Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) synthesizes several putative viral envelope proteins, including the spike (S), membrane (M), and small envelope (E) glycoproteins. Although these proteins likely are essential for viral replication, their specific roles in SARS-CoV entry have not been defined. In this report, we show that the SARS-CoV S glycoprotein mediates viral entry through pH-dependent endocytosis. Further, we define its cellular tropism and demonstrate that virus transmission occurs through cell-mediated transfer by dendritic cells. The S glycoprotein was used successfully to pseudotype replication-defective retroviral and lentiviral vectors that readily infected Vero cells as well as primary pulmonary and renal epithelial cells from human, nonhuman primate, and, to a lesser extent, feline species. The tropism of this reporter virus was similar to that of wild-type, replication-competent SARS-CoV, and binding of purified S to susceptible target cells was demonstrated by flow cytometry. Although myeloid dendritic cells were able to interact with S and to bind virus, these cells could not be infected by SARS-CoV. However, these cells were able to transfer the virus to susceptible target cells through a synapse-like structure. Both cell-mediated infection and direct infection were inhibited by anti-S antisera, indicating that strategies directed toward this gene product are likely to confer a therapeutic benefit for antiviral drugs or the development of a SARS vaccine.
The severe acute respiratory syndrome coronavirus (SARS-CoV) is the likely cause of an acute infectious respiratory disorder identified in highly lethal outbreaks during the past year (10, 18, 21, 32, 40). Infection is characterized by acute flu-like symptoms that progress to a severe febrile respiratory illness with significant mortality. Coronaviruses, comprising a genus of the Coronaviridae family, are enveloped positive-strand RNA viruses. In general, coronaviruses cause respiratory and enteric diseases in humans and domestic animals (15, 20). Two previously known human coronaviruses caused only mild upper respiratory infections (15, 20). In contrast, a highly pathogenic, severe respiratory disease is caused by the SARS-CoV, especially in the elderly (44). Coronaviruses can be divided into three serologically distinct groups (15). Phylogenetically, SARS-CoV is not closely related to any of the three groups (26), though it is most similar to the group II coronaviruses (33, 36).
Although the organization of the SARS-CoV genome is related to that of animal coronaviruses, its genetic sequence is unique, and the structure and function of its gene products are not known. At least 14 open reading frames (ORFs) can be identified in its genome (26, 34, 36). Among these, the replicase/transcriptase genes are located in the 5′ portion of the genome. At its 3′ end, the four major structural proteins (S, M, N, and E) are made through different subgenomic RNAs. Based on comparison to animal coronaviruses, three structural gene products are predicted to be present on the viral envelope: the spike (S), membrane (M), and small envelope (E) proteins (20, 26, 34). The structure of the SARS-CoV envelope differs in some respects from that of other enveloped viruses, such as retroviruses and lentiviruses, many of which contain one viral envelope protein.
Envelope or spike proteins from enveloped viruses have been used to pseudotype retroviral and lentiviral vectors for functional and gene transfer studies (29, 35, 43, 45); however, whether coronavirus glycoproteins could pseudotype these viruses was unknown. Here we report that replication-defective retroviral (Moloney murine leukemia virus) and lentiviral (human immunodeficiency virus type 1 [HIV-1]) vectors can be pseudotyped with the SARS-CoV S protein, and the properties of S related to entry have been defined. Using these pseudoviruses, we were able to determine the relative contributions of SARS-CoV envelope proteins to viral entry and fusion and to examine the roles of these different viral envelope gene products with respect to entry, cell specificity, and potential inhibition of viral replication.
MATERIALS AND METHODS
Antibodies, mouse immune serum, and media.
A human serum from a recovered SARS patient was kindly provided by William Bellini (Centers for Disease Control and Prevention [CDC], Atlanta, Ga.). Antibodies to CD11c, CD14, CD40, CD80, CD86, and HLA-DR were purchased from BD Pharmingen. A fluorescein isothiocyanate (FITC)-conjugated mouse antibody against the C-terminal His tag was purchased from Invitrogen (Carlsbad, Calif.). Media for human primary cell culture were purchased from Cambrex (East Rutherford, N.J.). RPMI 1640 medium and Dulbecco's modified Eagle medium were purchased from Invitrogen. A mouse immune serum against the SARS-CoV S protein was generated by vaccinating 10-week-old BALB/c mice with CMV/R plasmid DNA expression vectors, described below, encoding the S protein (mice were vaccinated with 25 μg, three times, at 3-week intervals, and were bled after 2 months). Negative-control antisera were obtained in a similar fashion by injecting the same plasmids with no insert.
Cell lines.
Human primary cell lines from renal proximal tubule epithelial cells (RPTEC), renal epithelial cells (HRE), renal cortex epithelial cells (HRCE), small airway epithelial cells (SAEC), bronchial epithelial cells (NHBE), lung fibroblasts (NHLF), lung microvascular endothelial cells (HMVEC-L), umbilical vein endothelial cells (HUVEC), microvascular endothelial cells (HMVEC), mammary epithelial cells (NHMEC), and keratinocytes (NHEK), as well as hepatocytes, were purchased from Cambrex. The following human and animal cell lines were purchased from the American Type Culture Collection: ACHN (human kidney adenocarcinoma), 293 (human embryonic kidney cells), 786-O (human kidney adenocarcinoma), A549 (human lung carcinoma), HeLa (human cervical adenocarcinoma), Colo205 (human colon adenocarcinoma), Jurkat (human T cells), CEM (human acute lymphoblast leukemia), M8166 (human CD4+ lymphoid cells), HL60 (human promyelocytic leukemia cells), THP-1 (human acute monocytic leukemia), Vero (African green monkey kidney epithelial cells), CRFK (cat kidney cortex epithelial cells), OK (opossum kidney cortex epithelial cells), M-1 (mouse kidney cortex epithelial cells), FC2.Lu and FC28.lu (cat lung fibroblasts), AK-D (cat lung epithelial cells), MLE12 (mouse lung epithelial cells), MM14.lu (mouse lung), LA-4 (mouse lung adenoma), LH4 (guinea pig lung fibroblasts), and CHL-11 (Chinese hamster lung fibroblasts). Human peripheral blood mononuclear cells were prepared from whole blood by Ficoll gradient centrifugation. The THP-1, THP-DC-SIGN (THP-1 cells expressing human DC-SIGN), and THP-DC-SIGNΔ35 (THP-1 cells expressing DC-SIGN with a 35-amino-acid [35-aa] cytoplasmic domain deleted) cell lines were kindly provided by D. R. Littman (19). Human T-cell leukemia cell lines A3R5 (a subline of CEM expressing both CCR5 and CXCR4), MT-2 (expressing CXCR4), and 293T were gifts from John Mascola.
Gene synthesis and construction of expression vectors.
Genes encoding the SARS-CoV S, M, and E proteins were synthesized by using human-preferred codons. To synthesize these genes, protein sequences obtained from GenBank (SARS-CoV strain Urbani, accession no. AY278741) were reverse translated by using human-preferred codons. Sets of 75-bp oligonucleotides with 25-bp overlaps covering each fragment were synthesized and gel purified. The oligonucleotides were assembled into DNA fragments by using Pfu Turbo Hotstart DNA polymerase (Stratagene, La Jolla, Calif.) at a 50-to-65°C gradient annealing temperature. DNA fragments were cloned into the pCR-Blunt II-Topo vector (Invitrogen) and sequenced. Clones with the fewest mutations were picked and were further corrected by using the QuikChange kit (Stratagene) according to the manufacturer's protocol. Fully corrected DNA fragments for each gene were finally cloned into the mammalian expression vector CMV/R-mcs. COOH-terminal deletion mutants were generated by using the QuikChange kit (Stratagene) and were cloned into the CMV/R-mcs expression vector, which contains the cytomegalovirus (CMV) enhancer/promoter and splice donor and the human T-cell leukemia virus type 1 R region (W. Akahata, Z.-Y. Yang, and G. J. Nabel, unpublished data). These mutants include (i) SΔCD, in which the cytoplasmic domain was truncated (terminated at aa 1229), (ii) SΔTM2, in which the transmembrane and cytoplasmic domains were deleted (terminated at aa 1190), and (iii) SΔHR1, in which the transmembrane, cytoplasmic, and heptad-2 domains were removed (terminated at aa 1153). For S(1190)-Myc-His, the S protein was truncated at aa 1190 to remove the transmembrane and cytoplasmic domains and was tagged with a Myc and a His epitope at the COOH-terminus. The expression vectors were sequenced on both strands to ensure that each gene was correct and were further confirmed by Western blot analysis.
Purification and differentiation of human mDC.
Myeloid dendritic cells (mDC) were purified from elutriated monocytes from healthy adult donors by a two-step procedure consisting of automated leukapheresis and counterflow centrifugal elutriation at the Transfusion Medicine Department of the Warren Grant Magnuson Clinical Center, National Institutes of Health, Bethesda, Md. (1). mDC were isolated from the elutriated monocyte fraction with negative selection by removing cells expressing BDCA-4 and CD9 with microbeads (Miltenyi Biotec, Auburn, Calif.), followed by positive selection using antibodies to CD1c (Miltenyi Biotec). mDC were then cultured in a medium containing granulocyte-macrophage colony-stimulating factor (10 ng/ml; PeproTech) and induced to differentiate to mature mDC by using poly(I · C) (50 ng/ml; Sigma, St. Louis, Mo.) for 48 h (5). Antibodies to CD11c and CD14 (BD Pharmingen) were used to assess the purity of DC, and antibodies to CD40, CD80, CD86, and HLA-DR (BD Pharmingen) were used to characterize the differentiation of DC by flow cytometry.
Production of pseudotyped lentiviruses and retroviruses.
Recombinant lentiviruses and retroviruses expressing a luciferase reporter gene were produced as described previously (17, 29). Briefly, 5 × 106 293T cells were plated in 10-cm-diameter tissue culture dishes the day before transfection. The cells were transfected the next day by using calcium phosphate reagent (Invitrogen). The amount of plasmid DNA used for making different pseudotyped vectors was as follows: for lentiviral vectors, 7 μg of pCMVΔR8.2 plus 7 μg of pHR′CMV-Luc and either 400 ng of CMV/R-SARS-S or 2 μg of pNGVL-4070A (Ampho); for retroviral vectors, 7 μg of pNGVL-GagPol (MLV) plus 7 μg of pLZR-Luc and 400 ng of CMV/R-SARS-S or 2 μg of pNGVL-4070A (amphotroic MLV gp70), respectively. Cells were transfected overnight, washed, and replenished with fresh medium. Forty-eight hours later, supernatants were harvested, filtered through a 0.45-μm-pore-size syringe filter, and stored in aliquots at −80°C. p24 levels were measured from different viral stocks (4) by using the Coulter HIV-1 p24 Antigen Assay kit (Beckman Coulter, Somerset, N.J.). One-tenth the amount of plasmids CMV/R-M and CMV/R-E were used with CMV/R-S for making combinational pseudoviruses.
Production of a GFP-Vpr-labeled SARS-CoV S-pseudotyped lentivirus.
A green fluorescent protein (GFP)-Vpr-labeled SARS-CoV S-pseudotyped lentivirus was produced by transfection of human embryonic kidney 293T cells with a pLAI provirus from which env had been deleted (10 μg), CMV/R-SARS-S (1 μg), and plasmid pEGFP-C3 (Clontech, Palo Alto, Calif.), containing the entire Vpr coding region fused to the carboxy terminus of eGFP (GFP-Vpr; 15 μg) (27). Cells were washed at 16 to 20 h posttransfection and replenished with fresh medium. Forty-eight hours later, supernatants were harvested, filtered through a 0.45-μm-pore-size syringe filter, and concentrated. Briefly, 32 ml of supernatant was layered onto 5 ml of Optiprep (Iodoxinal) medium (Invitrogen) and centrifuged at 50,000 × g for 1.5 h with a Surespin 630 rotor (Sorvall, Newtown, Conn.). The last 3 ml of supernatant remaining above the Optiprep interface was collected and frozen at −80°C in 500-μl aliquots.
Infection of cells with SARS-CoV and titration of SARS-CoV.
Cells in six-well dishes were infected with 100 μl of a 1:10 dilution of SARS-CoV strain Urbani (106.25 50% tissue culture infective doses/ml; CDC) per well under appropriate containment in a BSL3 laboratory. After 1 h of adsorption, the cells were washed three times with medium, replenished with 3 ml of fresh medium, and maintained at 37°C in a 5% CO2 incubator. Seventy-two hours after infection, 0.5 ml of tissue culture medium was harvested and incubated with Vero cells in a 96-well plate; viral titers in the medium were calculated in 50% tissue culture infective doses per milliliter 4 days after infection of Vero cells (38). The viral cytopathic effect was determined on days 3 and 4. Mature and immature mDC were infected in 96-well plates (10,000 cells/well) with 50 μl of 1:10-diluted viral stock and were titered similarly 3 days later.
Infection of cells with pseudovirus.
A total of 30,000 cells were plated into each well of a 48-well dish the day before infection. Cells were infected with 150 μl of viral supernatant for 16 to 18 h for SARS-CoV S-pseudotyped viruses and for 3 to 4 h for Ampho Env and Ebola glycoprotein-pseudotyped viruses. The viral supernatant was replaced with fresh medium at the end of infection. Forty-eight hours after infection, cells were lysed in “mammalian cell lysis buffer” (Promega, Madison, Wis.). The same amount of cell lysate was used in a luciferase assay with “Luciferase assay reagent” (Promega) according to the manufacturer's suggestions.
Transfection and Western blot analysis.
293T cells were transfected by using calcium phosphate (Invitrogen). Transfected cells were harvested 48 h after transfection. Cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.). The membrane was incubated with convalescent-phase human sera from a SARS patient (dilution, 1:2,500; kindly provided by William Bellini of the CDC) for 1 h at room temperature in blocking buffer (Tris-buffered saline, 1% bovine serum albumin, 5% skim milk, 0.3% Tween 20), followed by three washes in washing buffer (Tris-buffered saline-0.3% Tween 20). The blot was further incubated in blocking buffer with horseradish peroxidase-conjugated donkey anti-human immunoglobulin G (IgG) (dilution, 1:5,000; Chemicon, Temecula, Calif.) for 30 min and then washed four times in washing buffer. Detection was performed with the ECL reagent (Amersham, Piscataway, N.J.).
Cell staining with S(1190)-Myc-His.
A 500-ng portion of purified S(1190)-Myc-His glycoprotein was incubated with 106 cells in 100 μl of phosphate-buffered saline (PBS) containing 2% fetal bovine serum for 20 min on ice and then washed once with 1 ml of cold PBS. Each cell line was split into two aliquots, which were stained either with an FITC-conjugated mouse anti-His tag monoclonal antibody (dilution, 1:100; Invitrogen) or with an FITC-labeled isotype control, followed by flow cytometric analysis.
Confocal microscopy.
mDC (105) isolated from human elutriated monocytes were plated onto a 12-well-dish. Twenty-four to 48 h later, the cells were infected with 100 μl of a Vpr-GFP-labeled SARS-CoV S pseudolentivirus for 30 min. Cells were washed, detached with trypsin-EDTA, washed again, and added to human renal epithelial cells (786-O; 3 × 104 cells/well) plated onto 8-well coverslip slides (Nalge Nunc, Naperville, Ill.). Sequential images of live cells were recorded every 3 min by confocal microscopy (SP2-AOBS; Leica Microsystems), and uptake, polarization, and transfer were assessed with representative cells.
pH-dependent entry of SARS-CoV-S-pseudotyped lentiviral vectors.
Vero cells were plated in a 48-well-dish (30,000 cells/well) the day before infection. Cells were preincubated with the indicated amounts of ammonium chloride or bafilomycin A (Sigma) for 1 h. Pseudoviruses were mixed with the same concentrations of reagents in tubes and added to cells. Eight hours later, viruses were removed and replaced with fresh medium. Cells were harvested 48 h after infection, and a luciferase assay was performed.
Cell-mediated transfer of SARS-CoV and the SARS-CoV S pseudotyped lentiviral vector.
THP, THP-DC-SIGN, or THP-DC-SIGNΔ35 (30,000 cells/well) was incubated with the SARS-CoV S-pseudotyped lentiviral vector for 2 h and then washed three times with tissue culture medium. Cells were then added to Vero, A3R5, or MT2 cells (30,000 cells/well) plated in 24-well dishes. These cells were harvested 72 h later for a luciferase assay to assess THP, THP-DC-SIGN, or THP-DC-SIGNΔ35 cell-mediated transfer of pseudovirus to the respective cells.
To measure the transfer of SARS-CoV by mature mDC and to assess whether an anti-SARS-CoV S immune serum can block the transfer of SARS-CoV by mDC, mature mDC were incubated with SARS-CoV for 1 h, washed, detached with trypsin, and replated onto Vero cells in a 96-well-plate (10,000 cells/well) in the presence of a control or an S-specific mouse antiserum (dilution, 1:100). Cell culture medium (Dulbecco's modified Eagle medium-10% FBS) was collected 72 h later, and SARS-CoV titers in the cell culture medium were measured as described above.
RESULTS
Pseudotyping.
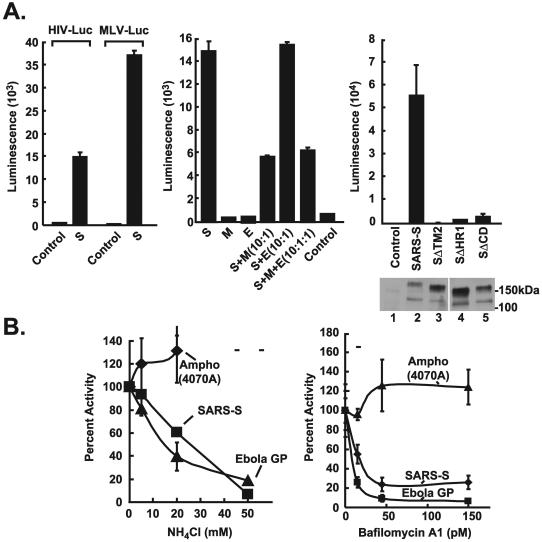
SARS-CoV envelope proteins were analyzed by cotransfection of expression vectors encoding either the S, M, or E glycoprotein with packaging plasmids for retroviral or lentiviral vectors into human 293T cells (17, 29). For comparison, envelope glycoproteins from amphotropic murine leukemia virus or Ebola virus (45) were substituted for the SARS-CoV envelope proteins. Vero cells, which support SARS-CoV replication (18, 32), were initially analyzed as target cells. Among the SARS-CoV gene products, only the S protein mediated entry into target cells, and it did so with both murine retroviral and human lentiviral vectors (Fig. 1A, left). Because both vectors could be pseudotyped with the S glycoprotein, further analyses were performed with the lentiviral vector, which can transduce and express recombinant genes in nondividing cells. Neither M nor E alone was able to support viral entry in the absence of S, suggesting that these glycoproteins serve other functions in the virus. Cotransfection of M together with the S glycoprotein inhibited the generation of functional lentiviral vector, in contrast to E, which did not alter the efficacy of gene transfer (Fig. 1A, center). To confirm the specificity of this effect and to demonstrate that full-length S was required for gene transfer, deletion mutants of S with various COOH-terminal deletions were prepared (Fig. 1A, upper right). Although these mutants showed comparable levels of cellular gene expression, the progressive COOH-terminal deletions showed markedly lower levels of recombinant gene transfer (Fig. 1A, lower and upper right, respectively), demonstrating that the cytoplasmic domain of S is required for viral entry.
FIG. 1.
Infection of Vero cells by S-pseudotyped retroviral and lentiviral vectors. Amphotropic (Ampho), S, and no-envelope control vectors were prepared as described in Materials and Methods. Viruses were used at similar multiplicities of infection, standardized by p24 protein levels. Viral pseudotypes were prepared by cotransfection of the indicated combinations of S, M, and E (center). 293T cell supernatants were used to infect Vero cell lines, and luciferase activity was analyzed as previously described (45). (A) (Left) Infection of the Vero cell line with the S-pseudotyped lentiviral or retroviral vector expressing luciferase (42). MLV, murine leukemia virus. (Center) The S glycoprotein, but not the M and E glycoproteins, mediates viral entry by the S-pseudotyped lentiviral vector. (Right) (Top) The requirement for the cytoplasmic domain of S was analyzed by generation of pseudotyped virus using full-length S proteins (S) or S proteins from which the COOH-terminal end was deleted. (Bottom) Expression of these S variants was confirmed by Western blot analysis. (B) pH-dependent entry of SARS-CoV S-pseudotyped lentiviral vectors. Pseudolentiviruses were incubated in the presence of increasing amounts of ammonium chloride (left) or bafilomycin (Sigma) (right). The experiment was performed in triplicate. Data are presented as the percentage of activity at the indicated dose relative to activity with no drug treatment. GP, glycoprotein.
pH-dependent fusion mediated by the SARS-CoV S protein.
Viral glycoproteins typically mediate attachment, fusion, and entry by one of two mechanisms. Viruses such as HIV or murine amphotropic retroviruses infect through a pH-independent cell fusion and entry process (30, 37). In contrast, influenza and Ebola viruses are prototypes for viruses that utilize a pH-dependent endocytotic pathway (43). To determine the pathway utilized by the SARS-CoV, the pH dependence of the SARS-CoV S-pseudotyped lentiviral vector was analyzed. Addition of ammonium chloride, which prevents acidification of the endosome, caused a dose-dependent reduction in viral entry (Fig. 1B, left) at concentrations similar to those described for other pH-dependent viral glycoproteins (3, 11, 43). This effect was also observed with another inhibitor of endosomal acidification, bafilomycin, also in a dose-dependent fashion (Fig. 1B, right).
Susceptibilities of human and animal cells to infection.
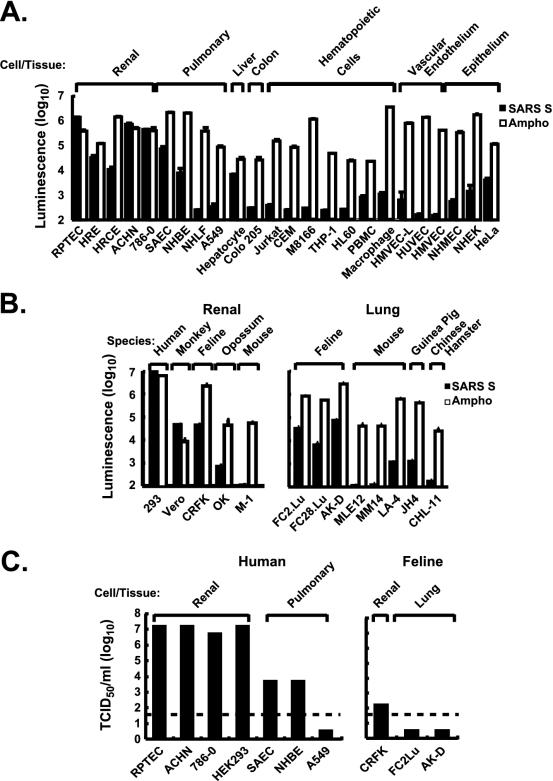
The specificity of the SARS-CoV S-pseudotyped virus was analyzed by transducing different human cell types, including epithelial, endothelial, and hematopoietic cells, and lung and renal cells from different species. Like Vero cells, a renal epithelial cell line derived from African green monkeys, human renal epithelial cells (HPTRC, HRE, HRCE, ACHN, and 786-O) were highly susceptible to infection compared to a known positive control with a broad host range, the 4070A amphotropic murine retroviral envelope. Respiratory tract epithelial cells were also readily transduced. In contrast, a number of cell types, including hepatocytes, lower airway fibroblasts, breast or colonic epithelial cells, vascular endothelial cells, or hematopoietic cells, were relatively resistant to transduction (Fig. 2A). Interestingly, renal epithelial cell lines from humans, nonhuman primates, and felines, and, to a lesser extent, lung cell lines from felines were susceptible to transduction, while similar cells from rodents were relatively resistant (Fig. 2B). The specificity of pseudotyped-virus transduction in these cell lines was confirmed by the susceptibility to infection by SARS-CoV. A number of human cell lines, particularly those of renal and pulmonary origin, that had not previously been recognized as susceptible to infection by the virus, because they did not show cytopathic effect, yielded high titers of virus (Fig. 2C). The susceptibilities of these cells to pseudovirus transduction correlated well with their abilities to support SARS-CoV replication. It therefore appears that a range of cell types and species are susceptible to infection mediated by the SARS-CoV S glycoprotein.
FIG. 2.
Tropism of a SARS-CoV S-pseudotyped lentiviral vector for human and animal cells and correlation with SARS-CoV infectibility. (A) Tropism of a SARS-CoV S pseudolentivirus for different types of human cells. All infections were performed in triplicate. Data are presented as averages ± standard deviations. Results from one of two independent experiments are shown. (B) Infectibility of renal cells from different species. Cells were infected and analyzed in triplicate. Data are presented as averages ± standard deviations. Results from one of two independent experiments are shown. (C) Infection of selected susceptible and resistant cells from panel A by SARS-CoV strain Urbani, titered as previously described (38) on Vero cells. Dashed line indicates the detection limit of infectivity of SARS-CoV.
Binding of the SARS-CoV S protein to DC-SIGN.
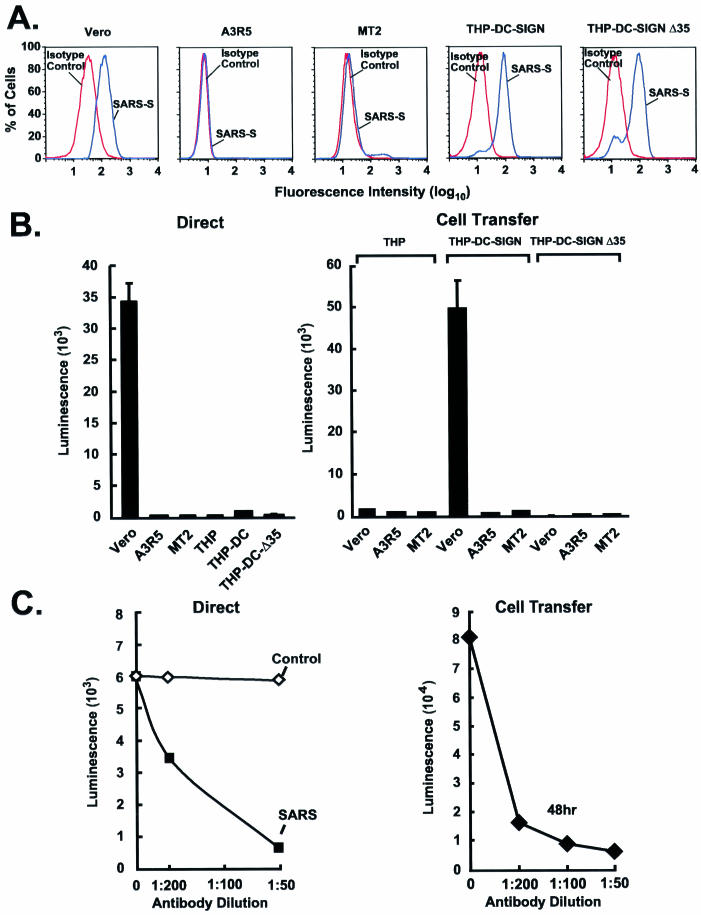
The S glycoprotein contains a number of N-linked glycosylation sites, which have been shown to affect binding to the DC-SIGN receptor on DC. This receptor regulates cell-mediated transmission for a number of viruses, including HIV, dengue virus, and CMV (2, 12, 14, 25, 39). To determine whether the SARS-CoV S protein could bind to DC-SIGN, THP-DC-SIGN or THP-DC-SIGNΔ35 (expressing a mutant form of DC-SIGN lacking the cytoplasmic domain required for internalization and transfer) (19) cells were incubated with purified His-tagged S. As expected, binding was readily detected in the permissive Vero cells by use of flow cytometry, in contrast to two nonpermissive T-cell leukemia cell lines, A3R5 and MT2 (Fig. 3A), or THP-1 cells lacking DC-SIGN (data not shown). In contrast, THP-DC-SIGN and Δ35 cells interacted with purified S glycoprotein (Fig. 3A, right panels), but unlike Vero cells, they could not be infected by the S-pseudotyped lentiviral vector (Fig. 3B, left). To determine whether DC-SIGN could nonetheless promote cell-mediated transfer of virus, the abilities of these cells to transfer the SARS-CoV S-pseudotyped lentiviral vector to Vero cells were analyzed. THP-DC-SIGN cells, but not THP or THP-DC-SIGNΔ35 cells, which are unable to internalize viruses (19), readily transferred virus to Vero cells (Fig. 3B, right), indicating that DC-SIGN or a related lectin on DC might facilitate cell-mediated transfer of virus. Both direct infection and DC-SIGN-mediated transfer were inhibited by a SARS-CoV S-specific mouse immune serum (Fig. 3C), confirming that S was necessary and sufficient for infection in both cases.
FIG. 3.
DC-SIGN-dependent uptake of the SARS-CoV S-pseudotyped lentiviral vector, and cell-mediated transfer and infection of target cells. (A) Binding of purified SARS-CoV S glycoprotein to cell lines. A total of 106 African green monkey kidney cells (Vero), human T-cell leukemia cells (A3R5 and MT2), or THP-1 myelomonocytic leukemia cells expressing wild-type or mutant forms of DC-SIGN (THP-DC-SIGN or THP-DC-SIGNΔ35, respectively) were incubated with purified S(1190)-Myc-His glycoprotein for 20 min on ice. Binding of S protein to the cells was detected by using an FITC-labeled anti-His (COOH-terminal) antibody (blue) (dilution, 1:100; Invitrogen). A FITC-labeled IgG isotype was used as a control (red). Data were analyzed by flow cytometry. (B) Direct viral entry (left) and cell-mediated virus transfer (right) of the SARS-CoVS-pseudotyped lentiviral vector from THP-1, THP-DC-SIGN, and THP-DC-SIGNΔ35 cells. (Left) Susceptibilities of Vero, A3R5, MT2, THP-1, THP-DC-SIGN, and THP-DC-SIGNΔ35 cells to SARS-CoV S-pseudotyped lentiviral vector infection were measured after transduction by use of the luciferase reporter. (Right) Cell-mediated pseudoviral transfer by THP-1, THP-DC-SIGN, or THP-DC-SIGNΔ35 cells (3 × 104) was also assessed by incubating the cells with the SARS-CoV S-pseudotyped lentiviral vector for 2 h at 37°C, followed by three washes before addition of the respective cells to the indicated target Vero cells at a 1:1 ratio. Cells were collected 72 h later for luciferase assay. (C) Inhibition of direct infection and cell-mediated transfer of SARS-CoV S pseudolentivirus by a mouse anti-SARS-CoV S protein antiserum. (Left) The SARS-CoV S-pseudotyped lentiviral vector was exposed to a mouse control or anti-S specific antiserum at the indicated dilutions for 60 min at 37°C before being added to Vero cells. (Right) For cell-mediated transfer, THP-DC-SIGN cells were incubated with pseudoviruses as described in the legend to panel B, followed by incubation with Vero cells in the presence of a control or anti-SARS-CoV S specific mouse antiserum for 48 h. After 48 h, cells were collected for luciferase assays.
Cell-mediated transfer of GFP-Vpr-labeled pseudotyped lentivirus by human mDC.
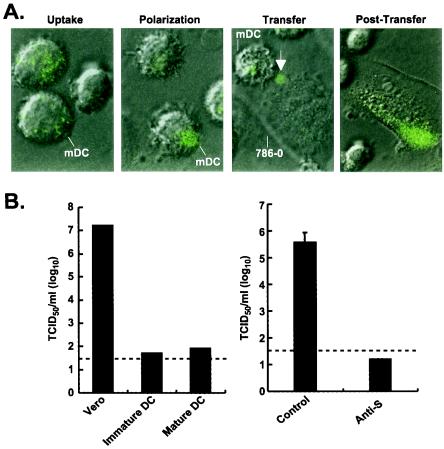
To determine whether cell-mediated transfer could be mediated by primary human DC, mature mDC were isolated and incubated with a GFP-Vpr-labeled SARS-CoV S-pseudotyped lentivirus (27). A similar vector pseudotyped with HIV gp160 was shown to mediate the formation of an “infectious synapse” that facilitates HIV infection (28), but it was not known whether similar structures could be formed by an unrelated virus whose target cell is nonlymphoid. mDC were incubated with the virus for 30 min, trypsinized, and transferred to fresh 786-O human renal cell cultures. Initially, the virus was distributed evenly throughout the DC (Fig. 4A, uptake), but within minutes, immunofluorescent foci had begun to form at the site of contact with Vero cells (Fig. 4A, polarization). The virus was observed to be transferred to the target cells through a structure analogous to the “synapse” previously described between mDC and lymphoid cells (Fig. 4A, transfer) (28). After transfer, a characteristic streak of fluorescence was seen at the site of entry, suggesting specific channeling of viral contents into cells (Fig. 4A, post-transfer). This effect was seen consistently and was not caused by tunneling of DC beneath the 786-O epithelial cell in culture. To confirm that mDC mediate infection by virus, immature and poly(I · C)-treated mDC were incubated with SARS-CoV. No direct infection was observed (Fig. 4B, left); however, mature mDC readily transferred virus that infected Vero cells (Fig. 4B, right). Transfer was inhibited by a specific anti-S mouse antiserum (Fig. 4B, right), documenting that cell-mediated transfer of SARS-CoV is mediated by mDC and is dependent on the interaction of the S glycoprotein with mDC.
FIG. 4.
Uptake and transfer of a GFP-Vpr-labeled SARS-CoV S-pseudotyped lentiviral vector and SARS-CoV by mature human mDC. (A) Uptake of a GFP-Vpr-labeled SARS-CoV S-pseudotyped lentiviral vector by mature mDC and subsequent transfer to renal epithelial cells by mDC, as detected by confocal microscopy. mDC were infected with a GFP-Vpr-labeled SARS-CoV S-pseudotyped lentivirus for 30 min at 37°C and were then added to human renal epithelial cells (786-O; 3 × 104 cells/well, plated 1 day before) in 8-well coverslip slides (Nalge Nunc) at a 1:1 ratio. Uptake, polarization, and transfer were assessed by confocal microscopy with representative cells. Arrow indicates transfer of labeled virus from DC to 786-O cells. (B) Human mature mDC are not directly infected by SARS-CoV (strain Urbani) but instead promote cell-mediated infection of susceptible target cells. (Left) Vero cells, immature mDC, or mature mDC were infected with SARS-CoV (strain Urbani) for 1 h in 96-well-dishes (2 × 104 cells/well), washed three times, and maintained in cell culture medium. (Right) Mature mDC were also infected for 1 h with SARS-CoV, washed, detached with trypsin, and replated onto 96-well-dishes with Vero cells (2 × 104 cells/well; 1:1 ratio) in the presence of a control or anti-SARS-CoV S specific mouse antiserum at a dilution of 1:100. Cell culture supernatants were collected 72 h later, and viral titers were measured as described previously (38). Virus yield is expressed as 50% tissue culture infective doses (TCID50) per milliliter. Dashed line indicates the detection limit of SARS-CoV.
DISCUSSION
In this study, we have examined the function of the SARS-CoV S protein and the contribution of other viral membrane proteins to virus fusion and entry. We suggest that, of these gene products, the S glycoprotein is necessary and sufficient for viral gene delivery. Although the origin of SARS-CoV is unknown, recent studies have suggested that it may have been transmitted from animals to humans (6, 7, 13). During the course of the SARS-CoV epidemic, molecular evolution throughout the SARS-CoV genome was detected. In addition to mutations in several regions of SARS-CoV, specific changes that involved the deletion of a 29-nucleotide region or insertion of an 82-nucleotide region in ORF8 were observed, and it is hypothesized that these changes may have facilitated the spread of the disease (7). It is possible that SARS-CoV has arisen from recombination between different coronaviruses, although the direct antecedents of the present strain are not yet defined (15, 20, 33, 36). While SARS-CoV is not closely related phylogenetically to any of three previously known subgroups of coronaviruses (26, 33, 34, 36), the recent discovery of human ACE2 protein as a cellular receptor that mediated SARS-CoV infection (23) suggests a parallel to hCD13 (or APN), utilized by the human coronavirus HCoV-229E (16, 23, 46); both ACE2 and hCD13 are peptidases.
The present study demonstrates that retroviral and lentiviral vectors can be pseudotyped with the SARS-CoV S protein. Using the pseudoviruses, we were able to demonstrate that S protein alone is necessary and sufficient for mediating viral attachment and entry. The cytoplasmic domain of the S protein is also required for viral entry. Interestingly, cotransfection of an M expression vector with S inhibited pseudovirus entry, indicating that an M-S interaction, as described for other coronaviruses (9, 24), might prevent S incorporation into the lentivirus virion. SARS-CoV S mediates viral entry in a pH-dependent manner, like Ebola virus and vesicular stomatitis virus (41, 43), suggesting that the S glycoprotein contains two structures predicted to be helical coiled-coils, analogous to those that have been described for avian, murine, and human enveloped viruses (11).
The tropism of SARS-CoV was studied by transducing or infecting different human and animal cell lines with SARS-CoV S pseudoviruses or SARS-CoV, respectively. Transduction of these cells by pseudovirions correlated well with their ability to support SARS-CoV replication. The infectibility of human respiratory tract cells, e.g., SAEC or NHBE (Fig. 2A and C) by SARS-CoV-S pseudovirions also correlates well with the primary clinical finding for this disease (10, 18). The susceptibility of human renal epithelial cells suggests that other cell types may contribute to the pathogenesis of SARS-CoV infection. This finding could explain the isolation of SARS-CoV from kidney tissue of a SARS patient (18) and the high incidence of acute renal failure requiring continuous dialysis in a cohort of critically ill SARS patients in Singapore (22).
Although envelope glycoproteins of coronaviruses have not been shown previously to pseudotype unrelated viruses, such as retroviral or lentiviral vectors, the findings here suggest that the S glycoprotein of the SARS-CoV is fusion competent and able to function in viral entry through a pH-dependent mechanism. Immune antisera from mice or convalescent patients can inactivate this function of S, suggesting that this approach may be used to screen for neutralizing antibodies without the use of live, infectious virus. It is noteworthy that such antibody intervention would be best able to contain viral infection, with concomitant generation of cell-mediated immunity. Previous studies using animal coronaviruses have suggested that antibody responses, in the absence of cell-mediated immunity, either temporarily contain (15) or in some cases may exacerbate (8, 31) infection, though there is no evidence to date that antibody-dependent enhancement occurs in SARS-CoV infection. This study has shown that in vitro neutralization can be achieved with antibodies and provides encouragement for immune therapy.
The recognition in this study that DC can take up SARS-CoV and transfer it to susceptible target cells has important implications for the pathogenesis of SARS-CoV in vivo. These cells can serve as a reservoir that may provide more-continuous exposure to the virus and contribute to the persistence and chronicity of infection. They could also affect antigen presentation and antibody-dependent clearance of SARS-CoV. The finding that mDC can promote cell-mediated transfer of virus could theoretically pose a barrier to effective neutralization by antibodies, but the findings reported here suggest that mDC-associated virus remains susceptible to neutralization. This study thus provides insight into the mechanism of viral entry, reveals a previously unrecognized mode of dissemination, and demonstrates that the SARS-CoV S glycoprotein is vulnerable to immune interventions for the prevention and treatment of SARS.
Acknowledgments
We thank Ati Tislerics and Tina Suhana for assistance with preparation of the manuscript, Karen Stroud and Toni Garrison for help with graphics, William Bellini and colleagues at the CDC for providing genetic sequence information and convalescent-phase antisera from patients infected with SARS-CoV, and Brian Murphy and Anjeanette Roberts of the Laboratory of Infectious Diseases for assistance and discussions.
REFERENCES
- 1.Abrahamsen, T. G., C. S. Carter, E. J. Read, M. Rubin, H. G. Goetzman, E. F. Lizzio, Y. L. Lee, P. A. Pizzo, and T. Hoffman. 1991. Stimulatory effect of counterflow centrifugal elutriation in large-scale separation of peripheral blood monocytes can be reversed by storing the cells at 37 degrees C. J. Clin. Apheresis 6:48-53. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, C. P., F. Lasala, J. Carrillo, O. Muniz, A. L. Corbi, and R. Delgado. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76:6841-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 4.Burgard, M., M. J. Mayaux, S. Blanche, A. Ferroni, M. L. Guihard-Moscato, M. C. Allemon, N. Ciraru-Vigneron, G. Firtion, C. Floch, F. Guillot, et al. 1992. The use of viral culture and p24 antigen testing to diagnose human immunodeficiency virus infection in neonates. N. Engl. J. Med. 327:1192-1197. [DOI] [PubMed] [Google Scholar]
- 5.Cella, M., M. Salio, Y. Sakakibara, H. Langen, I. Julkunen, and A. Lanzavecchia. 1999. Maturation, activation and protection of dendritic cells induced by double-stranded RNA. J. Exp. Med. 189:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2003. Prevalence of IgG antibody to SARS-associated coronavirus in animal traders—Guangdong Province, China, 2003. Morb. Mortal. Wkly. Rep. 52:986-987. [PubMed] [Google Scholar]
- 7.Chinese SARS Molecular Epidemiology Consortium. 30 January 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 10.1126/science.1092002. [DOI] [PubMed]
- 8.Corapi, W. V., R. J. Darteil, J. C. Audonnet, and G. E. Chappuis. 1995. Localization of antigenic sites of the S glycoprotein of feline infectious peritonitis virus involved in neutralization and antibody-dependent enhancement. J. Virol. 69:2858-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Haan, C. A., M. Smeets, F. Vernooij, H. Vennema, and P. J. Rottier. 1999. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 73:7441-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 12.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 13.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 14.Halary, F., A. Amara, H. Lortat-Jacob, M. Messerle, T. Delaunay, C. Houles, F. Fieschi, F. Arenzana-Seisdedos, J. F. Moreau, and J. Dechanet-Merville. 2002. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 17:653-664. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, K. V. 2001. Coronaviruses, p. 1187-1203. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 16.Holmes, K. V., G. Dveksler, S. Gagneten, C. Yeager, S. H. Lin, N. Beauchemin, A. T. Look, R. Ashmun, and C. Dieffenbach. 1993. Coronavirus receptor specificity. Adv. Exp. Med. Biol. 342:261-266. [DOI] [PubMed] [Google Scholar]
- 17.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 18.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 19.Kwon, D. S., G. Gregorio, N. Bitton, W. A. Hendrickson, and D. R. Littman. 2002. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 16:135-144. [DOI] [PubMed] [Google Scholar]
- 20.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 21.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 22.Lew, T. W., T. K. Kwek, D. Tai, A. Earnest, S. Loo, K. Singh, K. M. Kwan, Y. Chan, C. F. Yim, S. L. Bek, A. C. Kor, W. S. Yap, Y. R. Chelliah, Y. C. Lai, and S. K. Goh. 2003. Acute respiratory distress syndrome in critically ill patients with severe acute respiratory syndrome. JAMA 290:374-380. [DOI] [PubMed] [Google Scholar]
- 23.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim, K. P., and D. X. Liu. 2001. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 276:17515-17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, G., G. Simmons, S. Pohlmann, F. Baribaud, H. Ni, G. J. Leslie, B. S. Haggarty, P. Bates, D. Weissman, J. A. Hoxie, and R. W. Doms. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J. Virol. 77:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 27.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald, D., L. Wu, S. M. Bohks, V. N. KewalRamani, D. Unutmaz, and T. J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295-1297. [DOI] [PubMed] [Google Scholar]
- 29.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nussbaum, O., A. Roop, and W. F. Anderson. 1993. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J. Virol. 67:7402-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perlman, S. 1998. Pathogenesis of coronavirus-induced infections. Review of pathological and immunological aspects. Adv. Exp. Med. Biol. 440:503-513. [PubMed] [Google Scholar]
- 32.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 33.Rest, J. S., and D. P. Mindell. 2003. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 3:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 35.Sanders, D. A. 2002. No false start for novel pseudotyped vectors. Curr. Opin. Biotechnol. 13:437-442. [DOI] [PubMed] [Google Scholar]
- 36.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein, B. S., S. D. Gowda, J. D. Lifson, R. C. Penhallow, K. G. Bensch, and E. G. Engleman. 1987. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell 49:659-668. [DOI] [PubMed] [Google Scholar]
- 38.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W.-J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tassaneetrithep, B., T. H. Burgess, A. Granelli-Piperno, C. Trumpfheller, J. Finke, W. Sun, M. A. Eller, K. Pattanapanyasat, S. Sarasombath, D. L. Birx, R. M. Steinman, S. Schlesinger, and M. A. Marovich. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1977-1985. [DOI] [PubMed] [Google Scholar]
- 41.White, J., K. Matlin, and A. Helenius. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood, K. V., J. R. de Wet, N. Dewji, and M. DeLuca. 1984. Synthesis of active firefly luciferase by in vitro translation of RNA obtained from adult lanterns. Biochem. Biophys. Res. Commun. 124:592-596. [DOI] [PubMed] [Google Scholar]
- 43.Wool-Lewis, R. J., and P. Bates. 1998. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 72:3155-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organization. 2003. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS). [Online.] http://www.who.int/csr/sars/en/WHOconsensus.pdf.
- 45.Yang, Z., R. Delgado, L. Xu, R. F. Todd, E. G. Nabel, A. Sanchez, and G. J. Nabel. 1998. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 279:1034-1037. [DOI] [PubMed] [Google Scholar]
- 46.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]