.913/0.087 |
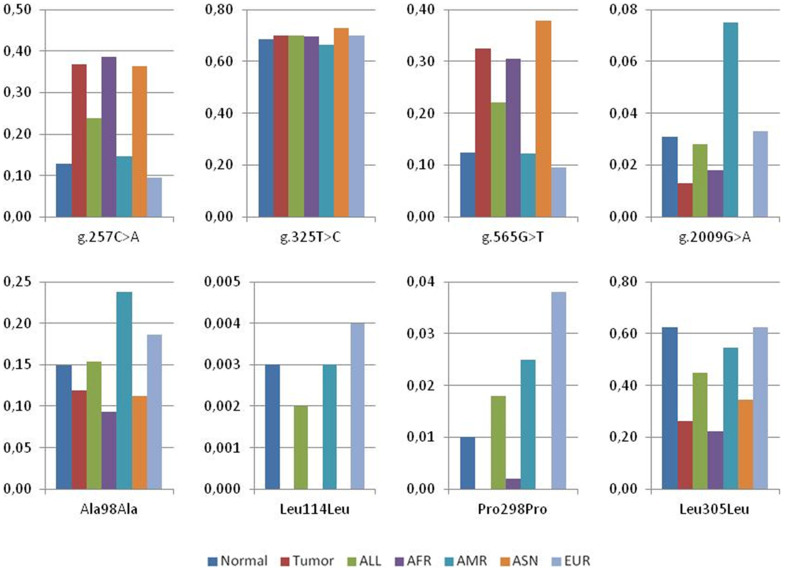
Figure 1. Allele frequencies of NAPRT1 genetic variants.
The graphs show the frequencies calculated from our results, both in normal and in tumor samples, for four non-coding and four silent variants. Data from 1000 Genomes reflecting the frequencies in different populations is also included. The novel polymorphisms g.2013A>G and g.676C>G as well as the two missense mutations Val106Met and Arg332Cys were not included since no data was available in the Ensembl Genome Browser. ALL: all individuals from phase 1 of the 1000 Genomes Project; AFR: African; AMR: American; ASN: East Asian; EUR: European.
To evaluate whether the variants have different prevalence in pathological conditions and normal tissues, 80 DNA samples from colon and gastric tumors were also analyzed. Apart from two silent alterations, which were also detected in normal samples (g.468C>T and g.1803C>T, at Ala98 and Leu305, respectively), no missense mutations were found in NAPRT1. Curiously, the g.1803C>T variant was more frequent in the normal population than in tumor samples (63% and 26%, respectively). From the five intronic variants found, g.257C>A and g.565G>T were more frequent in tumor than in normal samples (around 30% and 13%, respectively) (Table 2 and Figure 1). In the 1000 Genomes data, these are also more frequent in the Asiatic population, consistent with the fact that the tumor samples analyzed were from Asian origin, while control samples were mostly Caucasian. Thus, these results may be explained by different frequencies between populations rather than differences observed between control and tumor samples. In addition, we identified a polymorphism which was not previously described in intron 7 (g.2013A>G). The remaining variants showed similar frequencies between populations (Table 2 and Figure 1). To validate the population effect on the allelic differences, we used re-sampling statistics25,26 and compared NAPRT1 bootstrap confidence intervals (95%) for mean allele frequencies found in our data (control and tumor samples) with the 1000 Genomes allele frequencies (European and Asian populations). For most variants, we observed a similar trend between normal and European population frequencies, as well as between tumor and Asian population frequencies (Supplementary Table S3).
Impact of missense variants in the protein structures
Since there is no human NAPRT1 structure available, models of the protein were predicted by homology modeling and by ab initio modeling, based on Saccharomyces cerevisae27 and Enterococcus faecalis NAPRTases. After assessing quality (Supplementary Figure S3) the model with the lowest z-score was chosen (Supplementary Figure S4) to evaluate the structural impact of the NAPRT1 missense mutations found in our samples (Figure 2A).
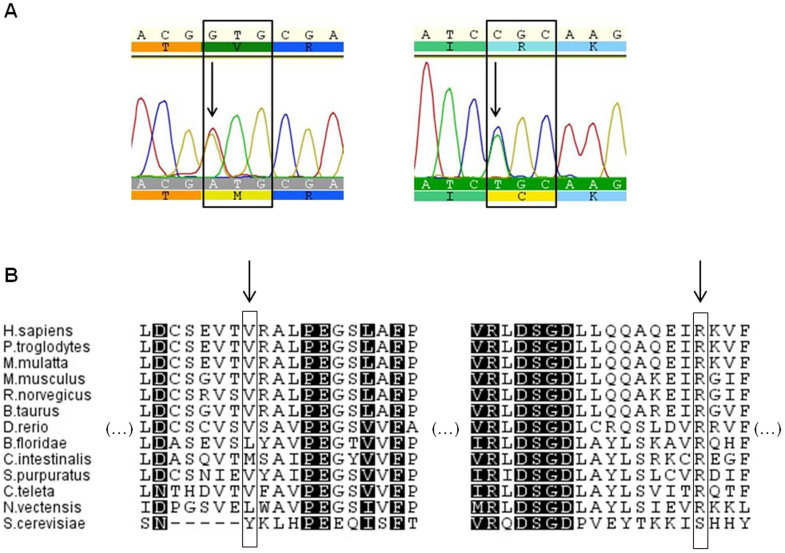
Figure 2. Missense mutations found in human NAPRT1 gene.
(A). Electropherograms showing 341C>T (left) and 1019C>T (right) replacements in heterozygosity that lead to Val106Met and Arg332Cys substitutions, respectively. Geneious software was used to visualize the sequences. (B). Localization of the amino acids in a multi-species alignment. V106 (left arrow) is conserved in vertebrates, and is substituted by interchangeable residues (Val/Leu) in invertebrate species. R332 (right arrow) is highly conserved, except in the yeast protein.
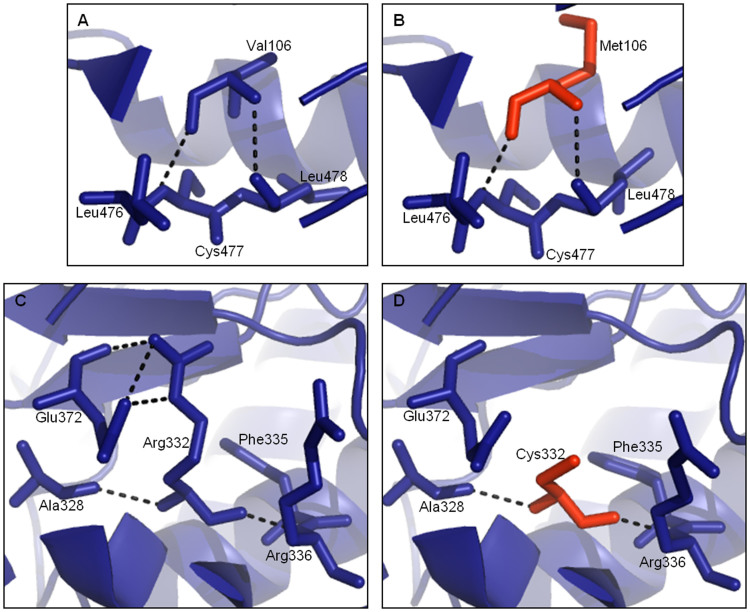
Taking into consideration residue conservation, Arg332 is invariant throughout all Metazoan species analyzed whereas Val106 is substituted by interchangeable residues (valine and leucine, except for yeast), and even methionine in Ciona intestinalis (Figure 2B). Accordingly, the SIFT28 and PolyPhen29 predictions considered the Val106Met substitution tolerated/benign and the Arg332Cys mutation as deleterious/probably damaging, respectively (Supplementary Table S4). In the structural models, the Val106Met replacement does not cause an apparent modification in the protein structure (Figure 3A and B) however, in the Arg332Cys replacement a distinct network of H-bond contacts is observed when a cysteine residue replaces the ancestral arginine (Figure 3C and D). Specifically, Arg332 establishes polar contacts with Ala328, Arg336, Phe335 and Glu372, and the polar contact with Glu372 is lost with the Cys332.
Figure 3. Structural impact of the human NAPRT1 missense mutations.
Val106 (A) interacts with Leu476, Cys477 and Leu478 and in the model with Met106 (B – red) the same interactions are maintained. Arg332 (C) establishes contacts with Ala328, Arg336, Phe335 and Glu372. Cys332 (D – red) loses polar contact with Glu372.
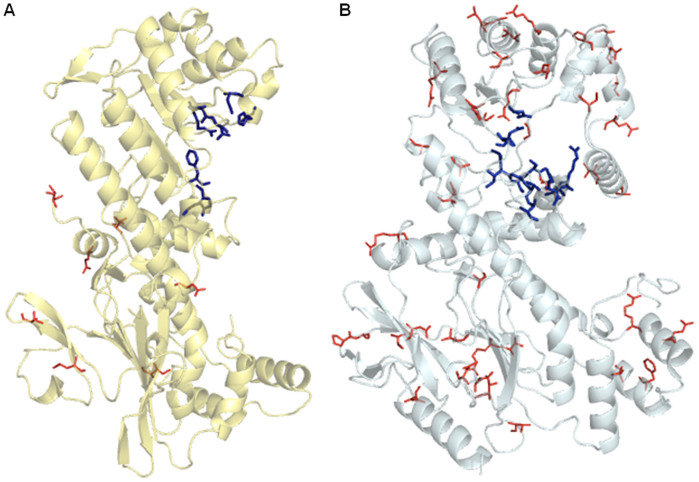
Using the NAMPT structure30 and our model for NAPRT1, we located all the missense mutations described so far as well as the predicted active sites in both proteins (Figure 4). Missense mutations found in NAMPT are far from the active sites or the dimer interface (Figure 4A), thus, their potential impact on the enzyme structure and/or function should be limited. On the other hand, NAPRT1 missense mutations, which are abundant, are spread all over the molecule (Figure 4B). This is consistent with SIFT28and PolyPhen29 predictions as well (Supplementary Table S4).
Figure 4. NAMPT and NAPRT1 missense spectra.
(A). The PDB structure of human NAMPT (PDB id: 3DKJ) representing only one of the two chains of the dimeric molecule. The active site residues (blue sticks) are Phe193, Arg311, Asp313, Asp279, His247; from chain 2 (not represented) the residues involved are Tyr18, Lys415, Lys42330. The missense mutations are shown in red lines. (B). The structure of the human NAPRT1 was predicted with I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) and represents one chain of the predicted dimer. The blue sticks represent the active sites as predicted by The Conserved Domain Database (CDD:29617), from NCBI43. The red lines illustrate the missense mutation residue location, which are all over the molecule.
Discussion
NAMPT and NAPRT1 are important enzymes in NAD metabolism, both in normal and in pathological conditions. In this study, we targeted the regions where most missense mutations were located. Variation data from Ensembl Genome Browser (Dec2010 - Jan2013) indicated an increasing number of coding region alterations in both genes, although more evident for NAPRT1 (Supplementary Figure S5). For instance, in the same period, 40 missense and 20 silent new mutations were described in NAPRT1, compared to 8 and 15, respectively, in NAMPT. This shows that NAPRT1 is more permissive to mutations and, thus, much more polymorphic than NAMPT, in which a strong purifying selection must be acting on.
Our results confirmed that NAMPT has a lower genetic diversity than NAPRT1. We found a smaller number of alterations and observed that missense mutations are rare in the human gene. In fact, we detected only intronic and synonymous variants. Intronic SNPs in NAMPT are associated with disease phenotypes related to glucose and lipid metabolism and other metabolic and vascular traits (see31 and32 for review), possibly due to alterations in splicing, protein binding or methylation sites that will affect the levels of protein expression. As for synonymous mutations, it has been recently shown that rs2302559 correlates with NAMPT serum levels33, thus, the impact of these novel variants is yet to be determined.
In NAPRT1 we detected two missense and five silent variants in samples from healthy individuals. One of these, g.676C>G, is an unreported alteration in exon 3 (Val142Val). Although commonly considered silent, synonymous mutations can have a phenotypic effect and be implicated in human disease26,33,34. Protein expression or conformation can be affected by speeding up or slowing down protein synthesis or by affecting splicing.
Using a novel methodology to estimate the pathogenicity of human genetic variants35, we observed that many NAPRT1 variants here described are associated with splicing sites (Supplementary Table S5). Given the location of the Val142 near the exon border, we used the NetGene2 online server, which predicts human splice sites36, to evaluate the impact of this new polymorphism. We observed that an alternative splice site near the mutated g.676C>G is predicted with higher confidence than the splice site that determines the exon 3-intron 3 boundary in the reference sequence, resulting in an alternatively spliced transcript. As a recent study suggests that synonymous mutations change the sequences that regulate splicing in oncogenes, and are frequently associated with cancer26, the impact of this, and other, synonymous mutations should be considered in further studies.
The absence of NAPRT1 expression has been reported in different types of cancer14,37, thus, we expected to find a higher number of alterations in NAPRT1 that could affect protein expression. This is of particular importance to validate NA as a chemoprotectant in the treatment of cancer patients with NAMPT inhibitors, which would be effective in NAPRT1-negative tumors only38. Curiously, no missense mutations were detected in the tumor samples studied, further supporting the role of intronic and synonymous mutations in promoting changes in protein expression, as discussed above. Recent work also shows that epistasis influences NAPRT1 gene expression39, suggesting that further studies will be necessary to establish which SNP or SNP associations are preponderant in defining NAPRT1-negative phenotypes.
Although the human NAMPT enzyme is well characterized, with known active site and dimer interface residues30,40,41, for human NAPRT1 no protein structure is yet available. A recent study42 on the kinetic characterization of this enzyme identified residues involved in activity, and used the Thermoplasma acidophilum NAPRTase to infer structural information. To study the structural impact of missense variants, we also built models for human NAPRT1, and evaluated Val106Met and Arg332Cys mutations considering residue conservation and changes in bond contacts. We further located known missense mutations of NAPRT1 and NAMPT in their respective structures, as well as residues involved in the active center30,43. For NAMPT, none of the missense mutations would interfere directly with the active center of the enzyme or with the dimer interface, whereas in NAPRT1, the missense mutations are located all over the molecule. Despite no direct bond to active center residues was detected, these mutations may influence assembly of the active dimer form, resulting in a dysfunctional or inactive protein.
The growing number of mutations described in NAPRT1 that occur all over the molecule is somewhat puzzling. Although it could be secondary in tissues that express NAMPT, NAPRT1 is useful for cells that lack NAMPT, such as neuron, and for cells in the digestive tract due to abundance of NA from bacterial metabolism44. Moreover, as NA is a more efficient NAD precursor4, situations with high NAD demand or NAMPT impairment would benefit from the activity of NAPRT1. Further studies are required to elucidate if the variants found in this study are correlated with protein expression levels, not only in physiological normal conditions but also in a disease context.
Methods
Samples
All samples used in this study are commercially available and were acquired as extracted DNA. Donors have given written, informed consent for their samples to be used for research purposes. DNA samples (108 healthy UK Caucasian blood donors) of the ECACC (European Collection of Cell Cultures) Human Random Control DNA Panel were purchased from Sigma-Aldrich (St. Louis, MO, USA). Human Adult Genomic DNA panels were obtained from AMS Biotechnology (Abigdon, U.K.), and included DNA from blood (25 samples, from 15 Caucasian, 2 Asian, 3 African American, 1 Center/S American, 3 Hispanic and 1 N/A individuals), colon and stomach tissues (16 samples, from 6 Caucasian and 10 Asian individuals) and colon and gastric tumors (80 samples, all of Asian origin).
Polymerase chain reactions
Polymerase chain reactions (PCR) were prepared using HotStarTaq® Master Mix Kit (Qiagen, Germantown, MD, USA), 0.2 µM (final concentration) of each primer and 10% Q solution (Qiagen). Primer sequences to NAMPT and NAPRT1 fragments, containing relevant mutation sites described in public databases (NCBI, Ensembl) and the literature, are provided in Supplementary Table S1.
PCR amplification was as follows: 95°C (15 min), 35 cycles at 94°C (30 sec), 58–64°C (1 min) and 72°C (1 min), and final extension at 72°C (10 min). Amplification products were separated by horizontal polyacrylamide gel electrophoresis and visualized by silver staining.
Sequencing analysis
PCR products were purified with ExoSAP-IT (USB Corporation, Santa Clara, CA, USA) according to the manufacturer's instructions and sequenced with the ABI Big Dye Terminator Cycle Sequencing Ready Reaction kit v3.1 (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA). Fragments were analyzed in an ABI PRISM 3130xl (Applied Biosystems). Sequences were aligned using Geneious v.5.5 (created by Biomatters, available from http://www.geneious.com/).
Protein alignments
Human NAMPT (NP_005737) and NAPRT1 (NP_660202) amino acid sequences were aligned with homologue proteins. Sequences were retrieved from the NCBI (http://www.ncbi.nlm.nih.gov/) and Joint Genome Institute (JGI) Genome Portal (genome.jgi.doe.gov/) databases (Supplementary Table S2). Alignments were performed using MUSCLE45 incorporated in Geneious v.5.5 and are shown in Supplementary Figures S1 and S2.
Homology modeling and structure visualization
The human NAPRT1 protein sequence was used as template to search for the best E-value PDB using the NCBI BlastP analysis46 This search identified structures from Enterococcus faecalis, Thermoplasma acidophilum and Saccharomyces cerevisiae as optimal structural templates. E. faecalis putative nicotinate phosphoribosyltransferase and S. cerevisiae Npt1p structures (PDB id: 2F7F and 1VLP) were used as templates in MODELLER47 to build human NAPRT1 structural models by homology modeling. Additionally, the I-TASSER online server48,49 was used to predict NAPRT1 structure by ab initio modeling. Accuracy of the models (Supplementary Figure S3) was estimated using ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php), as previously described50,51. Models of the NAPRT1 variants Val106Met and Arg332Cys were built in MODELLER, using as template the I-TASSER predicted structure (Supplementary Figure S4). The human NAMPT structure used corresponds to the PDB id: 3DKJ30. All structures were visualized in Pymol v1.1r152.
Mutation analyses
Re-sampling statistics were performed by bootstrap analysis using MATLAB (R2011a, MathWorks, Natick, MA, USA). The method used for the construction of the bootstrap confidence intervals (95%) was the accelerated bias-correction (Supplementary Table S3).
Pathogenicity prediction of the variants was performed as follows. Available information of the SIFT28 and PolyPhen29 predictions, for all dbSNP missense mutations in the human NAMPT and NAPRT1 reference transcripts (ENST00000222553 and ENST00000449291), were retrieved from Ensembl53 (release 75 - February 2014) (Supplementary Table S4).
For all the variants found in our study, the pathogenicity was also scored according to CADD (http://cadd.gs.washington.edu/score)35. Variants were retrieved from the 1000 Genomes Browser54 (http://browser.1000genomes.org/index.html) as vcf files, corresponding to the coordinates of NAMPT and NAPRT1 genes (chr7:105888731-105926772 and chr8:144656955-144660819). Variants were selected according to the position on the chromosome and scores are shown in Supplementary Table S5.
Author Contributions
R.M.S. designed and supervised the study. S.D.P. and S.S.S. performed the experiments. L.A., L.C., A.A. and R.M.S. analyzed the data. S.D.P. and R.M.S. wrote the paper. All authors revised and approved the manuscript.
Supplementary Material
SUPPLEMENTARY INFO
Acknowledgments
The authors are thankful to Joana Gonçalves and Filipe Pereira for kindly providing samples. IPATIMUP is an Associate Laboratory partially supported by FCT, the Portuguese Foundation for Science and Technology. This work was supported by the project PTDC/BIA-PRO/099888/2008, co-financed by FEDER through POFC/QREN (COMPETE FCOMP-01-0124-FEDER-009029). LA and RMS were supported by FCT Ciência2007 (Hiring of PhDs for the SCTN - financed by POPH - QREN - Typology 4.2 - Promoting Scientific Employment, co-financed by the MES national funding and The European Social Fund). LC and RMS are currently supported by the project Neuropath (CENTRO-07-ST24-FEDER-002034), co-funded by QREN “Mais Centro” program and the EU.
References
- Okumura S., Sasaki T., Minami Y. & Ohsaki Y. Nicotinamide phosphoribosyltransferase: a potent therapeutic target in non-small cell lung cancer with epidermal growth factor receptor-gene mutation. J. Thorac. Oncol. 7, 49–56 (2012). [DOI] [PubMed] [Google Scholar]
- Magni G. et al. Enzymology of NAD+ homeostasis in man. Cell. Mol. Life Sci. 61, 19–34 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel W. et al. Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 5, e263 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara N. et al. Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J. Biol. Chem. 282, 24574–24582 (2007). [DOI] [PubMed] [Google Scholar]
- Chiarugi A., Dolle C., Felici R. & Ziegler M. The NAD metabolome - a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 (2012). [DOI] [PubMed] [Google Scholar]
- Belenky P., Bogan K. L. & Brenner C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19 (2007). [DOI] [PubMed] [Google Scholar]
- Ocampo A., Liu J. & Barrientos A. NAD+ salvage pathway proteins suppress proteotoxicity in yeast models of neurodegeneration by promoting the clearance of misfolded/oligomerized proteins. Hum. Mol. Genet. 22, 1699–1708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford R. et al. Nicotinamide phosphoribosyltransferase and SIRT3 expression are increased in well-differentiated thyroid carcinomas. Anticancer Res. 33, 3047–3052 (2013). [PubMed] [Google Scholar]
- Samal B. et al. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol. Cell. Biol. 14, 1431–1437 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara A. et al. Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 307, 426–430 (2005). [DOI] [PubMed] [Google Scholar]
- Rongvaux A. et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32, 3225–3234 (2002). [DOI] [PubMed] [Google Scholar]
- Dahl T. B., Holm S., Aukrust P. & Halvorsen B. Visfatin/NAMPT: a multifaceted molecule with diverse roles in physiology and pathophysiology. Annu. Rev. Nutr. 32, 229–243 (2012). [DOI] [PubMed] [Google Scholar]
- Olesen U. H., Thougaard A. V., Jensen P. B. & Sehested M. A preclinical study on the rescue of normal tissue by nicotinic acid in high-dose treatment with APO866, a specific nicotinamide phosphoribosyltransferase inhibitor. Mol. Cancer Ther. 9, 1609–1617 (2010). [DOI] [PubMed] [Google Scholar]
- Watson M. et al. The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol. Cell. Biol. 29, 5872–5888 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames D. S. et al. Loss of NAPRT1 Expression by Tumor-Specific Promoter Methylation Provides a Novel Predictive Biomarker for NAMPT Inhibitors. Clin. Cancer Res. 19, 6912–6923 (2013). [DOI] [PubMed] [Google Scholar]
- Bottcher Y. et al. Genetic variation in the visfatin gene (PBEF1) and its relation to glucose metabolism and fat-depot-specific messenger ribonucleic acid expression in humans. J. Clin. Endocrinol. Metab. 91, 2725–2731 (2006). [DOI] [PubMed] [Google Scholar]
- Jian W. X. et al. The visfatin gene is associated with glucose and lipid metabolism in a Chinese population. Diabet. Med. 23, 967–973 (2006). [DOI] [PubMed] [Google Scholar]
- Reddy P. S. et al. PBEF1/NAmPRTase/Visfatin: a potential malignant astrocytoma/glioblastoma serum marker with prognostic value. Cancer Biol. Ther. 7, 663–668 (2008). [DOI] [PubMed] [Google Scholar]
- Wang L. S. et al. A polymorphism in the visfatin gene promoter is related to decreased plasma levels of inflammatory markers in patients with coronary artery disease. Mol. Biol. Rep. 38, 819–825 (2011). [DOI] [PubMed] [Google Scholar]
- Korner A. et al. Effects of genetic variation in the visfatin gene (PBEF1) on obesity, glucose metabolism, and blood pressure in children. Metabolism 56, 772–777 (2007). [DOI] [PubMed] [Google Scholar]
- Bailey S. D. et al. Common polymorphisms in the promoter of the visfatin gene (PBEF1) influence plasma insulin levels in a French-Canadian population. Diabetes 55, 2896–2902 (2006). [DOI] [PubMed] [Google Scholar]
- Johansson L. M., Johansson L. E. & Ridderstrale M. The visfatin (PBEF1) G-948T gene polymorphism is associated with increased high-density lipoprotein cholesterol in obese subjects. Metabolism 57, 1558–1562 (2008). [DOI] [PubMed] [Google Scholar]
- Lasky-Su J. et al. Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 1355–1358 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A. et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum. Mol. Genet. 19, 4745–4757 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat. Commun. 1, 34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Minana B., Valcarcel J., Gabaldon T. & Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 (2014). [DOI] [PubMed] [Google Scholar]
- Chappie J. S. et al. The structure of a eukaryotic nicotinic acid phosphoribosyltransferase reveals structural heterogeneity among type II PRTases. Structure 13, 1385–1396 (2005). [DOI] [PubMed] [Google Scholar]
- Ng P. C. & Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos E. S., Ho M. C., Almo S. C. & Schramm V. L. A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT. Proc. Natl. Acad. Sci. U S A 106, 13748–13753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddi-Rosa P., Oliveira C. S., Giuffrida F. M. & Reis A. F. Visfatin, glucose metabolism and vascular disease: a review of evidence. Diabetol. Metab. Syndr. 2, 21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Q., Heruth D. P. & Ye S. Q. Nicotinamide Phosphoribosyltransferase in Human Diseases. J. Bioanal. Biomed. 3, 13–25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stastny J. et al. Association of genetic variability in selected regions in visfatin (NAMPT) gene with anthropometric parameters and dietary composition in obese and non-obese Central-European population. Diabetes Metab. Syndr. 7, 166–171 (2013). [DOI] [PubMed] [Google Scholar]
- Sauna Z. E. & Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12, 683–691 (2011). [DOI] [PubMed] [Google Scholar]
- Kircher M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J. & Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 220, 49–65 (1991). [DOI] [PubMed] [Google Scholar]
- Olesen U. H., Hastrup N. & Sehested M. Expression patterns of nicotinamide phosphoribosyltransferase and nicotinic acid phosphoribosyltransferase in human malignant lymphomas. APMIS 119, 296–303 (2011). [DOI] [PubMed] [Google Scholar]
- Cerna D. et al. Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) activity by small molecule GMX1778 regulates reactive oxygen species (ROS)-mediated cytotoxicity in a p53- and nicotinic acid phosphoribosyltransferase1 (NAPRT1)-dependent manner. J. Biol. Chem. 287, 22408–22417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G. et al. Detection and replication of epistasis influencing transcription in humans. Nature 508, 249–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang T. et al. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat. Struct. Mol. Biol. 13, 661–662 (2006). [DOI] [PubMed] [Google Scholar]
- Takahashi R. et al. Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase. J. Biochem. 147, 95–107 (2010). [DOI] [PubMed] [Google Scholar]
- Galassi L. et al. Characterization of human nicotinate phosphoribosyltransferase: Kinetic studies, structure prediction and functional analysis by site-directed mutagenesis. Biochimie 94, 300–309 (2012). [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan K. L. & Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130 (2008). [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Potterton L., Yuan F., van Vlijmen H. & Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins 23, 318–326 (1995). [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A. & Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl M. J. Recognition of errors in three-dimensional structures of proteins. Proteins 17, 355–362 (1993). [DOI] [PubMed] [Google Scholar]
- Wiederstein M. & Sippl M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–410 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. L. C. The PyMOL Molecular Graphics System, Version 1.3r1 (2010). [Google Scholar]
- Flicek P. et al. Ensembl 2014. Nucleic Acids Res. 42, D749–755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY INFO