Abstract
Domestic dogs are unique from other animal models of cancer in that they generally experience spontaneous disease. In addition, most types of cancer observed in humans are found in dogs, suggesting that canines may be an informative system for the study of cancer genetics. Domestic dogs are divided into over 175 breeds, with members of each breed sharing significant phenotypes. The breed barrier enhances the utility of the model, especially for genetic studies where small numbers of genes are hypothesized to account for the breed cancer susceptibility. These facts, combined with recent advances in high-throughput sequencing technologies allows for an unrivaled ability to use pet dog populations to find often subtle mutations that promote cancer susceptibility and progression in dogs as a whole. The meticulous record keeping associated with dog breeding makes the model still more powerful, as it facilitates both association analysis and family-based linkage studies. Key to the success of these studies is their cooperative nature, with owners, scientists, veterinarians and breed clubs working together to avoid the cost and unpopularity of developing captive populations. In this article we explore these principals and advocate for colony-free, genetic studies that will enhance our ability to diagnose and treat cancer in dogs and humans alike.
Keywords: canine cancer; chip-seq, dog colony; exome; histiocytic sarcoma; osteosarcoma; RNA-seq; squamous cell carcinoma of the digit; transitional cell carcinoma; tumor sequencing; whole genome
Introduction
According to a 2009–2010 National Pet Owners Survey reported by the Pet Products Manufacturers Association, approximately 39% of American homes own at least one dog and 24% have two dogs (NumberofNet.com 2014). Thus, there are approximately 77.4 million pure-bred and mixed-breed dogs living in the United States (Texas Veterinary Cancer Registry 2012). Cancer is the leading cause of death in dogs over 10 years, with 50% of older dogs developing the disease and approximately one in four dogs eventually dying from it (Adams et al. 2010; Animal Cancer Foundation 2014; Bronson 1982; Dobson 2013; Vail and MacEwen 2000). Not surprisingly, dogs are diagnosed with many of the same cancers as humans (Khanna et al. 2006; Merlo et al. 2008), with an underlying presentation, clinical pathology, and treatment response mirroring that observed in humans (Cadieu and Ostrander 2007; Dorn 1976). This suggests that similar genetic mechanisms cause human and canine cancers and that genetic studies of canine disease may be a powerful way to advance our understanding of cancer in humans and companion animals alike (Cadieu and Ostrander 2007; Khanna et al. 2006; Ostrander 2012).
It is unclear whether cancer incidence in dogs is stabilized or increasing. Improved health care for pets now extends their lifespan, permitting the diagnosis of late-in-life diseases such as cancer. Also, as diagnostic tests improve in the veterinary community, dogs receive more accurate diagnoses than were available even a decade ago and, consequently, more effective therapy. In addition, owners are increasingly willing to pay for expensive diagnostic tests. Finally, as veterinary epidemiologists improve their ability to track canine cancer, scientists are increasingly able to predict which breeds are at an increased risk for each type of cancer. This allows diligent veterinarians to monitor individuals from at-risk breeds, leading to earlier diagnoses and more effective treatment.
In this article we explore our current understanding of canine cancer genetics. We argue that the days of maintaining dog colonies at veterinary schools, started with limited founders for the purpose of studying a single cancer type, are past. Rather, geneticists, veterinarians, and owners can work together to design highly accurate studies using pet dog populations (Karlsson and Lindblad-Toh 2008; Rowell et al. 2011; Shearin and Ostrander 2010). Typically, for any given cancer, the number of deleterious alleles segregating in a single dog breed is likely to be limited because dog fanciers use closed breeding programs to develop breeds with specific phenotypic traits (Karlsson and Lindblad-Toh 2008; Ostrander 2012; Ostrander and Kruglyak 2000; Parker et al. 2010). As a result, cancer genetic studies in pet dog populations presents a mechanism to circumvent the small families, outbred population structure, and locus heterogeneity that plague human cancer gene mapping (Karlsson and Lindblad-Toh 2008; Shearin and Ostrander 2010), while allowing scientists to avoid setting up breeding colonies at veterinary schools. These facts, combined with the ability to easily collect and sequence DNA and tissue-specific RNA from dogs underscores the notion that pet dogs are uniquely positioned to change our view of both human and canine cancer (Khanna et al. 2006; Ostrander 2012; Rowell et al. 2011).
At the heart of the proposition is the fact that by using multiple, recently developed genomic technologies, we can thoroughly scan any single dog's genome for variants as simple as a single nucleotide change or as complex as a gene family expansion. We can ascertain both allele-specific gene and noncoding RNA expression profiles. We can also develop somatic mutation profiles from individual tumors (Figure 1). Indeed, with the arrival of high-throughput sequencing (HTS), the first breed-specific whole genomes, together with catalogues of breed-specific variants, are now emerging in the scientific literature (Kim et al. 2012; Owczarek-Lipska et al. 2013), as is data from wild canids. These data will facilitate the detection of both rare and common disease alleles, and the resulting databases will aid clinical researchers in advancing their knowledge regarding cancer, as well as other complex genetic diseases.
Figure 1.
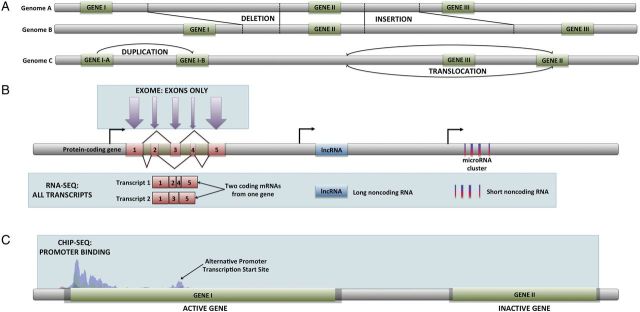
Graphic representation of the most commonly used HTS technologies. (A) Whole genome sequencing allows the detection of changes in genome structure such as insertion/deletion events, gene duplications, and translocations. (B) Whole exome sequencing produces only sequences within exons from genomic DNA. RNA-Seq generates the complete sequences of every RNA transcript, including alternative transcripts, and noncoding RNAs. (C) ChIP-Seq is able to identify the DNA-binding sites for specific transcription factors, as well as any other protein capable of binding to DNA.
Cancer Registries
The Surveillance, Epidemiology and End Results registry of the National Cancer Institute collects information on incidence, prevalence, and survival from a set of predetermined geographic regions of the United States, allowing researchers to assemble statistics regarding cancer mortality and trends for the entire country. Unfortunately, no such organization exists in veterinary science. However, individual registries have emerged in recent years, such as the National Veterinary Cancer Registry (http://nationalveterinarycancerregistry.org/about-nvcr/naturally-occuring-models), which, among other tasks, collects data for a national registry while developing a network of veterinary oncologists. The National Veterinary Cancer Registry is a strong advocate of pets as naturally occurring animal models for cancer studies. As such, they work to connect researchers with oncologists who share interests and to inform owners regarding ongoing clinical trials. Other similar organizations exist, including the Texas Veterinary Cancer Registry and the Veterinary Cancer Society. Although useful, such organizations provide little publically available data on cancer trends, breed-specific incidence of cancers, and treatment response. In addition, they often lack the resources to link breeders of at-risk populations with geneticists or epidemiologists studying a particular cancer type. Thus, a need for this type of research exists and cannot be done by colonies.
The best resources for establishing breed-specific trends in cancer research are highly targeted academic studies, some of which are discussed below, and selected veterinary school studies. The first such set of studies, done in the late 1960s, focused exclusively on Alameda and Contra Costa Counties in California (Dorn 1976; Dorn et al. 1968) and then on data from veterinary schools (Priester and Mantel 1971). Subsequent reviews were among the first to collate breed excesses of several of types of cancer (Madewell 1981), which had previously been reported as just single cancers or as case–case studies. These were also the first studies to suggest that spontaneous dog tumors could be informative for learning about human cancers (Schneider 1970; Schneider et al. 1968).
In recent years, American studies have been dwarfed by those from Europe, which often use registry data provided by pet insurance companies. For instance, the Animal Tumor Registry of Genoa, Italy, established in 1985, reported that cancer incidence is threefold higher in female dogs then male dogs (Merlo et al. 2008), with mammary cancer the most frequently diagnosed malignancy (incidence-rate [IR] = 191.8; 95% confidence interval [CI] = 182.2–201.4), followed closely by non-Hodgkin's lymphoma (IR = 22.9; 95% CI = 19.7–26.5) in bitches, and non-Hodgkin's lymphoma (IR = 19.9; 95% CI = 17.4–22.7) and skin cancer (IR = 19.1; 95% CI = 16.6–21.8) in male dogs. Operating since 1990, the Norwegian Cancer Registry also reports that mammary tumors are the most common (Arnesen et al. 1995). As expected, risk increased with age, with 10 years being the critical cut-off (Merlo et al. 2008). Breeds at highest risk include Leonbergers, Irish wolfhounds, Bernese mountain dogs, and great Danes, among others (Jitpean et al. 2012; Schneider 1970).
Although all studies seem to agree on at-risk breeds, there is a lack of agreement regarding specific rankings. The commonly cited study of UK Kennel Club–recognized breeds reports that overall cancer incidence is highest in the Irish water spaniel, followed by the flat-coated retriever, Hungarian wirehaired vizsla, Bernese mountain dog, Rottweiler, Italian spinone, Leonberger, Staffordshire bull terrier, Welsh terrier, and giant schnauzer (Adams et al. 2010). In Sweden, the list is slightly different, with the Bernese mountain dog topping the chart, followed by the Irish wolfhound, flat-coated retriever, boxer, and Saint Bernard (Bonnett et al. 1997).
Jane Dobson, a longtime expert in the field, reports that cutaneous histiocytoma is the most common canine tumor type reported overall in the United Kingdom, followed by lipoma, adenoma, soft tissue sarcomas, mast cell tumor, and lymphomas (Dobson 2013). This list obviously includes both malignant and benign tumors. A study from the Danish Veterinary Cancer Registry reported that the frequency of benign and malignant tumors is similar in their country, with the most commonly reported malignant neoplasms being adenocarcinomas (21%), followed by mast cell tumors (19%) and lymphomas (17%) (Brønden et al. 2010).
Breed-Specific Cancers
When multiple dog breeds are at an elevated risk for the same type of cancer, it is possible, even probable, that the breeds share an underlying genetic predisposition (e.g., all at-risk breeds segregate the cancer because they shared a common founder during breed development) (Goldstein et al. 2006; Karlsson et al. 2007; Ostrander 2012; Parker et al. 2007) (Figure 2). To investigate specific cancers, DNA from blood or cheek swab samples are collected from cases and aged-unaffected controls of one or more related at-risk breeds. The genomes of the two populations are then compared using arrays of single nucleotide polymorphisms (SNPs). The practice of comparing whole genomes of several dogs simultaneously as a predetermined set of SNPs is called a genome-wide association study (GWAS), and it remains a powerful tool for finding loci associated with any disease. The most commonly used commercially available SNP chips contain approximately 170,000 SNPs, allowing interrogation of every chromosome, and costing approximately $250 per individual to perform. A variety of statistical tools can be applied to find loci that distinguish cases from controls. This is followed by fine-mapping experiments that narrow the region of interest and DNA sequencing to identify the precise disease-associated mutation.
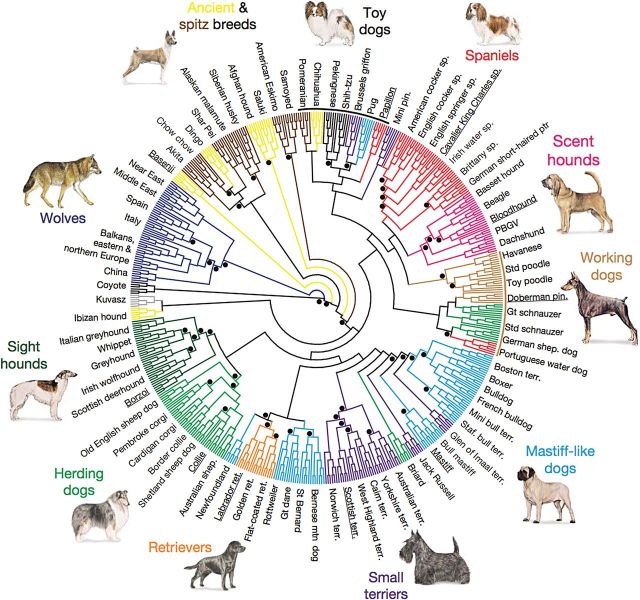
Figure 2.
Cladogram from (vonHoldt et al. 2010) depicting the structure of domestic dog breeds as ascertained from 48,036 autosomal SNP loci. Relationship between breeds was determined by haplotype-sharing for 10-SNP windows with at least 6 members representing each breed.
Increasingly, because of greater efficiency, scientists are circumventing GWASs and proceeding directly to whole-genome sequencing, which requires no fine-mapping or follow-up sequencing. This method produces the entire repertoire of variation within an individual's genome at once, including SNPs, structural variants such as insertion–deletion events, translocations, and copy number changes (Figure 1A). One Illumina HiSeq 2500 machine, which is typically used for such experiments, is able to generate 600 billion sites of genomic sequence in a matter of hours (Eisenstein 2012). The data produced is sufficient to scan the 2.5-billion base-pair genome of 10 dogs 10 times. Although the cost of DNA sequencing is about an order of magnitude more per individual than that of one SNP chip, the price of these methods continues to drop, with the expectation that sub-$1000 genomes will soon be within reach. These data are increasingly used also for the examination of a variety of morphologic, behavioral, or disease traits. In the interim, as scientists wait for the price to drop, some are focusing on studying only the 1% of the genome that codes for proteins and generating whole-exome sequences at a very low cost (Figure 1A). Our lab has tended to avoid such an approach because disease variants are not exclusive to coding genes and may reside within noncoding regulatory regions.
Regardless of how the data is collected, such experiments require a number of a priori considerations (Karlsson and Lindblad-Toh 2008; Karlsson et al. 2007; Shearin and Ostrander 2010). For instance, the number of cases and controls to be collected should be carefully considered. Ideally, the more cases assayed for a GWAS, the better. Obtaining unrelated samples from at least the grandparent level for both cases and controls is important to minimize false positives (Shearin and Ostrander 2010). Data from breeders can be extremely useful at this point. Controls are preferentially dogs from the same or a closely related breed that have passed the median age at which the disease usually presents (Shearin and Ostrander 2010). However, the older the control, the better the choice. Although it is likely that any set of controls will contain at least a few individuals with the risk allele, that number will be dwarfed by the number who lack it. Data are readily available regarding how breeds relate one to another and should be used in experimental design (vonHoldt et al. 2010).
Lindblad-Toh has done an elegant series of calculations to aid scientists in optimizing the number of samples needed for their GWASs (Lindblad-Toh et al. 2005). Because the linkage disequilibrium observed in dog breeds can be extensive, the number of SNPs needed for a canine GWAS is much smaller than that required for a comparable human study (Karlsson and Lindblad-Toh 2008; Lindblad-Toh et al. 2005; Sutter et al. 2004). By taking advantage of breed structure and selecting maximally unrelated dogs, the required number of samples can also be minimized. Thus, for a hypothetic trait with dominant inheritance, high penetrance, and no phenocopies, less then 20,000 SNPs would be needed to reach a confidence level of greater than 99% using data from 100 cases and 100 controls. Of course, no such ideal trait exists, and as genetic complexity increases, the number of samples must increase as well (Lindblad-Toh et al. 2005). Still, many complex disorders in dogs have been mapped using modest numbers of samples (e.g., Ahonen et al. 2013; Forman et al. 2013; Frischknecht et al. 2013; Pfahler and Distl 2012; Yokoyama et al. 2012), including cancer (Karyadi et al. 2013; Shearin et al. 2012).
For many of the above principles, squamous cell carcinoma of the digit is a particularly demonstrative case (Karyadi et al. 2013). Squamous cell carcinoma of the digit, the most frequent cutaneous squamous cell carcinoma in dogs, is found in giant schnauzers (odds ratio [OR] = 22.7), Gordon setters (OR = 11.1), Briards (OR = 10.4), the Kerry blue terrier (OR = 7.7), and black standard poodles (OR = 5.9; 95% CI = 4.8–7.2) (Goldschmidt and Shoufer 1998). Our GWAS, initially based on 31 standard poodle cases and 34 controls, identified a locus on canine chromosome 15 with a high level of significance that spanned a little more than 1 million base pairs. Additional mapping using 85 standard poodle cases as well as a small number of Gordon setters and Briards resolved the region to 24 kilobases. Sequencing revealed that all affected dogs, regardless of breed, carry the same founder mutation, a copy number variant that likely affects expression of KITL (Karyadi et al. 2013). This is an example of a regulatory mutation playing a role in disease susceptibility and highlights how data from a small number of dogs can contribute to our understanding of a disease that is important in canine and human health (Chung and Chanock 2011).
Breed-Specific Cancer Susceptibility
There are many other examples of breeds with either a predisposition for, or apparent protection from particular cancers. For instance, when considering brain and central nervous system cancers, a 2013 study showed that the boxer, golden retriever, French bulldog, and Boston and rat terriers were at a significantly increased risk, whereas the cocker spaniel and Doberman pinscher were at a low risk (Song et al. 2013). Interestingly, meningiomas were seen more frequently in dolichocephalic breeds (those with elongated muzzles), whereas glial tumors were observed more in brachycephalic breeds (short, upturned muzzles).
One frequently discussed breed-enriched cancer is osteosarcoma, which predominates in the long-limbed breeds such as the Irish wolfhound (Urfer et al. 2007), Scottish deerhound (Phillips et al. 2007) and great Dane, as well as other large breeds (Dobson 2013; Phillips et al. 2010). It has a standardized incidence rate of about 52 per 100,000. The dog is generally considered a good model for human osteosarcoma (Angstadt et al. 2011; Mueller et al. 2007; Rankin et al. 2012; Rowell et al. 2011; Withrow and Wilkins 2010). Particularly compelling are reports that IL-8 and SLC1A3, which are frequently overexpressed in canine osteosarcoma, are associated with poor outcome in human osteosarcoma (Paoloni et al. 2009). This knowledge will expand in the near future because researchers have used an HTS technique called RNA-Seq to detect the misexpression of these and many other genes between human osteosarcoma tumors and normal bone tissue (Märtson et al. 2013).
Histiocytic Sarcoma
Not every case in which multiple cancers are found in the same breed is there is a common founder. Rather, in some cases, the same rare cancer is seems to have arisen independently in distinct breeds. Histiocytic cancer covers a broad range of clinical presentations, from benign cutaneous histiocytoma to highly aggressive histiocytic sarcoma (HS) (Affolter and Moore 2000, 2002), which is an aggressive and lethal disorder of dendritic cell origin. Localized HS most commonly develops in the skin or subcutis of an extremity, although it can be found in other organs. Disseminated HS is a multisystem disease with tumors appearing in numerous organs simultaneously.
Both flat-coated retrievers (FCR) and Bernese mountain dogs (BMDs) are at high risk for HS, which affects approximately 20% of FCRs and 25% of BMDs (Abadie et al. 2009; Affolter and Moore 2002; Moore et al. 2006; Proschowsky et al. 2003). BMDs tend to present with a disseminated or visceral form of the disease, generally around age 6 to 7 years, with tumors appearing in the spleen, liver, and lungs (Abadie et al. 2009; Affolter and Moore 2002; Moore et al. 2006; Proschowsky et al. 2003). By comparison, approximately two-thirds of FCRs present with a mass in a joint and/or the surrounding muscle. Earlier literature referred to these as localized and disseminated malignant histiocytosis, respectively (Affolter and Moore 2002). However, they are now more correctly referred to as periarticular and visceral forms (Boerkamp et al. 2013).
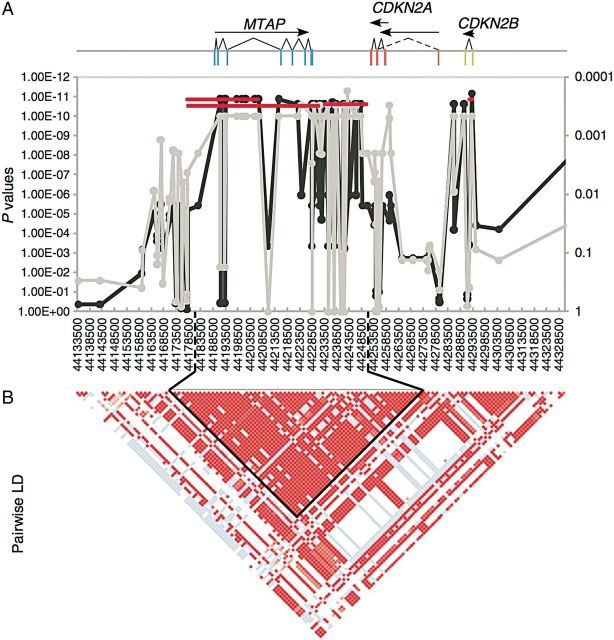
In 2012, we reported the first GWAS results in the BMD, showing an association between the MTAP-CDNK2A locus and increased susceptibility to HS (Figure 3). This is one of what is likely to be a growing number of cases where canine studies have preceded human studies. Interestingly, genetic studies done using both microsatellites and SNPs suggest no recent common ancestor to the FCR and BMD (Parker et al. 2004; vonHoldt et al. 2010). Thus, it is not surprising that their clinical presentation is different and that studies of copy number variation in BMD and FCR tumors highlights differences between breeds as well (Hedan et al. 2011). Ongoing studies include both GWASs as well as direct sequencing of affected and unaffected dogs from at-risk breeds.
Figure 3.
The region encompassing MTAP and CDK genes shown to be highly associated with histiocytic sarcoma from (Shearin et al. 2012). A 195-kb region on CFA11 is shown with X-axes for all plots listing SNPs in proximal-distal order. (A) 3 genes are shown with exons indicated as rectangles, introns as lines, and transcripts as arrows. Fisher exact association of HS with allele frequency is plotted along the Y-axis. The light gray line denotes association in a discovery cohort of 24 cases and 20 controls with P values on the right Y-axis. The black line indicates association in the complete cohort after imputation of genotypes (P values on left Y-axis). Horizontal lines depict haplotype association (P values on left Y-axis). (B) Pairwise LD plot with solid blocks indicating D′ = 1 and LOD score of 2. The black outline shows a haplotype block with 28 of 30 equally associated SNPs in this region.
Nearly all human cancer studies are being expanded using HTS in lieu of GWAS-based SNP arrays. HTS provides the ability to detect all variants within the genome rather than focusing on predetermined loci and, as described, obviates the need for later fine-mapping studies. Also, as the importance of rare variation becomes increasingly evident, it can be expected that more and more studies will use HTS rather then GWAS to find the mutation of origin (Cirulli and Goldstein 2010). Finally, the ability to characterize both common and rare structural variation is essential for the study of a complex disease such as cancer, where a considerable component of susceptibility may result from insertion–deletion events, copy number variation (Demichelis et al. 2012) and retrotransposion events (Lee et al. 2012).
Gene expression studies have also been illuminating in the case of HS, where fresh-frozen tissues from FCRs with either periarticular or visceral disease reveal changes in genes from 24 distinct pathways that the authors argue are involved in tumor development. Interestingly, most of the implicated pathways were important in either DNA repair or replication. Nine genes in particular, not previously implicated in HS, including ITGAD, SpiC, VCAM1, PPBP, and ENPEP, were observed to be downregulated in tumors, whereas four others were upregulated (Boerkamp et al. 2013).
One problem with this type of data is that it is based on an analysis of known cancer genes and is inherently less comprehensive than the exhaustive technique of RNA-Seq, which requires no a priori information about the transcriptome, and thus data are acquired on every transcript that is produced within the sample (Wang et al. 2009). Because it is inherently unbiased, RNA-Seq also permits the discovery of new genes, changes in exon usage based on cell type (Trapnell 2010), noncoding transcripts including long noncoding RNAs (lincRNAs) (Pasquinelli 2012), and microRNAs (Figure 1B) (Ryan et al. 2010), and, of obvious interest to cancer, gene fusions (Maher et al. 2009). To date, no GWAS or RNA-Seq study of FCR HS has been published. But when they are, it will be interesting to see if the same loci or an overlapping set of loci contributes to at least the visceral form of the disease that is observed more commonly in the BMD.
Bladder Cancer
One final example of a cancer with extraordinary breed specificity is that of transitional carcinoma of the bladder (TCC), which accounts for 20,000 to 30,000 new cases of dog cancer each year (Knapp et al. 2000). TCC is an especially challenging cancer to treat (Boria et al. 2005; Knapp 2001; Mohammed et al. 2003; Mohammed et al. 2004) because the typical trigonal location precludes surgical excision and complete cystectomy is not a viable option in pet dogs. Tumor growth within the bladder, urethra, and ureters often leads to urinary obstruction, and there is typically spread to distant organs. Chemotherapy is only partially effective, and as a result, most dogs with TCC ultimately die from the disease (Knapp et al. 2000; Mutsaers et al. 2003).
TCC occurs at the highest frequency in Scottish terriers, (18–20-fold relative risk compared with mixed breed dogs), followed by the West Highland white terrier (fivefold increase), and Shetland sheep dogs (4.5-fold increase) (Knapp 2001; Knapp 2007; Mutsaers et al. 2003). Other terrier breeds, collies, and beagles are also at an increased risk. TCC appears to arise from a combination of genetic and environmental factors, making it a particularly poor choice for colony-based studies (Glickman et al. 2004; Knapp 2001).
TCC is one cancer where the incorporation of HTS technologies is particularly advantageous. Using both whole-genome HTS and the lower-cost whole-exome sequencing, numerous high-frequency genome alterations have been identified in human TCC tumors (Guo et al. 2013). However, even at the whole-genome/exome level, this study was unable to detect gene fusion transcripts without additional RNA-Seq. Although not yet observed in human TCC studies, the potent combination of genome-level sequencing with RNA-Seq has been shown to yield unrivaled levels of information, including how tumor-dependent structural variation is linked to the allele-specific changes in expression (Tuch et al. 2010).
Cancer and Hormonal Pathways
When considering TCC, we made the point that environment can play a key role in the onset of some cancers, hence making it a poor cancer to study in a breeding colony. For other cancers, factors such as hormone levels are equally important and equally hard to study in the colony setting. Consider, for example, mammary cancer. It is well established that in addition to breed specificity, spaying and neutering, which is practiced on most American dogs, but only approximately 50% of European dogs (Trevejo et al. 2011), affects cancer risk. For instance, a study of golden retrievers done by University of California at Davis investigators revealed that nearly 10% of male dogs neutered before 1 year of age were eventually diagnosed with lymphosarcoma (Torres de la Riva et al. 2013). This is three times the rate observed by the same study for intact male dogs. Also significant was the fact that 8% of female dogs who were spayed after 1 year of age developed hemangiosarcoma (Torres de la Riva et al. 2013), four times the rate observed in intact female dogs and those spayed before age one.
This is true across breeds; other studies suggest an increase of approximately twofold to fivefold for spayed versus intact female dogs for hemangiosarcoma (Prymak et al. 1985). Even osteosarcoma rates increase with neutering (Cooley et al. 2002; Ru et al. 1998). Hormone-responsive cancers offer a unique avenue to leverage the combinatorial power of two comparative transcriptome-oriented techniques: RNA-Seq and chromatin immunoprecipitation sequencing (Figure 1C). Chromatin immunoprecipitation sequencing identifies modifications that modulate transcriptional activation given varying conditions, including presence and quantity of hormonal inducers. Coupling these methods has been used to great effect in identifying the activation of hormonal binding sites in responsive cancers and the resulting changes to the total transcriptional landscape (Prokesch and Lazar 2011; Ross-Innes et al. 2012).
Prostate Cancer and Dogs?
As a final consideration, it is important to keep in mind that for some rare cancers the dog is not the perfect genetic model but can nevertheless provide useful information. Prostate cancer is extremely rare in dogs, but curiously it occurs more frequently in neutered male dogs (Bryan et al. 2007). Neither GWASs nor family-based linkage studies are likely to be informative. However, aside from humans, the dog is the only animal to present with spontaneous prostate cancer, with clinical features, including late age at onset, and metastatic patterns similar to what is observed in humans (Cornell et al. 2000; Waters and Bostwick 1997a, 1997b).
Interestingly, polysomy of canine chromosome 13 has been observed in canine prostate tumors (Reimann-Berg et al. 2011; Winkler et al. 2006). This region is syntenic with human chromosomes 4q and 8q, the latter of which has been suggested as containing multiple prostate cancer loci (Barros-Silva et al. 2011; El Gammal et al. 2010). Recently, PCAT-1, a novel lincRNA believed to influence prostate cancer progression, was discovered on human chromosome 8q using RNA-Seq (Prensner et al. 2011). It will be interesting to see whether studies of canine tumors reveal similar or unique findings. In any case, the similarity in architecture and presentation makes the dog a unique model for studies of prostate tumor growth.
Summary
Technology moves rapidly and inexorably forward. Our need to improve our own health and the health of our family members, including our dogs, remains. For the past several years there has existed in public health the concept of One Health, which is a guiding principle acknowledging that the health of humans, animals, and the environment is highly intertwined (http://www.cdc.gov/onehealth/index.html). Although often focused on infections disease, the concept can be expanded to guide the way in which we conduct canine research in the future.
Gone are the days when captive colonies of dogs are easily justifiable for genetic studies. The rapid and striking advances in the quality, volume, and specificity of genomic information brought about by nascent technologies has ushered in an undeniable impetus for researchers to shift to other paradigms when tackling questions of animal health and their concomitant parallels to human health. Although we can debate the utility of such resources for other areas of study, for studies aimed at finding genetic loci that cause or protect from disease, the work can, as we have demonstrated here, be accomplished through collaboration and by embracing genomic technologies.
For humans, an era of personalized medicine, which uses genomic data to determine disease risk, select specific and effective treatment regimens, and predict relapse is rapidly becoming state of the art, particularly in the field of cancer (Chute and Kohane 2013). Although the scope and depth of available canine genomic data pale compared with what is currently collected on humans, we predict that directed or personalized treatments will soon become a reality for our pets (Khanna and Gordon 2009; Rankin et al. 2012; Rowell et al. 2011; Shearin and Ostrander 2010). Hence we seek to set forth principles that will guide researchers to responsibly work on improving the health and well-being of humans and animals, advocating that the complete compliment of modern genomic technologies be a part of every genetic researcher's tool kit.
Acknowledgments
We gratefully acknowledge the AKC Canine Health Foundation and the Intramural Program of the National Human Genome Research Institute.
References
- Abadie J, Hedan B, Cadieu E, De Brito C, Devauchelle P, Bourgain C, Parker HG, Vaysse A, Margaritte-Jeannin P, Galibert F, Ostrander EA, André C. Epidemiology, pathology, and genetics of histiocytic sarcoma in the Bernese mountain dog breed. J Hered. 2009;100:S19–S27. doi: 10.1093/jhered/esp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams VJ, Evans KM, Sampson J, Wood JLN. Methods and mortality results of a health survey of purebred dogs in the UK. J Small Anim Pract. 2010;51:512–524. doi: 10.1111/j.1748-5827.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- Affolter VK, Moore PF. Canine cutaneous and systemic histiocytosis: Reactive histiocytosis of dermal dendritic cells. Am J Dermatopathol. 2000;22:40–48. doi: 10.1097/00000372-200002000-00009. [DOI] [PubMed] [Google Scholar]
- Affolter VK, Moore PF. Localized and disseminated histiocytic sarcoma of dendritic cell origin in dogs. Vet Pathol. 2002;39:74–83. doi: 10.1354/vp.39-1-74. [DOI] [PubMed] [Google Scholar]
- Ahonen SJ, Pietilä E, Mellersh CS, Tiira K, Hansen L, Johnson GS, Lohi H. Genome-wide association study identifies a novel canine glaucoma locus. PLoS One. 2013;8:e70903. doi: 10.1371/journal.pone.0070903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angstadt AY, Motsinger-Reif A, Thomas R, Kisseberth WC, Guillermo Couto C, Duval DL, Nielsen DM, Modiano JF, Breen M. Characterization of canine osteosarcoma by array comparative genomic hybridization and RT-qPCR: Signatures of genomic imbalance in canine osteosarcoma parallel the human counterpart. Genes Chromosomes Cancer. 2011;50:859–874. doi: 10.1002/gcc.20908. [DOI] [PubMed] [Google Scholar]
- Animal Cancer Foundation. Home page. 2014 Available online (http://www.acfoundation.org. ), accessed on April 1, 2014. [Google Scholar]
- Arnesen K, Gamlem H, Glattre E, Grøndalen J, Moe L, Nordstoga K. Registraion of canine cancer. Tidsskr Nor Laegeforen. 1995;115:714–717. [PubMed] [Google Scholar]
- Barros-Silva JD, Ribeiro FR, Rodrigues A, Cruz R, Martins AT, Jerónimo C, Henrique R, Teixeira MR. Relative 8q gain predicts disease-specific survival irrespective of the TMPRSS2-ERG fusion status in diagnostic biopsies of prostate cancer. Genes Chromosomes Cancer. 2011;50:662–671. doi: 10.1002/gcc.20888. [DOI] [PubMed] [Google Scholar]
- Boerkamp KM, van der Kooij M, van Steenbeek FG, van Wolferen ME, Groot Koerkamp MJ, van Leenen D, Grinwis GC, Penning LC, Wiemer EA, Rutteman GR. Gene expression profiling of histiocytic sarcomas in a canine model: The predisposed flatcoated retriever dog. PLoS ONE. 2013;8:e71094. doi: 10.1371/journal.pone.0071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnett BN, Egenvall A, Olson P, Hedhammar A. Mortality in insured Swedish dogs: Rates and causes of death in various breeds, Vet Rec. 1997;141:S40–S44. doi: 10.1136/vr.141.2.40. [DOI] [PubMed] [Google Scholar]
- Boria PA, Glickman NW, Schmidt BR, Widmer WR, Mutsaers AJ, Adams LG, Snyder PW, DiBernardi L, de Gortari AE, Bonney PL, Knapp DW. Carboplatin and piroxicam in 31 dogs with transitional cell carcinoma of the urinary bladder. Vet Comp Oncol. 2005;3:73–80. doi: 10.1111/j.1476-5810.2005.00070.x. [DOI] [PubMed] [Google Scholar]
- Brønden LB, Nielsen SS, Toft N, Kristensen AT. Data from the Danish Veterinary Cancer Registry on the occurrence and distribution of neoplasms in dogs in Denmark. Vet Rec. 2010;166:586–590. doi: 10.1136/vr.b4808. [DOI] [PubMed] [Google Scholar]
- Bronson RT. Variation in age at death of dogs of different sexes and breeds. Am J Vet Res. 1982;43:2057–2059. [PubMed] [Google Scholar]
- Bryan JN, Keeler MR, Henry CJ, Bryan ME, Hahn AW, Caldwell CW. A population study of neutering status as a risk factor for canine prostate cancer. Prostate. 2007;67:1174–1181. doi: 10.1002/pros.20590. [DOI] [PubMed] [Google Scholar]
- Cadieu E, Ostrander EA. Canine genetics offers new mechanisms for the study of human cancer. Cancer Epidemiol Biomarker Prev. 2007;16:2181–2183. doi: 10.1158/1055-9965.EPI-07-2667. [DOI] [PubMed] [Google Scholar]
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Human Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- Chute CG, Kohane IS. Genomic medicine, health information technology, and patient care. JAMA. 2013;309:1467–1468. doi: 10.1001/jama.2013.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- Cooley DM, Beranek BC, Schlittler DL, Glickman MW, Glickman LT, Waters DL. Endogenous gonadal hormone exposure and bone sarcoma risk. Cancer Epidemiol Biomarkers Prev 11:1434. 2002 [PubMed] [Google Scholar]
- Cornell KK, Bostwick DG, Cooley DM, Hall G, Harvey HJ, Hendrick MJ, Pauli BU, Render JA, Stoica G, Sweet DC, Waters DJ. Clinical and pathological aspects of spontaneous canine prostate carcinoma: A retrospective analysis of 76 cases. Prostate. 2000;45:173–183. doi: 10.1002/1097-0045(20001001)45:2<173::aid-pros12>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Demichelis F, Setlur SR, Banerjee S, Chakravarty D, Chen JYH, Chen CX, Huang J, Beltran H, Oldridge DA, Kitabayashi N. Identification of functionally active, low frequency copy number variants at 15q21. 3 and 12q21. 31 associated with prostate cancer risk . Proc Natl Acad Sci U S A. 2012;109:6686–6691. doi: 10.1073/pnas.1117405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson JM. Breed-predispositions to cancer in pedigree dogs. ISRN Vet Sci 2013:doi:10.1155/2013/941275. 2013 doi: 10.1155/2013/941275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn CR. Epidemiology of canine and feline tumors. Comp Cont Educ Pract Vet. 1976;12:307–312. [Google Scholar]
- Dorn CR, Taylor DO, Frye FL, Hibbard HH. Survey of animal neoplasms in Alameda and Contra Costa Counties, California. I. Methodology and description of cases. J Natl Cancer Inst. 1968;40:295–305. [PubMed] [Google Scholar]
- Eisenstein M. The battle for sequencing supremacy. Nat Biotechnol. 2012;30:1023–1026. doi: 10.1038/nbt.2412. [DOI] [PubMed] [Google Scholar]
- El Gammal AT, Brüchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Köllermann J, Sauter G, Simon R, Wilczak W, Schwarz J, Bokemeyer C, Brümmendorf TH, Izbicki JR, Yekebas E, Fisch M, Huland H, Graefen M, Schlomm T. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- Forman OP, De Risio L, Mellersh CS. Missense mutation in CAPN1 is associated with spinocerebellar ataxia in the Parson Russell Terrier dog breed. PLOS One. 2013;8:e64627. doi: 10.1371/journal.pone.0064627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht M, Niehof-Oellers H, Jagannathan V, Owczarek-Lipska M, Drögemüller C, Dietschi E, Dolf G, Tellhelm B, Lang J, Tiira K, Lohi H, Leeb T. A COL11A2 mutation in Labrador retrievers with mild disproportionate dwarfism. PLoS One. 2013;8:e60149. doi: 10.1371/journal.pone.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman LT, Raghavan M, Knapp DW, Bonney PL, Dawson MH. Herbicide exposure and the risk of transitional cell carcinoma of the urinary bladder in Scottish terriers. J Am Vet Med Assoc. 2004;224:1290–1297. doi: 10.2460/javma.2004.224.1290. [DOI] [PubMed] [Google Scholar]
- Goldschmidt MH, Shofer FS. 1998. Skin tumors of the dog and cat. Woburn, MA: Butterworth-Heinemann. [Google Scholar]
- Goldstein O, Zangerl B, Pearce-Kelling S, Sidjanin D, Kijas J, Felix J, Acland G, Aguirre G. Linkage disequilibrium mapping in domestic dog breeds narrows the progressive rod-cone degeneration interval and identifies ancestral disease-transmitting chromosome. Genomics. 2006;88:541–550. doi: 10.1016/j.ygeno.2006.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, Dean M, Huang Y, Jia W, Zhou Q, Tang A, Yang Z, Li X, Song P, Zhao X, Ye R, Zhang S, Lin Z, Qi M, Wan S, Xie L, Fan F, Nickerson ML, Zou X, Hu X, Xing L, Lv Z, Mei H, Gao S, Liang C, Gao Z, Lu J, Yu Y, Liu C, Li L, Fang X, Jiang Z, Yang J, Li C, Zhao X, Chen J, Zhang F, Lai Y, Lin Z, Zhou F, Chen H, Chan HC, Tsang S, Theodorescu D, Li Y, Zhang X, Wang J, Yang H, Gui Y, Wang J, Cai Z. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genett. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedan B, Thomas R, Motsinger-Reif A, Abadie J, Andre C, Cullen J, Breen M. Molecular cytogenetic characterization of canine histiocytic sarcoma: A spontaneous model for human histiocytic cancer identifies deletion of tumor suppressor genes and highlights influence of genetic background on tumor behavior. BMC Cancer. 2011;11:201. doi: 10.1186/1471-2407-11-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitpean S, Hagman R, Ström Holst B, Höglund OV, Pettersson A, Egenvall A. Breed variations in the incidence of pyometra and mammary tumours in Swedish dogs. Reprod Domest Anim. 2012;6:347–350. doi: 10.1111/rda.12103. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NHC, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, Comstock KE, Keller ET, Mesirov JP, von Euler H, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Lindblad-Toh K. Leader of the pack: Gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9:713–724. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- Karyadi D, Karlins E, Decker B, vonHoldt BM, Carpintero-Ramirez G, Parker HG, Wayne RK, Ostrander EA. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS Genet. 2013;9:e1003409. doi: 10.1371/journal.pgen.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna C, Gordon I. Catching cancer by the tail: New perspectives on the use of kinase inhibitors. Clin Cancer Res. 2009;15:3645–3647. doi: 10.1158/1078-0432.CCR-09-0132. [DOI] [PubMed] [Google Scholar]
- Khanna C, Lindblad-Toh K, Vail D, London C, Bergman P, Barber L, Breen M, Kitchell B, McNeil E, Modiano JF, Modiano JF, Niemi S, Comstock KE, Ostrander E, Westmoreland S, Withrow S. The dog as a cancer model. Nat Biotechnol. 2006;24:1065–1066. doi: 10.1038/nbt0906-1065b. [DOI] [PubMed] [Google Scholar]
- Kim RN, Kim D-S, Choi S-H, Yoon B-H, Kang A, Nam S-H, Kim D-W, Kim J-J, Ha J-H, Toyoda A, Fujiyama A, Kim A, Kim MY, Park KH, Lee KS, Park HS. Genome analysis of the domestic dog (Korean Jindo) by massively parallel sequencing. DNA Res. 2012;19:275–287. doi: 10.1093/dnares/dss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp D, Glickman N, DeNicola D, Bonney P, Lin T, Glickman L. Naturally-occurring canine transitional cell carcinoma of the urinary bladder: A relevant model of human invasive bladder cancer. Urol Oncol. 2000;5:47–59. doi: 10.1016/s1078-1439(99)00006-x. [DOI] [PubMed] [Google Scholar]
- Knapp DW. Tumors of the urinary system. In: Withrow SJ, MacEwen EG, editors. Small Animal Clinical Oncology. 3rd ed. Philadelphia PA: WB Saunders; 2001. 490–499. [Google Scholar]
- Knapp DW. Tumors of the urinary system. In: Withrow SJ, Vail DM, editors. Withrow and MacEwen's Small Animal Clinical Oncology. 4th ed. St. Louis MO: Saunders; 2007. 655–657. [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, deJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin C, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli K, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Madewell BR. Neoplasms in dometic animals: A review of experimental and spontaneous carcinogenesis. Yale J Biol Med. 1981;54:111–125. [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Märtson A, Kõks S, Reimann E, Prans E, Erm T, Maasalu K. Transcriptome analysis of osteosarcoma identifies suppression of wnt pathway and up-regulation of adiponectin as potential biomarker. Genom Disc. 2013;1:3. [Google Scholar]
- Merlo DF, Rossi L, Pellegrino C, Ceppi M, Cardellino U, Capurro C, Ratto A, Sambucco PL, Sestito V, Tanara G, Bocchini V. Cancer incidence in pet dogs: Findings of the Animal Tumor Registry of Genoa, Italy. J Vet Intern Med. 2008;22:976–984. doi: 10.1111/j.1939-1676.2008.0133.x. [DOI] [PubMed] [Google Scholar]
- Mohammed SI, Craig BA, Mutsaers AJ, Glickman NW, Snyder PW, deGortari AE, Schlittler DL, Coffman KT, Bonney PL, Knapp DW. Effects of the cyclooxygenase inhibitor, piroxicam, in combination with chemotherapy on tumor response, apoptosis, and angiogenesis in a canine model of human invasive urinary bladder cancer. Mol Cancer Ther. 2003;2:183–188. [PubMed] [Google Scholar]
- Mohammed SI, Khan KN, Sellers RS, Hayek MG, DeNicola DB, Wu L, Bonney PL, Knapp DW. Expression of cyclooxygenase-1 and 2 in naturally-occurring canine cancer. Prostaglandins Leukot Essent Fatty Acids. 2004;70:479–483. doi: 10.1016/j.plefa.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Moore PF, Affolter VK, Vernau W. Canine hemophagocytic histiocytic sarcoma: A proliferative disorder of CD11d+ macrophages. Vet Pathol. 2006;43:632–645. doi: 10.1354/vp.43-5-632. [DOI] [PubMed] [Google Scholar]
- Mueller F, Fuchs B, Kaser-Hotz B. Comparative biology of human and canine osteosarcoma. Anticancer Res. 2007;27:155–164. [PubMed] [Google Scholar]
- Mutsaers AJ, Widmer WR, Knapp DW. Canine transitional cell carcinoma. J Vet Intern Med. 2003;17:136–144. doi: 10.1892/0891-6640(2003)017<0136:ctcc>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- NumberofNet.com. Number of dogs in America. 2014 Available online (www.numberof.net/number-of-dogs-in-america. ), accessed on April 1, 2014. [Google Scholar]
- Ostrander EA. Franklin H. Epstein Lecture. Both ends of the leash—the human links to good dogs with bad genes . New Engl J Med. 2012;367:636–646. doi: 10.1056/NEJMra1204453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Kruglyak L. Unleashing the canine genome. Genome Res. 2000;10:1271–1274. doi: 10.1101/gr.155900. [DOI] [PubMed] [Google Scholar]
- Owczarek-Lipska M, Jagannathan V, Drögemüller C, Lutz S, Glanemann B, Leeb T, Kook PH. A frameshift mutation in the cubilin gene (CUBN) in border collies with Imerslund-Gräsbeck Syndrome (selective cobalamin malabsorption) PloS One. 2013;8:e61144. doi: 10.1371/journal.pone.0061144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoloni M, Davis S, Lana S, Withrow S, Sangiorgi L, Picci P, Hewitt S, Triche T, Meltzer P, Khanna C. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics. 2009;10:625. doi: 10.1186/1471-2164-10-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H, Kukekova AV, Akey DT, Goldstein O, Kirkness EF, Baysa KC, Mosher DS, Aguirre GD, Acland GM, Ostrander EA. Breed relationships facilitate fine-mapping studies: A 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17:1652–1571. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H, Shearin AL, Ostrander EA. Man's best friend becomes biology's best in show: Genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–336. doi: 10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Pfahler S, Distl O. Identification of quantitative trait loci (QTL) for canine hip dysplasia and canine elbow dysplasia in Bernese mountain dogs. PLoS One. 2012;7((11)):e49782. doi: 10.1371/journal.pone.0049782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Lembcke L, Chamberlin T. A novel locus for canine osteosarcoma (OSA1) maps to CFA34, the canine orthologue of human 3q26. Genomics. 2010;96:220–227. doi: 10.1016/j.ygeno.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Stephenson B, Hauck M, Dillberger J. Heritability and segregation analysis of osteosarcoma in the Scottish deerhound. Genomics. 2007;90:354–363. doi: 10.1016/j.ygeno.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotech. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priester WA, Mantel N. Occurrence of tumors in domestic animals. Data from 12 United States and Canadian colleges of veterinary medicine . J Natl Cancer Inst. 1971;47:1333–1344. [PubMed] [Google Scholar]
- Prokesch A, Lazar MA. A hormone sends instant messages to the genome. Cell. 2011;145:499–501. doi: 10.1016/j.cell.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Proschowsky HF, Rugbjerg H, Ersbøll AK. Mortality of purebred and mixed-breed dogs in Denmark. Prev Vet Med. 2003;58:63–74. doi: 10.1016/s0167-5877(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Prymak C, McKee LJ, Goldschmidt MH, Glickman LT. Epidemiologic, clinical, pathologic, and prognostic characteristics of splenic hemangiosarcoma and splenic hematoma in dogs: 217 cases. J Am Vet Assoc. 1985;193:706–712. [PubMed] [Google Scholar]
- Rankin KS, Starkey M, Lunec J, Gerrand CH, Murphy S, Biswas S. Of dogs and men: Comparative biology as a tool for the discovery of novel biomarkers and drug development targets in osteosarcoma. Pediatr Blood Cancer. 2012;58:327–333. doi: 10.1002/pbc.23341. [DOI] [PubMed] [Google Scholar]
- Reimann-Berg N, Willenbrock S, Murua Escobar H, Eberle N, Gerhauser I, Mischke R, Bullerdiek J, Nolte I. Two new cases of polysomy 13 in canine prostate cancer. Cytogenet Genome Res. 2011;132:16–21. doi: 10.1159/000317077. [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell JL, McCarthy DO, Alvarez CE. Dog models of naturally occurring cancer. Trends Mol Med. 2011;17:380–388. doi: 10.1016/j.molmed.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru G, Terracini B, Glickman LT. Host related risk factors for canine osteosarcoma. Vet J. 1998;156:31–39. doi: 10.1016/s1090-0233(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Can. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R. Comparison of age, sex, and incidence rates in human and canine breast cancer. Cancer. 1970;26:419–426. doi: 10.1002/1097-0142(197008)26:2<419::aid-cncr2820260225>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Schneider R, Dorn CR, Klauber MR. Cancer in households: A human–canine retrospective study. J Natl Cancer Inst. 1968;41:1285–1292. [PubMed] [Google Scholar]
- Shearin A, Hedan B, Cadieu E, Erich SA, Schmidt EV, Faden DL, Cullen J, Abadie J, Kwon EM, Gröne A, Devauchelle P, Rimbault M, Karyadi DM, Lynch M, Galibert F, Breen M, Rutteman GR, André C, Parker HG, Ostrander EA. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidmiol Biomarkers Prev. 2012;21:1019–1027. doi: 10.1158/1055-9965.EPI-12-0190-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin A, Ostrander E. Leading the way: Canine models of genomics and disease. Dis Model Mech. 2010;3:27–34. doi: 10.1242/dmm.004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RB, Vite CH, Bradley CW, Cross JR. Postmortem evaluation of 435 cases of intracranial neoplasia in dogs and relationship of neoplasm with breed, age, and body weight. J Vet Intern Med. 2013;27:1143–1152. doi: 10.1111/jvim.12136. [DOI] [PubMed] [Google Scholar]
- Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, Kruglyak L, Ostrander EA. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texas Veterinary Cancer Registry. Man's best friend may conquer man's most feared illnesses, say Texas A&M veterinarians. 2012 Available online (http://texasvetcancerregistry.com/category/news/ ), accessed on April 1, 2014. [Google Scholar]
- Torres de la Riva G, Hart BL, Farver TB, Oberbauer AM, Messam LL, Willits N, Hart LA. Neutering dogs: Effects on joint disorders and cancers in golden retrievers. PLoS One. 2013;8:e55937. doi: 10.1371/journal.pone.0055937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevejo R, Yang M, Lund EM. Epidemiology of surgical castration of dogs and cats in the United States. J Am Vet Med Assoc. 2011;238:898–904. doi: 10.2460/javma.238.7.898. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Laborde RR, Xu X, Gu J, Chung CB, Monighetti CK, Stanley SJ, Olsen KD, Kasperbauerm JL, Moorem EJ, Broomerm AJ, Tanm R, Brzoskam PM, Mullerm MW, Siddiquim AS, Asmannm YW, Sunm Y, Kuersten S, Barker MA, De La Vega FM, Smith DI. Tumor transcriptome sequencing reveals allelic expression imbalances associated with copy number alterations. PLoS One. 2010;5:e9317. doi: 10.1371/journal.pone.0009317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urfer SR, Gaillard C, Steiger A. Lifespan and disease predispositions in the Irish wolfhound: A review. Vet Q. 2007;29:102–111. doi: 10.1080/01652176.2007.9695233. [DOI] [PubMed] [Google Scholar]
- Vail D, MacEwen E. Spontaneously occurring tumors of companion animals as models for human cancer. Cancer Invest. 2000;18:781–792. doi: 10.3109/07357900009012210. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A, Bryc K, Brisbin A, Knowles JC, Mosher DS, Spady TC, Elkahloun A, Geffen E, Pilot M, Jedrzejewski W, Greco C, Randi E, Bannasch D, Wilton A, Shearman J, Musiani M, Cargill M, Jones PG, Qian Z, Huang W, Ding Z, Zhang Y, Bustamante CD, Ostrander EA, Novembre J, Wayne RK. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Gen. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters DJ, Bostwick DG. The canine prostate is a spontaneous model of intraepithelial neoplasia and prostate cancer progression. Anticancer Res. 1997a;17:1467–1470. [PubMed] [Google Scholar]
- Waters DJ, Bostwick DG. Prostatic intraepithelial neoplasia occurs spontaneously in the canine prostate. J Urol. 1997b;157:713–716. [PubMed] [Google Scholar]
- Winkler S, Reimann-Berg N, Murua Escobar H, Loeschke S, Eberle N. Polysomy 13 in a canine prostate carcinoma underlining its significance in the development of prostate cancer. Cancer Genet Cytogenet. 2006;169:154–158. doi: 10.1016/j.cancergencyto.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Withrow SJ, Wilkins RM. Cross talk from pets to people: Translational osteosarcoma treatments. ILAR J. 2010;51:208–213. doi: 10.1093/ilar.51.3.208. [DOI] [PubMed] [Google Scholar]
- Yokoyama JS, Lam ET, Ruhe AL, Erdmanm CA, Robertson KR, Webb AA, Williams DC, Chang ML, Hytönen MK, Lohi H, Hamilton SP, Neff MW. Variation in genes related to cochlear biology is strongly associated with adult-onset deafness in border collies. PLoS Genet. 2012;8:e1002898. doi: 10.1371/journal.pgen.1002898. [DOI] [PMC free article] [PubMed] [Google Scholar]