Abstract
A major difference between lentiviruses such as human immunodeficiency virus (HIV) and most other retroviruses is their ability to productively infect nondividing cells. We present here genetic evidence for involvement of the capsid protein (CA) in the infectious phenotype in nondividing cells. A chimeric HIV type 1 (HIV-1) in which the MA and CA of HIV-1 are replaced with the MA, p12, and CA encoding sequences from murine leukemia virus (MLV) loses the ability to efficiently infect nondividing cells. Analysis of the accumulation of two-long-terminal-repeat circles implies that the impairment of nuclear transport of preintegration complexes is responsible for the restricted infection of this chimeric virus in nondividing cells. Incorporation of MLV MA and MLV p12 into HIV virions alone does not exert any adverse effects on viral infection in interphase cells. These results suggest that CA is the dominant determinant for the difference between HIV and MLV in the ability to transduce nondividing cells.
One unexpected aspect of human immunodeficiency virus (HIV) biology is the ability of this virus to infect nondividing cells (38, 62). Infection of nondividing T cells and macrophages by HIV is thought to play a major role in AIDS pathogenesis (33) and may also lead to the generation of latent reservoirs for HIV (15). The ability of HIV to productively infect nonproliferating cells is shared by other immunodeficiency viruses belonging to the lentivirus genus, but not by all other retroviruses. For example, retroviruses such as murine leukemia virus (MLV) and avian sarcoma virus cannot productively infect nondividing cells (27).
Upon entry into the cytoplasm, retroviruses undergo an uncoating and reverse transcription process that results in a large nucleoprotein complex called the preintegration complex (PIC) that contains all the components necessary for integration (6). The PIC is composed of viral cDNA and viral proteins, as well as some host cell proteins. Transfer of such large complexes across the nuclear membrane requires active transport mechanisms, a process often mediated by nuclear localizing signals (NLS) on imported proteins. A fundamental difference between HIV and MLV is that MLV PICs do not enter the nucleus until mitosis occurs (38, 55), whereas HIV PICs can enter the nucleus at any stage of the cell cycle (except in G0 of quiescent cells) (11, 65). Thus, an attractive model to explain the different infectious phenotypes in nondividing cells between HIV and MLV is that HIV PICs contain an NLS(s) that allows them to be translocated across the nuclear membrane of interphase cells, while MLV PICs do not have an NLS and, hence, only access the nucleus at mitosis, when the nuclear envelope breaks down (27, 58).
Based on this model, extensive studies have been carried out to identify various candidate proteins with signals that allow the HIV PIC to translocate into the nucleus. The viral matrix protein (MA) and Vpr both contain an NLS, and although mutations in these proteins reduce the efficiency of infection of HIV in macrophages (2, 16, 19, 60), they do not eliminate the ability of HIV to infect nondividing cells (28, 29, 54). Integrase (IN) and the central polypurine tract (5, 30, 66) have also been reported to play an important role in translocation of HIV PICs to the nucleus, although subsequent studies showed that viruses with mutations in multiple combinations of these elements still retained a significant ability to infect nondividing cells (18, 41, 42, 52). Therefore, it remains unclear how HIV PICs enter the nucleus in nondividing cells. However, the use of packaging constructs for lentiviral vectors that have the same phenotype as HIV in nondividing cells suggests strongly that the essential component is in the Gag and/or Pol gene products (49, 67).
Because of the inability to find mutations in candidate NLS-containing proteins that eliminate HIV entry into the nucleus, a recent publication conjectured that viral nucleic acids could be imported into the nucleus without the aid of virus-derived signals and that MLV was somehow prevented from using the same pathway (18). One of the major differences in the composition of the PICs of HIV and MLV is that MLV contains a large amount of the capsid (CA) protein encoded by the gag gene (6, 23), while CA protein is not strongly associated with the PIC of HIV (10, 21, 22, 34). In this study, we found that CA is the major determinant of infection in growth-arrested cells.
MATERIALS AND METHODS
Nomenclature and construction of chimeric proviruses.
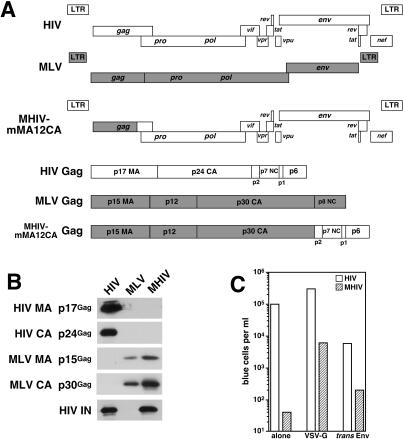
Chimeric proviruses were named as follows: MHIV-mMA12CA indicates that the MA and CA proteins of an HIV provirus have been replaced by the MA, p12, and CA proteins of MLV while the rest of the provirus is HIV. MHIV-mMA is an HIV provirus except for MA, which has been replaced by the MLV MA. Likewise, MHIV-mMA12 is an HIV-based chimera where MA has been replaced by the MLV MA and p12. HIV and MLV genes were taken from the infectious proviruses pLai (51) and pAMS (46), respectively. All the chimeras were made by precisely replacing HIV sequences with MLV sequences. The sequences at junctions between MLV and HIV in the chimeras are exactly the same as those of the parental viruses. Namely, in MHIV-mMA, the last four amino acids of the MLV MA (SSLY) are followed by the first four amino acids of the HIV CA (PIVQ). In MHIV-mMA12, the last four amino acids of the MLV p12 (SQAF) are followed by the first four amino acids of the HIV CA (PIVQ). In the MHIV-mMA12CA, the last four amino acids of the MLV CA (SKLL) are followed by the first four amino acids of the HIV type 1 (HIV-1) p2 (AEAM).
To perform single-cycle replication assays, we made reporter virus constructs which express either green fluorescent protein (GFP) or luciferase. The HIV-GFP construct, pLai3ΔenvGFP3, is based on an env-deficient molecular clone of HIV (56) in which the HIV nef gene is replaced by the enhanced GFP (EGFP) coding sequence following the Kozak sequence, as described previously (36). The junction between the env gene and EGFP open reading frame is as follows: . . . TAA G GTC GCC ACC ATG GTG AGC. . . (TAA, translational stop codon of the env gene; underlined portion, Kozak sequence; ATG, initiation codon of the EGFP gene). The termination codon of the EGFP gene was immediately followed by the unique XhoI site within the (defective) nef gene. The modified version of firefly luciferase was amplified from the pGL3-Basic vector (Promega, Madison, Wis.) with a unique XhoI site immediately downstream of the termination codon of the luciferase and then cloned into NcoI (within Kozak sequence) and XhoI of pLai3ΔenvGFP3, yielding pLai3ΔenvLuc2. The Gag coding sequences of these HIV reporter constructs were replaced with those of MLV to make the MHIV-based reporter constructs, MHIV-GFP and MHIV-Luc. The luciferase gene was also cloned into an HIV-1-based lentiviral vector (49) to create pHR′cmvLuc2.
A mammalian expression vector for MLV gag-pol expression was produced for the single-cycle infectivity assay. Briefly, long PCR was carried out to amplify the MLV gag-pol coding sequence using pAMS as template. The amplified PCR products were digested with EcoRI and NheI and cloned into the large fragment of EcoRI/XbaI-digested pCS2, a mammalian expression vector driven by the simian cytomegalovirus (CMV) IE94 promoter, yielding pCS2-mGP. A modified version of the luciferase gene of pGL3 Basic vector was cloned into the murine retrovirus-based vector pLNCXm1 (derivative of pLNCX [47]) at ApaI and HindIII sites to produce pLNCLuc which, upon infection, allows the high expression of luciferase under the control of strong activity of the human CMV immediate early promoter.
Virus stocks.
High-titer vesicular stomatitis virus G protein (VSV-G)-pseudotyped viruses were prepared by transient transfections of 293T cells performed either with the standard calcium phosphate precipitation method or with the FuGene 6 reagent (Roche, Indianapolis, Ind.). HIV and MHIV expression plasmids were cotransfected with VSV-G expression vectors (pL-VSV-G [4] or pMD-G [49]) in addition to pCMV-tat to express VSV-G for pL-VSV-G and to augment the expression of MHIV. In experiments using lentiviral vectors, the HIV-1 Gag/Pol expression vector (pΔR8.2) and MHIV expression plasmid were cotransfected with VSV-G expression vectors and with lentiviral vectors. gag-pol expression plasmids (pCS2-mGP and pJK3 [4]) were used for the production of VSV-G-pseudotyped MLV along with the murine retrovirus-based vectors (47) (pLtatSN for tat expression [3], pLNCluc for luciferase expression, and pLXCG for GFP expression) as well as the VSV-G constructs. MLV/tat was also made by infection of amphotropic Moloney MLV. Briefly, NIH 3T3 cells were transfected with pAMS and pLtatSN and culture supernatants were frozen as virus stocks.
Western blot analysis.
To examine the synthesis and processing of virion proteins by the chimeric viruses, molecular clones of wild-type HIV-1, MLV, and chimeric viruses were transfected to 293T cells by the standard calcium phosphate method or by using FuGene 6 reagent. On day 2 after transfection, the cells were washed with phosphate-buffered saline (PBS) and lysed in 200 μl of 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate lysis buffer. The lysates were centrifuged at 12,000 × g for 10 min to remove cell debris and kept on ice or frozen. Culture supernatant was collected, and cell debris was cleared by centrifugation at 500 × g for 5 min. Viral pellets were spun down by ultracentrifugation at 64,000 × g for 90 min in an SW28 rotor. The cell lysates and viral pellets were heated at 90°C for 5 min in the presence of sample buffer. Lysates with equal amounts of protein as measured by the optical density at 595 nm were loaded on sodium dodecyl sulfate-polyacrylamide gels. Following electrophoresis, the proteins were transferred to polyvinylidene difluoride membrane. The membranes were blocked for 60 min at room temperature with PBS containing 0.5% Tween 20 and 5% nonfat milk powder (Carnation) and incubated with primary antibodies at a 1:1,000 dilution unless otherwise noted for 1 h at room temperature. The blots were probed with the following antibodies: sheep anti-HIV-1 p17 MA (through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, from Michael Phelan); mouse monoclonal anti-HIV-1 p17 MA (Shu-lok Hu); mouse monoclonal anti-HIV-1 p24 CA (Shu-lok Hu), mouse monoclonal anti-HIV-1 gp120 Env (Yves Rivere); human anti-HIV-1 pooled antiserum; goat anti-MLV p15 MA (Quality Biotech Inc); rat monoclonal anti-MLV p30 CA (American Type Culture Collection); and goat serum reactive with MLV virion proteins (Quality Biotech Inc). The membranes were washed for 30 min in wash buffer (PBS containing 0.2% Tween 20) and then incubated with a 1:5,000 dilution of horseradish peroxidase-conjugated monoclonal antibodies that matched with the primary antibody for 60 min at room temperature. The membranes were washed three times for 30 min, and the bound antibody was detected with ECL Western blotting detection reagents (Amersham). In some cases, membranes were stripped and reprobed with another primary antibody.
Electron microscopy.
293T cells were transfected with MHIV-mMA12CA, HIV, or MLV Gag-Pol expression vectors. Two days after transfection, cells were washed once with PBS and then fixed in a solution of 2% paraformaldehyde and 2.5% glutaraldehyde. The samples were then dehydrated, embedded, and sectioned.
Infectivity assays.
Single-cycle infectivity of HIV, MLV/tat, and MHIV was measured by challenging MAGI cells with a serial dilution of virus and staining for β-galactosidase expression (35). GHOST cells (48) were also used for the infectivity assay; percentages of GFP-expressing GHOST cells were determined by flow cytometry 48 h after challenge. Growth-arrested MAGI and GHOST cells were prepared by treatment with 2 μg of aphidicolin/ml and by irradiation with 3,500 rads from a 137cesium source, respectively. In all the infection experiments, virus binding was enhanced either by spinoculation (50) or by addition of either 10 μg of Polybrene/ml or 20 μg of DEAE-dextran/ml.
Peripheral blood mononuclear cells were prepared by back-washing cells from a leukofilter (obtained from Puget Sound Blood Center, Seattle, Wash.) with PBS plus 1 mM EDTA. Peripheral blood mononuclear cells were separated from erythrocytes by Ficoll centrifugation. Cells were plated in 12-well plates, and nonadherent cells were aspirated 3 days following separation. Adherent monocyte-derived macrophages (MDM) were cultured in RPMI 1640 with 10% fetal bovine serum and 5% human serum for 10 to 14 days to allow MDM to differentiate completely before infection. The MDM, along with HeLa cells, were inoculated with luciferase-expressing virus stocks. Two to 3 days after infection, the cells were lysed with luciferase cell culture lysis buffer (Promega) and the lysis was tested for luciferase activity by adding luciferase assay substrate (Promega). Luciferase activity in MDM infected with different virus stocks was expressed after normalization by the titer in HeLa cells.
Real-time PCR.
One day before infection, HeLa cells were plated at 250,000 cells per well in 12-well plates in the presence or absence of aphidicolin at 2 μg/ml. Infections were performed in a similar manner as described above. One day after infection, extrachromosomal DNA (Hirt DNA) was isolated from infected cells by using the High Pure plasmid isolation kit (Roche), as classical bacterial plasmid miniprep methods can be applied to isolation of episomal DNA from mammalian cells (7; G. Siebenkolten, H. Leyendeckers, R. Christine, and A. Radbruch, QIAGEN News 2:11-12, 1995). We observed higher yields of DNA isolated from the miniprep method than DNA isolated from the standard Hirt protocol (data not shown). Genomic DNA was extracted with the DNeasy kit (Qiagen, Valencia, Calif.). Real-time PCR for HIV and MHIV was based on a previously published protocol (12). Nested PCR was performed to amplify two-long-terminal-repeat (2-LTR) circles, which were cloned into the TA vector (Promega). The resulting plasmid DNA, pLai3-2LTR, was used as the standard for real-time PCR of 2-LTR circles. Primers and probes for HIV and MHIV were prepared as described previously (12), except that the probes were labeled with the reporter fluorochrome 6-carboxyfluorescein at the 5′ termini and with a nonfluorescent quencher at the 3′ termini (Applied Biosystems, Foster City, Calif.). Reactions, including 25,000 cell equivalents, were carried out in a 20-μl volume with the Brilliant QPCR core reagent kit (Stratagene, Cedar Creek, Tex.) on an ABI 7900 HT kinetic PCR instrument (Applied Biosystems). Sequence detection software (version 2.0; Applied Biosystems) was used to analyze the quantitative PCR amplification data.
RESULTS
Generation of an infectious chimeric HIV-1 with MLV MA, p12, and CA.
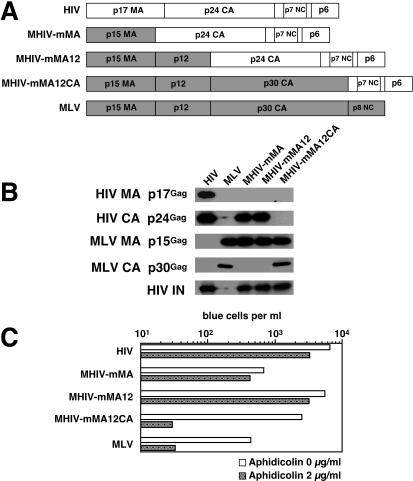
Previously, our group and others demonstrated that it is possible to make infectious chimeric viruses in which MA of MLV is replaced by HIV MA and vice versa (14, 17, 53). These studies prompted us to construct additional chimeric viruses between HIV-1 and MLV to identify the viral component(s) responsible for the infectious phenotypes in nondividing cells. Thus, the coding sequences of HIV-1 MA (p17gag) and CA (p24gag) were replaced with that of MLV MA (p15gag), p12 (p12gag), and CA (p30gag), yielding the HIV-based chimeric clone pMHIV-mMA12CA (Fig. 1A). Western blotting performed on virions pelleted from cell-free supernatants of cells transfected with the chimeric provirus showed that chimeric Gag polyproteins of MHIV-mMA12CA were correctly processed, as revealed by the presence of mature forms of MA (p15gag) and CA (p30gag) that comigrated with their counterparts of wild-type MLV (Fig. 1B). As expected, no reactivity against HIV p17 MA and p24 CA was found in MHIV-transfected supernatant (Fig. 1B). These results indicate that MHIV-mMA12CA is able to produce chimeric virions composed mostly of MLV Gag in the context of an HIV provirus. We assessed the infectivity of MHIV-mMA12CA by transfection of the provirus alone or by cotransfection with a VSV-G expression construct. The resulting viruses were tested for infectivity in the indicator cell line HeLa-CD4-LTR/β-galactosidase (MAGI). Although we observed minimal infection of MAGI cells by MHIV-mMA12CA (Fig. 1C), a VSV-G-pseudotyped version of MHIV-mMA12CA was infectious, yielding a titer of approximately 104 blue cells per ml (Fig. 1C). Expression of a truncated HIV envelope (44, 57) in trans could also rescue some of the infectivity (Fig. 1C), indicating that at least part of the problem with the infectivity of MHIV-mMA12CA was due to expression of the HIV envelope from the chimeric provirus (the same env truncation could not rescue infectivity in cis [data not shown]). Infectivity was sensitive to reverse transcriptase inhibitors (data not shown). Thus, the chimeric HIV with MA, p12, and CA of MLV Gag is infectious in single-cycle replication assays.
FIG. 1.
Generation of the infectious chimeric virus MHIV-mMA12CA. (A) Schematic representation of genomic organization of HIV-1, MLV, and MHIV (upper). Protein structure of the gag gene of these viruses is also shown (lower). MHIV-mMAp12CA encodes the MLV MA, p12, and CA and all the HIV-1 genes other than the HIV MA and CA. (B) Western blot analysis of purified virus particles of MHIV-mMA12CA together with HIV-1 and MLV. Polyprotein processing was tested for HIV-1 MA (p17gag) and CA (p24gag) as well as MLV MA (p15gag) and CA (p30gag) in addition to HIV-1 IN. Some unprocessed Gag and Gag-Pol proteins remain in the lane of MHIV-mMA12CA, but the anti-IN profile of MHIV-mMA12CA is the same as that of parental HIV-1 (not shown in this figure). (C) Single-cycle infectivity of MHIV-mMA12CA. Single-cycle infections of MHIV-mMA12CA were tested either alone or with envelope expression of VSV-G or a truncated HIV-1 Env in trans. Infectivity was measured with the MAGI assay by counting β-galactosidase-positive cells 2 days postinfection using viruses harvested after transfection of proviral clones with or without env expression vectors into 293T cells. These data are representative of three independent experiments.
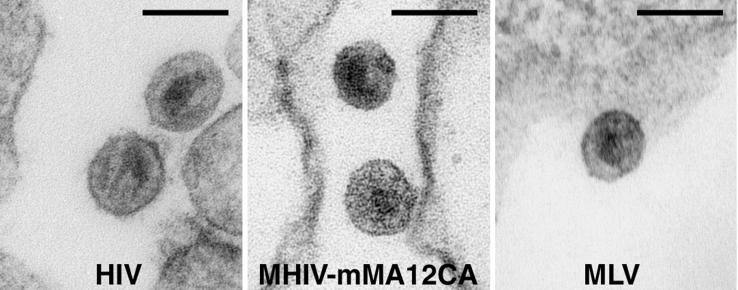
We also examined the viruses produced from 293T cells transfected with the MHIV-mMA12CA chimeric provirus by electron microscopy (Fig. 2). HIV and MLV proviruses were also transfected in parallel as controls. The core structures of lentiviruses differ subtly from those of the gammaretroviruses in that the lentiviruses contain conical core structures after maturation, while those of the gammaretroviruses tend to be more spherical (61). Virion particles released from 293T cells transfected with MHIV-Mma12CA exhibited dense round cores, which are more similar to those of MLV (Fig. 2). On the other hand, the core shape of the chimeric virus differed from that of the conical core structure of mature HIV(Fig. 2) These data indicate that the MHIV-mMA12CA core structure retains MLV-like cores and suggests that the shape of the cores is determined by the origin of the CA proteins (although we cannot rule out that the MA also plays a role).
FIG. 2.
Electron microscopy analysis of virions of HIV (left), MHIV-mMA12CA (center), and MLV (right) produced from 293T cells transfected with the respective proviruses. Bar, 100 nm.
The infectivity of HIV with most of Gag from MLV is restricted to dividing cells.
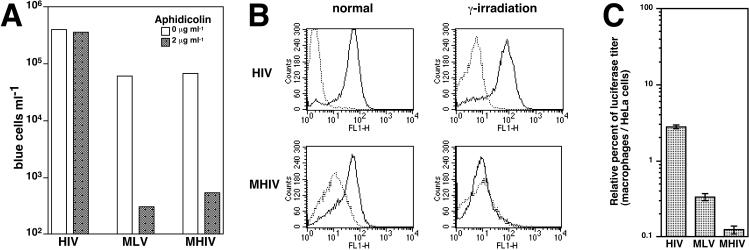
We next evaluated the infectivity of nondividing cells by the novel chimeric virus MHIV-mMA12CA using several different methods. First, nondividing cells were prepared by treatment of MAGI cells with aphidicolin, which arrests these cells in the S phase of the cell cycle. As expected, the parental HIV was capable of infecting nondividing cells as efficiently as dividing cells (Fig. 3A). In contrast, the titers of both MLV and the chimeric virus of HIV with most of MLV Gag were consistently reduced by 100-fold relative to the titers in dividing cells. Second, GHOST cells were irradiated from a 137cesium source to arrest cells in the G2 phase of the cell cycle and subsequently exposed to either HIV-1 or MHIV-mMA12CA (concentrated by 100-fold). Infection of GHOST cells was measured by fluorescence-activated cell sorter analysis of GFP-positive cells 2 days after infection. HIV infection of GHOST cells was not affected by γ-irradiation: the same change in GFP-positive peaks was seen in both treated and untreated cells (Fig. 3B, top). However, the GFP-positive population after infection of dividing GHOST cells with MHIV-mMA12CA was no longer present when the cells were nondividing (Fig. 3B, bottom). Third, we infected dividing and nondividing HeLa cells with wild-type and the MHIV-mMA12CA chimeric virus that encoded GFP in place of the nef gene and found that the chimeric virus could transduce GFP only in dividing cells (data not shown). Finally, we extended the above observations to terminally differentiated macrophages, a primary nondividing target for HIV. We determined macrophage infection by using luciferase reporter viruses. Briefly, the luciferase gene was cloned into the molecular infectious clone of HIV and MHIV-mMA12CA in place of the nef gene, or the luciferase gene was supplied by cotransfection of a lentiviral vector encoding the luciferase gene. MLV luciferase reporter viruses were made from the MLV-based target vector pLNCLuc encoding luciferase under the control of the human CMV IE promoter. We compared the amount of luciferase activity of each virus stock in a dividing cell type (HeLa) versus nondividing macrophages (Fig. 3C). Thus, compared with HIV infection of macrophages, the infection efficiency in macrophages with MHIV-mMA12CA was decreased by ∼20-fold, similar to the decrease observed with MLV. Our data showed that luciferase titers in macrophages varied among preparations of different donors. However, reductions in macrophage infection by MHIV-mMA12CA and MLV were consistent throughout several independent experiments. These results corroborate those obtained by artificially prepared nondividing cells and demonstrate that HIV with a core composed mostly of MLV Gag proteins is as defective as MLV itself in its ability to mediate infection in growth-arrested cells.
FIG. 3.
MHIV-mMA12CA does not establish efficient infection in nondividing cells. (A) Infectivity of MHIV in aphidicolin-treated cells. MAGI cells were treated in the presence or absence of aphidicolin and challenged with virus stocks. Virus titers are expressed as the number of β-galactosidase-expressing blue cells per milliliter. These data are representative of five independent experiments. (B) Infectivity of MHIV in γ-irradiated cells. GHOST cells were treated with 3,500 rads from a 137cesium source. HIV and concentrated MHIV, all of which were pseudotyped with the VSV-G envelope protein, were spinoculated into normal (dividing) or γ-irradiated (nondividing) GHOST cells. Two days after infection, the cells were fixed and analyzed with flow cytometry for GFP-expressing GHOST cells. Dotted lines indicate zidovudine-treated control cells. (C) Macrophage infection with MHIV-mMA12CA. Monocyte-derived macrophages were prepared by adherence to the bottom of wells and maintained for 10 to 14 days before infection. Infectivity is shown as the relative percentage of luciferase titer in HeLa cells per that in macrophages. Error bars indicate standard deviations of duplicate cultures. The luciferase titers of these particular data are as follows: 534,677 relative light units (RLU) for HeLa cells infected with HIV; 680,954 RLU for HeLa cells infected with MLV; 137,651 RLU for HeLa cells infected with MHIV; 14,669 RLU for macrophages infected with HIV; 2,281 RLU for macrophages infected with MLV; 178 RLU for macrophages infected with MHIV. These values are averages of duplicate wells. The data presented here are representative of at least four different independent experiments using different blood donors.
The defect of an HIV chimeric virus containing most of Gag from MLV is independent of both viral titer and of cis-acting sequences from HIV.
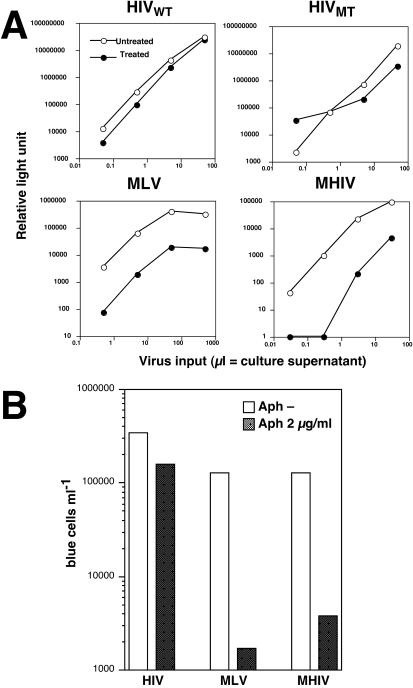
It has previously been pointed out that differences among studies examining the role of HIV MA and Vpr as genetic determinants of nuclear transport of PICs may be explained by the possibility that MA- and Vpr-mutated viruses exhibit their effect only when a low initial inoculum is used (59). In the MAGI assay, comparison of infectivity was made at a low titer. Therefore, we used the luciferase-encoding viruses described above, which are quantitative over a greater range of input virus, to measure infections at both high and low titers. Virus stocks for the luciferase reporter assay were serially diluted into either normal or aphidicolin-treated HeLa cells, and luciferase activity of the lysate was measured 2 days after infection. The results showed that MHIV-mMA12CA behaved similar to MLV, with decreased infectivity in nondividing cells regardless of viral input (Fig. 4A). For both viruses, we measured about a ∼1.5 log decrease of infectivity in nonproliferating cells when compared with that in proliferating cells at all dilutions of virus. Notably, the phenotype of the chimeric virus was much stronger than that of the previously described double mutant (MA− Vpr−) of HIV, which had marginal effects on infections in nondividing cells in this assay (Fig. 4A). Instead, the reduction of the chimeric virus was comparable to the reduction of MLV infectivity in nondividing cells.
FIG. 4.
(A) Dose-independent restriction of MHIV-mMA12CA in nondividing cells. Aphidicolin-treated HeLa cells were infected with increasing amount of luciferase-encoding viruses. Culture supernatants of transfected cells were used as inocula. Virus infectivity was judged by measuring luciferase titers of infected cell lysates 2 days after infection. (B) Transduction of the lacZ gene into dividing and nondividing HeLa cells. MAGI cells were treated in the presence or absence of aphidicolin and challenged with virus stocks by using retroviral vectors encoding the lacZ gene. Virus titers are expressed as the number of β-galactosidase-expressing blue cells per milliliter. These data are representative of two independent experiments.
It was possible that a region of HIV cDNA acts in a cis-dependent manner to affect transduction of nondividing cells. The chimeric virus MHIV-mMA12CA differs from the parental HIV-1 not only in the protein encoded by gag, but also in the DNA or RNA sequences in this region of the genome. Thus, to determine if the proteins encoded by this chimeric virus or the nucleic acid structures present on the viral cDNA were important, we used the chimeric virus to package a lentiviral vector encoding the β-galactosidase gene (wild-type HIV Gag/Pol was also used to package the same vector). Thus, resulting virions would have the same viral RNA but differ from each other in the contents of core structures (made from either HIV Gag proteins or MLV Gag proteins). Dividing and nondividing cells were challenged with these lentiviral vectors, and the efficiency of transduction was measured by counting β-galactosidase-positive blue cells. The results showed that transduction of the lentiviral target vectors by MHIV-mMA12CA was severely impaired in growth-arrested cells compared with that in control cells, in contrast to wild-type HIV-1, which transduced the lentiviral vector into nondividing cells at a level as high as in dividing cells (Fig. 4B). This observation was reproducible when we used another lentiviral vector that encodes the luciferase gene (data not shown). Thus, these results indicate that proteins are responsible for the different phenotypes of HIV and MHIV-mMA12CA in nondividing cells rather than cis-acting sequences.
HIV CA is required for efficient infection in nondividing cells.
MHIV-mMA12CA retains MLV MA, p12, and CA in place of HIV MA and CA. To narrow down the region responsible for the attenuated infectivity of the chimeric virus in nondividing cells, we genetically engineered several additional chimeric viruses. HIV MA was replaced by either MLV MA (resulting in the proviral clone pMHIV-mMA) or by MLV MA and MLV p12 (pMHIV-mMA12), as shown in Fig. 5A. Western blotting analysis of cell-free supernatants collected after transfection of these two proviral DNAs showed released virion particles with mature Gag proteins, MLV MA, and HIV CA (Fig. 5B). We analyzed infectivity of these chimeric viruses in both dividing and nondividing cells and found, like HIV, that MHIV-mMA and MHIV-mMA12 transduced nondividing cells almost as efficiently as dividing cells (Fig. 5C). Thus, neither MA nor MLV p12 is responsible for the inability of MHIV-mMA12CA to infect nondividing cells. We also attempted to construct an HIV with only the CA sequences of MLV, but such viruses were not infectious (data not shown). Nonetheless, the results with all of the chimeric viruses demonstrate that CA plays a crucial role in the infectivity of nondividing cells.
FIG. 5.
CA determines infectivity in nondividing cells. (A) Schematic illustration of the protein structure of Gag of new chimeric viruses. Note that two new chimeric viruses as well as MHIV-mMA12CA were created based on the HIV-1 infectious clone. (B) Western blot analysis of purified virus particles of new MHIV chimeras. See the legend to Fig. 1B for details. (C) Single-cycle infectivity of MHIV as well as parental MLV and HIV. For details, see the legend to Fig. 1C.
CA participates in nuclear import of PICs.
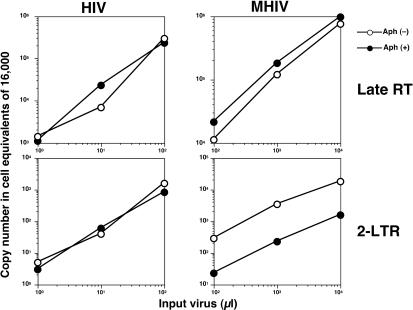
The restriction of MLV infection in nondividing cells is believed to occur at a postentry level, since MLV virions in nondividing cells give rise to similar amounts of reverse-transcribed viral cDNA copies in nondividing cells as in dividing cells, yet do not give rise to nuclear forms of MLV cDNA in nondividing cells (38, 55). We asked if this model also holds true for the infection phenotype of MHIV-mMA12CA in growth-arrested cells by exploiting a quantitative real-time PCR system to measure copy numbers of reverse-transcribed products in both dividing and nondividing cells infected with HIV and MHIV-mMA12CA. Equivalent amounts of reverse transcription products were detected 24 h postinfection for HIV as well as MHIV-mMA12CA in both nondividing cells as well as in dividing cells (Fig. 6). However, while the copy number of 2-LTR circles of wild-type HIV-1 (2-LTR circles are dead-end products for integration that have been used as a marker for nuclear entry of PICs) was similar in infections for both dividing and nondividing cells (Fig. 6), the level of 2-LTR circles formed by MHIV-mMA12CA diminished in aphidicolin-treated cells compared to the level in normal cells (Fig. 6). Similar results were obtained when cells were growth arrested by γ-irradiation (data not shown). These results suggest that, as is the case for MLV, a block in nuclear import of PICs is responsible for restriction of MHIV-mMA12CA in nondividing cells.
FIG. 6.
Infection of MHIV-mMA12CA is blocked at nuclear transport. Reverse transcription and nuclear transport of MHIV-mMA12CA and parental HIV-1 were monitored by measuring late reverse transcription products (upper) and 2-LTR circles (lower) as markers, respectively. Increasing amounts of virus inocula (x axis) were challenged with either normal or nondividing cells. The y axis indicates the copy number of each DNA analyzed with real-time quantitative PCR.
DISCUSSION
We present here genetic evidence for involvement of CA with the infectious phenotype of HIV in nondividing cells. A chimeric HIV-1 in which most of the Gag proteins were replaced with their counterparts from MLV lost the ability to efficiently infect nondividing cells, with a corresponding loss of 2-LTR circle formation as a marker of nuclear entry. The phenotype of this chimeric virus (MHIV-mMA12CA) is more similar to MLV than previously reported HIV mutants defective in NLS-containing proteins and is independent of the multiplicity of infection. The chimera also differs from other described mutants for HIV translocation into the nucleus that are defective in both in dividing and nondividing cells (5, 66). Therefore, this is the first identification of an HIV-1-based virus that lacks infectivity specifically in nondividing cells to the same extent as MLV.
The chimeric virus MHIV-mMA12CA still encodes C-terminal HIV-1 Gag proteins (p2, NC, p1, and p6), but it has lost the ability to efficiently infect nondividing cells. This suggests that those C-terminal HIV-1 Gag proteins as well as Pol proteins do not possess dominant positive effects on HIV infection in nondividing cells. In addition, our data rule out the possibility that a cis-acting element is involved in nuclear transport of HIV, since packaged lentivirus vectors showed the phenotype of the Gag proteins used to package them (Fig. 4B). Thus, we have shown that the Gag proteins (MA, p12, and CA) are important for infectivity in interphase cells. However, an involvement of MA in this phenotype is not supported by the observation that MHIV-mMA infects nondividing cells as efficiently as dividing cells. Finally, the data on MHIV-mMA12 make it unlikely that p12 negatively regulates infection in nondividing cells, because MHIV-mMA12 replicated well in growth-arrested cells. These observations suggest that CA is the major determinant of the infectious phenotype in nonproliferating cells.
Previous reports have shown that the CA protein not only functions in virus assembly and virion maturation but plays a role in postentry steps. For instance, there are HIV CA mutants capable of completing reverse transcription but unable to facilitate the nuclear transport of PICs (13, 25, 26), suggesting direct participation of HIV CA in a postentry step. Involvement of CA in the postentry step seems to be true for MLV as well (1). More importantly, MLV CA is involved in the Fv1 restriction that takes place at nuclear transport of viral cDNA (32). Thus, these observations are consistent with our finding that CA is important for nuclear transport of PICs. Although a definitive test of the importance of CA in infection of nondividing cells would be the transfer of the nondividing cell phenotype of HIV to MLV, we have not yet been successful in constructing MLV-based chimeric proviruses that produce virions (data not shown).
The nuclear transport of HIV PICs is mediated by an active transport mechanism using NLS (reviewed in references 27 and 58). This model led to the assumption of NLS present on viral proteins that are part of the PIC-mediated nuclear entry. Indeed, NLS of viral proteins MA, Vpr, and IN that are biochemically associated with PICs have been identified. In contrast, HIV CA does not meet the definition of the NLS in this model: HIV CA is not strongly associated with the PICs, based on biochemical data (10, 21, 22, 34), and HIV CA does not seem to have an NLS activity, as it is fractionated exclusively in cytoplasm (8, 31). Therefore, we do not think that HIV CA acts as an NLS to transport the HIV PIC through the nucleus of interphase cells.
So, how does CA show its effect on nuclear transport of PICs? Our data are consistent with a model postulating that MLV is prevented from accessing the nucleus because of the presence of CA in the PIC of MLV. In this regard, it is interesting that MLV CA differs from HIV CA in that it is closely associated with MLV PICs (6, 23) as opposed to HIV CA, which dissociates from HIV PICs soon after viral fusion with cellular membrane (9, 21, 22, 34) (although there may be some particles with CA in the cytoplasm [45]). It is tempting to speculate that such tight association of MLV CA with PICs prevents cellular transport machinery from interacting with putative NLS on MLV PICs, thereby retaining PICs within the cytoplasm of interphase cells. On the other hand, HIV CA, by dissociating from PICs of HIV-1, might expose the viral nucleic acids (or other components of the PIC) to cellular transport machinery so that HIV PIC can be transported to the nucleus of nondividing cells (24). Therefore, our working model is that CA alters the accessibility of the PICs to cellular or viral factors that determine nuclear entry and thereby determines the fate of nuclear transport in nondividing cells.
One prediction of this model is that MLV CA will remain associated with PIC in the chimeric MHIV-mMA12CA virus. We have not yet been able to test this prediction, because of the low titers of our virus. Nonetheless, we do not feel that the low titers are an explanation for the phenotypes we observed, since pseudotyping the MHIV-mMA12CA with VSV-G allowed us to use nonconcentrated virus (Fig. 2), and we found that the phenotype was independent of viral dose (Fig. 4).
What factors function as an NLS after disruption of the CA, either soon after entry into the cytoplasm in the case of HIV or presumably near mitosis in the case of MLV? For HIV, redundant and multiple portions of viral proteins might serve as NLS, as previously pointed out (reviewed in references 27 and 58). MLV PICs were presumed to migrate into the nucleus of mitotic cells passively via disrupted nuclear membrane rather than via nuclear pores by NLS. However, given the identification of MLV mutants that cannot bring their PICs into the nucleus of dividing cells, nuclear transport of MLV PICs might be actively regulated in mitotic cells (63, 64). Thus, MLV as well might encode a possible NLS factor in viral proteins that is masked by CA until near mitosis. As opposed to these ideas about the involvement of viral proteins in nuclear transport, host factors associated with PICs might supply an NLS activity to PIC. Indeed, several host factors identified as a component of PIC (HMGa1, Ku, and BAF) of both HIV and MLV have NLS activities (20, 37, 39, 40, 43). Thus, it is not yet clear whether it is viral or cellular components that are ultimately recognized by the cellular import machinery that was recently characterized (24). Although many aspects of this model remain to be tested, our data suggest that nuclear transport of retroviruses is both positively and negatively influenced by the CA composition of the incoming virion.
Acknowledgments
We acknowledge the assistance of the FHCRC flow cytometry and EM core facilities. We are grateful to Tim Dellit, Maxine Linial, Mark Rogel, and Yegor Voronin for critical reading of the manuscript and to the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIH, for the anti-HIV-1 MA antibody (contributed by Michael Phelan).
M.Y. was partially supported by the Uehara Memorial Foundation. This work was supported by NIH grants R01 AI 51153 and R37 AI 30927 to M. Emerman and equipment purchased by the James B. Pendelton Foundation.
REFERENCES
- 1.Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. [DOI] [PubMed] [Google Scholar]
- 2.Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623-631. [DOI] [PubMed] [Google Scholar]
- 3.Bartz, S. R., and M. Emerman. 1999. Human immunodeficiency virus type 1 Tat induces apoptosis and increases sensitivity to apoptotic signals by up-regulating FLICE/caspase-8. J. Virol. 73:1956-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. [DOI] [PubMed] [Google Scholar]
- 5.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 6.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 7.Bowers, M. T., S. Ramachandran, and B. W. Yu. 1999. Modified miniprep method for the rapid recovery of episomes from transfected breast epithelial cells. BioTechniques 26:276-280, 282. [DOI] [PubMed] [Google Scholar]
- 8.Bukrinskaya, A. G., A. Ghorpade, N. K. Heinzinger, T. E. Smithgall, R. E. Lewis, and M. Stevenson. 1996. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc. Natl. Acad. Sci. USA 93:367-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky, M., N. Sharova, and M. Stevenson. 1993. Human immunodeficiency virus type 1 2-LTR circles reside in a nucleoprotein complex which is different from the preintegration complex. J. Virol. 67:6863-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 13.Cartier, C., P. Sivard, C. Tranchat, D. Decimo, C. Desgranges, and V. Boyer. 1999. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J. Biol. Chem. 274:19434-19440. [DOI] [PubMed] [Google Scholar]
- 14.Chen, B. K., I. Rousso, S. Shim, and P. S. Kim. 2001. Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells. Proc. Natl. Acad. Sci. USA 98:15239-15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chun, T. W., and A. S. Fauci. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc. Natl. Acad. Sci. USA 96:10958-10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 17.Deminie, C. A., and M. Emerman. 1994. Functional exchange of an oncoretrovirus and a lentivirus matrix protein. J. Virol. 68:4442-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckstein, D. A., M. P. Sherman, M. L. Penn, P. S. Chin, C. M. De Noronha, W. C. Greene, and M. A. Goldsmith. 2001. HIV-1 Vpr enhances viral burden by facilitating infection of tissue macrophages but not nondividing CD4+ T cells. J. Exp. Med. 194:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 21.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 26.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fouchier, R. A., and M. H. Malim. 1999. Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv. Virus Res. 52:275-299. [DOI] [PubMed] [Google Scholar]
- 28.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freed, E. O., G. Englund, F. Maldarelli, and M. A. Martin. 1997. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell 88:171-173. [DOI] [PubMed] [Google Scholar]
- 30.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. [DOI] [PubMed] [Google Scholar]
- 32.Goff, S. P. 1996. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell 86:691-693. [DOI] [PubMed] [Google Scholar]
- 33.Hirsch, V. M., M. E. Sharkey, C. R. Brown, B. Brichacek, S. Goldstein, J. Wakefield, R. Byrum, W. R. Elkins, B. H. Hahn, J. D. Lifson, and M. Stevenson. 1998. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat. Med. 4:1401-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karageorgos, L., P. Li, and C. Burrell. 1993. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res. Hum. Retrovir. 9:817-823. [DOI] [PubMed] [Google Scholar]
- 35.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, L., K. Yoder, M. S. Hansen, J. Olvera, M. D. Miller, and F. D. Bushman. 2000. Retroviral cDNA integration: stimulation by HMG I family proteins. J. Virol. 74:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Limon, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Limon, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, C. W., and A. Engelman. 2003. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 77:5030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mammano, F., F. Salvatori, S. Indraccolo, A. De Rossi, L. Chieco-Bianchi, and H. G. Gottlinger. 1997. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J. Virol. 71:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, A. D., M. F. Law, and I. M. Verma. 1985. Generation of helper-free amphotropic retroviruses that transduce a dominant-acting, methotrexate-resistant dihydrofolate reductase gene. Mol. Cell. Biol. 5:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-982, 984-986, 989-990. [PMC free article] [PubMed] [Google Scholar]
- 48.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 50.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 52.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reed, M., R. Mariani, L. Sheppard, K. Pekrun, N. R. Landau, and N. W. Soong. 2002. Chimeric human immunodeficiency virus type 1 containing murine leukemia virus matrix assembles in murine cells. J. Virol. 76:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogel, M. E., L. I. Wu, and M. Emerman. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 69:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnierle, B. S., J. Stitz, V. Bosch, F. Nocken, H. Merget-Millitzer, M. Engelstadter, R. Kurth, B. Groner, and K. Cichutek. 1997. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc. Natl. Acad. Sci. USA 94:8640-8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevenson, M. 2002. Molecular biology of lentivirus-mediated gene transfer. Curr. Top. Microbiol. Immunol. 261:1-30. [DOI] [PubMed] [Google Scholar]
- 59.Trono, D., and P. Gallay. 1997. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell 88:171-174. [DOI] [PubMed] [Google Scholar]
- 60.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27-70. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 62.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan, B., A. Fassati, A. Yueh, and S. P. Goff. 2002. Characterization of Moloney murine leukemia virus p12 mutants blocked during early events of infection. J. Virol. 76:10801-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 66.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 67.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]