Abstract
We previously described the adaptation of the neutralization-sensitive human immunodeficiency virus type 1 (HIV-1) strain IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker. During long-term propagation of this resistant isolate, designated FF3346, on primary peripheral blood leukocytes in vitro, an HIV-1 variant appeared that had regained sensitivity to neutralization by soluble CD4 (sCD4) and the broadly neutralizing monoclonal antibody b12. When an early passage of FF3346 was subjected to limiting-dilution culture in peripheral blood mononuclear cells, eight virus variants with various degrees of neutralization resistance were isolated. Two of them, the sCD4 neutralization-resistant variant LW_H8res and the sCD4 neutralization-sensitive variant LW_G9sens, were selected for further study. Interestingly, these two viruses were equally resistant to neutralization by agents that recognize domains other than the CD4 binding site. Site-directed mutagenesis revealed that the increased neutralization sensitivity of variant LW_G9sens resulted from only two changes, an Asn-to-Ser substitution at position 164 in the V2 loop and an Ala-to-Glu substitution at position 370 in the C3 domain of gp120. In agreement with this notion, the affinity of b12 for monomeric gp120 containing the N164S and A370E substitutions in the background of the molecular clone LW_H8res was higher than its affinity for the parental gp120. Surprisingly, no correlation was observed between CD4 binding affinity for monomeric gp120 and the level of neutralization resistance, suggesting that differences in sCD4 neutralization sensitivity between these viruses are only manifested in the context of the tertiary or quaternary structure of gp120 on the viral surface. The results obtained here indicate that the neutralization-sensitive strain IIIB can become neutralization resistant in vivo under selective pressure by neutralizing antibodies but that this resistance may be easily reversed in the absence of immunological pressure.
Resistance to antibody neutralization is a hallmark of primary human immunodeficiency virus type 1 (HIV-1) isolates that have not been passaged through T-cell lines in vitro. This property allows these viruses to persist in vivo and, as such, has significant implications for vaccine strategies aimed at inducing HIV-neutralizing antibodies. Although it is known that HIV-1 becomes neutralization sensitive when cultured in vitro in permanent T-cell lines (9, 21, 42, 48), recent studies have shown that experimental introduction of a chimeric T-cell-line-adapted, neutralization-sensitive simian-human immunodeficiency virus in a rhesus macaque resulted in adaptation to a neutralization-resistant phenotype (6, 13, 36).
We previously reported that HIV-1 also has the capacity to revert from a neutralization-sensitive to a neutralization-resistant phenotype in vivo. The virus variant that was reisolated 7 years after an accidental infection of a laboratory worker in the United States (27, 28, 31, 37, 46), designated FF3346, had a neutralization resistance that was comparable to that generally observed for primary HIV-1 isolates (1). These results suggested that neutralization-sensitive HIV-1 variants lack the capacity to persist in vivo or, at least, lack the capacity to dominate the viral quasispecies in vivo in the presence of neutralizing antibodies.
The increased neutralization resistance of isolate FF3346 was accompanied by 38 amino acid substitutions in the gp120 envelope; three of these involved residues in the CD4 binding site (CD4bs) of gp120, A281 (replaced with Val), E370 (replaced with Ala), and K429 (replaced with Glu). E370 is important for CD4 binding (24, 25, 39, 40) and contributes up to 57% of the interatomic contacts between gp120 and CD4 (17). We hypothesized that the striking E370A mutation may have been the only substitution allowed in the background of FF3346 that was compatible with replication competence and, at the same time, provided an escape from CD4bs-directed neutralizing antibodies in vivo; somewhat similar escape configurations have been described in vitro (19, 20, 44). As neutralization resistance is likely to be dispensable when neutralizing antibodies are absent from the environment in which the virus is replicating, we reasoned that the resistance of isolate FF3346 might be lost during propagation in the absence of neutralizing antibodies, even when only primary lymphocytes, which are typically used for the culturing of primary virus isolates, would be used as target cells.
Here, we report that in vitro culturing of the neutralization-resistant isolate FF3346 on primary lymphocytes indeed resulted in progeny virus with increased sensitivity to neutralization by soluble CD4 (sCD4) and the broadly neutralizing human monoclonal antibody (MAb) b12. This phenotypic change could be attributed primarily to a dual amino acid reversion: an Asn → Ser substitution at position 164 in the V2 loop and an Ala → Glu substitution at position 370 in the C3 region of gp120. The results obtained in this study strongly support the notion that the neutralization resistance of HIV-1 strains is a dynamic process that can easily be modulated depending on the milieu of the replicating virus (32, 45).
MATERIALS AND METHODS
PBMCs.
For virus propagation, experiments were performed with phytohemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMCs) with a wild-type CCR5 genotype (CCR5+/+) pooled from at least five healthy blood donors according to previously described procedures (34). The CCR5 genotype was determined by PCR as described elsewhere (12). PBMCs were isolated from buffy coats after Ficoll density gradient centrifugation. For PHA stimulation, 5 × 106 cells/ml were cultured for 3 days in Iscove's modified Dulbecco's medium (BioWhittaker, Verviers, Belgium) supplemented with 10% fetal calf serum (HyClone, Logan, Utah), penicillin (10 U/ml), streptomycin (10 μg/ml), cyproxin (5 μg/ml), and PHA (5 μg/ml; Murex Biotech, Dartford, England). Subsequently, cells (106/ml) were grown in the absence of PHA in medium supplemented with 10 U of recombinant interleukin 2 (Chiron Benelux BV, Amsterdam, The Netherlands) per ml.
Virus isolation.
The neutralization-resistant variant FF3346 was reisolated by cocultivation of PBMCs from healthy donors with PBMCs from an accidentally infected laboratory worker (designated LW-F) in 1992, approximately 7 years after the assumed time of infection (46). Recently, biological virus clones were isolated from the quasispecies of the second PBMC passage of the FF3346 isolate by limiting-dilution coculture of FF3346-infected PBMCs (50 cells/well) with PHA-stimulated PBMCs (105 cells/well) in 96-well microtiter plates (34). Each week, culture supernatants were tested for the presence of p24 in an in-house p24 antigen capture enzyme-linked immunosorbent assay (ELISA). Simultaneously, half of the cells were transferred to empty microtiter plates, and 105 fresh PHA-stimulated PBMCs were added to propagate the culture. Viruses were considered clonal when at least one third of the microcultures were positive. From these p24-positive cultures, virus stocks were grown, and 50% tissue culture infectious doses (TCID50s) were determined by endpoint dilution.
Molecular cloning and expression of replication-competent viruses.
Biological virus clones LW_H8res and LW_G9sens, resistant and sensitive to sCD4 and MAb b12 neutralization, respectively, were obtained from the quasispecies of the FF3346 isolate by limiting-dilution coculture, as described above. Genomic DNA was isolated from PBMCs infected with either LW_H8res or LW_G9sens. The entire envelope of strain LW_H8res was amplified by PCR with Taq and Pwo DNA polymerases (Expand High Fidelity; Boehringer Mannheim, Germany) with the primer combinations and conditions described previously (1, 43). PCR products were purified with the GFX purification kit (Amersham Pharmacia) and inserted into the pGEM-T Easy vector (Promega, Madison, Wis.). To ensure proper ligation of the inserts, the resulting plasmids were sequenced with a BigDye terminator cycle sequencing reaction kit (ABI Prism; Applied Biosystems, Warrington, United Kingdom), according to the manufacturer's protocol, on an ABI Prism 373S automated sequencer.
To generate the full-length molecular clone of LW_H8res, the pGEM-T vector containing the LW_H8res envelope gene was digested with SalI and BamHI, and the resulting fragment (positions 5786 to 8480 of the envelope gene; numbers relative to the HXB2 reference sequence) was ligated into SalI- and BamHI-digested pLAI-2, which contains the molecular clone of HIV-1 IIIB (see Fig. 3A) (26). Recombinant viruses were obtained by transfection of 70% confluent 293T cells with 10 μg of plasmid DNA and Lipofectamine (Gibco BRL, Breda, The Netherlands) as described by the manufacturer. Two days after transfection, the supernatants were used to infect PHA-stimulated PBMCs.
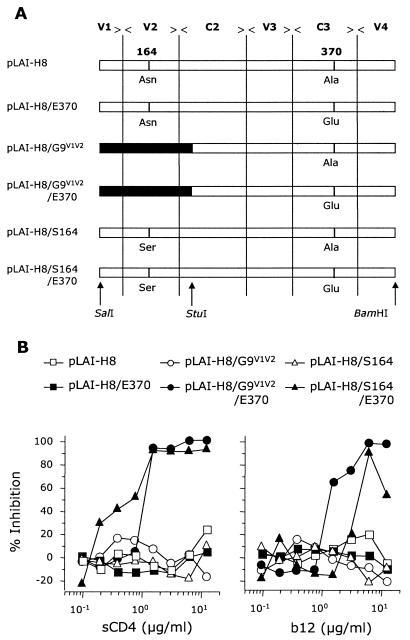
FIG. 3.
Construction of chimeric virus clones and analysis of their sensitivity to neutralization by sCD4 and MAb b12. (A) The SalI/BamHI fragment of the LW_H8res gp120 envelope was inserted into the pLAI-2 vector, which contains the molecular clone of HIV-1 strain IIIB, to yield pLAI-H8. Site-directed mutagenesis was used to engineer three different variants of pLAI-H8 with amino acid substitutions at positions 164 and 370, pLAI-H8/E370, pLAI-H8/S164, and pLAI-H8/S164/E370 (the changes of A370 to Glu and N164 to Ser in the background of pLAI-H8 are depicted). In addition, the V1/V2 fragment of strain LW_G9res was placed into pLAI-H8, to yield pLAI-H8/G9V1V2, or into pLAI-H8/E370, to yield pLAI-H8/E370/G9V1V2. (B) Neutralization sensitivity of the chimeric molecular virus clones pLAI-H8 (open squares), pLAI-H8/G9V1V2 (open circles), pLAI-H8/E370/G9V1V2 (solid circles), pLAI-H8/E370 (solid squares), pLAI-H8/S164 (open triangles), and pLAI-H8/S164/E370 (solid triangles) for sCD4 and MAb b12. Virus stocks of the chimeric molecular clones were obtained by infection of PHA-stimulated PBMCs with supernatant from transfected 293T cells. Neutralization experiments were performed as described for Fig. 1.
To insert the V1/V2 domain of LW_G9sens into the LW_H8res background, gp120 of LW_G9sens was digested with SalI and StuI (positions 5786 to 6833 relative to HXB2). The resulting 1047-bp fragment was then ligated to the SalI- and StuI-digested pGEM-T Easy vector containing the LW_H8res envelope with a rapid ligation kit (Hoffman-La Roche, Basel, Switzerland). The vector was then digested with SalI and BamHI, and the resulting fragment was cloned into the background of pLAI-2. By site-directed mutagenesis, the Ala residue at position 370 and the Asn at position 164 were replaced with Glu and Ser, respectively. These mutations were introduced into the LW_H8res envelope in the pGEM-T vector. Therefore, primers containing sequences that encoded the 370Glu or 164Ser residue and one or two silent mutations resulting in an XbaI or SpeI restriction site, respectively, were designed. For the introduction of both 370Glu and 164Ser, two PCR products were generated. The two PCR products with the 370Glu sequence were digested with XbaI and then ligated to each other. The products with the 164Ser-encoding sequence were ligated to each other after digestion with SpeI. The envelopes generated containing either the 370Glu or 164Ser mutations were ligated into the pGEM-T Easy vector, after which a SalI/BamHI envelope fragment was cloned into the pLAI-2 background (Fig. 3A).
Neutralizing agents and neutralization sensitivity of HIV-1 variants.
All viruses were tested for neutralization sensitivity to increasing concentrations of sCD4, b12, HIVIg (a preparation of purified polyclonal immunoglobulin derived from HIV-infected donors), Amshps (pooled sera from 34 patients from the Amsterdam Cohort) (50), MAbs gp13, gp68, and F105 (which, like b12, recognize epitopes overlapping the CD4bs on gp120) (30, 35), and MAb 902 (which binds to the V3 loop of gp120) (7). From each virus isolate, an inoculum of 100 TCID50/ml in a 100-μl final volume was incubated for 2 h at 37°C with increasing concentrations of sCD4, serum, or antibody. Subsequently, 105 PHA-stimulated human PBMCs were added to the mixtures in 96-well microtiter plates.
To avoid interference of anti-p24 antibodies in patient sera in the subsequent p24 ELISA, plates incubated with HIVIg or Amshps were washed the following day. On days 7 and 14, virus production in supernatants was analyzed in triplicate with an in-house p24 antigen capture ELISA. The percent viral inhibition was calculated as the mean reduction in p24 production of cultures in the presence of the neutralizing agent compared to that in viral cultures in the absence of neutralizing ligand. In general, 50% inhibitory concentrations (IC50) were determined by linear regression.
Binding of monomeric gp120 of HIV-1 variants to MAbs b12 and CD4-IgG2.
To determine the relative binding affinities of MAbs b12 and CD4-IgG2, which was used in this assay as a surrogate for sCD4, a gp120 ELISA was performed as described recently (21, 25). HIVIg, at 1 μg/ml, was used to ensure that equivalent amounts of gp120 were captured onto microtiter plates coated with a polyclonal antibody to the C5 region of gp120 (Aalto Bioreagents). Bound antibody was detected with a peroxidase-conjugated anti-human immunoglobulin secondary antibody in conjunction with tetramethylbenzidene (TMB; Pierce) as the substrate. Color development was stopped by adding sulfuric acid (2 M), and absorbance was measured at 450 nm. Apparent binding affinities were calculated as the antibody concentration at half-maximal binding.
RESULTS
Isolation and characterization of HIV variants with various neutralization sensitivities that are present in the quasispecies of the FF3346 isolate.
Seven years after the presumed time of accidental infection of an American laboratory worker with the neutralization-sensitive HIV-1 strain IIIB, a neutralization-resistant variant, denoted FF3346, that was highly resistant to neutralization by sCD4 and MAb b12 was isolated. Although FF3346 was only propagated in PBMCs in vitro, virus progeny that had regained sensitivity to neutralization by sCD4 and MAb b12 emerged after eight successive PBMC passages spanning a period of 6 years of in vitro cultivation. Upon performing limiting dilution of cryopreserved PBMCs that were infected with the second passage of the FF3346 isolate, we obtained eight virus variants that were either sensitive or resistant to neutralization by sCD4 (data not shown). Analysis of the gp120 envelope sequences from these eight virus variants showed that the presence of a Glu residue at position 370 correlated with a neutralization-sensitive viral phenotype, whereas an Ala at position 370 was present in the neutralization-resistant virus variants (data not shown).
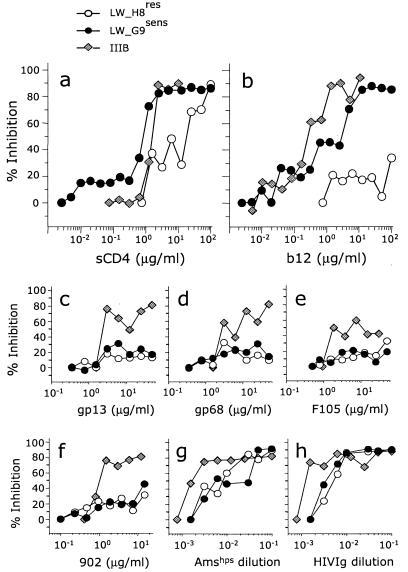
We selected two virus variants, LW_H8res and LW_G9sens, with opposite neutralization sensitivities for sCD4 and MAb b12. LW_G9sens differs from LW_H8res at only 10 positions, including the residue at position 370 (Ala for LW_H8res and Glu for LW_G9sens; Fig. 1). For the LW_H8res virus variant, the IC50s of sCD4 and MAb b12 were 12 and >100 μg/ml, respectively, compared to 0.8 and 1.6 μg/ml, respectively, for the LW_G9sens virus variant (Fig. 2a and b).
FIG. 1.
Amino acid sequences of virus clones LW_H8res and LW_G9sens in comparison to IIIB. The viruses were isolated by limiting dilution from PBMCs infected with an isolate (FF3346) obtained 7 years after accidental infection of a laboratory worker. The predicted amino acid sequences of the envelope glycoproteins were derived from the consensus sequences of the envelope fragments ranging from V1 to V5, which were amplified and sequenced from DNA isolated from infected PBMC cultures. Variable and constant domains are indicated, and the numbering of amino acid positions is relative to HXB2; dots indicate residues identical to those in the reference sequence, and dashes indicate amino acid deletions relative to HXB2. The location of the restriction site for StuI is also depicted.
FIG. 2.
Neutralization sensitivity of LW_H8res (open circles), LW_G9sens (solid circles), and IIIB (gray diamonds). We incubated 100 TCID50 of each virus inoculum with increasing concentrations of (a) sCD4, (b) MAb b12, (c) MAb gp13, (d) MAb gp68, (e) MAb F105, (f) MAb 902, (g) pooled polyclonal serum Amshps, and (h) pooled polyclonal serum HIVIg. MAbs b12, gp13, gp68, and F105 are directed to epitopes overlapping the CD4bs of gp120, whereas MAb 902 is directed to an epitope in the V3 loop of gp120. Incubation of virus with sCD4, MAbs, or sera was performed for 2 h at 37°C before PHA-stimulated PBMCs were added. The concentration of p24 in culture supernatants was measured, and the percent viral inhibition was calculated as the mean reduction in p24 level in triplicate cultures in the presence of the neutralizing ligand compared to that in cultures with virus only. IC50s were determined by linear regression when possible. Neutralization assays were performed at least twice.
Compared to the T-cell-line-adapted strain IIIB, LW_H8res and LW_G9sens were both relatively resistant to neutralization by MAbs gp13, gp68, and F105, which recognize epitopes overlapping the CD4bs of gp120, and MAb 902, which binds to the V3 loop of gp120 (Fig. 2c to f). The two viruses were also less sensitive than IIIB to neutralization by two polyclonal immunoglobulin preparations derived from HIV-infected donors (HIVIg and Amshps; Fig. 2g and h).
Molecular determinant for neutralization sensitivity of FF3346-derived HIV-1 variants.
To gain insight into the molecular basis for sCD4 and MAb b12 neutralization sensitivity, a molecular clone of LW_H8res was generated by inserting the envelope gene of LW_H8res into the vector pLAI-2, which contains the molecular clone of IIIB. The resulting plasmid, pLAI-H8, was then used as a template to generate chimeric viruses and for site-directed mutagenesis (Fig. 3A). We first focused on the role of residue 370 in the neutralization sensitivity of the FF3346-derived virus variants, as this residue is highly conserved among HIV-1 isolates and directly involved in CD4 binding. Moreover, the presence of an Ala residue at position 370 always correlated with resistance to sCD4 and MAb b12 neutralization in the eight variants isolated from the FF3346 quasispecies.
By site-directed mutagenesis, an Ala → Glu substitution was introduced at position 370 in pLAI-H8 to yield pLAI-H8/E370. Furthermore, chimeric viruses were generated in which a SalI-StuI fragment containing the V1/V2 region of the LW_G9sens envelope was cloned into pLAI-H8 and pLAI-H8/E370 to yield pLAI-H8/G9V1V2 and pLAI-H8/G9V1V2/E370, respectively (Fig. 3A). Resistance to neutralization by sCD4 and MAb b12 was observed for viruses derived from pLAI-H8, pLAI-H8/E370, and pLAI-H8/G9V1V2 (Fig. 3B), indicating that the presence of a Glu residue at position 370 or of the V1/V2 domain of LW_G9sens in the background of LW_H8res did not result in virus with a neutralization-sensitive phenotype. In contrast, virus derived from pLAI-H8/G9V1V2/E370 was highly sensitive to neutralization by sCD4 and MAb b12 (Fig. 3B). In fact, the neutralization sensitivity was comparable to that of virus variant LW_G9sens.
Considering that the V1/V2 fragment of LW_G9sens is different from that of LW_H8res only at positions 141, 144, and 164, we compared the residues at these positions with the observed sensitivity of all eight virus variants that were isolated from the quasispecies of FF3346 to neutralization by sCD4 and b12. For all eight clones, the presence of a Ser residue at position 164 in the V2 loop of gp120 correlated with a neutralization-sensitive phenotype, whereas residues at the other positions varied irrespective of the neutralization sensitivity of the virus variant (data not shown). To confirm that a combined N164S/A370E substitution was indeed responsible for the neutralization sensitivity of LW_G9sens, we performed site-directed mutagenesis with pLAI-H8 and pLAI-H8/E370 as templates to replace the Asn at position 164 with Ser. The resulting plasmids were designated pLAI-H8/S164 and pLAI-H8/S164/E370, respectively (Fig. 3A).
Virus derived from pLAI-H8/S164 was resistant to neutralization by sCD4 and MAb b12 (IC50, >12.5 μg/ml for both sCD4 and MAb b12; Fig. 3B). However, virus derived from pLAI-H8/S164/E370, which contains the dual N164S and A370E substitutions, was sensitive to neutralization by sCD4 and b12. The sensitivity to neutralization was equivalent to that observed for virus variant LW_G9sens (0.4 and 4.0 μg/ml, respectively).
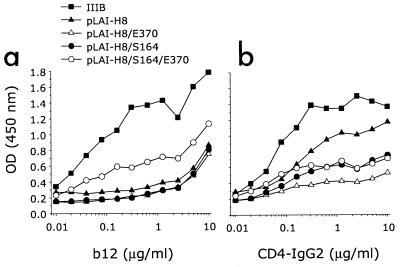
To determine whether the differences in neutralization sensitivity correlated with changes in binding affinity for gp120, we subsequently performed an ELISA with monomeric gp120 from the different mutant viruses after NP-40 lysis of virus-containing culture supernatant from infected PBMCs. Although we cannot be absolutely certain that this preparation contains only monomeric gp120, previous studies have demonstrated that the antibody preparation used in this assay for antigen capture does not bind or binds very weakly to oligomeric forms of gp120 and gp120-gp41 complexes, indicating that most likely only binding to monomeric gp120 is being measured here (14, 21-23). In agreement with its neutralization sensitivity for MAb b12, monomeric gp120 from virus derived from pLAI-H8/S164/E370 bound b12 with relatively better affinity than gp120 from neutralization-resistant variants (Fig. 4a). The relative antibody binding affinity for gp120 from virus derived from pLAI-H8/S164/E370 was lower than that for wild-type virus IIIB, which also agrees with the higher sensitivity of IIIB to b12 neutralization (Fig. 2). For CD4-IgG2, we did not observe a correlation between binding affinity for monomeric gp120 and the degree of sCD4 neutralization resistance for the corresponding virus (Fig. 4b). Thus, it appears that changes in the binding affinity of CD4 for monomeric gp120 from the neutralization-resistant and -sensitive viruses do not correlate with changes in affinity for functional oligomeric gp120 present on the surface of the virus.
FIG. 4.
Binding of MAbs b12 and CD4-IgG2 to monomeric gp120 of IIIB (solid squares) and gp120 of virus derived from pLAI-H8 (solid triangles), pLAI-H8/E370 (open triangles), pLAI-H8/S164 (solid circles), and pLAI-H8/S164/E370 (open circles). gp120, standardized to equivalent amounts by using HIVIg (1 μg/ml), was captured on microtiter plate wells with an anti-C5 antibody and then probed with antibody at the concentrations indicated. Absorbance was measured at 450 nm after 30 min.
DISCUSSION
Broad resistance of primary HIV-1 isolates to neutralizing antibodies is typically lost when the viruses are adapted to growth in permanent T-cell lines. The underlying mechanism for this phenotypic change is not fully understood but is most likely driven by either the absence of neutralizing antibodies, which results in removal of antibody-imposed selective pressure, or the limited availability of CD4 on permanent T-cell lines. In the latter case, the low expression level of CD4 on T-cell lines may select for a more open quartenary conformation of the viral envelope glycoproteins (29), which, coincidentally, results in better accessibility for antibodies and other neutralizing agents.
The increased neutralization sensitivity of resistant primary HIV-1 isolates after passage on primary lymphocytes has not been reported previously, and it may be argued that the number of passages may have been too small to allow the accumulation of mutations required for the development of a neutralization-sensitive phenotype. In our present study, the prolonged successive propagation in primary lymphocytes of the neutralization-resistant isolate FF3346, which was recovered from a laboratory worker who was accidentally infected with the neutralization-sensitive strain IIIB (46), led to the emergence of progeny virus variants, some of which had regained sensitivity to neutralization by sCD4 and MAb b12. This observation emphasizes that not the propagation in T-cell lines, but merely the absence of neutralizing antibodies in vitro may allow the selection of virus variants that have lost their resistance to neutralization. This most likely occurs in return for a higher efficiency of binding to the cellular CD4 entry complex, which has been reported to be typically inefficient in primary isolates (22). However, although sensitivity to neutralization by sCD4 and b12 was increased, resistance to neutralization by other MAbs against the CD4bs as well as an MAb against the V3 loop remained the same. Progeny virus variants that were sensitive to neutralization by sCD4 and b12 were as sensitive to neutralization by two polyclonal sera as resistant progeny variants.
Our observations also reemphasize the importance of limiting the number of passages in vitro to preserve the phenotype of primary HIV isolates. Although we first observed a large population of HIV-1 variants with increased neutralization sensitivity after a 6-year period during which the virus was passaged eight times on primary lymphocytes, our findings do imply that changes in neutralization sensitivity may occur at even earlier time points during short-term culture on primary lymphocytes, as evidenced by the isolation of neutralization-sensitive progeny even after the second passage of the resistant parental isolate FF3346. Whether this will also occur with more typical primary HIV-1 isolates remains to be determined. However, one way to avoid the possibility of introducing a phenotypically mixed viral quasispecies when determining the neutralization sensitivity of primary virus isolates would be the use of single-round neutralization assays with envelope sequences that have been cloned directly from the primary viruses (4, 10, 11, 15, 32, 45).
The HIV-1 isolate FF3346 had 38 amino acid substitutions in its gp120 envelope gene in comparison to that of the neutralization-sensitive virus IIIB with which the laboratory worker was accidentally infected. Among these substitutions was a Glu → Ala mutation at position 370. This substitution is striking, considering the highly conserved nature of this residue (http://hiv-web.lanl.gov/content/hiv-db/mainpage.html). Interestingly, in the neutralization-sensitive progeny of FF3346 designated LW_G9sens, the Ala at position 370 was re-replaced with Glu. However, site-directed mutagenesis showed that the difference between FF3346 and its neutralization-sensitive progeny in susceptibility to neutralization by sCD4 and MAb b12 was determined not only by a Glu or Ala residue at position 370, but also by the replacement of the Asn residue at position 164 in the V2 loop with Ser, which is also found in the neutralization-sensitive strain IIIB. Considering that the interaction of the conserved E370 residue with CD4 is assumed to be essential for virus binding to and entry into the target cell (24, 38), we speculate that the Ala substitution at position 370 in the LW_H8res strain may have been selected to escape from CD4bs-directed antibodies that were induced upon infection of the laboratory worker. This is supported by previous studies, which have shown that substitution of the E370 residue in either HXB2 (X4 dependent) or YU2 (R5 dependent) results in diminished gp120 binding to sCD4, CD4bs antibodies, and antibodies to CD4-induced epitopes (24, 25, 33, 41, 49). Comparable escape mechanisms have also been described by others (18-20, 44, 45).
The presence of a virus variant in which the Ala residue at position 370 had been re-replaced with Glu suggested that having this residue at position 370 in the background of the IIIB gp120 envelope may provide the virus with better replication fitness. However, short-term replication kinetics were indistinguishable between LW_H8res and LW_G9sens in PBMCs in vitro, with comparable maximum p24 antigen levels (data not shown). In further agreement, neither LW_H8res nor LW_G9sens became the dominant virus variant in a direct 4-week competition experiment, implicating that fitness differences under our in vitro conditions, if any, are marginal (data not shown). Thus, merely having a Glu at position 370 does not appear to endow LW_G9sens with increased viral fitness over LW_H8res. Reincorporation of the Glu residue may therefore only be advantageous in the context of other amino acid substitutions that differ between the two variants.
Conversely, the preservation of fitness of the LW_H8res variant, with the Ala at position 370, may have been established by compensatory mutations elsewhere in gp120 (Fig. 1a) (1). Of note here is the N164S substitution in the V2 loop, which has previously been reported to compensate for the altered replication competence of a b12-neutralization escape mutant of JR-CSF (20). How this mutation influences exposure of the CD4bs on the virus remains to be established, but it is likely to have an influence on the efficiency of CD4 binding to target cells. Our finding that a neutralization-sensitive phenotype may emerge and be selected on primary cells may also apply in vivo. With progression of disease, neutralizing antibody titers may drop when CD4+-T-cell counts decline (3, 5, 8, 47). Under these conditions, HIV variants with reduced neutralization resistance but increased replication kinetics may emerge. We have indeed demonstrated the presence of more rapidly replicating viruses with progression of disease (2, 16, 43). This implies that neutralizing antibodies contribute to the control of even neutralization-resistant HIV by continuously selecting virus variants with a neutralization-resistant phenotype and coinciding impaired replicative capacity.
Based on the in vivo readaptation of the T-cell-line-adapted strain IIIB to neutralization resistance, we previously concluded that the neutralization resistance of primary HIV can indeed be considered an escape mechanism (45). This idea was supported by the observation that despite cellular and humoral immune responses elicited in the accidentally infected laboratory worker, virus replication could not be suppressed (27, 28, 31, 37). The in vitro reversal towards a neutralization-sensitive phenotype during propagation on primary cells in the absence of neutralizing antibodies, as shown here, further supports this hypothesis. Moreover, the observation that only two substitutions in gp120, N164S and A370E, are sufficient for this phenotypic change not only indicates that previously assumed deleterious mutations in gp120 may be compatible with replication competence, but also demonstrates that such simple mutations may offer the virus a mechanism to adapt rapidly when faced with inadequate antibody responses (18, 19, 32, 45).
Acknowledgments
We thank Ray Sweet (Smithkline Beecham) for kindly providing four-domain sCD4, Alfred Prince (Lindsey F. Kimball Research Institute) for HIVIg, and Jaap Goudsmit (University of Amsterdam) for Amshps. The human MAbs gp13 and gp68 were a kind gift from Martin Schutten (Erasmus University, Rotterdam). We also thank James Robinson (Tulane University) for providing MAb 17b. MAbs F105, 902, and 48d were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH; contributed by Marshall Posner, Bruce Chesebro, and James Robinson, respectively). We thank Dennis Burton for providing us with antibody IgG1 b12, for critical reading of the manuscript, and for helpful suggestions. We are also thankful to Ben Berkhout (University of Amsterdam) for providing the full-length sequence of pLAI-2 and to Ronald van Rij and Frank Miedema for critiquing the manuscript and constructive discussions.
This work was supported by Dutch AIDS Foundation grants 1304 and 7009.
REFERENCES
- 1.Beaumont, T., A. van Nuenen, S. Broersen, W. A. Blattner, V. V. Lukashov, and H. Schuitemaker. 2001. Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J. Virol. 75:2246-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaak, H., M. Brouwer, L. J. Ran, F. de Wolf, and H. Schuitemaker. 1998. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J. Infect. Dis. 177:600-610. [DOI] [PubMed] [Google Scholar]
- 3.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 4.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 5.Cecilia, D., C. Kleeberger, A. Munoz, J. V. Giorgi, and S. Zolla-Pazner. 1999. A longitudinal study of neutralizing antibodies and disease progression in HIV-1-infected subjects. J. Infect. Dis. 179:1365-1374. [DOI] [PubMed] [Google Scholar]
- 6.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro, B., and K. Wehrly. 1988. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J. Virol. 62:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuenchitra, T., C. Wasi, S. Louisirirojchanakul, S. Nitayaphan, R. Sutthent, J. H. Cox, M. S. De Souza, A. E. Brown, D. L. Birx, and V. R. Polonis. 2003. Longitudinal study of humoral immune responses in HIV type 1 subtype CRF01_AE (E)-infected Thai patients with different rates of disease progression. AIDS Res. Hum. Retroviruses 19:293-305. [DOI] [PubMed] [Google Scholar]
- 9.Clapham, P. R., and A. McKnight. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809-1829. [DOI] [PubMed] [Google Scholar]
- 10.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., K. E. Sheridan, C. Lai, L. Zhang, and D. D. Ho. 1996. Characterization of the functional properties of env genes from long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 70:5306-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Roda Husman, A. M., M. Koot, M. Cornelissen, I. P. Keet, M. Brouwer, S. M. Broersen, M. Bakker, M. T. Roos, M. Prins, F. de Wolf, R. A. Coutinho, F. Miedema, J. Goudsmit, and H. Schuitemaker. 1997. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann. Intern. Med. 127:882-890. [DOI] [PubMed] [Google Scholar]
- 13.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, M. Fernandes, K. Liou, R. Gomila, J. Lee, and J. Sodroski. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J. Virol. 74:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwa, D., J. Vingerhoed, B. Boeser, and H. Schuitemaker. 2003. Increased in vitro cytopathicity of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J. Infect. Dis. 187:1397-1403. [DOI] [PubMed] [Google Scholar]
- 17.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis, J., P. Balfe, C. Arnold, S. Kaye, R. S. Tedder, and J. A. McKeating. 1998. Development of a neutralizing antibody response during acute primary human immunodeficiency virus type 1 infection and the emergence of antigenic variants. J. Virol. 72:8943-8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKeating, J. A., J. Bennett, S. Zolla-Pazner, M. Schutten, S. Ashelford, A. L. Brown, and P. Balfe. 1993. Resistance of a human serum-selected human immunodeficiency virus type 1 escape mutant to neutralization by CD4 binding site monoclonal antibodies is conferred by a single amino acid change in gp120. J. Virol. 67:5216-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo, H., L. Stamatatos, J. E. Ip, C. F. Barbas, P. W. Parren, D. R. Burton, J. P. Moore, and D. D. Ho. 1997. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. J. Virol. 71:6869-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas, 3rd, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore, J. P., J. A. McKeating, Y. X. Huang, A. Ashkenazi, and D. D. Ho. 1992. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J. Virol. 66:235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, J. P., J. A. McKeating, W. A. Norton, and Q. J. Sattentau. 1991. Direct measurement of soluble CD4 binding to human immunodeficiency virus type 1 virions: gp120 dissociation and its implications for virus-cell binding and fusion reactions and their neutralization by soluble CD4. J. Virol. 65:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantophlet, R., E. Ollmann Saphire, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 27.Pincus, S. H., K. G. Messer, P. L. Nara, W. A. Blattner, G. Colclough, and M. Reitz. 1994. Temporal analysis of the antibody response to HIV envelope protein in HIV-infected laboratory workers. J. Clin. Investig. 93:2505-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pincus, S. H., K. G. Messer, D. H. Schwartz, G. K. Lewis, B. S. Graham, W. A. Blattner, and G. Fisher. 1993. Differences in the antibody response to human immunodeficiency virus-1 envelope glycoprotein (gp160) in infected laboratory workers and vaccinees. J. Clin. Investig. 91:1987-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 30.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 31.Reitz, M. S., Jr., L. Hall, M. Robert-Guroff, J. Lautenberger, B. M. Hahn, G. M. Shaw, L. I. Kong, S. H. Weiss, D. Waters, R. C. Gallo, et al. 1994. Viral variability and serum antibody response in a laboratory worker infected with HIV type 1 (HTLV type IIIB). AIDS Res. Hum. Retroviruses 10:1143-1155. [DOI] [PubMed] [Google Scholar]
- 32.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 34.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schutten, M., A. McKnight, R. C. Huisman, M. Thali, J. A. McKeating, J. Sodroski, J. Goudsmit, and A. D. Osterhaus. 1993. Further characterization of an antigenic site of HIV-1 gp120 recognized by virus neutralizing human monoclonal antibodies. AIDS 7:919-923. [DOI] [PubMed] [Google Scholar]
- 36.Si, Z., M. Cayabyab, and J. Sodroski. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J. Virol. 75:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sipsas, N. V., S. A. Kalams, A. Trocha, S. He, W. A. Blattner, B. D. Walker, and R. P. Johnson. 1997. Identification of type-specific cytotoxic T lymphocyte responses to homologous viral proteins in laboratory workers accidentally infected with HIV-1. J. Clin. Investig. 99:752-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas, 3rd, P. W. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, J. Li, and J. Sodroski. 1991. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 65:5007-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner, S., R. Tizard, J. DeMarinis, R. B. Pepinsky, J. Zullo, R. Schooley, and R. Fisher. 1992. Resistance of primary isolates of human immunodeficiency virus type 1 to neutralization by soluble CD4 is not due to lower affinity with the viral envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 72:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watkins, B. A., M. S. Reitz, Jr., C. A. Wilson, K. Aldrich, A. E. Davis, and M. Robert-Guroff. 1993. Immune escape by human immunodeficiency virus type 1 from neutralizing antibodies: evidence for multiple pathways. J. Virol. 67:7493-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 46.Weiss, S. H., J. J. Goedert, S. Gartner, M. Popovic, D. Waters, P. Markham, F. di Marzo Veronese, M. H. Gail, W. E. Barkley, J. Gibbons, et al. 1988. Risk of human immunodeficiency virus (HIV-1) infection among laboratory workers. Science 239:68-71. [DOI] [PubMed] [Google Scholar]
- 47.Wong, M. T., R. Q. Warren, S. A. Anderson, M. J. Dolan, C. W. Hendrix, S. P. Blatt, G. P. Melcher, R. N. Boswell, and R. C. Kennedy. 1993. Longitudinal analysis of the humoral immune response to human immunodeficiency virus type 1 (HIV-1) gp160 epitopes in rapidly progressing and nonprogressing HIV-1-infected subjects. J. Infect. Dis. 168:1523-1527. [DOI] [PubMed] [Google Scholar]
- 48.Wrin, T., T. P. Loh, J. C. Vennari, H. Schuitemaker, and J. H. Nunberg. 1995. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J. Virol. 69:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 50.Zwart, G., L. van der Hoek, M. Valk, M. T. Cornelissen, E. Baan, J. Dekker, M. Koot, C. L. Kuiken, and J. Goudsmit. 1994. Antibody responses to HIV-1 envelope and gag epitopes in HIV-1 seroconverters with rapid versus slow disease progression. Virology 201:285-293. [DOI] [PubMed] [Google Scholar]