Highlights
-
•
Bacteria are typically found within complex microbial communities in nature.
-
•
Molecular interactions between co-infecting bacteria can profoundly affect disease prognosis and treatment.
-
•
In vivo models and genomic tools are providing new insights into interbacterial behavior during infection.
-
•
There is potential to target interbacterial interactions as part of a therapeutic strategy.
Keywords: polybacterial disease, synergy, infection, metatranscriptomics, cell–cell signaling, polymicrobial infection
Abstract
Polybacterial diseases involve multiple organisms that act collectively to facilitate disease progression. Although this phenomenon was highlighted early in the 20th century, recent technological advances in diagnostics have led to the appreciation that many infections are far more complex than originally believed. Furthermore, it is apparent that although most treatments focus on the dominant bacterial species in an infection, other microbes, including commensals, can have a profound impact on both the response to therapy and virulence. Very little is known about the molecular mechanisms that underpin interactions between bacteria during such infections. Here, we discuss recent studies identifying and characterizing mechanisms of bacterial interaction and the biological processes they govern during certain diseases. We also highlight how possible strategies for targeting these interbacterial interactions may afford a route towards development of new therapies, with consequences for disease control.
A growing appreciation for the polymicrobial nature of infections
The earliest examination of human infection sites using rudimentary microscopic methods revealed the colonization of multiple bacterial species 1, 2. Despite this observation, the possibility that infections may be polymicrobial in nature was generally overlooked for many years. Potential interest in the topic was further stifled by the belief that disease was mediated by a single virulence factor produced by the dominant organism. Although this is true for some infections, immunization against single virulence factors has proven unsuccessful in many situations, suggesting far greater complexity than initially envisaged. We now appreciate that bacterial virulence is multifactorial and that the majority of infections result from colonization by more than one microbe 1, 2.
This re-evaluation has been driven in recent years by the improvement of diagnostic and genomic technologies; characterization of bacteria associated with many infections including chronic wounds, otitis media, and respiratory tract infections has revealed the presence of specific combinations of species (Table 1). Furthermore, other infections that were previously classified as being caused by single bacterial species have now been shown to be associated with a considerably more diverse bacterial population (Table 1). These findings have prompted a substantial effort aimed at understanding the nature of polymicrobial disease that is reflected in the escalation in publications on the topic, which have trebled in the past 5 years. This focus is certainly timely, given the health care and other socio-economic costs associated with many of these diseases and their treatment (Table 1).
Table 1.
Examples of polybacterial human diseases and the consequences of bacterial interactiona
| Disease | Dominant bacteria | Consequences of co-infection; likely interbacterial interactions | Healthcare costs | Refs |
|---|---|---|---|---|
| Cystic fibrosis lung | Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, Burkholderia cepacia complex, Stenotrophis maltophila | Increased antimicrobial tolerance, biofilm development, and biomass; metabolite perception, AI-2 and DSF-mediated signaling | $49 000 per patient per year USD | 15, 29, 62, 63 |
| Device-related infections | S. aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Proteus mirabilis, Providencia stuartii | Co-infection promotes progression to bacteremia; mixed biofilm formation | $2.3 billion USD | 64, 65 |
| Urinary tract infection | Escherichia coli (UPEC), Staphylococcus saprophyticus, Group B Streptococcus, Enterococcus faecalis, Klebsiella sp., Enterobacter sp., Proteus sp. | More persistent infection; modulation of host immunity by non-pathogen promotes infection by pathogen | $1.6 billion USD | 66, 67 |
| Pneumonia | Streptococcus pneumoniae, H. influenza | Increased biofilm formation and colonization of nasopharynx | $7.4 billion USD (Medicare only) | 68, 69 |
| Otitis media | H. influenzae, S. pneumoniae, Moraxella catarrhalis | Increase in biofilm, colonization of nasopharynx, antibiotic resistance and persistence of the infection; mixed biofilm formation and AI-2 signaling implicated | $2.88 billion USD | 9, 10, 70 |
| Periodontitis | Porphyromonas gingivalis, Treponema denticola, Veillonella atypica, Fusobacterium nucleatum, Streptococcus sp., Aggregatobacter actinomycetemcomitans | Biofilm formation, persistent infection and chronic inflammation; metabolite exchange, co-aggregation, interspecies signaling | £2.7 billion (UK, NHS) | 1, 22, 26, 66, 71, 72, 73 |
| Wound infections and diabetic ulcers | S. aureus, P. aeruginosa, Enterococcus sp., Bacteriodes fragilis, Corynebacterium sp. | Delayed wound healing, resistance to antibiotics; biofilm formation, peptidoglycan sensing, modulation of host immunity |
Diabetic foot ulcers: $9–13 billion USD | 21, 74, 75, 76, 77 |
| Inflammatory bowel disease | Correlated with presence of adherent-invasive E. coli, Fusobacterium sp., increase in Gammaproteobacteria and reduction in Firmicutes, Clostridium sp. and Bifidobacterium species | Direct interbacterial interactions unknown, immune modulation by one species may promote or suppress growth of another speciesb | Unknown | 78, 79 |
Abbreviations: NHS, National Health Service; UPEC, uropathogenic E. coli; USD, US dollar.
In a mouse model of colitis, prior infection by Helicobacter pylori was shown to reduce inflammation caused by subsequent infection with Salmonella Typhimurium through modulation of host immunity.
Recent microbial community surveys of infections have provided extensive information about microbiota diversity, gene content, and potential functions. There is as yet less understanding of the metabolic activities of these diverse organisms during the infection process and the molecular mechanisms that underpin interspecies interactions. A number of in vitro studies have given insight into interspecies interactions. This body of work suggests that a variety of molecules are exchanged between bacteria, enabling various competitive, synergistic, or neutral relationships to be conducted. More recently, physical contact between bacteria has been implicated in modulation of microbial behavior. Extrapolations of the findings from in vitro systems to the disease state need to be made cautiously of course. Nevertheless, both the detection of molecules during infection by metabolome analysis and mutational analysis of bacteria in model polymicrobial infections provide experimental support for some of the conclusions drawn. Here, we discuss the major types of interbacterial interaction that may govern disease, highlight recent advances in our understanding of interbacterial interactions in vivo, and discuss potential implications of targeting these interactions as a route towards control of bacterial disease. Interactions between bacteria and viruses, or bacteria and fungi, are also known to be important in several disease settings. We will focus on bacterial–bacterial interactions, but direct the reader to several recent reviews addressing other aspects of polymicrobial infections including interkingdom interactions 1, 2, 3, 4.
Exchange of chemicals between bacteria during disease
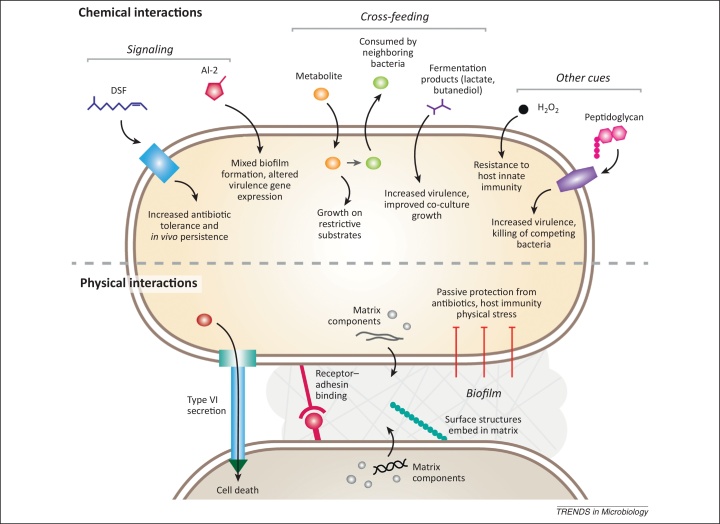
Bacteria produce many types of diffusible molecules that can affect their neighbors in different ways (Figure 1). Bacteria of the same species often use signal molecules to monitor aspects of their environment such as population density (a process that has been termed quorum sensing) and to modulate their behavior accordingly. Molecules are generally classed as ‘signals’ if they are produced by a dedicated pathway at a specific stage of growth, and elicit responses in the receiver organism that are distinct from those required for the processing of the molecule. Such effects usually occur following perception by a specific receptor and act to benefit both producer and receiver. The signal molecules synthesized by bacteria belong to a wide range of chemical classes and diverse mechanisms for signal perception and transduction have been described 5, 6. Some types of signal molecules produced for intracellular signaling can be recognized and influence other non-producing species through ‘eavesdropping’ 5, 6. Other interspecies interactions range from the symbiotic, such as the cross-feeding of intermediary metabolites during the enzymatic breakdown of complex carbon sources, to the strictly antagonistic, such as the production of lethal quantities of antibiotics or bacteriocins. Interactions can benefit one partner while the other is unaffected (where the molecules involved are termed ‘cues’) or allow subtle, non-lethal manipulation of the other species for the benefit of the producer (termed ‘coercion’). Many of these chemical interactions are contingent on the close proximity of bacteria, which can result from physical interactions such as formation of mixed biofilms (discussed below).
Figure 1.
Schematic illustrating potential bacterial interspecies interactions. Bacteria can influence other cells in a community through both chemical (top) and physical interactions (bottom). Chemical interactions include the production and perception of specific signal molecules such as diffusible signal factor (DSF) and autoinducer-2 (AI-2), which benefit both the producer and the receiver strain. The cross-feeding of metabolites between members of a community allows growth on complex carbon sources, and can also elicit responses distinct from metabolism, as a means of surveying the bacterial community. H2O2 and peptidoglycan are two examples of interspecies cues that elicit beneficial responses in the receiving species. Physical interactions between cells include those involved in formation of biofilms, which protect cells from stresses such as antibiotics and host immunity. Formation of mixed-species biofilms involves receptor and adhesin-mediated co-aggregation, interaction of other surface appendages such as pili and fimbriae, and the regulated secretion of an extracellular matrix in which surface structures become embedded. Contact-dependent interactions can also be antagonistic, as seen with type VI secretion in which toxins are translocated into neighboring cells.
Interspecies communication through diffusible signals
As outlined above, bacteria can sense signal molecules produced by other bacteria in their immediate environment during disease. We will illustrate this facet of interspecies communication in the context of mixed infections through consideration of autoinducer-2 (AI-2) and signals of the diffusible signal factor (DSF) family. It should be noted that other types of signal molecule including N-acyl homoserine lactones (AHLs), Pseudomonas quinolone signal (PQS), and 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) have been found to be produced during infection and to influence other bacteria through eavesdropping 5, 6.
AI-2
AI-2 is a ribose-derived signal molecule produced and perceived by multiple different bacteria [7]. The enzyme responsible for AI-2 synthesis is LuxS, which is also involved in central metabolism through a pathway responsible for recycling S-adenosyl methionine. AI-2 acts as an intraspecies quorum sensing signal in a number of Gram-positive and Gram-negative bacteria, where it influences biofilm formation and production of various virulence factors [8]. AI-2 may not have a role in quorum sensing in all bacteria but may nevertheless be secreted, because it can be toxic to cells. Studies of bacteria associated with otitis media have revealed synergy between the luxS-deficient species Moraxella catarrhalis and the AI-2 producers Haemophilus influenzae and Streptococcus pneumoniae 9, 10. AI-2 synthesis by H. influenzae promotes mixed biofilm formation, and development of a persistent M. catarrhalis infection in chinchillas [9], whereas AI-2 synthesis by S. pneumoniae increases M. catarrhalis colonization of the nasopharynx in co-infection of mice or chinchillas [10]. Interestingly, AI-2 alters the course of S. pneumoniae and M. catarrhalis co-infections in vivo but does not affect mixed biofilm formation in vitro [10]. Such findings highlight the need for both animal model studies and in vitro experiments to fully explore the consequences of different types of interaction (Box 1).
Box 1. Reconstruction of polybacterial infections using in vivo models.
Studies using in vivo models that closely mimic features of human polybacterial diseases are key in bridging the gap from the lab to the clinic. Modeling polybacterial infections presents specific challenges such as establishing a mixed infection and, in some cases, managing the effects of the native microbiota. When choosing an in vivo model it is important to know how closely the model infection reflects aspects of the human disease such as colonization, chronic infection, and interaction of the pathogen(s) with the immune system. Practical issues must also be taken into account, such as cost, ethics, manipulability, and options for controlling the native flora.
Several invertebrate models have already been used to examine interbacterial interactions that contribute to virulence, as well as some features of the innate immune response. Their ease of use makes them suited for high throughput experiments. For example, experiments using the fruit fly Drosophila melanogaster and the pathogen P. aeruginosa were used to demonstrate the virulence-enhancing effects of a subset of oropharangeal commensal organisms [80] and of peptidoglycan from Gram-positive bacteria [21].
Studies in mammals are needed when the research question requires reconstruction of more specific features of a disease, such as persistent infection or colonization of a specific organ. Different mammalian models offer distinct advantages and disadvantages in how accurately they represent the human disease and the degree of control they offer the scientist. For example, bacterial periodontitis can be modeled in the oral cavity of antibiotic-treated rats or in mouse skin wound infections. Although the former model is a closer representation of the disease, the wound infection model is easier to administer and monitor. It is also easier to exclude other bacteria in this model. Both models have been useful in revealing some of the interbacterial interactions that influence this disease 26, 81.
Developing accurate animal models of CF lung infection has been challenging. Various CFTR (cystic fibrosis transmembrane conductance regulator) mutant mice are available, however, these do not develop the chronic infections that characterize the human disease [82]. Chronic infection is mimicked by implanting agar beads with embedded bacteria directly into mouse or rat lungs, and this model has been used to investigate the synergy of P. aeruginosa with other CF pathogens [29]. New models in CFTR mutant pigs and ferrets may offer more options for modeling the development and progression of chronic CF lung infection [83].
Advances in sequencing and metabolomics approaches have made it possible to observe the events of human infections in detail. It is likely that these will be deployed in in vivo models to improve the resolution of our understanding of these infections.
AI-2 signaling may also influence the virulence of Pseudomonas aeruginosa in the cystic fibrosis (CF) lung. AI-2 is produced by certain bacteria found in CF sputum, such as Staphylococcus and Streptococcus species that are members of the oropharyngeal flora, but not by P. aeruginosa. Co-culture with either of two different AI-2 producing strains of Staphylococcus and Streptococcus species was shown to increase damage caused by P. aeruginosa in rat lung infections. Furthermore, CF sputum contains AI-2 at sufficient levels to alter P. aeruginosa gene expression in vitro, supporting the idea that interspecies AI-2 signaling is a contributing factor in the course of pathogenesis in CF [11].
DSF
DSF family signals are fatty acid molecules that share a characteristic cis-unsaturated double bond at position two of the fatty acid chain [12]. DSF was discovered in the plant pathogen Xanthomonas campestris but has since been shown to be produced by several clinically relevant bacteria including Stenotrophomonas maltophilia and Burkholderia species. Interspecies DSF signaling is implicated in CF lung infections involving P. aeruginosa, Burkholderia cenocepacia, and S. maltophilia. P. aeruginosa responds to DSF (cis-11-methyl-2-decenoic acid) produced by S. maltophilia and the related molecule BDSF (cis-2-dodecenoic acid) from B. cenocepacia, although it does not synthesize either of these molecules. Exposure of P. aeruginosa to DSF alters biofilm formation and polymyxin tolerance [13], whereas BDSF causes a modest decrease in quorum sensing signal production and type III secretion [14]. A recent study by Twomey and colleagues [15] presented several lines of evidence supporting the contention that DSF family signaling contributes to P. aeruginosa persistence and antibiotic resistance in CF lung infection. Specifically, DSF and BDSF are both found at physiologically relevant levels in CF sputum from adult patients, where their presence is correlated with B. cenocepacia and S. maltophilia colonization. Furthermore, DSF treatment increases the persistence of P. aeruginosa in a mouse model of CF lung infection. Together, these data provide evidence that interspecies DSF-mediated bacterial interactions occur in the CF lung and may influence the efficacy of antibiotic treatment.
Antibiotics, indole, and other cues
Bacteria can also respond to their neighbors in ways that fall outside strict definitions of either signaling or metabolite exchange (which we will examine below). A number of bacteria produce antibiotic compounds that may have a role in competition for niches or resources. Antibiotics at sub-lethal concentrations can alter the behavior of some clinically-relevant bacteria in ways that promote survival or infection [16]. For example, treatment of Staphylococcus aureus with beta-lactam antibiotics promotes biofilm formation [17]. Many bacteria produce the metabolite indole, which can modulate the virulence of some bacterial pathogens that do not produce the molecule [18]. For example, indole suppresses P. aeruginosa virulence through interference with AHL quorum sensing [19], and also suppresses virulence of S. aureus, where it acts to repress the expression of several virulence factors including α-hemolysin and enterotoxin [20]. Finally, peptidoglycan and hydrogen peroxide can function as interspecies cues. P. aeruginosa senses peptidoglycan from Gram-positive bacteria through a specific two-component system receptor; this triggers enhanced virulence and killing of Gram-positive commensals [21]. Hydrogen peroxide, which is produced by oral streptococci such as Streptococcus gordonii during fermentation of sugars, induces increased resistance to host innate immunity in the periodontitis-causing bacterium Aggregatibacter actinomycetemcomitans [22].
Metabolite cross-feeding
In addition to antagonistic interactions, different species of bacteria can also act in a cooperative fashion through metabolite cross-feeding, where one species utilizes an end-product of a metabolic pathway of another species. The metabolic abilities of the different bacteria in the community thereby complement each other. Nutritional cooperation is especially well-documented in the oral bacteria, and has been demonstrated for pairs of bacteria 17, 18, as well as certain consortia such as the set of five species from dental plaque that collectively degrade the salivary mucin glycoprotein MUC5B 23, 24. Metabolic relationships can be coupled to other types of communication, sometimes in a reciprocal fashion. An example of this is the mixed biofilm formed by Veillonella atypica and S. gordonii, in which V. atypica consumes lactate produced by S. gordonii, which itself produces amylase in response to a diffusible molecule from V. atypica [25].
Cross-feeding has consequences beyond supporting the growth of bacteria with limited metabolic capacities. Recently, some bacteria have been shown to preferentially consume the fermentation products of neighboring bacteria when faced with a choice of carbon sources. The choice of a ‘social’ carbon source has downstream effects on growth and virulence. A. actinomycetemcomitans shows better colonization and greater pathogenesis in mouse abscess infection when co-cultured with S. gordonii, and this advantage is dependent on its ability to metabolize lactate produced by S. gordonii [26]. In this case, cross-feeding appears to benefit A. actinomycetemcomitans by precluding nutrient competition with faster-growing resident streptococci at an early stage, allowing it to persist and cause infection later. The opportunistic pathogen P. aeruginosa also shows altered virulence phenotypes when grown on fermentation products produced by many of the bacteria that reside with P. aeruginosa in the CF airway, in comparison to glucose. P. aeruginosa grown on 2,3-butanediol shows increased biofilm formation, virulence factor production, and antimicrobial activity against various bacteria [27]. Thus, even though it is not necessary from a metabolic perspective, surveillance of the metabolites in the environment allows P. aeruginosa to assess which bacteria are co-residents and alter its behavior and lifestyle accordingly. The emergence of new metabolomics technologies has made it possible to explore the breadth and importance of metabolic interactions in polymicrobial infections by directly characterizing the content of infection sites (discussed further below).
Contact-dependent interactions between bacteria during infection
Although interactions between bacteria mediated by diffusible chemicals have received the bulk of attention, contact-dependent interactions are gaining increasing interest. Many insights into contact-dependent signaling have come from the study of biofilm formation, where physical interactions have been demonstrated to modulate community growth and behavior (Figure 1). In addition, the consequences of physical interaction are very different in polybacterial infections compared to single-strain models, suggesting that these interactions play a key role in enabling bacteria to adopt a polybacterial lifestyle.
Lessons from mixed-species biofilm infections
Biofilms are surface-attached bacterial communities, embedded in a matrix, which show radically different properties compared to free-living bacterial cultures. Studies of chronic clinical infections using in vivo and in vitro models have highlighted significant impacts of mixed-species biofilms in a range of chronic infections such as otitis media, wound infections, and CF lung infections (Table 1). In mixed-species biofilms, synergistic interactions often occur and result in beneficial effects such as greater biofilm biomass and enhanced antibiotic tolerance 10, 28, 29. Pili and fimbriae, which are hair-like adhesive structures on the bacterial cell surface, play a critical role in cell–cell interactions during mixed-species biofilm formation 30, 31. In addition, flagella have been shown to be important for the initial surface attachment of bacteria including Escherichia coli, Vibrio cholera, and P. aeruginosa during infection 31, 32, 33. Co-aggregation, the adhesion of genetically distinct bacteria to each other, is a process that has been most extensively investigated in oral polymicrobial biofilm formation. Oral biofilm formation occurs in an ordered, sequential process with the attachment of each bacterial species acting as a scaffold for the attachment of a subsequent species. Interactions are highly specific between pairs of bacterial species and are mediated by an adhesin on one cell type binding to a complementary receptor on another 34, 35.
Interbacterial physical interactions can also promote disease in ways that are independent of biofilm formation. Recent studies of enteric and oral pathogens have shown that co-aggregation of infecting bacteria promotes adherence to and invasion of eukaryotic cells 36, 37. Furthermore, physical interactions between co-infecting bacteria such as S. pneumoniae and H. influenzae have also been shown to result in altered virulence factor expression to promote colonization of an environmental niche [38].
Do other physical interactions between bacteria contribute to infection?
Several mechanisms of physical interaction between bacterial cells have been described that may play a role during infection. One of the most recently identified is the type VI secretion system (T6SS), which mediates interactions between many Gram-negative bacterial species. The T6SS was first identified in two human pathogens, V. cholerae and P. aeruginosa and was initially thought to be involved in virulence by directly targeting eukaryotic cells 39, 40. It has subsequently been discovered that most T6SSs actually mediate interbacterial competition by transporting toxins directly into competitor bacterial cells, whereas attacking cells possess cognate immunity proteins to prevent self-intoxication. T6SSs have now been identified in a vast array of pathogenic bacteria. It is thought that T6SS could play a role in a polybacterial infection by providing a competitive advantage for those bacteria with these functions. The antagonistic activity could aid initial colonization and enable a pathogen to defend a niche against invading competitors.
Russel et al. [41] recently hypothesized several non-antagonistic functions for T6SSs in polybacterial environments including toxin–immunity complexes that may function as signaling molecules in immune cells. A role for the T6SS in enhancing the extracellular matrix in biofilm populations via toxin-mediated cell lysis was also suggested. The presence of antibacterial T6SSs in bacteria associated with chronic polymicrobial infections such as P. aeruginosa, Serratia marcescens, and Burkholderia sp., indicates a potential role for the T6SS during infection 40, 42, 43. An important observation in support of this contention is that CF patients chronically infected with P. aeruginosa were found to possess antibodies that cross-react with Hcp1 (a structural protein from the P. aeruginosa antibacterial T6SS-1), suggesting that T6SS is active in the CF lung and may promote the survival of P. aeruginosa over other bacteria [40].
Other types of contact-dependent interaction are known to play roles in interbacterial interactions during environmental scenarios. Metal-reducing soil bacteria can express complex networks of electrically conductive pili known as nanowires [44], whereas contact-dependent inhibition through two-partner secretion systems that can deliver toxins to neighboring cells are encoded by many strains of pathogenic E. coli as well as many other pathogens [45]. Further work will uncover if these types of interactive systems play a role during infection.
Profiling and further understanding interbacterial interactions
Although great strides have been made in developing animal models to reproduce the etiology of human polymicrobial diseases (Box 1), these approaches still present considerable drawbacks and challenges. Complementary approaches that directly characterize the human infections are believed to deliver more meaningful insights into complex microbial infections and their activities. Genomic, proteomic, and metabolomics tools have been successful in probing the environment of polymicrobial infection sites. Whereas metagenomics surveys the metabolic potential and functional capabilities encoded by members of the microbial community, metatranscriptomics, metaproteomics, and metabolomics provide insights into the current metabolic activities during infection.
The potential of these tools was illustrated recently by Lim and colleagues [46], who used metagenomic and metatranscriptomic approaches to characterize the bacterial and viral communities present in the sputum of five CF patients with chronic lung infections. Each CF patient was found to possess a unique microbial profile with dominant infecting organisms persisting over time, although abundances varied during periods of exacerbation or antibiotic treatment. The metatranscriptome analyses showed the greatest variation both between patients and over time [45]. Lim and colleagues considered that this approach was best able to capture the dynamic nature of these complex communities [45]. A number of studies have reported metabolite profiling of sputum from CF patients that provide insight into the environment of bacterial community of the CF airway and alterations in the environment that occur during exacerbation [47]. These studies highlight the potential of these technologies to be effective tools to reveal the varied and dynamic nature of polymicrobial communities during infection.
New technologies to study metabolic exchange processes within polymicrobial communities are also being developed. For example, Moree and colleagues [48] recently described the use of matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) and MALDI-Fourier transform-ion cyclotron resonance (MALDI-FT-ICR) imaging mass spectrometry (MALDI-IMS) combined with tandem mass spectrometry (MS/MS) networking to provide insight into the metabolic interactions between CF lung pathogens. Using these technologies, they were able to visualize and identify the metabolites secreted by each organism and reveal a complex molecular interplay that involved suppression, increased production, and biotransformation of a range of molecules.
These techniques also allow predictions based upon studies of monospecies and multispecies communities in model systems to be tested in a real infection, and they can reciprocally inform the design of the model experiments so that the experimental conditions more closely mimic the infected state. An example of the first point is the assay of signal molecules in the sputum of CF patients that shows that they are present at levels that are physiologically relevant for interspecies signaling, as determined from model in vitro systems [15]. The second point is illustrated by the work of Jorth and colleagues [49] who explored the use of RNA sequencing (RNA-seq) to obtain a high-resolution transcriptome of A. actinomycetemcomitans during growth in vitro and in a murine abscess infection. A large number of metabolic genes were more highly expressed during infection, including those for formate dehydrogenase (fdhF1F2) and fumarate reductase (frdABCD), which are associated with fermentative and anaerobic metabolism, respectively. Overall, this experimental strategy provides an excellent outline for future investigations of bacterial physiology during infectious processes, and for the design of in vitro experiments under appropriate growth conditions.
Understanding interbacterial interactions may improve therapy
Exploitation of the wealth of metagenomic and metagenetic data on polybacterial disease is crucial for informed clinical judgment and appropriate treatment (Box 2). Currently the preferred treatment strategy for many polybacterial infections is still the use of broad-spectrum antibiotics 50, 51, 52, 53. Unfortunately such courses of antibiotics are generally ineffective or fail in many cases [54]. This problem is compounded by the continued emergence and increasing prevalence of multiple drug-resistant strains. Thus, alternative treatments strategies are required. By taking into account the complement of infecting organisms and the complex interactions between them, a new approach where therapy is personalized to each patient could be developed for treating such infections. Below we discuss some of the approaches that could be used in combating polybacterial infections and highlight initial evidence for their clinical potential.
Box 2. Analyzing the polybacterial nature of CF respiratory infections to inform clinical judgment and treatment.
The arrival of high-throughput parallel sequencing technologies has already revolutionized the study of microbiology within a short period 84, 85. Several studies have suggested that the throughput and quality of data being generated by these approaches bring personalized diagnostics within immediate reach 86, 87. Polymicrobial lung disease associated with CF is a well-documented case where this type of approach may be beneficial 86, 87.
Historically, a limited number of bacterial species including Pseudomonas aeruginosa, Burkholderia cenocepacia, Staphylococcus aureus, and Haemophilus influenzae were established as important CF pathogens. The ability to routinely identify these pathogens has for the most part informed antibiotic treatment in patients with CF presenting with pulmonary exacerbations [85]. However, it has been widely reported that treatment based on antibiotic susceptibilities of these specific CF pathogens does not correlate strongly with clinical outcome [88].
The appreciation that CF airway infections are more broadly polymicrobial than originally thought has offered potential explanations for the sometimes poor response to antibiotic therapy 85, 88. Molecular profiling of the CF-derived sputum has revealed that there are dozens of bacterial genera present, including Streptococcus and a variety of anaerobes including Prevotella, Fusobacterium, and Veillonella. Specific bacterial species have been revealed to be colocated in different regions of the airway [89]. The relative contribution by these newly identified members and their location within the CF airway to clinical status, disease progression, and the efficacy of antibiotic treatment against pathogens remains to be fully examined.
Improved patient outcomes may be achievable with the application of more focused antibiotic therapy that is based on comprehensive microbiological analyses of each patient. A current EU funded project CFMATTERS (Cystic Fibrosis Microbiome-determined Antibiotic Therapy Trial in Exacerbations: Results Stratified) aims to address this issue through a clinical trial comparing the use of microbiome-directed antibiotic treatment versus standard therapy (http://www.cfmatters.eu). The analysis should enhance scientific knowledge of the polymicrobial lung disease, based on bacterial molecular profiling of sputum, allowing the use of stratified targeted antibacterial therapy that can be compared with standard empirical antibacterial therapy currently used. If successful the approach may pave the way for more effective therapeutic regimes and ultimately contribute to personalized CF treatment and the development of personalized treatment for other polybacterial diseases.
Interference with interbacterial communication
Through work on individual bacterial species, several strategies have been developed to inhibit or interfere with bacterial cell–cell signaling systems that regulate virulence factor synthesis and gene expression 55, 56. There are several studies describing small molecules that are effective in interfering with these processes that were identified either through screening of chemical libraries or that are structural analogs of the signal molecules [51]. Other approaches to interfere with signaling have used enzymes that degrade the signal molecules 55, 56. For every class of bacterial cell–cell signal discovered thus far, a mechanism has been discovered that inhibits, destroys, or removes it. Work on the inhibition of these signaling systems in single species provides a platform for manipulation of interspecies interactions during polybacterial disease.
As discussed previously, there is evidence that interspecies signaling with DSF family molecules influences biofilm formation and increases polymyxin tolerance in P. aeruginosa, factors that may contribute to the refractory nature of polymicrobial lung infections to therapy. It remains to be seen whether interference with DSF signaling during infection may negatively affect P. aeruginosa persistence. Intriguingly, Deng et al. [57] have recently demonstrated that treatment with DSF or analogs make bacterial pathogens including Bacillus cereus and S. aureus more susceptible to antibiotic treatment in vitro. The mechanistic bases for these effects are as yet unknown.
Indole signaling is known to suppress virulence of some important pathogens and the potential for the therapeutic use of related molecules is being investigated. Indole derivatives have shown potential as post-biotics, because indole-3-propionic acid is a powerful antioxidant and 7-hydroxyindole diminishes virulence and colonization of P. aeruginosa [58]. In order to capitalize on these observations, screening for more natural and synthetic indole derivatives that are nontoxic and cannot be easily metabolized by other pathogens is now needed, in conjunction with further in vivo studies.
Disruption of interbacterial contact and attachment
Pili and fimbriae are adhesive surface structures required for biofilm formation and attachment to both the host and other bacteria; their loss often results in dramatically reduced virulence. Chemical targeting of these structures is being considered as an approach to neutralizing various infections. This has been exploited in recent efforts to target uropathogenic E. coli by inhibiting type I pili. Deploying pilicides (mimics of the normal pilin subunits) interfered with pilin export, leading to reduced cell aggregation and improved clearance of infection in various in vivo models 59, 60. Although pilicides have not been tested specifically in treating a mixed bacterial infection, effective use in several non-sterile mouse models, which most likely carry commensals, suggests the potential of this approach.
Adhesion inhibitors based on synthetic receptor analogs have also proved to be potent in disrupting adhesion of many bacteria [61]. For example, synthetic peptides and glycerophosphate derivatives mimicking fragments of fimbrial adhesins have been effective against several oral infection studies of mice [61]. Although these studies provide evidence of the potential of adhesion inhibitors, the use of these molecules in their present form is not desirable owing to their proinflammatory properties and potential cytotoxicity. Moreover, their potency against polybacterial infection still needs to be scrutinized using more appropriate in vivo models.
As evidenced in this section, substantial effort has been focused on the attenuation of interbacterial interactions, with promising results in many cases. Although still in its infancy, this analysis provides new tools which, in combination with other therapeutic measures including symbiotic therapy, phage therapy, and the use of polybacterial vaccines, could facilitate improved therapeutic approaches to prevent and treat polybacterial infection [1]. Although the strategies outlined above present attractive solutions, many challenges exist in developing effective treatments targeting interbacterial interactions exclusively, including obstacles in specificity and delivery of the treatment, potential negative implications of systemic inhibition, and knock-on effects on normal flora. Despite advances in developing animal models for polymicrobial disease, the in vivo efficacy of most of these approaches remains to be assessed (Box 1).
Concluding remarks
It is now clear that many infections are polybacterial in nature, and that interactions between different organisms can contribute to disease progress and clinical outcome. These interactions may involve commensal bacteria as well as pathogens and are mediated by a range of molecular mechanisms that include interspecies signaling, metabolite exchange, and cell–cell contact. The consequences of interaction are often modulation of pathogen behavior including alterations in virulence factor synthesis, biofilm formation, and antibiotic resistance or tolerance. More research is needed to provide a better mechanistic insight into the complex interplay between potential pathogenic agents, commensal organisms, and their eukaryotic hosts (Box 3). Understanding the molecular basis and biological effect of these interbacterial processes may lead to an improvement of treatment regimens and also define new targets and strategies for disease control.
Box 3. Outstanding questions.
-
•
How can existing diagnostic tools be improved and new tools be established to characterize the bacterial communities in polybacterial diseases and to identify the molecules involved in interspecies interactions?
-
•
Will targeted antibacterial therapy based on bacterial molecular profiling be more effective than standard empirical antibacterial therapy?
-
•
Non-pathogenic organisms coexist with pathogens in many sites of infection. What is the impact of these organisms on the virulence and antibiotic susceptibility of pathogens?
-
•
The molecular mechanisms that underpin interbacterial interactions during infection remain poorly understood. Will new mechanisms be uncovered?
-
•
The impact of polybacterial infections on the host immune response needs to be examined. How will this differ from the response to individual pathogens?
-
•
Development of biotherapeutics against polybacterial infection has begun with vaccine and phage therapy. Can an understanding of interbacterial interactions provide a platform for targeted interference strategies and improved care?
Acknowledgments
We thank Yvonne McCarthy and Max Dow for their helpful comments and reading of the manuscript. The work of the authors has been supported in part by grants awarded by the Wellcome Trust (WT100204AIA senior fellowship grant to R.P.R.) and the EU Seventh Framework Programme (Grant No. 603038 to R.P.R.).
References
- 1.Peters B.M. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brogden K.A. Human polymicrobial infections. Lancet. 2004;365:253–255. doi: 10.1016/S0140-6736(05)17745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCullers J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014;12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 4.Harriott M.M., Noverr M.C. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19:557–563. doi: 10.1016/j.tim.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riedel K. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology. 2001;147:3249–3262. doi: 10.1099/00221287-147-12-3249. [DOI] [PubMed] [Google Scholar]
- 6.Fugère A. Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS ONE. 2014;9:e86705. doi: 10.1371/journal.pone.0086705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardie K.R., Heurlier K. Establishing bacterial communities by “word of mouth”: LuxS and autoinducer 2 in biofilm development. Nat. Rev. Microbiol. 2008;6:635–643. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- 8.Pereira C.S. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 9.Armbruster C. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. MBio. 2010 doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez A.C. Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog. Dis. 2014;70:280–288. doi: 10.1111/2049-632X.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan K. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 12.Ryan R.P., Dow J.M. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011;19:145–152. doi: 10.1016/j.tim.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Ryan R.P. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 14.Deng Y. Cis-2-dodecenoic acid signal modulates virulence of Pseudomonas aeruginosa through interference with quorum sensing systems and T3SS. BMC Microbiol. 2013;13:231–242. doi: 10.1186/1471-2180-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Twomey K.B. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 2012;6:939–950. doi: 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernier S.P., Surette M.G. Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 2013;4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan J. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio. 2012;3:e00198–e212. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J., Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 19.Chu W. Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl. Environ. Microbiol. 2012;78:411–419. doi: 10.1128/AEM.06396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J. Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2013;97:4543–4552. doi: 10.1007/s00253-012-4674-z. [DOI] [PubMed] [Google Scholar]
- 21.Korgaonkar A. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsey M.M., Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickström C. Proteolytic degradation of human salivary MUC5B by dental biofilms. Microbiology. 2009;155:2866–2872. doi: 10.1099/mic.0.030536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickström C., Svensäter G. Salivary gel-forming mucin MUC5B – a nutrient for dental plaque bacteria. Oral Microbiol. Immunol. 2008;23:177–182. doi: 10.1111/j.1399-302X.2007.00407.x. [DOI] [PubMed] [Google Scholar]
- 25.Egland P.G. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey M.M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkataraman A., Rosenbaum M. Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa. ISME J. 2014;8:1210–1220. doi: 10.1038/ismej.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K.W.K. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bragonzi A. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS ONE. 2012;7:e52330. doi: 10.1371/journal.pone.0052330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barken K.B. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 31.Pratt L.A., Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 32.O’Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 33.Watnick P.I., Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright C.J. Microbial interactions in building of communities. Mol. Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang R. Bacterial interactions in dental biofilm. Virulence. 2011;2:435–444. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira A.L. Diarrhea-associated biofilm formed by enteroaggregative Escherichia coli and aggregative Citrobacter freundii: a consortium mediated by putative F pili. BMC Microbiol. 2010;10:57. doi: 10.1186/1471-2180-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards A.M. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect. Immun. 2006;74:654–662. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cope E.K. Regulation of virulence gene expression resulting from Streptococcus pneumoniae and nontypeable Haemophilus influenzae interactions in chronic disease. PLoS ONE. 2011;6:e28523. doi: 10.1371/journal.pone.0028523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pukatzki S. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mougous J.D. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell A.B. Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz S. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murdoch S.L. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 2011;193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malvankar N.S., Lovley D.R. Microbial nanowires: a new paradigm for biological electron transfer and bioelectronics. ChemSusChem. 2012;5:1039–1046. doi: 10.1002/cssc.201100733. [DOI] [PubMed] [Google Scholar]
- 45.Ruhe Z.C. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim Y.W. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J. Cyst. Fibros. 2012;12:154–164. doi: 10.1016/j.jcf.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Twomey K.B. Microbiota and metabolite profiling reveal specific alterations in bacterial community structure and environment in the cystic fibrosis airway during exacerbation. PLoS ONE. 2013;8:e82432. doi: 10.1371/journal.pone.0082432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moree W.J. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorth P. Probing bacterial metabolism during infection using high-resolution transcriptomics. J. Bacteriol. 2013;195:4991–4998. doi: 10.1128/JB.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talbot G.H. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 2006;42:657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 51.Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 52.Bjarnsholt T. Applying insights from biofilm biology to drug development – can a new approach be developed? Nat. Rev. Drug Discov. 2013;12:791–808. doi: 10.1038/nrd4000. [DOI] [PubMed] [Google Scholar]
- 53.Kostakioti M. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013;3 doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leekha S. General principles of antimicrobial therapy. Mayo Clin. Proc. 1991;86:156–167. doi: 10.4065/mcp.2010.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Njoroge J., Sperandio V. Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol. Med. 2009;1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rasko D., Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 57.Deng Y. Diffusible signal factor (DSF) quorum sensing signal and structurally related molecules enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens. BMC Microbiol. 2014;14:51. doi: 10.1186/1471-2180-14-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2009;2:75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chorell E., Pinkner J. Design and synthesis of C-2 substituted thiazolo and dihydrothiazolo ring-fused 2-pyridones: pilicides with increased antivirulence activity. J. Med. Chem. 2010;53:5690–5695. doi: 10.1021/jm100470t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chorell E. Synthesis and application of a bromomethyl substituted scaffold to be used for efficient optimization of anti-virulence activity. Eur. J. Med. Chem. 2011;46:1103–1116. doi: 10.1016/j.ejmech.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartmann M. Inhibition of bacterial adhesion to live human cells: activity and cytotoxicity of synthetic mannosides. FEBS Lett. 2012;586:1459–1465. doi: 10.1016/j.febslet.2012.03.059. [DOI] [PubMed] [Google Scholar]
- 62.Baldan R. Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS ONE. 2014;9:e89614. doi: 10.1371/journal.pone.0089614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Sullivan A.K. Health care utilization and costs for cystic fibrosis patients with pulmonary infections. Manag. Care. 2011;20:37–44. [PubMed] [Google Scholar]
- 64.Armbruster C.E. Increased incidence of urolithiasis and bacteremia during Proteus mirabilis and Providencia stuartii coinfection due to synergistic induction of urease activity. J. Infect. Dis. 2014;209:1524–1532. doi: 10.1093/infdis/jit663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Von Eiff C. Infections associated with medical devices: pathogenesis, management and prophylaxis. Drugs. 2005;65:179–214. doi: 10.2165/00003495-200565020-00003. [DOI] [PubMed] [Google Scholar]
- 66.Kline K.A. Immune modulation by group B Streptococcus influences host susceptibility to urinary tract infection by uropathogenic Escherichia coli. Infect. Immun. 2012;80:4186–4194. doi: 10.1128/IAI.00684-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 2003;49:53–70. doi: 10.1067/mda.2003.7. [DOI] [PubMed] [Google Scholar]
- 68.Thomas C.P. Incidence and cost of pneumonia in medicare beneficiaries. Chest. 2012;142:973–981. doi: 10.1378/chest.11-1160. [DOI] [PubMed] [Google Scholar]
- 69.Shak J.R. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol. 2013;21:129–135. doi: 10.1016/j.tim.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahmed S. Incremental health care utilization and costs for acute otitis media in children. Laryngoscope. 2014;124:301–305. doi: 10.1002/lary.24190. [DOI] [PubMed] [Google Scholar]
- 71.Grenier D., Mayrand D. Nutritional relationships between oral bacteria. Infect. Immun. 1986;53:616–620. doi: 10.1128/iai.53.3.616-620.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect. Immun. 1992;60:5298–5301. doi: 10.1128/iai.60.12.5298-5301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chapple I.L.C. Time to take periodontitis seriously. BMJ. 2014;348 doi: 10.1136/bmj.g2645. [DOI] [PubMed] [Google Scholar]
- 74.Karchmer A.W. In: The Diabetic Foot: Medical and Surgical Management. Veves A., editor. Humana Press; 2012. Microbiology and treatment of diabetic foot infections; pp. 331–346. [Google Scholar]
- 75.Pastar I. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE. 2013;8:e56846. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rice J.B. Burden of diabetic foot ulcers for medicare and private insurers. Diabetes Care. 2014;37:651–658. doi: 10.2337/dc13-2176. [DOI] [PubMed] [Google Scholar]
- 77.Dowd S.E. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS ONE. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Higgins P., Johnson L. Prior Helicobacter pylori infection ameliorates Salmonella Typhimurium-induced colitis: mucosal crosstalk between stomach and distal intestine. Inflamm. Bowel Dis. 2011;17:1398–1408. doi: 10.1002/ibd.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kostic A.D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sibley C.D. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kesavalu L. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect. Immun. 2007;75:1704–1712. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilke M. Mouse models of cystic fibrosis: phenotypic analysis and research applications. J. Cyst. Fibros. 2011;10(Suppl. 2):S152–S171. doi: 10.1016/S1569-1993(11)60020-9. [DOI] [PubMed] [Google Scholar]
- 83.Keiser N., Engelhardt J. New animal models of cystic fibrosis: what are they teaching us? Curr. Opin. Pulm. Med. 2011;17:478–483. doi: 10.1097/MCP.0b013e32834b14c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Febrer M. Advances in bacterial transcriptome and transposon insertion-site profiling using second-generation sequencing. Trends Biotechnol. 2011;29:586–594. doi: 10.1016/j.tibtech.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 85.Surette M.G. The cystic fibrosis lung microbiome. Ann. Am. Thorac. Soc. 2014;11(Suppl. 1):S61–S65. doi: 10.1513/AnnalsATS.201306-159MG. [DOI] [PubMed] [Google Scholar]
- 86.Rogers G.B. Interpreting infective microbiota: the importance of an ecological perspective. Trends Microbiol. 2013;21:271–276. doi: 10.1016/j.tim.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pattison S.H. Molecular detection of CF lung pathogens: current status and future potential. J. Cyst. Fibros. 2013;12:194–205. doi: 10.1016/j.jcf.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sibley C.D., Surette M.G. The polymicrobial nature of airway infections in cystic fibrosis: Cangene Gold Medal Lecture. Can. J. Microbiol. 2011;57:69–77. doi: 10.1139/w10-105. [DOI] [PubMed] [Google Scholar]
- 89.Willner D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012;6:471–474. doi: 10.1038/ismej.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]