Abstract
Viral immune evasion strategies are important for establishment and maintenance of infections. Many viruses are in possession of mechanisms to counteract the antiviral response raised by the infected host. Here we show that a herpes simplex virus type 1 (HSV-1) mutant lacking functional viral protein 16 (VP16)—a tegument protein promoting viral gene expression—induced significantly higher levels of proinflammatory cytokines than wild-type HSV-1. This was observed in several cell lines and primary murine macrophages, as well as in peritoneal cells harvested from mice infected in vivo. The enhanced ability to stimulate cytokine expression in the absence of VP16 was not mediated directly by VP16 but was dependent on the viral immediate-early genes for infected cell protein 4 (ICP4) and ICP27, which are expressed in a VP16-dependent manner during primary HSV infection. The virus appeared to target cellular factors other than interferon-induced double-stranded RNA-activated protein kinase R (PKR), since the virus mutants remained stronger inducers of cytokines in cells stably expressing a dominant-negative mutant form of PKR. Finally, mRNA stability assay revealed a significantly longer half-life for interleukin-6 mRNA after infection with the VP16 mutant than after infection with the wild-type virus. Thus, HSV is able to suppress expression of proinflammatory cytokines by decreasing the stability of mRNAs, thereby potentially impeding the antiviral host response to infection.
Elimination of infectious agents relies on the ability of the infected organism to mount an immune response capable of restricting the microbe. For viruses, efficient elimination of the infection requires a proinflammatory host response and development of type 1 immunity (14, 35). This type of response is characterized by activation of macrophages, natural killer (NK) cells, and cytotoxic T lymphocytes (CTLs) and production of proinflammatory cytokines and chemokines, including interferons (IFNs), tumor necrosis factor alpha (TNF-α), and various interleukins (ILs), such as IL-12, IL-18, and IL-23 (14, 35).
In order to establish an infection in such an environment, many pathogens have evolved mechanisms to evade or suppress the host immune response. For instance, a wealth of different strategies are used by viruses to inhibit NK cell activity, including viral expression of inhibitory receptors (24). Viruses also interfere with the early antiviral response mediated by IFN-α/β (8), as well as with antigen presentation to CTLs (48).
Herpes simplex virus type 1 (HSV-1) and HSV-2 are closely related DNA viruses causing infections manifested as dermatologic, immunologic, or neurologic disorders (47). Previous studies in this and other laboratories have demonstrated that macrophages, NK cells, and different T-lymphocyte populations all play important roles in control of HSV infections (3, 12, 20, 40). We have recently shown that HSV infection triggers production of cytokines and chemokines in two temporal waves, one independent of viral replication and one dependent on this activity (18, 26, 28). Furthermore, we have reported that HSV induces expression of the chemokine RANTES/CCL5 in the murine macrophage cells line RAW264.7 through a mechanism dependent on the viral gene encoding infected cell protein 0 (ICP0) (18). In addition, this laboratory has identified the cellular IFN-induced double-stranded RNA-activated protein kinase R (PKR) as a central component in induction of TNF-α, IL-6, and RANTES, since expression of a dominant-negative mutant form of PKR strongly inhibits production of these cytokines in HSV-infected macrophages (18, 26, 28).
Besides the immunostimulatory activity of HSV, this virus is also in possession of mechanisms capable of suppressing the antiviral host response. For instance, viral ICP34.5 counteracts the action of IFN-α/β by reversing the phosphorylation of eukaryotic initiation factor 2α mediated by PKR (9). Another viral protein, ICP47, associates with transporter in antigen processing, or TAP, and thus inhibits presentation of antigens to CD8+ T cells in the context of major histocompatibility complex class I (7, 10).
Here we report the finding of a novel mechanism of immune suppression by HSV. We demonstrate that the virus down-modulates the production of several proinflammatory cytokines in a number of different cell types by mediating instability of proinflammatory cytokine mRNA. Moreover, our data provide evidence that this phenomenon is also seen in vivo and hence suggest that HSV targets the proinflammatory host response as a means of immune evasion.
MATERIALS AND METHODS
Mice, cells, and viruses.
The mice used were eight-week-old female BALB/c mice (M&B Taconic). Activation of peritoneal cells (PCs) was induced by injection of 2 ml of 10% thioglycolate into the peritoneal cavity. Five days later, PCs were harvested by lavage of the peritoneal cavity with cold phosphate-buffered saline (PBS) supplemented with 2% fetal calf serum (FCS; BioWhittaker) and 20 IU of heparin (Leo Pharma) per ml. The cells were washed once in RPMI 1640 medium-5% FCS, seeded at a density of 3.5 × 105 per well in 96-well tissue culture plates, and left overnight before further treatment.
The murine macrophage cell lines RAW264.7 and J774A.1 and the murine fibroblast cell line NIH 3T3 were maintained in Dulbecco's minimal essential medium (DMEM) with 1% Glutamax I (Life Technologies) supplemented with antibiotics and FCS at a concentration of 5% (RAW264.7 and J774A.1) or 10% (NIH 3T3). The growth conditions for the cell lines pBKCMV and PKRM7, which are derived from RAW264.7 (16), further included 200 μg of G418 (Roche) per ml. For 1 to 4 h prior to virus infection, NIH 3T3 cells were deprived of serum. The human osteosarcoma cell line U2OS was kept in DMEM with 10% FCS. The African green monkey kidney cell line Vero and the murine fibroblast cell line L929 were grown in minimal essential medium containing 5% FCS.
The wild-type (wt) viruses used in this study were the KOS and 17+ strains of HSV-1. The mutant viruses used are shown in Table 1. The viruses were produced as previously described (6, 27). The virus was quantified by three methods. First, plaque titration on U2OS cells (17+, in1814, and dl1403), Vero cells (KOS and d22), or Vero-derived cell lines (gL-86, vi-13, and d27-1). Second, glycoprotein D levels in the virus preparations were estimated by Western blotting. Third, the preparations were examined by electron microscopy. Only preparations in which the ratio of the viral titer to the particle concentration resembled that of preparations of wt virus were used for experiments. Prior to use, the virus was thawed and used as infectious virus, subjected to heat inactivation at 56°C for 30 min, or inactivated by exposure to UV light for 15 min. Phosphonoacetic acid (PAA) was obtained from Sigma and used at a concentration of 500 μg/ml.
TABLE 1.
HSV-1 mutants used in the present study
| Gene deleted | Parental strain | Mutant designation | Kinetic class | Reference |
|---|---|---|---|---|
| VP16 | 17+ | in1814 | La | 1 |
| ICP0 | 17+ | dl1403 | IE | 43 |
| gL | KOS | gL86 | Lb | 23 |
| ICP4 | KOS | vi-13 | IE | 39 |
| ICP22 | KOS | d22 | IE | 13 |
| ICP27 | KOS | d27-1 | IE | 34 |
Tegument protein.
Virion glycoprotein.
In vivo virus infection and ex vivo cytokine production.
Mice were inoculated intraperitoneally with 106 PFU of either a mock virus preparation, infectious wt virus, or VP16 mutant HSV-1. At 24 h postinfection (p.i.), the mice were sacrificed and PCs were harvested as described above. To allow measurement of cytokine production by the isolated cells, PCs were cultured in RPMI 1640 medium supplemented with 5% FCS at a density of 3.5 × 105 per well in 96-well tissue culture plates. Twenty-four hours later, the supernatants were harvested and cytokine concentrations were measured.
Virus plaque titration.
Vero and U2OS cells were seeded at a density of 1.2 × 106/5-cm-diameter plate and left overnight to settle. Infection was performed by incubating cells with 100 μl of serial dilutions of the virus in a total volume of 500 μl. The tissue culture dishes were rocked every 15 min to ensure even distribution of the virus. After 1 h, supernatants were removed and 8 ml of preheated minimal essential medium supplemented with 2% FCS and 0.02% human immunoglobulin was added. The cells were incubated for 2 to 6 days, depending on the virus strain, and the cells were stained with 0.03% methylene blue to allow quantification of the plaques.
ELISA.
Murine IL-6, IL-12 p40, and RANTES/CCL5 were detected by enzyme-linked immunosorbent assay (ELISA). Maxisorp plates were coated by overnight incubation at 4°C with the primary antibody (2 μg of anti-IL-6 [Pharmingen] per ml, 6 μg of anti-IL-12 p40 [Pharmingen] per ml, or 0.5 μg of anti-RANTES [Peprotech] per ml) in coating buffer (15 mM Na2CO3; 35 mM NaHCO3; 0.02% sodium azide, pH 9.6). After blocking for 2 h at 37°C or 3 h at room temperature with PBS (pH 7.4) containing 1% (wt/vol) BSA and 0.05% sodium azide, samples and standard dilutions of cytokine (concentration range, 3.9 to 2,000 pg/ml) were added to the wells and the plates were incubated at room temperature for 2 h or overnight at 4°C. Subsequently, the wells were incubated for 2 h at room temperature with a biotin-labeled detection antibody (1 μg of anti-IL-6 [Pharmingen] per ml, 2 μg of anti-IL-12 p40 [Pharmingen] per ml, or 0.25 μg of anti-RANTES [Peprotech] per ml) in blocking buffer. Finally, horseradish peroxidase (HRP)-conjugated streptavidin diluted in blocking buffer was added, the mixture was incubated for 20 min at 20°C, and the result was visualized by the TMB system (R&D Systems). After 10 min, the color reaction was stopped by addition of 5% H2SO4. Between steps, the plates were washed three or four times with PBS containing 0.05% (vol/vol) Tween 20. The results were quantified by reading the A450.
TNF-α and IFN-α/β bioassays.
For measurement of TNF-α and IFN-α/β, L929 cells were seeded at a density of 2 × 104 per well in 96-well tissue culture plates and left for 16 to 24 h at 37°C in a humidified atmosphere with 5% CO2. Infectious virus in the supernatants to be analyzed for cytokine contents were inactivated by exposure to UV light for 15 min. The L929 cell culture supernatants were removed and replaced with the test samples in successive twofold dilutions. For detection of TNF-α, actinomycin D (ActD) was added to a final concentration of 1 μg/ml and the cells were incubated for 18 h at 38.5°C with 1 μg of ActD per ml. For detection of IFN-α/β, the cells were incubated overnight with the test samples and then infected with vesicular stomatitis virus. After an incubation period of 2 to 3 days at 37°C, cells were fixed in 10% formaldehyde and stained with crystal violet (1 mg/ml). Measurement of light A580 and comparison with TNF-α and IFN-α/β standard dilution series allowed assessment of cytokine bioactivity.
Isolation of nuclear extracts and electrophoretic mobility shift assay.
To isolate nuclear proteins, the cell monolayer was washed twice with ice-cold PBS, scraped off the plate, and centrifuged (2,000 × g for 1 min). The cells were resuspended in a hypotonic buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 0.2 mM leupeptin, 0.2 mM pepstatin A, 0.1 mM Na3VO4) and left on ice for 15 min. NP-40 was added to 0.6%, and the mixture was vortexed for 15 s and centrifuged at 10,000 × g for 1 min. The supernatants were removed, and pellets were washed once in the hypotonic buffer. Extraction buffer (20 mM HEPES [pH 7.9], 20% glycerol, 1.5 mM MgCl2, 420 mM NaCl, 0.5 mM DTT, 0.2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 0.2 mM leupeptin, 0.2 mM pepstatin A, 0.1 mM Na3VO4) was added to the nuclei, and the mixture was incubated for 30 min at 4°C with rocking. The samples were centrifuged at 10,000 × g for 15 min at 4°C, and supernatants containing nuclear proteins were harvested.
To assay for DNA-binding activity, 5 μg of protein in 3 μl of nuclear extraction buffer was mixed with 4 μg of poly(dI-dC) and 50,000 cpm of 32P-labeled probe in 18 μl. The final concentrations were 4 mM Tris-HCl, 23 mM HEPES (pH 7.9), 66 mM NaCl, 5 mM MgCl2, 0.7 mM EDTA, 1 mM DTT, and 14% glycerol. After 25 min of incubation at room temperature, the reaction mixture was subjected to electrophoresis on a nondenaturing 5% polyacrylamide gel in 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA). The gel was dried and analyzed by autoradiography. As a probe, we used the distal κB site of the murine NOS2 promoter (5′-TAG GGG GAT TTT CCC CTC-3′).
Whole-cell extracts and Western blotting.
Virus samples were denatured in sample buffer (140 mM Tris-HCl [pH 8.5], 10% glycerol, 2% sodium dodecyl sulfate, 1 mM EDTA, 0.019% Serva blue G250, 0.06% phenol red), heated to 80°C for 10 min, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were blotted onto a polyvinylidene difluoride membrane and blocked for 1 h with TBS (10 mM Tris, 140 mM NaCl) supplemented with 0.05% Tween 20 and 5% skim milk powder. Murine monoclonal anti-glycoprotein D (Virusys) was added for overnight incubation at 4°C. The membrane was washed four times for 10 min each in washing buffer (TBS with 0.05% Tween 20) and incubated for 1 h at room temperature with an HRP-conjugated polyclonal anti-mouse immunoglobulin (Transduction Laboratories). The membrane was washed as described above, and the HRP-conjugated antibody was visualized by enhanced chemiluminescence.
Transfections and reporter gene assay.
NIH 3T3 cells were transfected with LipofectAMINE (Invitrogen). Briefly, for each well, 2.9 μg of DNA (pGL2-220 RANTES promoter (5) and 7.7 μl of LipofectAMINE were mixed in a volume of 80 μl of serum-free DMEM. After 20 min of incubation at room temperature, 400 μl of serum-free medium was added to the mixture. The cells, seeded at a density of 106 per well in six-well tissue culture plates, were washed once with 5 ml of preheated serum-free medium, and the DNA-LipofectAMINE solution was gently added to the cells. Plates were gently rocked every 20 min for 3 to 4 h, at which point 2.5 ml of preheated medium supplemented with 12% FCS was added. After overnight incubation, the growth medium was exchanged with serum-free DMEM, and 2 h later, the cells were infected. After an 8- to 24-h incubation period, the cells were lysed and luciferase activity was measured with the Luciferase Assay System (Promega).
RT-PCR.
To isolate RNA, cells were lysed in Trizol (Invitrogen) and phase separated by addition of chloroform and centrifugation at 12,000 × g for 15 min (4°C). The aqueous phase was isolated, and RNA was precipitated with isopropanol and pelleted by centrifugation for 10 min at 10,000 × g (4°C). Two to 4 μg of RNA was subjected to reverse transcription (RT) with oligo(dT) as the primer and Expand reverse transcriptase. The cDNA was amplified by PCR with the following primers: RANTES, 5′-ATA TGG CTC GGA CAC CAC TC-3′ (sense) and 5′-GAT GCC GAT TTT CCC AGG AC-3′ (antisense); IL-6, 5′-TTC TGG AGT ACC ATA GCT AC-3′ (sense) and 5′-AGT TCT TCG TAG AGA ACA AC-3′ (antisense); β-actin, 5′-CCC ACT CCT AAG AGG AGG ATG-3′ (sense) and 5′-AGG GAG ACC AAA GCC TTC AT-3′ (antisense). The products for RANTES, IL-6, and β-actin spanned 330, 370, and 214 bp, respectively.
Statistical analysis.
The data are presented as means ± the standard errors of the means (SEM). Statistical significance was estimated with Student's t test for unpaired observations, and P values of <0.05 were considered significant.
RESULTS
Mutation of VP16 potentiates the cytokine-inducing potential of HSV-1.
Replication of HSV is initiated by entry of the virus into cells through fusion of the viral and cellular membranes (42). Following this event, a number of so-called tegument proteins present in the virion are released into the infected cell, and these tegument proteins facilitate viral replication through different mechanisms. One tegument protein, VP16, is a transcriptional activator that associates with cellular transcription factors and enhances transcription of viral immediate-early (IE) genes and hence production of progeny virus.
Previous studies in this laboratory have shown that production of many cytokines and chemokines during HSV infection relies on a functional viral genome (17, 18, 26), whereas TNF-α expression is induced partly independently of viral replication (28). In an attempt to identify the viral genes responsible for this phenomenon, we examined the ability of the HSV-1 mutant in1814 to induce expression of cytokines. This mutant carries a 12-bp insertion in the VP16 gene, which compromises the ability of VP16 to interact with cellular transcriptional activators (1), thereby impairing the replication competence of this virus.
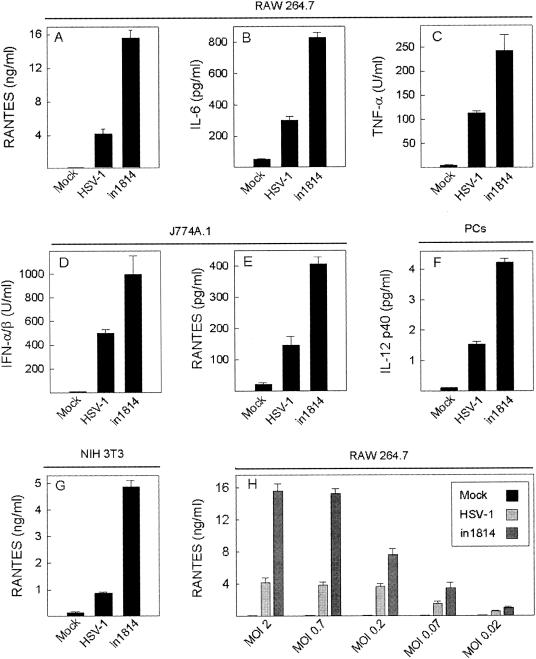
To our surprise, we found that uncoupling of VP16 strongly potentiated the ability of HSV-1 to trigger expression of IL-6, TNF-α, IFN-α/β, IL-12 p40, and RANTES in several different cell lines and primary cells (Fig. 1A to G). This finding was reproduced with three different virus preparations of both wt HSV-1 and in1814. To further characterize in1814-induced cytokine production, we performed a dose-response experiment, which revealed that the enhancement of cytokine production by in1814 versus wt HSV-1 was seen over a broad range of virus concentrations (Fig. 1H), hence adding more weight to the potential physiological relevance of our finding.
FIG. 1.
VP16 mutation potentiates HSV-1-induced cytokine expression. The cells (RAW264.7, A, B, C, and H; J774A.1, D and E; PCs, F; NIH 3T3, G) were seeded in triplicate cultures, left overnight to settle, and treated with a mock preparation or infected with wt HSV-1 or in1814 (A to F, MOI of 1; G, MOI of 2; H, MOI of 0.02 to 2) as indicated. At 8 (C) and 24 (A, B, and D to H) h later, the supernatants were harvested and RANTES, IL-6, and IL-12 p40 were measured by ELISA while IFN-α/β and TNF-α were measured by bioassays. The results are shown as means ± SEM. Essentially similar results were seen in more than five independent experiments for the results presented in panels A to E, G, and H and in two independent experiments for the result presented in panel F.
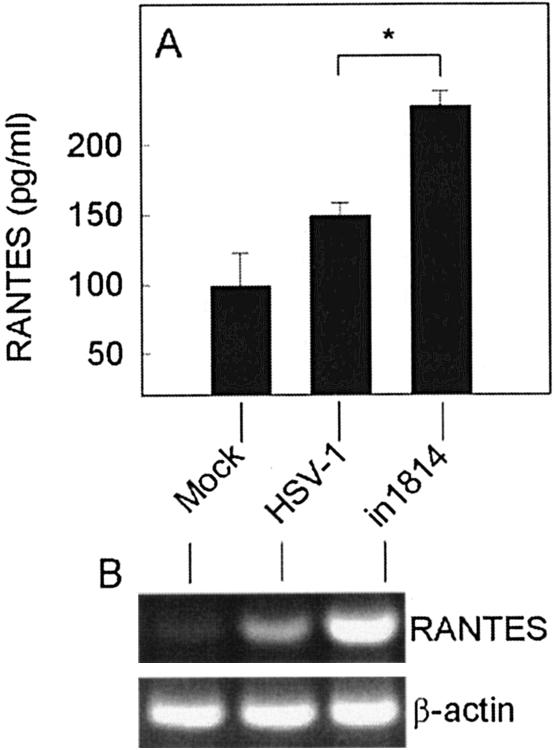
In order to examine if the same scenario was present during HSV-1 infection in vivo, we infected BALB/c mice intraperitoneally and harvested PCs 24 h later. The cells were cultured for 24 h, and RANTES levels in the supernatants were measured. As seen in Fig. 2A, PCs infected in vivo with wt virus did not produce RANTES expression to any major extent after 24 h of culture in vitro. However, PCs from mice infected with in1814 were induced to express RANTES and, importantly, produced significantly greater amounts than PCs from mice infected with wt virus. When mRNA was harvested from the PCs immediately after the mice had been sacrificed, wt HSV-1 did stimulate expression of RANTES but not to the same extent as in1814 (Fig. 2B). In summary, impaired VP16 activity led to hyperactivation of cytokine production during HSV-1 infection in vitro and in vivo.
FIG. 2.
VP16 mutation potentiates expression of RANTES during HSV-1 infection in vivo. Eight-week-old female BALB/c mice were infected with wt or VP16-deficient HSV-1 (106 PFU) or treated with a mock preparation. At 24 h later, PCs were harvested and either cultured for 24 h in RPMI 1640 medium supplemented with 5% FCS or lysed for subsequent RNA purification. (A) RANTES was measured in the cell culture supernatants by ELISA. The results are shown as means of five mice per group ± SEM. Essentially similar results were seen in two independent experiments. *, P < 0.05. (B) Total RNA was reverse transcribed and analyzed for the presence of RANTES and β-actin by PCR. Essentially similar results were seen in two independent experiments.
Suppression of cytokine expression by HSV-1 is dependent on the viral genes for ICP4 and ICP27 but is independent of post DNA replication events.
Since VP16 induces expression of IE genes, which in turn promote expression of the downstream early (E) and late (L) gene products, we wanted to examine if VP16 per se or VP16-induced genes were responsible for the observed hyperstimulatory potential of in1814 with respect to cytokine production. To this end, we used a panel of HSV-1 mutants each lacking a gene encoding one of the regulatory IE HSV proteins (Table 1). These are all transcribed through a VP16-dependent mechanism (4).
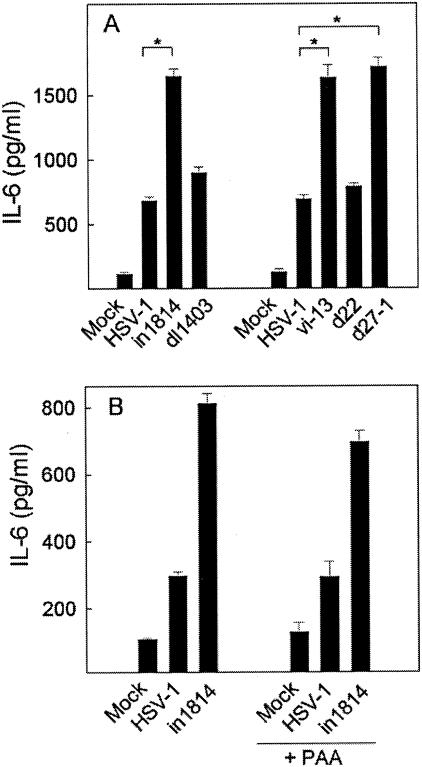
RAW264.7 cells were infected with the mutants, and IL-6 was measured in the supernatants harvested 24 h p.i. Lack of neither ICP0 (dl1403) nor ICP22 (d22) appeared to affect the IL-6-inducing potential of the virus (Fig. 3A), thus showing that the virus-cell interactions that trigger cytokine expression may differ between cytokines or groups of cytokines, since we have previously reported that ICP0 is required for induction of RANTES expression in RAW264.7 cells (18). Interestingly, mutants lacking either ICP4 (vi-13) or ICP27 (d27-1) closely resembled in1814 in being hyperpotent inducers of cytokine production (Fig. 3A). In NIH 3T3 cells, vi-13 and d27-1 induced RANTES expression to the same extent as did in1814 (data not shown). However, for reasons we do not understand, the ICP4 mutant induced RANTES expression only slightly more strongly than did the wt virus in RAW264.7 cells and the ICP27 deletion mutant resembled the wt virus with respect to induction of RANTES expression in this cell line (data not shown).
FIG. 3.
Down-regulation of cytokine expression by HSV-1 is dependent on viral IE and/or E genes. RAW264.7 cells were seeded in triplicate cultures and left overnight to settle. The cells received a mock preparation or wt or mutant HSV-1 (MOI of 1) as indicated. In panel B, some wells were treated with PAA (500 μg/ml) prior to infection. The cells were incubated for 24 h, and supernatants were harvested for measurement of IL-6 by ELISA. Essentially similar results were seen in three independent experiments. Results are shown as means ± SEM. *, P < 0.05.
To evaluate if the suppression of cytokine expression was mediated by HSV-1 L gene products, we infected RAW264.7 cells with wt HSV-1 or in1814 in the presence or absence of PAA, an inhibitor of viral DNA replication and hence L gene expression. As shown in Fig. 3B, the presence of PAA did not affect induction of IL-6 by either virus, thus suggesting that DNA replication and viral L genes are dispensable for suppression of cytokine expression. This result further demonstrated that the observed phenomenon was not simply a consequence of cytopathic effects (CPE) triggered by infection with wt virus but not in1814, vi-13, and d27-1, since PAA abolished any overt CPE caused by infection.
Collectively, these data suggested that HSV-1 suppressed expression of cytokines by a mechanism dependent on ICP4 and ICP27 and independent of post DNA replication events.
PKR is not the major target in HSV-1-mediated suppression of cytokine expression.
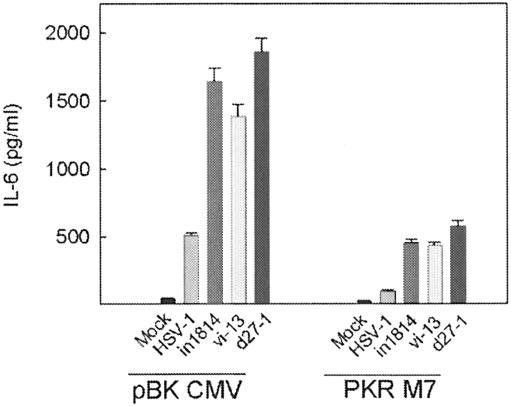
We have previously demonstrated that PKR is required for optimal HSV-induced expression of many cytokines, including IL-6, RANTES, and TNF-α (18, 26, 28). Thus, we hypothesized that viral targeting of PKR would lead to decreased expression of proinflammatory mediators. To address this question, we compared RAW264.7 cells stably transfected with a dominant-negative PKR construct (PKR M7) and those stably transfected with a control vector (pBK CMV) with respect to HSV-induced expression of IL-6. Confirming previous findings (26), lack of functional PKR strongly impaired IL-6 expression after HSV-1 infection (Fig. 4). Despite this, in1814, vi-13, and d27-1 remained significantly stronger inducers of IL-6 expression than wt HSV-1, even in the absence of a functional PKR system. Thus, PKR did not seem to be the major target in HSV-1-mediated down-regulation of proinflammatory cytokine production.
FIG. 4.
Suppression of cytokine expression by HSV-1 is independent of PKR. RAW264.7-derived cell lines stably transfected with control plasmid (pBK CMV) or a dominant-negative PKR construct (PKR M7) were serum starved for 5 h before treatment with a mock preparation or infection with wt HSV-1 or the mutant in1814 (VP16), vi-13 (ICP4), or d27-1 (ICP27) (MOI of 1). Supernatants were harvested 24 h later, and IL-6 was measured by ELISA. The results are shown as means of triplicate cultures ± SEM. Essentially similar results were observed in three independent experiments.
HSV-1-mediated suppression of cytokine expression is exerted at the level of mRNA stability.
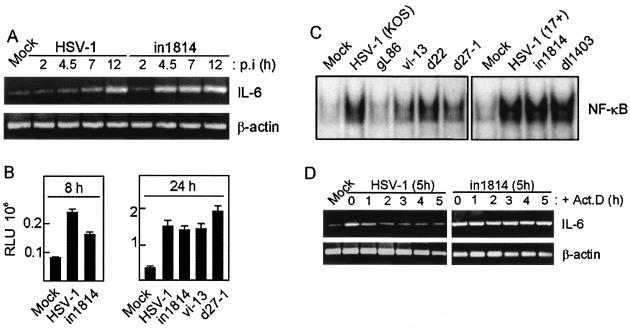
In order to address the mechanism underlying the observed phenomenon, we first did a time course experiment in which RAW264.7 cells were infected with wt HSV-1 or in1814, after which mRNA was harvested and IL-6 and β-actin were amplified by RT-PCR. As shown in Fig. 5A, IL-6 mRNA accumulated slowly after infection with wt HSV-1, with only modestly elevated levels at 4.5 and 7 h p.i. and strong induction seen only at 12 h p.i. By contrast, infection with in1814 already led to strong induction of IL-6 mRNA within 4.5 h of infection, which remained elevated through the 12 h of the experiment. The observed phenomenon was not attributable to the nonquantitative nature of the RT-PCR technique, since real-time PCR analysis of selected samples from the same RNA preparation gave the same result (data not shown).
FIG. 5.
HSV-1 down-modulates proinflammatory gene expression at the level of mRNA stability. (A) RAW264.7 cells were infected with HSV-1 or in1814, and RNA was harvested at the indicated time point p.i. IL-6 and β-actin mRNAs were amplified by RT-PCR and visualized by ethidium bromide staining of the agarose gel. Essentially similar results were seen in two independent experiments. (B) NIH 3T3 cells were transiently transfected with a RANTES promoter gene construct and treated as indicated. After 8 and 24 h, cells were lysed and luciferase activity was measured. The results are shown as means ± SEM. RLU, relative luciferase units. Essentially similar results were seen in two independent experiments. (C) J774A.1 cells were treated for 2 h with a mock virus preparation or infected with wt or mutant HSV-1 (MOI of 1) as indicated. Nuclear proteins were harvested, and NF-κB DNA-binding activity was measured by electrophoretic mobility shift assay. Essentially similar results were seen in two independent experiments. (D) RAW264.7 cells were infected with HSV-1 or in1814 for 5 h, at which point ActD was added. RNA was extracted from cells at various intervals after the addition of ActD, and IL-6 and β-actin mRNAs were amplified by RT-PCR and visualized by ethidium bromide staining of the agarose gel. Essentially similar results were seen in two independent experiments.
To examine if the differences in the kinetics of cytokine mRNA accumulation were attributable to transcriptional effects, we transfected NIH 3T3 cells with a RANTES promoter construct and infected cells with wt HSV-1 or in1814. This construct includes most of the known cis elements involved in activation of RANTES expression, including two NF-κB sites, one NF-IL-6 binding site, one IFN-stimulated response element, and one cyclic AMP response element (5). After 8 or 24 h of infection, cells were lysed and luciferase activity was measured. As seen in Fig. 5B, wt HSV-1 was at least as strong an activator of transcription from the RANTES promoter as in1814, vi-13, and d27-1, hence demonstrating that the suppression of cytokine expression by HSV-1 was not mediated at the level of transcription. This idea was further supported by the finding that wt HSV-1 did not suppress activation of the transcription factor NF-κB (Fig. 5C), which is essential for virus-induced expression of all of the cytokines examined in this work (21, 22, 27).
Another potential mechanism through which HSV-1 could inhibit production of cytokines is at the level of pre-mRNA splicing. This idea is particularly interesting here in light of the findings that ICP27 is known to be capable of inhibiting splicing of host pre-mRNA (38) and that the ICP27-deficient mutant d27-1 induced significantly higher levels of proinflammatory cytokines than the wt virus (Fig. 3B and 4B). However, since the genes encoding IFN-α and IFN-β are intronless, and since production of IFNs was elevated after infection with in1814 or d27-1 compared to that seen after infection with the wt virus (Fig. 1D and data not shown), it seemed that inhibition of pre-mRNA splicing was not the main mechanism through which HSV-1 inhibits proinflammatory cytokine expression.
With the aim of examining if infection with wt HSV-1 versus in1814 differentially affected the stability of mRNA for proinflammatory genes, we treated RAW264.7 cells with the two viruses for 5 h, at which point we added ActD to block de novo RNA synthesis. Total RNA was harvested at various time points following this treatment, and IL-6 and β-actin were amplified by RT-PCR. In cells treated with wt HSV-1, a modest but clear induction of IL-6 mRNA was seen at 5 h p.i. (Fig. 5D). Prevention of new RNA synthesis led to a rapid decrease in the levels of IL-6 mRNA with a clear decrease already 1 h after addition of ActD. Interestingly, in cells treated with in1814, the levels of IL-6 mRNA only decreased very slowly after addition of ActD, with no significant decrease observed even 5 h after addition of ActD. Notably, the levels of β-actin mRNA were not decreased at this time point regardless of the infection. Real-time PCR analysis of selected samples from the same RNA preparation gave essentially similar results (data not shown). These data thus suggested that wt HSV-1, but not in1814, destabilizes IL-6 mRNA.
DISCUSSION
The first line of defense against viral infections involves production of IFN-α/β (2) and activation of the innate immune response by recruitment of, e.g., NK cells (36) and antigen-presenting cells, notably, dendritic cells. The latter process is regulated by chemokines, including RANTES/CCL5 (15). The innate immune response also shapes the nature of the subsequent adaptive immune response, mainly through the cytokines produced by leukocytes of the innate immune system. For instance, IL-12 drives development of type 1 immunity, which is generally believed to promote defense against viral infections (32). In this work, we demonstrate that HSV-1 suppresses production of cytokines exerting their effects at many different stages of the antiviral response, including IFN-α/β, TNF-α, IL-6, IL-12, and RANTES. The virus thus hampers the ability of the host to mount an antiviral response and in this way promotes the establishment of infection.
We found that the HSV-1 mutant in1814, with a mutation in VP16 that strongly impairs the ability to activate IE gene transcription, was a hyperpotent inducer of expression of many cytokines and chemokines in a number of cell lines, as well as in primary PCs. Moreover, ex vivo production of RANTES by PCs after 24 h of infection in vivo was significantly augmented after infection with in1814 compared to wt HSV-1. However, the observed phenomenon did not appear to be mediated directly by VP16, since HSV-1 mutants lacking the viral IE gene for ICP4 or ICP27 (both expressed through a VP16-dependent mechanism [4]) mimicked in1814 with respect to cytokine-inducing potential. In addition, we found that the potentiated ability of the HSV-1 mutants to induce cytokine expression was not simply due to a reduced CPE of these viruses compared to that of wt virus, since treatment with PAA, which totally prevented the virus-induced CPE, did not affect cytokine expression. Finally, the lack of effect of PAA demonstrates that suppression of cytokine expression by HSV-1 is independent of DNA replication and viral L gene expression. The observation that replication-impaired mutants display a greater cytokine-inducing potential than the wt virus may appear contradictory to previous reports from this laboratory, showing that HSV-induced cytokine expression is dependent on viral gene expression (18, 26, 28). However, given the highly reproducible nature of both the previous and present findings, they ought to be reconciled. We favor the idea that the mutants are in fact weaker inducers of cytokine gene transcription than the wt virus because of reduced viral gene expression. However, the mechanism of mRNA destabilization, which is operative in wt HSV-1 but impaired in in1814, vi-13, and d27-1, gives the net outcome that cells infected with the mutants produce greater amounts of cytokines. This hypothesis is supported by the data presented in Fig. 5B, which show that in1814 is indeed a weaker activator of transcription from the RANTES promoter than is the wt virus at early time points p.i.
Our finding that suppression of cytokine expression was mediated by ICP4 and ICP27 suggested that the mechanism described in this paper could also be at play during reactivation of HSV from latency, since activation of HSV replication after latency is independent of VP16 but is initiated by accumulation of superthreshold levels of IE genes (31). It is interesting to speculate that these two viral proteins might delay cytokine production to an extent that would allow the virus to proceed with replication before facing the potent adaptive immune response during reactivation of HSV-1 from latency.
We have previously shown that HSV-induced expression of many cytokines is dependent on PKR (18, 26, 28). In one study, we showed that induction of RANTES, which is sensitive to UV treatment of the virus, correlates with accumulation of viral IE genes and is inhibited by dominant-negative PKR (18). We have subsequently shown that HSV-induced RANTES expression is dependent on NF-κB (19, 21), and others have reported that HSV-1 activates NF-κB through a PKR-dependent mechanism (46). Therefore, a viral mechanism of PKR inhibition would potentially reduce expression of proinflammatory cytokines. Others have demonstrated that HSV can reverse PKR phosphorylation of eukaryotic initiation factor 2α by an ICP34.5-dependent mechanism, and in an unusual mutant form, HSV can prevent PKR activation by a Us11-dependent mechanism (9, 29). However, the mechanism by which HSV-1 suppresses proinflammatory cytokine production in the present study appears to be mainly independent of PKR, since in1814 remained a significantly stronger inducer of cytokine expression than the wt virus in a macrophage cell line stably overexpressing a dominant-negative mutant form of PKR.
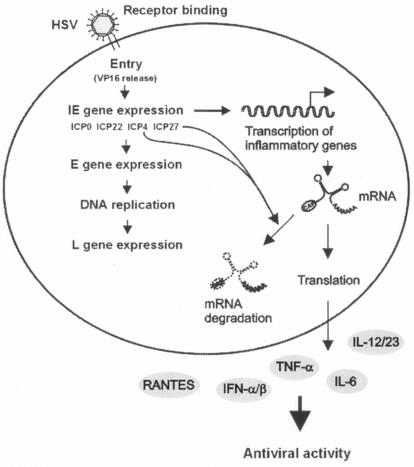
Many viruses are in possession of mechanisms to down-modulate the production of cytokines. For instance, African swine fever virus inhibits activation of NF-κB, thereby preventing establishment of a proinflammatory cytokine response (30, 33). Recently, it was demonstrated that measles virus counteracts cytokine expression by inhibiting signal transduction through the Jak-STAT pathway (25). Here, we found that HSV counteracted cytokine expression during the first hours of infection by destabilizing the mRNAs of proinflammatory genes. Although the molecular events governing this process still remain to be described, our present findings, together with previous work in this laboratory (18, 26-28), support the model shown in Fig. 6. HSV IE gene expression induces transcription of genes encoding proinflammatory cytokines (18, 26-28). However, to counteract this response, two IE proteins (ICP4 and ICP27) are capable of triggering mechanisms that destabilize virus-induced cellular mRNAs, with the net outcome being only modest production of proinflammatory cytokines. One potential mechanism could involve activation of RNase L by ICP4 and ICP27 mRNA, with subsequent nonselective degradation of RNA in the cell. However, since the stability of β-actin mRNA was not differentially affected by infection with wt HSV-1 versus in1814, RNase L appears not to be a likely candidate. Moreover, a recent study demonstrated that HSV-1 does not activate RNase L and that mRNA degradation in HSV-infected cells is independent of RNase L (41). The HSV tegument protein UL41 (or VHS), which promotes mRNA degradation, has previously been shown to decrease accumulation of some virus-induced cellular mRNAs (44), although the majority of cellular genes induced by HSV-1 infection are not affected by UL41 (45). However, the suppression of cytokine expression described here appears to be mediated by a distinct mechanism, since treatment of cells with UV-inactivated HSV-1, which did deliver tegument proteins to the cell, did not affect lipopolysaccharide-induced RANTES expression (data not shown). Also, the observation that β-actin mRNA levels were unaffected by virus infection argues for a more specific mechanism being involved.
FIG. 6.
Model of HSV-1-mediated suppression of cytokine expression. Accumulation of HSV IE genes correlates with induction of cytokines and chemokine expression. The data presented in this work demonstrate that two IE genes, ICP4 and ICP27, actively counteract the expression of many proinflammatory cytokines with antiviral potential by decreasing mRNA stability, a phenomenon that could potentially impede the host defense against HSV.
HSV uses a wealth of strategies to counteract the antiviral response of the infected host. It has previously been reported that the virus counteracts PKR-mediated events (9, 29) and inhibits maturation of dendritic cells (37), antigen presentation by major histocompatibility complex class I (7, 10), and killing of cells by CTLs (11). To this list we now add inhibition of cytokine expression. By preventing infected cells from producing antiviral and proinflammatory mediators, HSV-1 may hamper the ability of the host to mount a strong immune response, thus facilitating viral replication during the early stages of primary, as well as recurrent, HSV infection.
Acknowledgments
We thank Roger D. Everett, Chris M. Preston, Patricia G. Spear, Neal A. Deluca, Stephen Rice, and David M. Knipe for providing the virus mutants. The technical support of Birthe Søby and Elin Jakobsen is greatly appreciated.
This work was supported by grants from The Carlsberg Foundation, The Lundbeck Foundation, and The Danish Medical Research Council (grant 22-02-0144). J.M. and L.M. were supported by fellowships from the Faculty of Health Sciences, University of Aarhus.
REFERENCES
- 1.Ace, C. I., T. A. McKee, J. M. Ryan, J. M. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biron, C. A. 1999. Initial and innate responses to viral infections. Curr. Opin. Microbiol. 2:374-381. [DOI] [PubMed] [Google Scholar]
- 3.Biron, C. A., K. S. Byron, and J. L. Sullivan. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320:1731-1735. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 25:1-19. [DOI] [PubMed] [Google Scholar]
- 5.Casola, A., R. P. Garofalo, H. Haeberle, T. F. Elliott, R. Lin, M. Jamaluddin, and A. R. Brasier. 2001. Multiple cis regulatory elements control RANTES promoter activity in alveolar epithelial cells infected with respiratory syncytial virus. J. Virol. 75:6428-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellermann-Eriksen, S. 1993. Autocrine secretion of interferon-α/β and tumour necrosis factor-α synergistically activates mouse macrophages after infection with herpes simplex virus type 2. J. Gen. Virol. 74:2191-2199. [DOI] [PubMed] [Google Scholar]
- 7.Fruh, K., K. Ahn, H. Djaballah, R. Sempe, P. A. Peterson, and Y. Yang. 1995. A viral inhibitor of peptide transporter of antigen presentation. Nature 375:415-418. [DOI] [PubMed] [Google Scholar]
- 8.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15:259-267. [DOI] [PubMed] [Google Scholar]
- 9.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411-415. [DOI] [PubMed] [Google Scholar]
- 11.Jerome, K. R., J. F. Tait, D. M. Koelle, and L. Corey. 1998. Herpes simplex virus type 1 renders infected cells resistant to cytotoxic T-lymphocyte-induced apoptosis. J. Virol. 72:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodukula, P., T. Liu, N. van Rooijen, M. J. Jager, and R. L. Hendricks. 1999. Macrophage control of herpes simplex virus type 1 replication in the peripheral nervous system. J. Immunol. 162:2895-2905. [PubMed] [Google Scholar]
- 13.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucey, D. R., M. Clerici, and G. M. Shearer. 1996. Type 1 and type 2 cytokine dysregulation in human infectious, neoplastic, and inflammatory diseases. Clin. Microbiol. Rev. 9:532-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackay, C. R. 2001. Chemokines: immunology's high impact factors. Nat. Immunol. 2:95-101. [DOI] [PubMed] [Google Scholar]
- 16.Maggi, L. B., Jr., M. R. Heitmeier, D. Scheuner, R. J. Kaufman, R. M. Buller, and J. A. Corbett. 2000. Potential role of PKR in double-stranded RNA-induced macrophage activation. EMBO J. 19:3630-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malmgaard, L., S. R. Paludan, S. C. Mogensen, and S. Ellermann-Eriksen. 2000. Herpes simplex virus type 2 induces interleukin-12 in macrophages through a mechanism involving NF-κB. J. Gen. Virol. 81:3011-3020. [DOI] [PubMed] [Google Scholar]
- 18.Melchjorsen, J., F. S. Pedersen, S. C. Mogensen, and S. R. Paludan. 2002. Herpes simplex virus selectively induces expression of the CC chemokine RANTES/CCL5 in macrophages through a mechanism dependent on PKR and ICP0. J. Virol. 76:2780-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melchjorsen, J., and S. R. Paludan. 2003. Induction of RANTES/CCL5 by herpes simplex virus is regulated by nuclear factor κB and interferon regulatory factor. J. Gen. Virol. 84:2491-2495. [DOI] [PubMed] [Google Scholar]
- 20.Mogensen, S. C. 1978. Macrophages and age-dependent resistance to hepatitis induced by herpes simplex virus type 2 in mice. Infect. Immun. 19:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mogensen, T. H., J. Melchjorsen, P. Hollsberg, and S. R. Paludan. 2003. Activation of NF-κB in virus-infected macrophages is dependent on mitochondrial oxidative stress and intracellular calcium: downstream involvement of the kinases TGF-β-activated kinase 1, mitogen-activated kinase/extracellular signal-regulated kinase kinase 1, and IκB kinase. J. Immunol. 170:6224-6233. [DOI] [PubMed] [Google Scholar]
- 22.Mogensen, T. H., and S. R. Paludan. 2001. Molecular pathways in virus-induced cytokine production. Microbiol. Mol. Biol. Rev. 65:131-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 24.Orange, J. S., M. S. Fassett, L. A. Koopman, J. E. Boyson, and J. L. Strominger. 2002. Viral evasion of natural killer cells. Nat. Immunol. 3:1006-1012. [DOI] [PubMed] [Google Scholar]
- 25.Palosaari, H., J. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paludan, S. R. 2001. Requirements for the induction of interleukin-6 by herpes simplex virus-infected leukocytes. J. Virol. 75:8008-8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paludan, S. R., S. Ellermann-Eriksen, V. Kruys, and S. C. Mogensen. 2001. Expression of TNF-α by herpes simplex virus-infected macrophages is regulated by a dual mechanism: transcriptional regulation by NF-κB and activating transcription factor 2/Jun and translational regulation through the AU-rich region of the 3′ untranslated region. J. Immunol. 167:2202-2208. [DOI] [PubMed] [Google Scholar]
- 28.Paludan, S. R., and S. C. Mogensen. 2001. Virus-cell interactions regulating induction of tumor necrosis factor alpha production in macrophages infected with herpes simplex virus. J. Virol. 75:10170-10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell, P. P., L. K. Dixon, and R. M. E. Parkhouse. 1996. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 70:8527-8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 32.Ramshaw, I. A., A. J. Ramsay, G. Karupiah, M. S. Rolph, S. Mahalingam, and J. C. Ruby. 1997. Cytokines and immunity to viral infections. Immunol. Rev. 159:119-135. [DOI] [PubMed] [Google Scholar]
- 33.Revilla, Y., M. Callejo, J. M. Rodriguez, E. Culebras, M. L. Nogal, M. L. Salas, E. Vinuela, and M. Fresno. 1998. Inhibition of nuclear factor κB activation by a virus-encoded IκB-like protein. J. Biol. Chem. 273:5405-5411. [DOI] [PubMed] [Google Scholar]
- 34.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romagnani, S. 1997. The Th1/Th2 paradigm. Immunol. Today 18:263-266. [DOI] [PubMed] [Google Scholar]
- 36.Salazar-Mather, T. P., J. S. Orange, and C. A. Biron. 1998. Early murine cytomegalovirus (MCMV) infection induces liver natural killer (NK) cell inflammation and protection through macrophage inflammatory protein 1α (MIP-1α)-dependent pathways. J. Exp. Med. 187:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salio, M., M. Cella, M. Suter, and A. Lanzavecchia. 1999. Inhibition of dendritic cell maturation by herpes simplex virus. Eur. J. Immunol. 29:3245-3253. [DOI] [PubMed] [Google Scholar]
- 38.Sandri-Goldin, R. M. 2001. Nuclear export of herpes virus RNA. Curr. Top. Microbiol. Immunol. 259:2-23. [PubMed] [Google Scholar]
- 39.Shepard, A. A., and N. A. DeLuca. 1991. Activities of heterodimers composed of DNA-binding- and transactivation-deficient subunits of the herpes simplex virus regulatory protein ICP4. J. Virol. 65:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sin, J. I., J. J. Kim, C. Pachuk, C. Satishchandran, and D. B. Weiner. 2000. DNA vaccines encoding interleukin-8 and RANTES enhance antigen-specific Th1-type CD4+ T-cell-mediated protective immunity against herpes simplex virus type 2 in vivo. J. Virol. 74:11173-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, T. J., R. H. Silverman, and D. A. Leib. 2003. RNase L activity does not contribute to host RNA degradation induced by herpes simplex virus infection. J. Gen. Virol. 84:925-928. [DOI] [PubMed] [Google Scholar]
- 42.Spear, P. G. 2001. A first step toward understanding membrane fusion induced by herpes simplex virus. Mol. Cell 8:2-4. [DOI] [PubMed] [Google Scholar]
- 43.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 44.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyma, and M. Azuma. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 81:1763-1771. [DOI] [PubMed] [Google Scholar]
- 45.Taddeo, B., A. Esclatine, and B. Roizman. 2002. The pattern of accumulation of cellular RNAs in cells infected with a wild-type and a mutant herpes simplex virus 1 lacking the virion host shutoff gene. Proc. Natl. Acad. Sci. USA 99:17031-17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taddeo, B., T. R. Luo, W. Zhang, and B. Roizman. 2003. Activation of NF-κB in cells productively infected with HSV-1 depends on activated protein kinase R and plays no apparent role in blocking apoptosis. Proc. Natl. Acad. Sci. USA 100:12408-12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitley, R. J. 2001. Herpes simplex virus, p. 2461-2509. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 48.Yewdell, J. W., and A. B. Hill. 2002. Viral interference with antigen presentation. Nat. Immunol. 3:1019-1025. [DOI] [PubMed] [Google Scholar]