Abstract
In models of the replication of human hepatitis delta virus (HDV) RNA, it is generally assumed that circular RNAs are the only templates. However, noncircular HDV RNAs are also produced during replication, and it is known that replication can be initiated by transfection with noncircular RNAs. Therefore, strategies were devised to determine the relative ability of different HDV RNA species to initiate RNA replication. One strategy used in vivo intermolecular competition following cotransfection into cells, between two sequence-marked HDV RNA species. Circular RNA templates were found to be at least severalfold more efficient than a dimeric linear template. Unit-length linear species, that is, equivalent to circles opened at different sites, were in most cases but not always of efficiency comparable to that of each other. Greater-than-unit-length linear species were more efficient than unit-length species, presumably because of the increased opportunities for template switching. Genomic linear RNAs were generally of initiation ability comparable to that of antigenomic RNAs. A second strategy measured the ability of initiation to occur on different regions of HDV RNAs that were twice the unit length. In summary, results from these two experimental strategies make clear that linear HDV RNA species, as well as circles, can contribute to the overall process of HDV genome replication. In addition, the results from the two experimental strategies provided information on the impact of template switching during RNA-directed transcription.
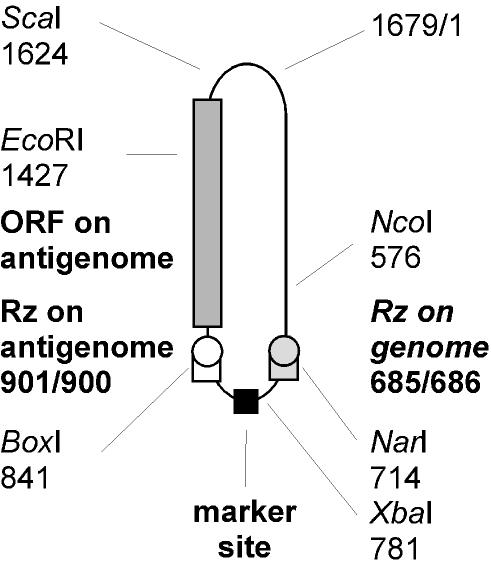
The RNA genome of hepatitis delta virus (HDV) is 1,679 nucleotides (nt) in length. This RNA and its exact complement, the antigenome, are single-stranded species with a circular conformation (26). Both RNAs can fold into an unbranched rodlike structure with extensive intramolecular base pairing (12, 26). Features of both the genome and antigenome are superimposed in Fig. 1. Each RNA contains a unique domain that acts as a site-specific ribozyme. Also, the antigenome contains an open reading frame for a 195-amino-acid protein known as the small delta protein. This protein, which is essential for genome replication (4), is not actually translated from the antigenome but from a less-than-full-length capped and polyadenylated mRNA (5, 8).
FIG. 1.
Superposition of sequence features on genomic and antigenomic RNA circles of HDV. The unit-length (1,679-nt) HDV RNA is indicated as a circular conformation with folding into an unbranched rodlike structure. The numbering is from the nucleotide sequence of Kuo et al. (12), with the origin indicated as 1679/1. Unique restriction enzyme sites relevant to this study are also indicated. Features from the genome and its exact complement, the antigenome, are superimposed on this figure. Note the locations of the genomic and antigenomic ribozymes, the open reading frame for the small delta protein, and, of particular relevance to this study, the location at the bottom of the rodlike structure of a marker site.
RNA-directed RNA synthesis and replication of HDV involve the transcription of RNA multimers that are processed by these ribozymes to release unit-length linear RNAs, which are subsequently ligated to produce RNA circles (13, 22). An additional and alternative form of processing occurs on nascent antigenomic RNAs, to produce the above-mentioned mRNA for the small delta antigen (11).
In models that have been proposed to explain HDV replication, the genomic and antigenomic RNA circles are considered to be the only templates. Hence, the models have been referred to as “double-rolling circle models” (15, 17, 25). However, what has not been properly addressed experimentally is the issue of whether or not these circles are actually the only templates. The nascent linear RNA multimers might act as templates even prior to posttranscriptional processing. Furthermore, many reported transfection studies have shown that linear HDV RNAs are actually capable of initiating HDV genome replication (6, 16). It has sometimes been assumed that such linear RNAs might have to be converted to circles before they could act as templates for initiation (14). However, this opinion is in contrast to data showing that linear RNAs that are only several nucleotides greater than unit length can initiate (6). Furthermore, in a more recent study it was shown that the polymerase used for transcription, presumably pol II, can carry out template switching when faced with a unit-length HDV RNA template (3).
Therefore, we undertook to determine the relative ability of various forms of HDV RNA to initiate RNA-directed RNA synthesis. We made use of two separate strategies. In the first we carried out an intermolecular competition between cotransfected sequence-marked HDV RNAs. In the second, we used an intramolecular approach, in which cells were transfected with linear dimers of the HDV RNA, where each monomer was specifically sequence marked. The results obtained from applications of these two strategies clarify important limitations of the current double-rolling circle model.
MATERIALS AND METHODS
Plasmids and RNA preparation.
All HDV monomeric and dimeric inserts were cloned into the vector pcDNA3 (Invitrogen). In most cases, the ends of the HDV inserts were blunted by a fill-in reaction before cloning into the unique EcoRV site of pcDNA3. As an exception, EcoRI-EcoRI dimers and monomers were cloned into the unique EcoRI site of pcDNA3. Following this cloning strategy, RNAs transcribed from these constructs will all have the same small flanking 5′ and 3′ non-HDV vector sequences.
Constructs randomized at positions 790 to 796, around the bottom region of the HDV rodlike structure, were achieved by using the strategy employed by Wu et al. (27) with minor modifications. Several resulting constructs were tested, and one with a neutral change was chosen for future experiments. Thus, the wild-type sequence 5′-CGAUAGA was replaced with 5′-UGAAGCA.
Prior to in vitro RNA transcription with T7 polymerase, the DNA constructs were linearized with NotI. Genomic and antigenomic 1.1-mer RNAs were transcribed by using the earlier-described plasmids pTW101 and pTW114, respectively (27). Both plasmids contain a T7 promoter and terminator. The in vitro RNA transcriptions were performed by using a RiboMAX large-scale RNA production system-T7 (Promega). mRNA for the delta protein was transcribed by expression PCR and an mMESSAGE mMACHINE high-yield capped RNA transcription T7 kit (Ambion) as previously described (18). All RNAs were gel purified by using the strategy described previously (10). For most intermolecular competition experiments, linear antigenomic unit-length RNA bearing a neutral mutation and opened at the NcoI site was used. The sources of HDV RNA circles used in Table 1 are described in Results.
TABLE 1.
Intermolecular competition between unmarked circular genomic templates and marked linear dimeric RNA
| Source of unmarked circular monomersa | Ratio of unmarked to marked RNAs before and after replication was initiatedb at day:
|
|||
|---|---|---|---|---|
| 0 | 4 | 6 | 9 | |
| Genomic RNA from HDV particles | 1.5 | 6.8 | 7.0 | 6.9 |
| Total RNA from transfected Huh7 cells | 0.16 | 1.0 | 0.6 | — |
The two sources of circles are described further in the text.
In Northern analyses of the RNAs from the cotransfected δ293 cells, two sequence-specific oligonucleotide hybridizations were carried out, as in Fig. 2. Quantitation for the marked and unmarked RNAs was carried out by using a bio-imager (Fuji) with results as shown. —, not detected.
Cell culture and transfection.
Huh7, a line of human hepatoma cells (19), was cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in the absence of added antibiotics. In some studies we also used δ293 cells. This is a line of human embryonic kidney cells (Invitrogen) that has been stably transfected to express the small delta protein under the control of tetracycline. δ293 is a derivative of the Flp-in T-Rex-293 cell line (Invitrogen), originally based on HEK 293 cells (7). Transfection of either Huh7 or δ293 cells with RNA or cDNA was performed by using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). In studies with Huh7 cells the HDV RNAs were cotransfected with an mRNA species to express the small form of the delta protein. In the case of δ293 cells, such mRNA was not used. Delta antigen expression was induced by tetracycline added at 4 h, the time when the transfection mixture was removed. For intermolecular competition experiments, cells were cotransfected with equal masses of the competing RNAs. Transfected cells were then split at day 1 and reseeded as smaller but identical cultures. Unless indicated otherwise, cells were harvested at days 2, 4, and 6 posttransfection and total RNA was analyzed by Northern assay.
Northern analysis.
Total RNA was extracted with Tri Reagent (Molecular Research Center), glyoxalated, and examined by using gels of 1.7% agarose. For hybridization we used short oligonucleotide probes that had been 5′ labeled by using T4 kinase and [γ-32P]ATP. For detection of the genomic wild-type sequence, we used antigenomic probe 5′-CACTTTTCTCTCGATTCTCTATCGGA, located at positions 813 to 788. To hybridize with the marked genome, we used another oligonucleotide probe, 5′-CACTTTTCTCTCGATTCTGCTTCAGA (positions 813 to 788 on marker). As the standard for hybridization and quantitation, we used an equimolar mixture of two cDNA fragments with and without the neutral marker mutation. Hybridization was performed at 50°C in Ekono Hybridization Solution (Research Products International). Membranes were washed first in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min and then two times for 10 min each in 2× SSC plus 1% sodium dodecyl sulfate at 50°C. Detection and quantitation of radioactivity were by bio-imager (Fuji). Membranes then were stripped by boiling for 10 min in 0.1% sodium dodecyl sulfate-0.1× SSC and were subsequently reprobed (1).
RESULTS
Definition of initiation of RNA replication.
For all the studies described here, we define the “initiation” of HDV RNA replication from a transfected RNA template in a particular way. We mean the summation of all events up until the time when the newly made RNA transcripts have become of sufficient length and have undergone processing to form unit-length HDV RNAs that in turn can become the templates for continued RNA-directed RNA synthesis. Therefore, within this definition, initiation may be controlled by the ability of the transfected RNA to survive nucleolytic degradation and to be transported to the site for recognition by a host polymerase and the starting of RNA-directed transcription. It may also include transcription events after synthesis is started, such as template switching. Finally, it may include posttranscriptional events, such as ribozyme cleavage and subsequent ligation, to form RNA circles.
Strategy for intermolecular competition between sequence-marked HDV RNA species.
The 1,679-nt unit-length HDV genomic RNA circle and its exact complement, the antigenome, share several features. In addition to identical lengths and circular conformation, they both are predicted to fold into an unbranched rodlike structure and each contains within its sequence a domain that facilitates site-specific ribozyme cleavage. Figure 1 shows a superposition of some of these features on a single circle. Also indicated, by using the nucleotide sequence numbering of Kuo et al. (12), are the sites at which cDNA copies of the sequence are cut with several restriction enzymes, the relevance of which will subsequently be explained.
The major objective of our studies was to measure the relative ability of different HDV RNA species to act as templates for the “initiation” of RNA-directed replication. Our strategy was to do this by using cotransfection of two RNA species that could be distinguished in hybridization assays. With this in mind, we exploited our studies, which have shown that at what we will refer to as the bottom end of the rodlike structure it is possible to make a small number of changes in the nucleotide sequence without producing any deleterious effects on the ability of the HDV RNA to undergo replication and accumulation (27). Therefore, for the present competition strategy, we created and tested HDV RNA sequences with and without such a neutral “marker sequence” at this location. Then, in Northern analyses of total RNA from transfected cells, we were able to use 5′-end-labeled oligonucleotide probes and to distinguish between and quantitate the amounts of replicating HDV RNAs with and without the marker sequence.
Several additional points need to be explained about the rationale of this competition assay. (i) In most cases, the original cotransfection used RNAs that were gel purified and were delivered in what was expected to be an equal mass ratio. (ii) When this transfection was into Huh7 cells, it was accompanied by a 10% amount, by mass, of gel-purified, capped, and polyadenylated mRNA to express transiently the small delta protein. In contrast, for transfection into δ293 cells able to express large amounts of this protein, cotransfection of mRNA for the small delta protein was not needed. (iii) We expected that the two transfected HDV RNA species, with and without the marker sequence, would compete only in terms of their ability to initiate HDV RNA replication. However, following this initiation, we expected that the newly produced marked and unmarked RNAs would be otherwise identical in size and conformation and so could act equally well as templates for additional rounds of RNA-directed transcription. Thus, at all times after the initiation, we expected no further changes in the ratio of marked to unmarked RNAs. However, if one of the competing RNAs underwent a deleterious nucleotide sequence change during initiation, then one would expect that, with increasing periods of time after transfection, this changed genome would show a compounding of this disadvantage relative to an unaffected RNA. Therefore, to guard against this possibility occurring and confounding our interpretations, within each study we measured the ratio of marked to unmarked genomes for at least three different times after the initiation of replication. (iv) Both in the initiation of replication and all subsequent steps, the two competing genomes should share the available essential small delta protein and all the required host factors. In summary, the proposed competition assay should specifically reveal the differences in the ability of cotransfected RNAs to initiate RNA-directed replication.
Competition between RNA templates with circular versus linear conformation.
As mentioned in the introduction, the key issue to be resolved was to determine the extent to which the conformation of the RNA template could affect the initiation ability. Therefore, we considered a competition between linear and circular RNAs.
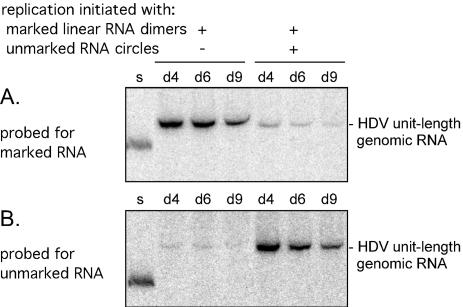
As the linear template we used in vitro-transcribed genomic dimeric RNA with ends located at the NcoI site (position 576), upstream of the genomic ribozyme (Fig. 1). Each monomer within this dimer contained the sequence marker (Fig. 1). In a preliminary experiment, just this linear dimer RNA was transfected into δ293 cells expressing the small delta protein. The cells were harvested at days 4, 6, and 9 after transfection, and the total RNA was examined by Northern blotting for the accumulation of unit-length HDV RNA. As shown in Fig. 2A, significant accumulation (per microgram of total RNA) was already maximal by day 4.
FIG. 2.
Northern assays for accumulation of unit-length genomic RNAs following transfection of δ293 cells with marked and unmarked HDV genomic RNAs. Replication was initiated with marked genomic linear RNA dimers, with or without unmarked genomic RNA circles (as obtained from purified HDV particles in serum). At days (d) 4, 6, and 9 after transfection, total RNA was extracted and assayed by Northern blotting to detect marked HDV RNA sequences (A). After quantitation the filter was stripped and rehybridized to detect unmarked HDV sequences (B). As a standard for quantitation, the Northern blot includes a sample, lane s, which contained an equimolar mixture of DNA fragments bearing the unmarked and marked sequences. Note that the signal detected in lanes 2 to 4 of panel B represents a low level of residual signal from the prior hybridization shown in panel A.
Next, we tested the initiation efficiency when the same template was cotransfected with an approximately equimolar amount (as judged by prior Northern analysis) of unmarked genomic RNA extracted from viral particles in the serum of a woodchuck superinfected with HDV. Consistent with previous studies, more than 50% of this virion RNA was in a circular conformation (data not shown). As shown in Fig. 2A, we found this cotransfection led to a >8-fold reduction in the accumulation of the marked RNAs.
Next, we stripped the Northern filter and rehybridized it to detect the accumulation of unmarked unit-length RNAs arising from the replication of the virion RNAs. As shown in Fig. 2B, there was a significant signal, consistent with the interpretation that, in a cotransfection, the virion RNAs were more efficient in “initiating” RNA replication than the linear dimeric RNA.
We also considered a quite different source of unmarked RNA circles. This source was achieved by transiently cotransfecting Huh7 cells with a DNA construct expressing 1.2 times the unit-length genomic RNA, along with a plasmid to express the large form of the delta protein. Under these conditions there is accumulation only of DNA-directed HDV RNAs processed to unit length. About 34% of these RNAs were circular in conformation and were not undergoing RNA-directed replication (data not shown). The total RNA from these cells was then used in a cotransfection into δ293 cells along with marked linear dimer RNA. Again we found that the RNA circles were more efficient in the initiation of RNA replication.
A quantitation of the two sets of competition data is summarized in Table 1. It can be seen that in both cases the ratio of unmarked RNA to marked RNA increased as a consequence of replication by about fivefold. Note that prior to transfection the circles obtained from the HDV particles would already have undergone RNA-directed replication, with associated changes such as those due to RNA editing and possibly nucleotide misincorporations (9, 20). Some of these changes could interfere with replication potential, such as via the ability to produce functional small delta protein. However, in the transfected δ293 cells there is provided an abundance of small delta protein.
Our interpretation is that RNA circles are at least five times more efficient than linear dimers at initiating replication. This could actually be an underestimate, as only 34 to 50% of the transfected unmarked unit-length RNA was actually circular in conformation. In Discussion we will consider further the possible reasons for this difference in efficiency.
These results made it clear that linear dimers have an efficiency of initiation less than that of circular monomers but still of potential significance to replication. Therefore, the following experiments were undertaken to apply the same basic competition assay to measure the relative initiation ability of different forms of linear RNA templates. As will be described, we tested the following features: (i) location of the opening site on monomeric RNA, (ii) various lengths of template ranging from monomers up to dimers, (iii) polarity, that is genomic versus antigenomic, and (iv) the number of ribozymes on either the template or nascent transcript. As will be seen, these variables also led to considerations of template switching during RNA-directed transcription.
Comparison of linear monomeric HDV RNA templates opened at different locations.
Competition studies were carried out between linear HDV RNA templates that were almost exactly unit length; that is, some RNAs have as much as a 6-nt terminal redundancy. It was thus almost as if these templates were monomeric RNA circles that had been opened up at different unique locations. One reason for these studies was the previous observation that, in the liver of an HDV-infected woodchuck, many of the accumulated HDV RNA species were not circles but were rather linear monomers opened up at locations other than the site of ribozyme cleavage (2).
As summarized in Table 2, we considered six examples of unmarked antigenomic linear templates and four of genomic linear templates. These were equivalent to circles opened at the six indicated locations. Each was tested in a competition assay along with a sequence marked unit-length antigenomic RNA opened up at position 576, as indicated in Fig. 1. Each of these RNAs was transcribed in vitro and gel purified, so that for the cotransfection into δ293 cells we were able to begin with equal amounts of both marked and unmarked RNA species. Then, at days 2, 4, and 6 after transfection, we determined for the replicating RNAs the ratio of unmarked RNA to marked RNA.
TABLE 2.
Intermolecular competition to assay initiation ability of different unit-length linear antigenomic and genomic templates
| Specifics of 5′ end of HDV sequence in RNA templatea | Ratio of unmarked to marked RNAs after replication initiated in:
|
|||||
|---|---|---|---|---|---|---|
| δ293 cellsb at day:
|
Huh7 cellsb at day:
|
|||||
| 2 | 4 | 6 | 2 | 4 | 6 | |
| Antigenomic | ||||||
| 576 (NcoI) | 1.3 | 1.1 | 1.1 | 1.6 | 1.2 | 1.2 |
| 714 (NarI) | ND | ND | ND | — | 0.04 | 0.04 |
| 781 (XbaI) | 1.7 | 1.5 | 1.5 | 0.8 | 1.4 | 1.4 |
| 841 (BoxI) | 1.2 | 1.2 | 1.2 | 0.8 | 0.7 | 0.8 |
| 1427 (EcoRI) | 0.3 | 0.3 | 0.3 | 0.4 | 0.4 | 0.4 |
| 1624 (ScaI) | 0.3 | 0.3 | 0.3 | ND | ND | ND |
| Genomic | ||||||
| 714 (NarI) | ND | ND | ND | — | 0.05 | 0.05 |
| 781 (XbaI) | ND | ND | ND | 2.3 | 1.6 | 1.8 |
| 841 (BoxI) | ND | ND | ND | 0.8 | 0.6 | 0.7 |
| 1427 (EcoRI) | ND | ND | ND | — | 0.07 | 0.05 |
The unit-length linear RNA templates of antigenomic or genomic polarity were transcribed in vitro by using T7 polymerase. The locations of the 5′ ends are as represented in Fig. 1. These unmarked RNAs were transfected into cells along with an equal mass of a unit-length antigenomic species with a 5′ end at position 576 (NcoI) and containing the marker sequence.
Transfections were either into the δ293 cells stably expressing induced small delta protein or along with a transient source of mRNA (10% by mass relative to the unit-length RNAs) for the delta protein, when using Huh7 cells. In each case, at 1 day after transfection, the cells were trypsinized and were reseeded as identical cultures by using wells of smaller area. At days 2, 4, and 6 after transfection, total RNA was harvested and analyzed by Northern assay to detect first the unit-length HDV RNA that was marked and then that which was unmarked. From the quantitation of these data, we deduced for each RNA sample the relative ratio of unmarked RNA to marked RNA. ND, not done; —, not detected.
Consider first the results for competition of the antigenomic RNAs. The first result with the RNA opened at position 576 is actually a control comparison between RNAs that differ only at the marker site. As expected, these two RNAs maintained the same ratio at all three time points and the value was about 1:1. We thus deduce that insertion of the marker sequence is neutral and has no detectable effect on HDV replication. Next we considered unmarked antigenomic RNAs opened at different locations. As can be seen, in each case the ratios were time independent. (We interpret this as evidence that during the “initiation” there was not the production of genetically altered HDV RNAs whose accumulation was less than optimal. That is, we deduce that our assay is measuring only differences in the ability to “initiate” replication.) It can also be seen that relative to the marked RNA standard the initiation efficiency for most species of linear templates was within about threefold of the marked standard. As an exception, the antigenomic RNA opened at position 714 was 25 times less able to initiate relative to standard. Two more exceptions were found when we considered genomic RNAs. Those opened at positions 714 and 1427 were >10-fold less efficient.
Also shown in the table are the results obtained in δ293 cells. The results with this stable source of small delta protein were not significantly different from those obtained with Huh7, where the initiation was dependent upon cotransfection with delta protein mRNA, as a transient template for the translation of the small delta protein.
From the results in Table 2 we make the following comments. (i) Most of the unit-length RNAs were of comparable ability to initiate RNA replication. (ii) There was no major difference between antigenomic and genomic RNAs. (iii) In only three cases was the position of the opening site associated with a significant loss in the ability to initiate replication. (iv) For the opening at position 714, there was an inhibition observed for both the genomic and antigenomic templates. This location is within the domain needed for a functional genomic ribozyme (Fig. 1). (v) However, it must be noted that the opening at position 841, within the antigenomic ribozyme domain, had no significant effect on initiation by either the genomic or antigenomic RNA template. (vi) The opening at position 1427, within the open reading frame for the small delta protein, had no effect on initiation by the antigenomic RNA but significantly interfered with initiation by the corresponding genomic RNA.
In a previous study with linear unit-length RNA templates, we were able to show that initiation of replication occurred in the absence of detectable levels of “repair” of the template RNA (3). That is, it was achieved via template switching during transcription of the transfected linear RNA. Therefore, in Table 2, since each of these templates was of unit length, it seems reasonable to deduce that initiation of replication was dependent upon transcription involving at least one template-switching event. In addition, for the genomic RNA opened at position 841, in order to produce a nascent antigenomic RNA with two ribozymes, there must have been two template switches. Despite this requirement, the RNA template was efficient in initiating replication. Thus, we would deduce that for the efficient templates one versus two template switches conferred no particular advantage or disadvantage, at least in this assay. That is, in this assay, template switching was not a rate-limiting event. In contrast, for those unit-length RNAs that were relatively inefficient at initiating replication, we would suggest that the limiting event was the ability to achieve even a single template switch.
Comparison of RNA templates that are greater than unit length.
Next, we moved to consider initiation of HDV replication by linear RNAs that ranged in length up to dimers, with either antigenomic or genomic polarity. As shown in Table 3, we indicate for these templates the positions of both the 5′ and 3′ ends, as well as the overall length of the template. We also indicate the number of ribozyme domains present in each of the RNA templates. As for Table 2, each of these unmarked RNA templates was tested in comparison with the same marked unit-length linear antigenomic RNA.
TABLE 3.
Intermolecular competition to assay initiation ability of linear antigenomic and genomic templates that are greater than unit length
| RNA templatea details
|
Ratio of unmarked to marked RNAs after replication was initiatedb at day:
|
|||||
|---|---|---|---|---|---|---|
| 5′ end | 3′ end | Length (no. of times unit length) | No. of ribozymes | 2 | 4 | 6 |
| Antigenomic | ||||||
| 962 (SalI)* | 781 (XbaI) | 1.1 | 2 | 6.0 | 5.6 | 5.2 |
| 781 (XbaI)* | 781 (XbaI) | 2.0 | 2 | 7.8 | 7.8 | 8.2 |
| 781 (XbaI) | 781 (XbaI) | 2.0 | 2 | 9.8 | 16.8 | 12.2 |
| 841 (BoxI)* | 841 (BoxI) | 2.0 | 1 | 5.0 | 5.6 | 5.0 |
| 841 (BoxI) | 841 (BoxI) | 2.0 | 1 | 2.9 | 5.5 | 6.6 |
| Genomic | ||||||
| 619 (StyI) | 781 (XbaI) | 1.1 | 2 | 7.3 | 4.9 | 4.9 |
| 781 (XbaI) | 781 (XbaI) | 2.0 | 2 | 18.8 | 28.6 | 15.6 |
| 841 (BoxI) | 841 (BoxI) | 2.0 | 2 | — | 2.6 | 2.9 |
| 1427 (EcoRI) | 1427 (EcoRI) | 2.0 | 2 | — | 6.2 | 7.7 |
Indicated for the template RNAs are the positions of the 5′ and 3′ ends, the overall length, and the number of ribozymes.
As in Table 2, the cotransfections were into either Huh7 cells or δ293 cells (indicated by *). In each experiment we used the same unit-length marked antigenomic RNA template. Also, the mass of competitor RNA was equal to that of the marked template. In the case of the dimeric constructs, we added a factor of two to convert the input mass ratio to a molar ratio. Quantitation was as in Table 2. —, not detected.
Consider the results, as summarized in Table 3, in terms of what they reveal about the influence of the template length on initiation ability. In addition, consider these data relative to the results for the monomeric linear RNAs, as summarized in Table 2.
Table 3 lists for each of the tested templates the number of (intact) ribozyme domains present. This number showed no correlation with the observed ability of these RNAs to achieve initiation.
For both genomic and antigenomic RNA templates, when the length was increased from monomer to dimer, there was at least a threefold increase of initiation ability. In the case of one dimer template (genomic dimer, EcoRI-EcoRI), the increase was at least 60-fold. It was more dramatic because the corresponding monomer was relatively poor at initiation. However, for some RNAs, even when the monomer was relatively efficient, there was still about a eightfold increase when the template was increased to dimeric size.
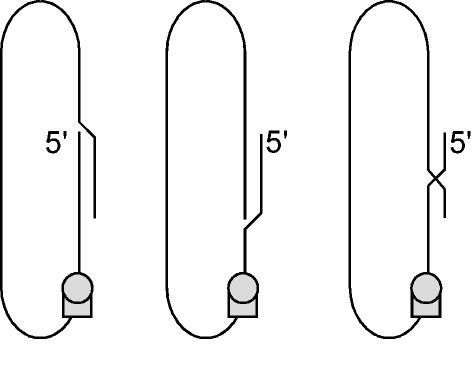
We were surprised to observe a similar increase in initiation efficiency for two RNA templates that were not dimers but only 1.1 times the unit length. Initiation from these 1.1× species, like the unit-length species, still required a template switch; however, as further considered in Fig. 4 and Discussion, we suggest that the advantage conferred by the terminal redundancies (162 and 181 nt) provided an enhanced ability to achieve template switching during transcription. Moreover, the advantage offered by this amount of terminal redundancy on the linear RNA appears to be just as good as for a dimeric RNA.
FIG. 4.
Model of possible template switching during transcription of a greater-than-unit-length linear genomic RNA template. The left panel shows the predicted folding into a rodlike structure of a 1.1× linear genomic RNA template. Note that the transcription of antigenomic RNA from this template might pause at the 5′ end of the template. However, what needs to be clarified here is that, in all our studies with in vitro transcribed linear RNA templates, there was present at the 5′ end short runs of non-HDV sequence. Thus, to avoid fatal misincorporation errors, transcription must have paused before the precise 5′ end of the template. We propose that the folding of the HDV sequences into the rodlike structure facilitated the pausing within the HDV sequences. Furthermore, as represented in the middle and right panels, we propose that, for a template of greater than unit length, there are multiple alternative foldings. This in turn would allow multiple choices at which template switching could occur. In contrast, for RNA templates of almost exactly unit length, there would only be a limited number of choices, which in some cases might preclude viable template switching.
Theoretically, dimeric RNA templates could have a real initiation advantage if the RNA transcribed from this template is greater than unit length and contains two ribozymes. The advantage could be that such a transcript could be achieved and processed to unit-length RNA circles without the need for any template switching. To test for this, one of the dimer RNAs (genomic dimer, BoxI-BoxI) was specifically designed as a template that could not produce the primary transcript with two ribozymes without need for the strand transfer. We found, as shown in Table 3, that this template was significantly poorer than all the other dimeric RNA templates. This result supports the interpretation that there could be an advantage associated with the possibility of transcription in the absence of template switching.
Earlier, we considered the unit-length linear RNAs of both genomic and antigenomic polarity and observed that in most cases they were of comparable ability to initiate replication (Table 2). Again, in these studies with greater-than-unit-length templates, we found that in most cases, genomic and antigenomic RNAs were equally good as templates for initiation of RNA-directed RNA synthesis. As a qualification, after consideration of the experimental errors in ratio determination, we would assert that any difference in initiation efficiency between genomic and antigenomic RNA templates is less than twofold.
Strategy of intramolecular competition for initiation from a multimeric RNA template.
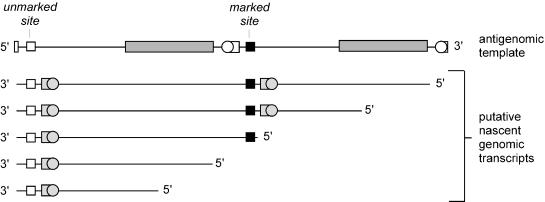
In all the studies described above we measured relative initiation abilities of various RNA templates by means of an intermolecular competition assay. For the following we used an alternative approach involving a single linear dimeric RNA as template and measured intramolecular competition.
As represented in Fig. 3, the linear dimeric antigenomic RNA template consisted of two almost identical unit-length monomers. The only difference was that the neutral change, the marker sequence, was present only on the 3′ monomer. Also shown in the figure are five potential nascent RNAs that might be initiated and transcribed from this template and then what processing features might be present on these transcripts (prior to any template switching). In the first two examples the nascent RNA will be able to be processed by ribozyme cleavage to produce unit-length HDV RNAs containing the marker sequence and capable of initiating further rounds of RNA-directed replication. In contrast, for the next three examples, the nascent RNAs will contain only one ribozyme domain. However, if template switching is achieved, it will be possible to generate transcripts that contain two ribozyme domains and can be processed to unit-length species. These species will be replication competent and will contain either the marked site or the unmarked site.
FIG. 3.
Initiation of transcription of genomic RNA from an antigenomic linear dimer RNA template. The upper line represents a dimer of antigenomic RNA whose 5′ and 3′ ends correspond to the middle of the antigenomic ribozyme. The other indicated features are as in Fig. 1. Next are shown five examples of putative nascent genomic RNAs transcribed from this template. Note that the first and fourth and the second and fifth transcripts begin at what correspond to equivalent sites on the two monomers.
Such a dimeric antigenomic RNA template was transfected into either δ293 cells or Huh7 cells. Total RNA was extracted at days 2, 4, and 6, and was subjected to Northern analyses to detect the ratio of marked to unmarked sequences in the replicating RNA. From the results summarized in Table 4, it can be seen that accumulation of unit-length marked sequences was greater than for the unmarked sequences. A similar experiment was performed in which the antigenomic RNA was replaced by a genomic dimer. Again the marked site was placed in the 3′ end of the template. Again, the results indicated that the marked sequences were more efficiently accumulated.
TABLE 4.
Intramolecular competition for initiation on linear dimeric templates
| Polarity of templatea | Ratio of marked RNA to unmarked RNA after replication was initiated inb:
|
|||||
|---|---|---|---|---|---|---|
| δ293 cells at day:
|
Huh7 cells at day:
|
|||||
| 2 | 4 | 6 | 2 | 4 | 6 | |
| Antigenomic | 12.0 | 15.4 | 15.5 | — | 7.2 | 10.1 |
| Genomic | 5.7 | 7.5 | 4.4 | — | 4.6 | 4.2 |
The antigenomic RNA template was a dimer with ends located at position 841 (BoxI). The 5′ monomer was unmarked and the 3′ monomer was marked, as indicated in Fig. 3. The genomic RNA template was a dimer with ends located at position 714 (NarI), with the 5′ monomer unmarked and the 3′ monomer marked.
Details of the transfection and subsequent quantitation are as given in Table 3. —, not detected.
Several comments can be made from these data. The very existence of replicating unmarked RNAs means that template switching did occur from some transcripts initiated on the dimer template. However, we cannot distinguish whether these initiations involved transcription events beginning within the 5′ and/or 3′ monomer of the template. Next, we note that the replicating marked RNAs were relatively much more abundant. Two sources of such RNAs can be envisaged. We might expect that most could have arisen from transcripts that begin using the 3′ monomer and that can be elongated and processed without the need for template switching. Theoretically possible is that others could have initiated within regions of either the 3′ or 5′ monomers of the template and later made use of template switching. To resolve this, we considered four templates in addition to the linear antigenomic dimer template shown in Fig. 3. These had deletions from the monomer at the 3′ end of the template of 1,267, 1,455, 1,551, and 1,618 nt. When these species were used to initiate replication, the observed ratios of marked to unmarked RNAs generated were 8.0, 6.6, 4.3 and 4.5, respectively. The last template had only 9 nt 3′ of the marker sequence, and so we would deduce that all of the marked RNAs generated from this template arose via template switching of transcripts initiated within the 5′ monomer of the template.
For the studies in Table 3, we observed that linear dimer templates were better than monomers. Now, with these studies summarized in Table 4, we can conclude that the increased efficiency of initiation is primarily from events that lead to transcription of the 3′ sequences of the template. We consider this to be further support for the interpretation that many of these additional transcription events involved nascent RNAs that did not need to undergo template switching to achieve lengths that contained two ribozymes and could be processed to unit-length RNA circles. Consistent with this explanation, the circular RNA templates as considered in Table 1 were even more efficient at initiation than was the linear dimer.
DISCUSSION
While it is clear that natural HDV infections are initiated by an RNA template that is a unit-length circular genomic RNA, in this study we have addressed the subsequent question of which HDV RNAs produced during genome replication can subsequently go on and act as templates for additional rounds of RNA-directed transcription. The previously described rolling circle models have assumed that only RNAs of unit-length and circular conformation can act as templates (15, 17, 25). Moreover, such models have not incorporated any information as to the relative template ability of genomic and antigenomic RNAs.
In the present study we used strategies involving RNA transfection and the use of sequence-marked HDV RNAs to measure what we have defined as the relative ability to “initiate” RNA replication. A major advantage of our strategies, whether using inter- or intramolecular competitions, is that there is a sharing between the RNA template competitors of components essential for RNA-directed RNA synthesis. Shared are all forms of the delta protein, both those provided by the transfection and those produced by the subsequent replication. In addition, all the essential host factors, including the RNA polymerase(s), are shared.
Under the experimental conditions used, we found that most genomic and antigenomic RNAs were of comparable initiating ability. This was true for both linear unit-length and greater-than-unit-length RNAs (Tables 2 and 3). These results are in contrast to reports that antigenomic RNAs are less efficient templates for RNA-directed synthesis and/or use different RNA polymerases (15, 17, 23, 24). Furthermore, from our results we would propose that the observed 10-fold excess of genomic to antigenomic RNA during HDV replication (5) is not due to differences in templating ability but to something else, such as differences in posttranscriptional RNA processing.
We also report that linear RNAs of greater than unit length were several times more efficient at initiation than unit-length RNAs (Tables 2 and 3). We demonstrated that this advantage was not linked to the number of ribozymes present in that template RNA and would propose that in some cases the advantage could be associated with the increased option of exploiting nucleotide homology during template switching. As explained in Fig. 4, we consider that it is the folding of the rodlike structure that both caused pausing in RNA transcription and which allows productive template switching. In addition, we suggest but have not established that, for some of the linear dimeric RNA templates tested, there was an advantage associated with the ability to transcribe, without any template switching, a greater-than-unit-length RNA that, because of the presence of at least two ribozyme domains, could go on to be processed to unit-length RNA circles.
Using our assay, we showed that circular RNA templates had at least fivefold more initiation potential than a linear dimer (Table 1). Several possible explanations might be suggested for this difference. First, circular templates might more readily fold into a conformation suitable for RNA polymerase recognition, binding, and initiation of RNA transcription. The second is based on the knowledge that RNA circles are more stable than linear RNAs (21). Thus, if the transfected RNA templates can be used for more than one round of transcription, then circles will be more efficient than linears. Finally, the explanation of possibly greatest relevance is that, for a circular RNA template, just one initiation event can produce, without the need for any template switching, a multimeric primary transcript. Furthermore, this transcript could proceed multiple times around the circular template, thereby producing multiple processed unit-length RNAs, each of which would be processed to new RNA circles, and every one of them would produce several circular templates during the new round of initiation.
Overall, our results provide evidence that a double-rolling circle model should be considered the preferred pathway for HDV replication. However, the other linear RNA species that arise during transcription and prior to RNA processing must now be considered alternative templates for the initiation of new rounds of RNA-directed RNA synthesis. That is, they do not represent merely intermediates or end products of replication but rather an additional source of RNA templates. Furthermore, their contribution to the initiation of new rounds of replication might be amplified by the fact that they, in turn, are able to produce transcripts that are processed to become new circular RNAs.
In addition, the present study confirms and extends our concept that template switching can occur during the initiation of HDV replication (3). Here, it was striking that some templates opened up in essential cis-acting sequences, such as the antigenomic ribozyme domain and the open reading frame for the small delta protein, were able to initiate, indicating successful and precise template switching (Table 2). Moreover, in our studies of intramolecular competition from greater-than-unit-length linear templates, we observed a significant preference for this switching to occur to HDV sequences at the 3′ end of the template (Fig. 3 and Table 4). Expressed in terms of the three template representations in Fig. 4, it was as if there was somehow a preference for the 3′ end of the template sequences to be folded into the rodlike structure, both to force the pause in transcription and to define the site at which template switching during transcription could be achieved.
Acknowledgments
Constructive comments on the manuscript were given by Glenn Rall, Richard Katz, and Chi Tarn. We thank Hans J. Netter and Chi Tarn for the particles purified from the serum of an infected woodchuck.
This work was supported by grants AI-26522 and CA-06927 from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Calin, G. A., C. D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich, and C. M. Croce. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 99:15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, J., G. Moraleda, S. Gudima, and J. Taylor. 2000. Efficient site-specific nonribozyme opening of hepatitis delta virus genomic RNA in infected liver. J. Virol. 74:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, J., and J. Taylor. 2002. In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J. 21:157-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, M., S.-Y. Hsieh, and J. Taylor. 1990. Role of two forms of the hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, P.-J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. Gerin, and J. Taylor. 1986. Structure and replication of the genome of hepatitis δ virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glenn, J. S., J. M. Taylor, and J. M. White. 1990. In vitro-synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J. Virol. 64:3104-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 8.Gudima, S., S.-Y. Wu, C.-M. Chiang, G. Moraleda, and J. Taylor. 2000. Origin of the hepatitis delta virus mRNA. J. Virol. 74:7204-7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudima, S. O., J. Chang, G. Moraleda, A. Azvolinsky, and J. Taylor. 2002. Parameters of human hepatitis delta virus replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J. Virol. 76:3709-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudima, S. O., E. G. Kazantseva, D. A. Kostyuk, I. L. Shchaveleva, O. I. Grishchenko, L. V. Memelova, and S. N. Kochetkov. 1997. Deoxyribonucleotide-containing RNAs: a novel class of templates for HIV-1 reverse transcriptase. Nucleic Acids Res. 25:4614-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh, S.-Y., M. Chao, L. Coates, and J. Taylor. 1990. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J. Virol. 64:3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo, M. Y.-P., J. Goldberg, L. Coates, W. Mason, J. Gerin, and J. Taylor. 1988. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 62:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo, M. Y. P., L. Sharmeen, G. Dinter-Gottlieb, and J. Taylor. 1988. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 62:4439-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazinski, D. W., and J. M. Taylor. 1994. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J. Virol. 68:2879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modahl, L. E., and M. M. C. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and regulation. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. C. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 20:6030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75:10161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 20.Netter, H. J., T.-T. Wu, M. Bockol, A. Cywinski, W.-S. Ryu, B. C. Tennant, and J. M. Taylor. 1995. Nucleotide sequence stability of the genome of hepatitis delta virus. J. Virol. 69:1687-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puttaraju, M., and M. Been. 1995. Generation of nuclease resistant circular RNA decoys for HIV-tat and HIV-rev by autocatalytic splicing. Nucleic Acids Res. 33:49-51. [PubMed] [Google Scholar]
- 22.Reid, C. E., and D. W. Lazinski. 2000. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. USA 97:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheu, G.-T. 2002. Initiation of hepatitis delta virus (HDV) replication: HDV RNA encoding the large delta antigen cannot replicate. J. Gen. Virol. 83:2507-2513. [DOI] [PubMed] [Google Scholar]
- 24.Sheu, G.-T., and M. M. C. Lai. 2000. Recombinant hepatitis delta antigen from E. coli promotes hepatitis delta virus RNA replication only from the genomic strand but not from the antigenomic strand. Virology 278:578-586. [DOI] [PubMed] [Google Scholar]
- 25.Taylor, J. M. 1999. Human hepatitis delta virus: structure and replication of the genome. Curr. Top. Microbiol. Immunol. 239:108-122. [Google Scholar]
- 26.Wang, K.-S., Q.-L. Choo, A. J. Weiner, J.-H. Ou, C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta viral genome. Nature 323:508-513. [DOI] [PubMed] [Google Scholar]
- 27.Wu, T.-T., H. J. Netter, D. W. Lazinski, and J. M. Taylor. 1997. Effects of nucleotide changes on the ability of hepatitis delta virus to transcribe, process, and accumulate unit-length, circular RNA. J. Virol. 71:5408-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]