Abstract
Human immunodeficiency virus type 1 particles form by budding at the surface of most cell types. In this process, a piece of the plasma membrane is modified into an enveloped virus particle. The process is driven by the internal viral protein Pr55gag. We have studied how host proteins in the membrane are dealt with by Pr55gag during budding. Are they included in or excluded from the particle? The question was approached by measuring the relative concentrations of host and viral proteins in the envelope of Pr55gag particles and in their donor membranes in the cell. We observed that the bulk of the host proteins, including actin and clathrin, were passively included into the virus-like Gag particles. This result suggests that budding by Pr55gag proceeds without significant alteration of the original host protein composition at the cell membrane. Nevertheless, some proteins were concentrated in the particles, and a few were excluded. The concentrated proteins included cyclophilin A and Tsg-101. These were recruited to the plasma membrane by Pr55gag. The membrane-bound cyclophilin A was concentrated into particles as efficiently as Pr55gag, whereas Tsg-101 was concentrated more efficiently. The latter finding is consistent with a role for Tsg-101 in Gag particle release.
Human immunodeficiency virus type 1 (HIV-1) particles assemble and bud at the surface of most cell types (14). The process can be reproduced by expressing the gene for internal viral protein Pr55gag only, implying that this protein carries the crucial functions for virus budding. Morphological studies of the assembly of such Pr55gag particles, also called Gag particles, and of HIV-1 suggest that budding is initiated by the formation of a small curved layer of Pr55gag below the plasma membrane (PM) of the host cell (14, 15). This structure grows into a particle-sized bud and further into an almost complete particle that is still tethered to the PM by a membranous neck. Particles are finally released by a pinching-off reaction in the neck.
The virus budding process is very similar to the process by which intracellular transport vesicles are formed. In the latter case, cytoplasmic proteins direct budding at cellular membranes, also called donor membranes (22). Like Pr55gag, they induce membrane curvature by forming a submembranous protein layer or coat. As this process is integrated with the sorting of, e.g., cargo proteins into the vesicles, we wondered how host proteins in the PM are dealt with during virus budding. Are they excluded from or included in the enveloped virus or Gag particles? This question is relevant because the structural proteins and the genome of HIV-1 are apparently not organized into a tightly packed structure; in the small icosahedral enveloped viruses, such a structure automatically excludes foreign components (5).
The existing models for the horizontal organization of HIV-1 glycoproteins and Pr55gag in the plane of the membrane leave significant free space for host protein “stowaways” (10, 14, 29). Furthermore, there seems to be additional space at the center of the budding particle, and this space apparently is not occupied by virus-specific components. So far there have been several reports about the inclusion of specific host proteins in highly purified HIV-1, but as their presence has not been related to their densities in the viral and donor membranes, it is not possible to conclude whether they passively enter the particles or whether they are actively included (concentrated) or excluded (diluted) (32). Although host proteins that become passively incorporated into the virus or even partially excluded from it during budding may be functionally significant for virus replication, the most important ones are likely to be those that become concentrated.
In order to monitor the fate of host proteins during virus budding, Hammarstedt et al. recently developed an assay to compare the protein components of the virus particle and the PM, i.e., the donor membrane, on the basis of membrane area (18). They analyzed the inclusion of host proteins in Pr65gag particles of Moloney murine leukemia virus (Mo-MuLV) with or without the viral envelope (Env) protein at different levels of production (18). They observed that the bulk of the host proteins present in the donor membrane were passively included in the Gag particles; only a few were found to be either actively included or excluded. In the present study, we analyzed the inclusion of host proteins in HIV-1 Gag particles. The host proteins cyclophilin A (CypA) and Tsg-101 were actively incorporated into the particles, but the majority of the PM proteins, including actin and clathrin, were included at densities similar to those found in the donor membrane. These findings confirm the earlier findings obtained with Pr65gag of Mo-MuLV and suggest that the retrovirus and lentivirus Gag proteins operate in budding without significant host protein exclusion.
MATERIALS AND METHODS
Cell cultures.
Baby hamster kidney cells (BHK-21; American Type Culture Collection, Manassas, Va.) were grown as described previously (38). The growth of 293T cells was also described previously (20). Jurkat cells (Sven Pettersson, Center for Genomic Research, Karolinska Institute, Stockholm, Sweden) were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 10 mM HEPES, 2 mM glutamine, 100 U of penicillin G sodium/ml, and 100 μg of streptomycin/ml. All cell culture media were obtained from Invitrogen (Paisley, United Kingdom).
Expression plasmids and viral vectors.
Infectious Semliki Forest virus (SFV) vectors were produced in BHK-21 cells transfected by electroporation with equal amounts of helper 1 and SFV vector RNAs (24). The replication-competent vector RNAs were transcribed in vitro from pSFV-helper 1 and pSFV-capsid (pSFV-C) constructs (37). The pSFV-C constructs used were pSFV-C/HIVgag, containing the Pr55gag gene from HIV-1 strain BH10 (20), and pSFV-C/NP, containing the A/PR/8 nucleoprotein gene from influenza virus (NPflu) (42). For expression analysis, nearly confluent cultures of BHK-21 cells in 6-cm dishes were infected with SFV vectors for 1 h at 37°C (24). Jurkat cells were infected with SFV vectors for 2 h at 37°C in RPMI 1640 medium supplemented with 2 mM glutamine, 10 mM HEPES, 0.2% bovine serum albumin, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Subconfluent 293T cells in 6-cm dishes were transfected with 10 μg of pCMV-HIVgag (20) or pRRLsin.cPPT.CMV.GFP.Wpre DNA (expressing enhanced green fluorescent protein; obtained from Luigi Naldini, Institute for Cancer Research and Treatment, University of Turin Medical School) by calcium phosphate precipitation (9).
Metabolic labeling.
Metabolic labeling of cells with [35S]methionine (Met) or [32P]orthophosphate (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) was done with low-phosphate, low-Met (low-phosphate/Met) medium (12.5 μg of phosphate/ml, 3.0 μg of Met/ml; i.e., 1/10 the regular concentrations) (medium no. 991303; National Veterinary Institute, Uppsala, Sweden) supplemented with 2 mM glutamine, 5% fetal calf serum, 20 mM HEPES, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. For labeling with [35S]Met, cells were grown in low-phosphate/Met medium supplemented with 30 μg of unlabeled Met/ml. After 24 h, the medium was replaced with low-phosphate/Met medium supplemented with 100 μCi of [35S]Met/ml (35S labeling medium), and the cells were incubated for 15 h before being infected with SFV-C vectors. Labeling was continued with fresh 35S labeling medium after infection. Particles were collected in fresh 35S labeling medium for 1 h from 5 to 6 h after infection, the last 15 min in the presence of an excess of unlabeled Met (300 μg/ml). For labeling with [32P]orthophosphate, cells were grown in low-phosphate/Met medium supplemented with 30 μg of unlabeled Met/ml and 25 μCi of [32P]orthophosphate/ml. After 24 h, the medium was replaced with low-phosphate/Met medium supplemented with 25 μCi of [32P]orthophosphate/ml (32P labeling medium), and incubation was continued for an additional 15 h before infection of the cells with SFV-C vectors. After infection, labeling was continued with fresh 32P labeling medium. Particles were collected in low-phosphate/Met medium for 1 h from 5 to 6 h after infection, without further labeling. Cultures of 293T cells were labeled for 21 h at 25.5 h after DNA transfection in low-Met medium (1/10 the regular Met concentration) supplemented with 100 μCi of [35S]Met/ml. Particles in medium were collected during the last 4 h of the labeling period.
Isolation of PM and Gag particles.
Homogenization of cells and isolation of Gag-enriched membranes (or corresponding membranes from uninfected control cells) by flotation in a sucrose step gradient were done as described previously (38) with some modifications. The postnuclear fraction of homogenized cells (800 μl) was mixed with 2.5 ml of 70% (wt/wt) sucrose in water; placed at the bottom of a Beckman (Palo Alto, Calif.) SW41 centrifuge tube; and overlaid with 1 ml of 50% (wt/wt) sucrose, 2 ml each of 40, 35, and 30% (wt/wt) sucrose, and 1.5 ml of 10% (wt/wt) sucrose in water. The gradient was run at 36,000 rpm (160,000 × g) for 16 h at 4°C. Fractions of 1 ml were collected from the top. Membranes in samples of each fraction were recovered by centrifugation in a Beckman JA18.1 rotor at 17,000 rpm (34,000 × g) for 1.5 h at 4°C after the samples were first diluted twofold with 10 mM Tris-HCl (pH 7.4). Gag-enriched membranes in pooled fractions 6 and 7 (or corresponding membranes from control cells) were recovered by centrifugation in a Beckman SW41 rotor at 35,000 rpm (151,260 × g) for 1 h at 4°C after fivefold dilution with 10 mM Tris-HCl (pH 7.4). Gag particles in medium were purified by sedimentation in a 5 to 20% (wt/vol) iodixanol gradient (Optiprep; Nycomed Pharma, Oslo, Norway) and concentrated from diluted gradient fractions as described previously (18). Medium with Gag particles from 293T cells was filtered through a 0.45-μm-pore-size filter (Schleicher & Schuell, Dassel, Germany) before particle purification by sedimentation.
Analysis by SDS-PAGE and quantification of phospholipids and proteins.
The [32P]phospholipid-labeled samples were taken up in 3% sodium dodecyl sulfate (SDS), heated for 2 min at 70°C, mixed with an equal volume of 2×-concentrated sample buffer (0.3 M Tris HCl [pH 8.8], 9.4 mM EDTA, 46.6% [vol/vol] glycerol, 0.04% [wt/vol] bromophenol blue), and analyzed by SDS-20% polyacrylamide gel electrophoresis (PAGE). The gel was dried and exposed to a Bas-III image plate (Fujifilm Sverige AB, Stockholm, Sweden). The [35S]Met-labeled samples were analyzed for proteins by SDS-PAGE with a 6 to 15% polyacrylamide gradient. The gel was fixed in 10% trichloroacetic acid-40% methanol for 30 min at room temperature (RT) before being dried and exposed to Fuji X-ray film or a Bas-III image plate. The radioactivity in protein or in mixed lipid-detergent-micelle bands was determined by using a Bas 2000 Bio-Image analyzer system equipped with Image Gauge version 3.3 software (Fujifilm Sverige AB).
Western blot analyses.
The primary antibodies used for Western blot analyses were a rabbit polyclonal anti-CypA antibody (PA3-021; Affinity Bioreagents, Inc., Golden, Colo.), a mouse monoclonal anti-actin antibody (MAB1501R; Chemicon International, Temecula, Calif.), and goat polyclonal anti-Tsg-101 (sc-6037) and anti-clathrin heavy chain (sc-6579) antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). The secondary antibodies used were horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (IgG) (NA934) and sheep anti-mouse IgG (NA931) (both from Amersham Pharmacia Biotech, Uppsala, Sweden) and donkey anti-goat IgG (sc-2020; Santa Cruz Biotechnology).
For Western blot analyses, samples were subjected to SDS-PAGE (6 to 15%) and blotted onto a pure nitrocellulose membrane (0.2-μm-pore size; Bio-Rad Laboratories, Hercules, Calif.) at 100 mA for 19 h. To avoid nonspecific antibody binding, the membrane was washed in a buffer that contained 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Tween 20, and 0.5% fat-free dry milk (blocking buffer) for 1 h 50 min at RT and then in blocking buffer containing 5% fat-free dry milk for 10 min. The membrane was washed for 10 min at RT with 20 mM Tris-HCl (pH 7.5)-150 mM NaCl-0.1% Tween 20 (binding buffer) before reaction with the primary antibody in binding buffer for 1 h at RT. The membrane was washed three times for 10 min each time at RT with binding buffer and once for 10 min with blocking buffer containing 5% fat-free dry milk. The reaction with the secondary antibody was carried out for 1 h at RT in blocking buffer. The membrane was washed three times for 10 min each time with binding buffer. Finally, proteins that reacted with antibodies were detected by an enhanced chemiluminescence reaction. For this, the membrane was developed for 1 min at RT in a mixture containing 10 ml of 1.25 mM luminol (5-amino-2,3-dihydro-1,4-phthalazine-dione), 0.1 M Tris-HCl (pH 8.5)-100 μl of 68 mM p-coumaric acid in dimethyl sulfoxide, and 30 μl of 3% H2O2 and then exposed to Fuji X-ray film for a short time (15 s to 15 min). Quantification of reactive protein bands from the Fuji X-ray film was done by using a luminiscence image analyzer equipped with a Las-1000 camera and Image Gauge version 3.0 software (Fujifilm Sverige AB). A dilution series of samples was used to define an interval of values that were linear relative to the concentrations of samples. Only values in the interval were used for quantification. The membrane was also used for the detection of [35S]Met-labeled proteins by longer exposure to Fuji X-ray film.
Transmission electron microscopy (EM).
BHK-21 cells infected with SFV-C vectors expressing Pr55gag (SFV-C/Pr55gag vectors) were fixed at 6 h postinfection in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer containing 0.1 M sucrose and 3 mM CaCl2 (pH 7.4). The cells were scraped with a wooden stick, transferred to an Eppendorf tube, and further fixed overnight in a refrigerator. The cells were rinsed in 0.15 M sodium cacodylate buffer containing 3 mM CaCl2 (pH 7.4) and centrifuged. The pellets were postfixed in 2% osmium tetroxide in 0.07 M sodium cacodylate buffer containing 1.5 mM CaCl2 (pH 7.4) at 4°C for 2 h, dehydrated in ethanol followed by acetone, and embedded in LX-112 (Ladd, Burlington, Vermont). Sections were contrasted with uranyl acetate followed by lead citrate. Pr55gag-enriched cell membranes, pooled and concentrated by centrifugation from sucrose step gradient fractions, were fixed and embedded as described above. Gag particles, pooled and concentrated by centrifugation from iodixanol gradient fractions, were fixed in the same fixative as that described above. The pellets were rinsed in buffer (0.15 M sodium cacodylate-3 mM CaCl2 [pH 7.4]), embedded in warm 10% gelatin (37°C) for 10 min, placed in a refrigerator for 20 min, and fixed in the same fixative overnight. The gelatin pellet was removed from the tube and cut into small pieces. The specimens were postfixed and processed as described above. The contrasted thin sections were examined with a Leo 906 (Zeiss, Oberkochen, Germany) transmission electron microscope at 80 kV.
RESULTS
Separation of Gag particles from cell-derived membrane vesicles.
The production of HIV-1 or the corresponding Gag particles in cultured cells is compromised by the constitutive release of membrane vesicles from the cells (16) (2). When virus or Gag particles are isolated by sedimentation, portions of these vesicles cofractionate with the Gag particles because of similar physical features. In a previous study of Mo-MuLV Gag particles, Hammarstedt et al. largely succeeded in overcoming this problem in two steps (18). First, SFV-C vectors were used for the efficient production of Gag particles in the cells (24, 37). This strategy facilitated a short collection period for Gag particles and hence minimized the time available for the constitutive release of cell vesicles. Second, Gag particles were separated from cell vesicles in a 5 to 20% iodixanol gradient by centrifugation at 160,000 × g for 90 min. A similar approach was used here for the isolation of HIV-1 Gag particles.
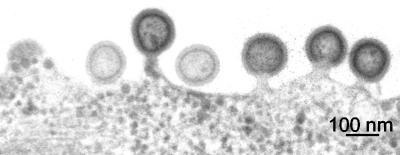
The Pr55gag gene was expressed in BHK-21 cells by using SFV-C/Pr55gag vectors (20). The initial translation product is an SFV capsid (CSFV)-Pr55gag fusion protein which is cotranslationally cleaved by an autoprotease activity of CSFV into CSFV and Pr55gag. The latter becomes myristoylated at its N terminus and is used for the assembly of Gag particles at the PM, whereas CSFV forms nonbudding SFV nucleocapsids in the cytoplasm. Figure 1 shows the budding of HIV-1 Gag particles at the surface of BHK-21 cells infected with SFV-C/Pr55gag vectors. Gag particles are seen as rather homogeneously sized spheres (diameter, 100 to 130 nm) with a characteristic layer of Pr55gag beneath the lipid bilayer in the section view, as shown before (14, 15).
FIG. 1.
Budding of Gag particles at the cell surface. BHK-21 cells were infected with SFV-C/Pr55gag vectors and fixed after 6 h. Thin sections of the cells were prepared and analyzed by EM.
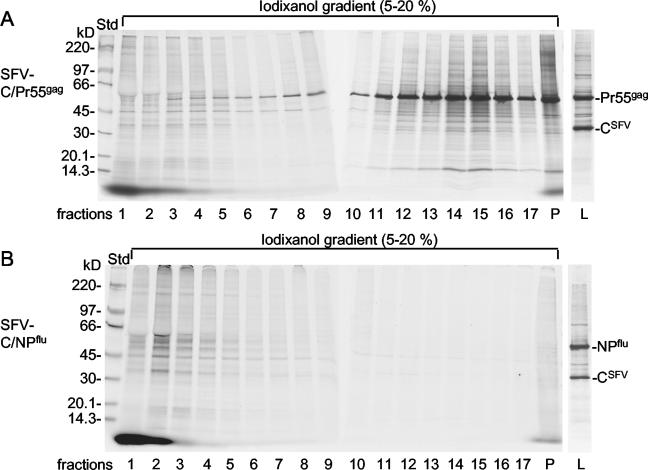
Figure 2A shows the sedimentation of Gag particles in a 5 to 20% iodixanol gradient. In this experiment, BHK-21 cells were labeled with [35S]Met for 15 h before infection with the vectors (prelabeling) and for 6 h after infection (postlabeling). Prelabeling was necessary for host protein detection because the SFV-C vectors inhibit host mRNA translation. The released particles in culture supernatants were collected for 1 h between 5 and 6 h after infection and subjected to centrifugation. It was apparent that the Gag particles, characterized by the prominent Pr55gag band, sedimented to the bottom part of the gradient, with a concentration peak in fractions 14 and 15. About half of total Pr55gag was found in fractions 13 to 15. A large number of apparent host proteins cofractionated with Pr55gag. These could have represented components of comigrating cell vesicles or of the Gag particles themselves. Apparent host proteins also were observed at the top of the gradient (fractions 1 to 5). In earlier work with Mo-MuLV Gag particles, it was demonstrated that the latter material represented protein components of putative cell vesicles. Specifically, slowly sedimenting particles, and not rapidly sedimenting particles, were released by BHK-21 cells infected with SFV-C vectors expressing NPflu (SFV-C/NPflu vectors). This control was repeated, and the previous results were confirmed (Fig. 2B). Thus, the apparent host proteins observed in the Gag particle fractions of the iodixanol gradient were carried by the Gag particles themselves rather than by contaminating cell vesicles. However, it remained possible that protein-poor cell vesicles contaminated the Gag particles. Such contamination would go undetected in protein analyses and would complicate subsequent lipid-based quantification of Gag particle membranes (see below).
FIG. 2.
Purification of 35S-labeled Gag particles. (A) Sedimentation analyses of 35S-labeled particles released from HIV-1 gag-expressing cells. BHK-21 cells were labeled with [35S]Met before and after infection with SFV-C/Pr55gag vectors. Gag particles in the medium were isolated by sedimentation in a 5 to 20% iodixanol gradient. After fractionation and dilution of the iodixanol, particles were recovered from each fraction by centrifugation and analyzed by SDS-PAGE (6 to 15%). Autoradiographs of the gels are shown. Lanes: 1, top fraction; P, pellet in the tube with the iodixanol gradient; L, NP-40 extract of vector-infected cells. (B) Sedimentation analyses of 35S-labeled particles released from NPflu-expressing cells. Labeling of cells that had been infected with SFV-C/NPflu vectors and analyses of particles were as described for panel A.
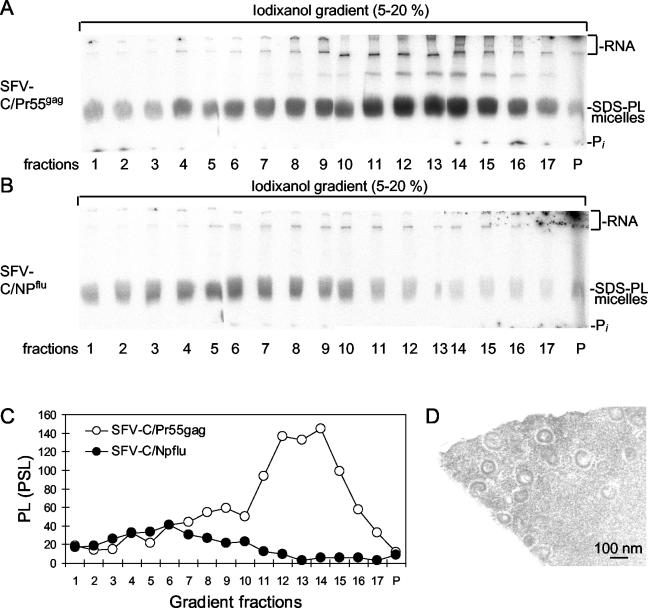
To study this possibility, we analyzed [32P]phospholipid-labeled particles released from cells infected with SFV-C/Pr55gag and SFV-C/NPflu vectors by sedimentation in 5 to 20% iodixanol gradients. The results are shown in Fig.3A and B. In this experiment, vector-infected cells were prelabeled and postlabeled for 40 and 5 h, respectively, with [32P]orthophosphate, and particles in culture supernatants were collected between 5 and 6 h after infection. Particles recovered from each fraction of the iodixanol gradient were taken up in hot SDS, mixed with sample buffer, and analyzed by SDS-PAGE (20%). In this analysis, [32P]lipids migrate as a broad band in the form of SDS-lipid mixed micelles (18). We found that the SFV-C/Pr55gag vector-infected cells produced a major group of membrane particles corresponding to the Gag particles and a minor group of slowly sedimenting particles. In contrast, the culture medium of cells infected with the SFV-C/NPflu vectors contained predominantly slowly sedimenting particles. Quantification of the rapidly sedimenting membrane particles in fractions 11 to 15 from the two sedimentation analyses suggested that less than 5% of the Gag particle preparations represented contamination (Fig. 3C). Therefore, we concluded that there was no significant contamination of isolated Gag particles with cell vesicles.
FIG. 3.
Purification of 32P-labeled Gag particles. (A) Sedimentation analyses of 32P-labeled particles released from HIV-1 gag-expressing cells. BHK-21 cells were labeled with [32P]orthophosphate for 40 h and then infected with SFV-C/Pr55gag vectors. After these steps, labeling was continued, and released particles were collected and separated as described in the legend to Fig. 2A. The isolated particles were solubilized in hot SDS and analyzed for 32P-labeled SDS-phospholipid (SDS-PL) mixed micelles by SDS-PAGE (20%). Labeled RNA at the top of the gels and free orthophoshate (Pi) at the bottom are indicated. (B) Sedimentation analyses of 32P-labeled particles released from NPflu-expressing cells. Labeling of cells and analyses of particles were as described for panel A. (C) Quantification of 32P-labeled SDS-lipid mixed micelles from panels A and B. PL, phospholipids; PSL, photostimulated luminescence. (D) EM analysis of isolated particles. Particles in fractions 13 to 15 of the separation procedure shown in Fig. 2A were pooled, concentrated by centrifugation at the tip of a centrifuge tube, embedded in gelatin, processed for sectioning, and analyzed by EM. Note the ∼100-nm particles with an apparent Gag layer.
We also considered the possibility that the apparent host proteins carried by the Gag particles represented proteins that had been secreted by the cell and nonspecifically adsorbed to the Gag particles. This possibility was studied by mixing a culture supernatant containing unlabeled Gag particles with a supernatant from prelabeled cells infected with SFV-C/NPflu vectors. After the mixture was incubated for 1 h at 37°C, the particles were sedimented in an iodixanol gradient. However, no cofractionation of host proteins was detected (data not shown), suggesting that the apparent host proteins of the Gag particles were incorporated during and not after budding. The purity of the Gag particles isolated by sedimentation was analyzed by sectioning and EM after the particles were concentrated at the bottom of a centrifuge tube. To facilitate processing of the small particle pellet during sectioning, we first embedded it in gelatin. Although this step increased the level of background staining, we could identify numerous particles which all appeared to have the characteristic features of Gag particles (Fig. 3D).
Isolation of Pr55gag-enriched cell membranes.
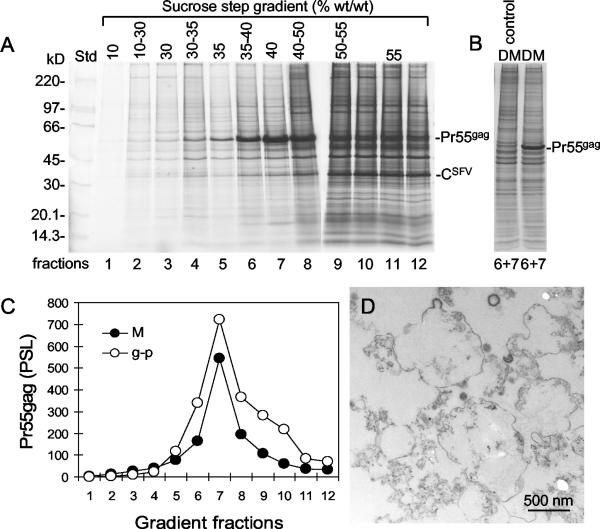
To isolate donor membranes from which Gag particles bud, microsomes were prepared from SFV-C/Pr55gag-infected BHK-21 cells that had been prelabeled and postlabeled with [35S]Met for 15 and 6 h, respectively. Pr55gag-enriched cell membranes were purified by flotation centrifugation with a previously described 10, 30, 35, 40, 50, and 55% sucrose step gradient (38). Pr55gag-containing microsomes floated from the dense loading zone and accumulated in 40% sucrose (Fig. 4A and C). The density of the Pr55gag-containing microsomes corresponded to that of released Gag particles when analyzed in the step gradient (Fig. 4C). This density is higher than that of the previously studied Mo-MuLV Pr65gag-enriched cell membranes, which accumulated in 35% sucrose (18). It is also higher than the densities of the Golgi apparatus and typical PMs, the bulk of which accumulated in 30% sucrose and 30 to 35% sucrose, respectively, but lower than the density of endoplasmic reticulum membranes, which floated to the 40%-50% sucrose interphase in corresponding analyses (38). SFV nucleocapsids, characterized by the intense CSFV band, remained predominantly in the loading zone (Fig. 4A, fractions 10 to 12). The Pr55gag-enriched membranes in fractions 6 and 7 were pooled, diluted, and concentrated by sedimentation at the bottom of a centrifuge tube. Analysis by EM showed that the isolated microsomes contained membrane structures with occasional budding profiles of apparent Gag particles (Fig. 4D). Therefore, the Pr55gag-enriched microsomes in fractions 6 and 7 of the step gradient most likely corresponded to the membranes where the Gag particles were formed. In these donor membranes, Pr55gag apparently was present mostly as prebudding structures.
FIG. 4.
Purification of donor membranes. (A) Flotation centrifugation analyses of microsomes from HIV-1 gag-expressing cells. BHK-21 cells were labeled with [35S]Met and infected with SFV-C/Pr55gag vectors as described in the legend to Fig. 2A. The cells were homogenized, and a postnuclear supernatant was prepared and subjected to flotation centrifugation in a 10, 30, 35, 40, 50, and 55% sucrose step gradient. The gradient was fractionated, and membranes were recovered from each fraction, after dilution, by centrifugation and analyzed by SDS-PAGE (6 to 15%). Autoradiographs of the gels are shown. Std, standard. (B) Comparison of protein profiles of Gag-enriched membranes (donor membranes [DM]) from SFV-C/Pr55gag vector-infected cells and corresponding membranes from uninfected cells. Analyses were done as described for panel A. Lanes show protein analyses for pooled fractions 6 and 7. (C) Quantification of Pr55gag in the floating microsomes (M) from panel A. Included is a quantification of Pr55gag in Gag particles (g-p) that were subjected to a similar flotation analysis. PSL, photostimulated luminescence. (D) Morphological analysis of donor membranes. Membranes in fractions 6 and 7 were pooled, concentrated by centrifugation at the tip of a centrifuge tube, sectioned, and analyzed by EM. Note the apparent budding Gag particles in some of the vesicles.
One might argue that Gag insertion, patching, and budding in donor membranes might alter the original host protein composition, i.e., that the donor membranes already represent modified, envelope-like membranes. To study this possibility, we compared the protein compositions in membrane fractions from uninfected cells and cells infected with SFV-C/Pr55gag vectors (Fig. 4B). We found that the host protein compositions were very similar in both preparations. This result suggests that Gag does not significantly alter host protein compositions in donor membranes.
Host protein sorting in donor membranes during the budding of Gag particles.
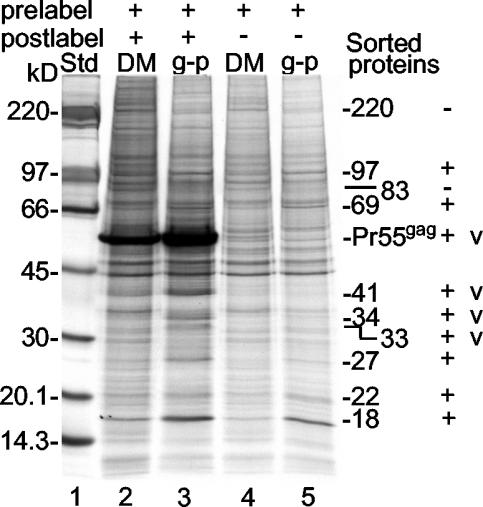
Parallel cultures of SFV-C/Pr55gag vector-infected BHK-21 cells that had been labeled before and after infection with [32P]orthophosphate and [35S]Met, respectively, or only prelabeled with [35S]Met were used for the preparation of Pr55gag-enriched membranes (donor membranes; pooled fractions 6 and 7 in Fig. 4A) and Gag particles. The relative phospholipid contents of the donor membrane and Gag particle preparations were determined by separating SDS-lipid mixed micelles of solubilized samples of the 32P-labeled donor membranes and Gag particles by SDS-20% PAGE and measuring the corresponding 32P radioactivity (18). This strategy allowed analysis of equal portions, based on phospholipid contents, of 35S-labeled donor membrane and Gag particle preparations by SDS-PAGE (6 to 15%) and thereby determination of the relative membrane surface densities of their proteins. An analysis of such equalized donor membrane and Gag particle samples from pre- and postlabeled cells revealed an intense Pr55gag band and several additional bands of weaker intensity (Fig. 5, lanes 2 and 3). The fact that almost all of the additional bands were also seen in the corresponding samples from cells that were only prelabeled demonstrated unequivocally that they were host proteins (Fig. 5, lanes 4 and 5). The exceptions were the 33-, 34-, and 41-kDa proteins that, together with Pr55gag, were found only in donor membranes and Gag particles from pre- and postlabeled cells. These probably represent the CSFV (33-kDa) and Pr55gag fragments.
FIG. 5.
Host protein sorting in the donor membrane during the budding of Gag particles. Samples of 35S-labeled Gag particles (g-p) and donor membranes (DM) were adjusted to contain equal amounts of membranes and then analyzed by SDS-PAGE (6 to 15%). The donor membrane and Gag particle preparations were made in cells that were either labeled both before (prelabeled) and after (postlabeled) vector infection (lanes 2 and 3) or only prelabeled (lanes 4 and 5). In the latter case, only host proteins were labeled and detected. Std, standard. The molecular masses of vector proteins (v) as well as those of host proteins that were either concentrated or diluted during budding are indicated. The latter proteins are marked as positively (+) or negatively (−) sorted proteins.
The most striking observation made when the protein landscape of the donor membranes was compared with that of the Gag particles was their overall similarity. This finding implies that the bulk of the host proteins in the donor membranes are included in the Gag particles; i.e., there is no significant sorting of host proteins during Pr55gag-driven budding. Nevertheless, we observed a few proteins in the donor membranes that appeared at lower concentrations in the Gag particles. Two examples were the 220- and 83-kDa proteins (Fig. 5). We also found a few proteins that were concentrated in the Gag particles along with Pr55gag and its putative fragments. Examples were the 18-, 22-, 27-, 69-, and 97-kDa proteins (Fig. 5). The concentration factors were 3.3 ± 1.4 (mean and standard deviation) for Pr55gag and in the range of 2 to 3 for the host proteins.
Passive inclusion of host proteins in Gag particles made in Jurkat T lymphocytes by SFV-C vectors or in 293T cells by nuclear gene expression.
Although the Pr55gag gene can be expressed with SFV-C vectors in BHK-21 cells and Gag particles can be produced, the cells cannot function as host cells for HIV-1. Therefore, we studied the host protein incorporation into Gag particles produced in Jurkat cells, which originate from T lymphocytes, i.e., natural HIV-1 host cells. We first compared Gag particle production in SFV-C/Pr55gag vector-infected BHK-21 and Jurkat cells by using [35S]Met pulse-labeling of Pr55gag and subsequent chase analysis. We found that 11.4% ± 2.1% of the labeled Pr55gag was released as Gag particles during a 4-h chase in Jurkat cells and that 7.8% ± 2.5% was released in BHK-21 cells. Longer chase times did not result in any significant increase in the yield of labeled particles (data not shown).
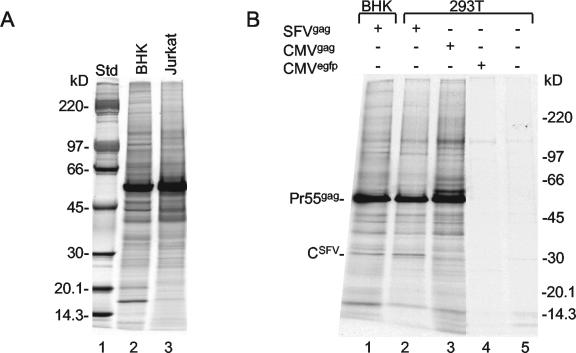
To monitor host protein incorporation into Gag particles, we pre- and postlabeled vector-infected BHK-21 and Jurkat cells with [35S]Met, collected released particles in the culture medium, and purified the Gag particles by sedimentation in iodixanol gradients as described above. In this experiment, donor membranes were not isolated, but Gag particles from BHK-21 and Jurkat cells were compared with each other on an equal-lipid basis. The comparison showed that the Gag particles produced in Jurkat cells contained a plethora of bands in addition to that of Pr55gag (Fig. 6A, lanes 2 and 3). As the overall loads of host proteins in the two preparations were rather similar, we conclude that the bulk of the host proteins in the donor membranes of Jurkat cells are also included passively in the Gag particles, when these are formed by budding.
FIG. 6.
Inclusion of host proteins in Gag particles made in Jurkat T lymphocytes by SFV expression and in 293T cells by nuclear gene expression. (A) BHK-21 cells and Jurkat cells were prelabeled with [35S]Met and infected with SFV-C/Pr55gag vectors. The cells then were postlabeled, and Gag particles were collected between 5 and 6 h postinfection. Samples of iodixanol gradient-purified Gag particles were adjusted to contain equal amounts of membranes and then analyzed by SDS-PAGE (6 to 15%). Particles produced in BHK-21 cells are shown in lane 2, and particles produced in Jurkat cells are shown in lane 3. Std, standard. (B) BHK-21 cells were infected with SFV-C/Pr55gag vectors (lane 1), and 293T cells were infected with SFV-C/Pr55gag vectors (lane 2). 293T cells were additionally transfected with pCMV-HIVgag DNA (lane 3) or pRRLsin.cPPT.CMV.GFP.Wpre DNA (lane 4) or mock transfected (lane 5). The infected BHK-21 and 293T cells were labeled with [35S]Met before and after infection for 17 and 6 h, respectively. Particles produced during the last 1 h of the labeling period were collected and purified by sedimentation in iodixanol gradients. The transfected and mock-transfected cells were incubated for 25.5 h and then labeled for 21 h. In these cases, the particles produced during the last 4 h of the labeling period were collected and purified. SDS-PAGE (6 to 15%) analysis of the various particle preparations is shown. Note that 1.25 and 1.07 times more Gag particles were analyzed in lanes 3 and 1, respectively, than in lane 2. Note also the apparent SFV capsid protein that is seen only in the particles produced by SFV-C vector-infected cells.
One might argue that the rather indiscriminate host protein incorporation into Gag particles might result from the high-level expression of the Pr55gag gene that is obtained with the SFV-C vectors. Therefore, we studied Gag particles produced in 293T cells by nuclear cytomegalovirus promoter-driven Pr55gag gene expression. To this end, we transfected 293T cells with pCMV-HIVgag DNA by using Ca2PO4 coprecipitation. The cells were labeled with [35S]Met, and Gag particles released into the medium were purified by sedimentation in an iodixanol gradient. The protein profiles of these particles were compared with those of Gag particles produced in SFV-C/Pr55gag vector-infected 293T and BHK-21 cells and with similarly sedimenting microvesicles of pRRLsin.cPPT.CMV.GFP.WpreDNA-transfected and mock-transfected 293T cells (Fig. 6B). We observed very similar loads of apparent host proteins in the different Gag particle preparations (Fig. 6B, lanes 1 to 3). These loads were significantly higher than that of the microvesicle background of the control cells (Fig. 6B, lanes 4 and 5), suggesting that the apparent host proteins in the Gag particles produced in 293T cells were most likely carried by the Gag particles themselves. Therefore, we conclude that the rather indiscriminate host protein incorporation into Gag particles is not an artifact of the SFV expression system.
CypA is concentrated in Gag particles.
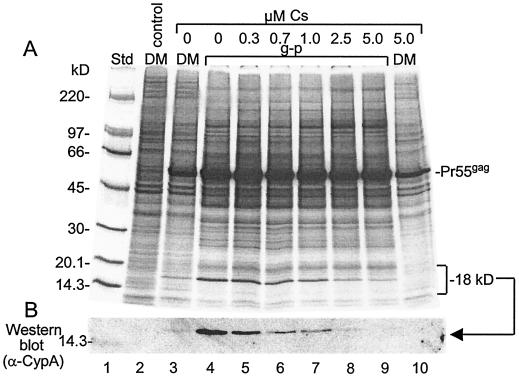
The prominent 18-kDa protein which was concentrated in the Gag particles produced in BHK-21 cells (Fig. 5) corresponded in size to CypA. This host protein has peptidyl-prolyl cis-trans isomerase activity and was found earlier in HIV-1 (11, 26, 39). CypA binds via its isomerase domain to a proline-rich stretch of the HIV-1 CA protein (12). The binding can be outcompeted by the drug cyclosporine, which binds to the isomerase domain of CypA (11, 26, 39). CypA is required for the efficient infection of cells with HIV-1 (3, 4). Recently, it was shown to modulate viral sensitivity to host restriction factors (40). To find out whether the 18-kDa protein represented CypA, we tested its incorporation into Gag particles when produced in SFV-C/Pr55gag vector-infected cells in the presence of 0 to 5 μM cyclosporine. The experiment showed that the 18-kDa protein was clearly outcompeted from the Gag particles by cyclosporine in a dose-dependent manner (Fig. 7A, lanes 4 to 9). The incorporation of the protein was almost totally blocked by 5 μM cyclosporine. The identity of the 18-kDa protein was further investigated by Western blot analysis with a polyclonal anti-CypA antibody, and we concluded that the 18-kDa protein was CypA (Fig. 7B). Quantification of Pr55gag and CypA in the Gag particles from untreated cells, taking into account the number of methionines in each protein, yielded a molar ratio of 1 CypA to 11.5 ± 2.7 Pr55gag. This ratio is very similar to the 1:10 ratio found earlier for HIV-1 (11, 39).
FIG. 7.
Concentration of CypA into Gag particles. Donor membranes (DM) and Gag particles (g-p) were produced by using SFV-C/Pr55gag vector-infected BHK-21 cells that were pre- and postlabeled with [35S]Met but with cyclosporine (Cs) added at the indicated concentrations at 2 h postinfection. Gag particles (lanes 4 to 9) and donor membranes (lanes 3 and 10) were adjusted to contain equal amounts of membranes and then analyzed by SDS-PAGE (6 to 15%) in duplicate. One gel was processed for autoradiography (A), and the other one was used for the detection of CypA by Western blotting (B). Donor membranes from uninfected cells were analyzed in parallel (lane 2). Std, standard.
In the experiment shown in Fig. 7, we also monitored the recruitment of CypA to donor membranes. We found that membranes of uninfected control cells did not contain CypA (Fig. 7, lane 2). However, CypA was recruited to donor membranes of vector-infected cells and became concentrated 3.0 ± 0.6 times in Gag particles during budding (Fig. 7, lanes 3 and 4). CypA was outcompeted from the donor membranes in the presence of 5 μM cyclosporine (Fig. 7, lane 10). Altogether, these results suggest that CypA forms a complex with a fraction of membrane-bound Pr55gag before assembly into budding structures and that the Pr55gag-CypA complexes become concentrated in Gag particles like CypA-free Pr55gag, i.e., at a concentration factor of about 3.
Incorporation of Tsg-101 into Gag particles.
Tsg-101 is a host protein required for HIV-1 budding (6, 13, 27). In the cell, Tsg-101 is involved in the sorting of proteins into multivesicular bodies (21). These structures are formed by luminal budding of the membrane of late endosomes. Tsg-101 binds to a PTAP motif in the p6 domain of Pr55gag (35). If p6 cannot interact with functional Tsg-101, then HIV-1 appears to be arrested at a late stage of its budding process (6, 13).
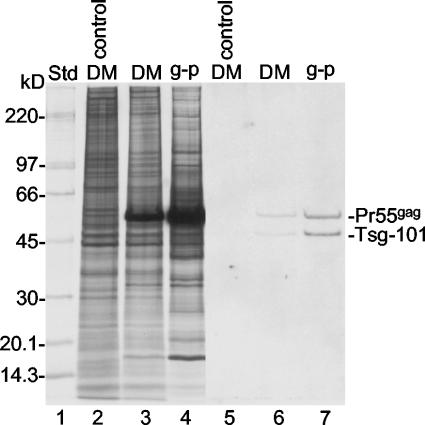
To find out how Tsg-101 was dealt with at the PM during the budding of Gag particles, we isolated donor membranes and Gag particles from SFV-C/Pr55gag vector-infected BHK-21 cells that had been pre- and postlabeled with [35S]Met. Samples of the two preparations were equalized on the basis of lipid contents and subjected to SDS-PAGE on duplicate gels. One was used for Western blotting with an anti-Tsg-101 antibody, and the other was used for autoradiography. We found that 55- and 47-kDa proteins were detected in both preparations on the Western blot (Fig. 8, lanes 6 and 7). The 55-kDa protein apparently represented nonspecific staining of Pr55gag because it corresponded in size to the intensively 35S-labeled Pr55gag band (Fig. 8, lanes 3 and 4) and was stained when only the secondary antibody was used for Western blotting (data not shown). Furthermore, it was not found in membranes of uninfected cells (Fig. 8, lane 5). In contrast, the 47-kDa protein probably represented Tsg-101. The apparent mass of this protein was close to that reported for Tsg-101 (45 kDa; see references mentioned above), and this protein was not observed when the anti-Tsg-101 antibody was excluded from staining (data not shown). The minor size discrepancy was probably due to differences in the gel systems used for analysis. A comparison between the blot and the autoradiograph showed that the stained Tsg-101 band corresponded to a 35S-labeled host protein migrating in front of Pr55gag. However, as this band was equally abundant in the membranes of vector-infected and control cells, most of the protein in this band must have represented another host protein comigrating with Tsg-101 (Fig. 8, lanes 2 and 3). Quantification of stained Tsg-101 and Pr55gag and of 35S-labeled Pr55gag in the donor membranes and the Gag particles demonstrated that Tsg-101 became more concentrated during budding than Pr55gag (concentration factors of 5.4 ± 2.4 and 3.3 ± 1.4, respectively). This result suggests that Tsg-101 forms complexes with a small fraction of membrane-bound Pr55gag and that these complexes are preferentially incorporated into Gag particles.
FIG. 8.
Concentration of Tsg-101 into Gag particles. Donor membranes (DM) and Gag particles (g-p) from BHK-21 cells infected with SFV-C/Pr55gag vectors and pre- and postlabeled with [35S]Met as well as donor membranes from uninfected control cells were analyzed by SDS-PAGE on an equal-lipid basis. The proteins in the gel were transferred to a filter, and this was used for the detection of Tsg-101 by Western blotting (lanes 5 to 7). Lanes 2 to 4 represent an autoradiograph of 35S-labeled proteins separated on a duplicate gel. Note that abundant Pr55gag was stained nonspecifically in the blot. Std, standard.
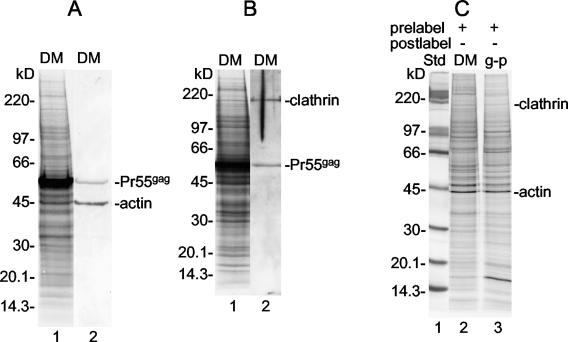
Fate of cytoskeletal proteins and clathrin during the budding of Gag particles.
The dynamic processes of the PM are controlled by the underlying cell cortex. This is composed of interconnected and membrane-anchored actin filaments (28). At the cytoplasmic side of the PM are coat proteins, such as clathrin, that direct the invagination and vesicularization of the PM (36). Therefore, an interesting question was what happens to these proteins during the budding of Gag particles. Using Western blotting, we were able to assign specific 35S-labeled protein bands appearing in the SDS-PAGE analysis of isolated donor membranes to actin and clathrin (Fig. 9A and B). We used protein analyses of the Gag particle and donor membrane samples of prelabeled cells (Fig. 5, lanes 4 and 5) to monitor the fate of these proteins during Gag particle budding (Fig. 9C). From a comparison of the intensities of the bands in the two samples, it was evident that there was no significant change in the concentration of either of the proteins. This result suggests that these proteins are incorporated in an undiluted form from donor membranes into Gag particles. However, this conclusion is based on the assumption that the bands assigned to the proteins do not contain significant amounts of a comigrating protein(s). Quantification of radioactivity in the bands showed that the amounts of actin and clathrin in the Gag particles were about 1.6 and 0.2% those in Pr55gag, respectively.
FIG. 9.
Passive inclusion of actin and clathrin in Gag particles. (A and B) Donor membranes (DM) from SFV-C/Pr55gag vector-infected BHK-21 cells were analyzed by multiple SDS-PAGE (6 to 15%) analyses. The proteins in the gels were transferred to filters, and these were used for the detection of actin (panel A, lane 2) and clathrin (panel B, lane 2). Lanes 1 in panels A and B represent autoradiographs of the respective filters. Note that Pr55gag was recognized nonspecifically in all Western blot analyses. (C) Actin and clathrin proteins from earlier protein analyses of membrane-equalized donor membrane and Gag particle (g-p) samples derived from prelabeled SFV-C/Pr55gag vector-infected BHK-21 cells (Fig. 5, lanes 4 and 5). Note that actin and clathrin are present at corresponding concentrations in the donor membranes and the Gag particles. Std, standard.
DISCUSSION
The budding of HIV-1 and corresponding Gag particles at the PM has apparent similarities to that of CopI, CopII, and clathrin-coated transport vesicles in the cell (14, 22). In both cases, cytoplasmic proteins form a layer below the cytoplasmic leaflet of the cell or the organelle membrane, inducing membrane curvature, budding, and pinching off of a membrane vesicle. For intracellular transport vesicles, the coat rapidly dissociates after vesicle formation to facilitate subsequent vesicle fusion with the target membrane. Similarly, for HIV-1, the Pr55gag layer matures through autoproteolytic processing into an internal capsid and a submembraneous matrix layer (41). The apparent difference between the viral and the cellular budding events is topological, the virus budding outward from and the cellular vesicles budding into the cytoplasm. However, cellular outward budding does also exist, e.g., during multivesicular body formation in the late endosome (21). The purpose of cellular transport vesicles is to effect vectorial transport of specific proteins in the cell. This transport is achieved through the selective incorporation of cargo proteins into the transport vesicles at the donor membrane and their release at the target membrane. Other proteins of the donor membrane, in particular, cargo receptors, are also incorporated but are not released and therefore return to the donor membrane when the transport vesicles are recycled.
So far, it has remained unclear how the Pr55gag-driven budding process affects host proteins in the cell membrane. To find out whether an individual host protein becomes concentrated (sorted in), diluted (sorted out), or passively included in Gag particles during budding at the cell membrane, we compared the protein contents of Gag particle and donor membrane preparations on an equal-lipid (membrane) basis. This study required reliable techniques for Gag particle and donor membrane isolation and sensitive assays to determine their protein compositions and lipid contents. Our solution to the analytical demands was solubilization of the Gag particle and donor membrane preparations in hot SDS and subsequent SDS-PAGE. Hammarstedt et al. recently showed that this simple protein analysis system can also be used to separate the lipids of an SDS-solubilized membrane from its other components (18). The lipids form mixed micelles with SDS; these migrate as a band on a 20% gel. This band can conveniently be detected and quantified in samples with [32P]phospholipids.
The major problem with particle isolation is contamination with cell-derived vesicles that accumulate in the culture medium over time (2, 16). In the present study, we succeeded in reducing the contamination to an insignificant level by using a high-level production system based on the SFV-C expression vectors and purification of particles by sedimentation in an iodixanol gradient (7, 37). The efficient production made it possible to collect sufficient Gag particles for analyses in a short period. Based on lipid quantification, the host vesicle contamination amounted to less than 5% of the Gag particle preparation (Fig. 3). Ott et al. (34) found that the proteins of contaminating vesicles in an HIV-1 preparation can be removed by digestion with subtilisin. However, as this treatment probably leaves the lipid membranes of the host vesicles and digests the external proteins of the Gag particles, we did not find it useful for our studies of protein concentrations in membranes. In the present work, we purified the donor membranes for the Gag particles by flotation of microsomes from producer cells in a sucrose step gradient. The donor membranes were primarily identified by their high Pr55gag content. Although the Pr55gag-enriched donor membranes had a density that corresponded to that of Gag particles, morphological analysis demonstrated that Pr55gag in the membranes was present mostly in prebudding structures. The most striking result of the lipid-normalized protein comparison of the Gag particles and the donor membranes was that the bulk of the host proteins in the donor membranes were incorporated into the Gag particles in an undiluted fashion. This result suggests that the formation of the Pr55gag layer below the PM and its induction of the virus bud proceed without significant alteration of the preexisting protein composition of the PM. As similar host protein inclusion in Mo-MuLV Gag particles was found earlier, this feature seems to be a general one for Gag particles of retroviruses and lentiviruses. How is this possible?
EM analyses of the cytoplasmic side of the PM of cells producing Gag particles have shown that Pr55gag proteins are organized as a hexameric lattice below the membrane (28). Model building suggests that the lattice is formed by Pr55gag trimers organized such that each Pr55gag protein is shared by two hexameric rings (10). The center-to-center distance between adjacent hexamers is 7 nm, and the openings of the rings are 4 nm. Only 1 out of 12 openings is occupied by glycoprotein. This fenestrated layer of Pr55gag proteins should accept a significant inclusion of donor membrane host proteins, as was found in our study. Why, then, has this feature not been observed before? The obvious explanation is that the majority of the host proteins appear in the particles in amounts that represent less than 1% of Pr55gag. Together with their large size variations, this property makes them difficult to differentiate from a general background signal. Nevertheless, analyses by HPLC in particular have identified a respectable number of host proteins in HIV-1 (1, 32, 33). In view of our present results, we believe that the bulk of the host proteins are incorporated passively into the particles.
Our results also demonstrated that a few proteins were concentrated into Gag particles during budding. Among these, we identified two, CypA and Tsg-101. These proteins were shown earlier to interact with Pr55gag and also were identified in released virus (6, 11, 12, 26, 35, 39). Our experiments suggested that CypA-Pr55gag and Tsg-101-Pr55gag complexes have already been formed in prebudding Pr55gag structures in the donor membrane and then become concentrated into budding particles. The CypA/Pr55gag ratio of ∼1:12 in the particles and in the donor membrane is consistent with the existence of a homooligomeric Pr55gag assembly intermediate that interacts with one molecule of CypA. Tsg-101 was present at a much lower level, a property inconsistent with a structural role. On the other hand, Tsg-101 moves from the donor membrane into Gag particles more efficiently than Pr55gag. This finding might reflect the fact that Tsg-101 is required for the release of Gag particles from the donor membrane (13, 17). It is possible that Tsg-101 alone or together with other host factors induces the clustering of Tsg-101-Pr55gag complexes in the membrane. Such a cluster then might associate with the Pr55gag layer of a “mature” Gag particle bud through Pr55gag-Pr55gag interactions and facilitate the pinching-off reaction.
It remains to be discovered whether the Pr55gag interaction represents the sole way of concentrating host proteins into Gag particles. Another possibility is raft-lipid association. Rafts have been suggested to facilitate Pr55gag-driven budding, and some “raft proteins” of the host PM have been identified in Gag particles and HIV-1 (20, 25, 30, 31). However, so far there is no information about whether these raft proteins are concentrated into the particles or passively included (30).
Our study indicated that some proteins in the donor membrane, e.g., 220- and 83-kDa proteins (Fig. 5), were partially excluded from the Gag particles during budding. While these proteins were not identified, others have reported that, e.g., CD45, HLA-DQ, HLA-DP, and the HIV-1 receptor and coreceptors are not found in virions but are present at the PM (1, 8, 19, 23). Exclusion of host proteins could result from the Pr55gag lattice functioning as a filter for transmembrane PM proteins with bulky cytoplasmic domains, such as CD45, or multiple transmembrane segments, such as the coreceptors. Alternatively, excluded proteins might form large oligomeric structures, such as CD4 with p56lck proteins, or associate with lipid microdomains that are left out of the budding particles.
At present, we cannot exclude the possibility that the assembly process for HIV-1 in naturally infected cells is more selective in host protein incorporation than what we observed here for the Gag particles produced at high levels by the SFV-C expression vectors in BHK-21 cells. However, several facts suggest that our system might be relevant to the way in which HIV-1 sorts host proteins during budding. First, we showed that similar levels of host proteins were incorporated into Gag particles produced in pCMV-HIVgag DNA-transfected 293T cells, SFV-C/Pr55gag vector-infected 293T cells, SFV-C/Pr55gag vector-infected Jurkat cells, and SFV-C/Pr55gag vector-infected BHK-21 cells (Fig. 6). This finding suggests that the rather indiscriminate host protein incorporation into Gag particles is not specific for SFV vector-directed Pr55gag expression in BHK-21 cells but also occurs in natural HIV-1 host cells and by nuclear gene expression. Second, the Gag particles produced by the SFV vector system in BHK-21 cells corresponded in size, shape, and morphology to immature HIV-1 particles (14, 15). Third, the Gag particles incorporated CypA and Tsg-101 in the same manner as HIV-1 (6, 11, 26, 39). Fourth, like HIV-1 release, SFV-C/Pr55gag vector-directed production of Gag particles is dependent on Tsg-101, as it can be inhibited by Tsg-101 small interfering RNA (13; M. Suomalainen, unpublished data). Fifth, earlier studies in which SFV-C/Pr65gag vectors were used to produce Mo-MuLV Gag particles showed a similar passive incorporation of most PM proteins into the Gag particles (18). Those studies also showed that host protein incorporation was not affected by decreasing the gag gene expression level ∼8-fold by using SFV-1/Pr65gag vectors (24) instead of SFV-C/Pr65gag vectors (37) or by incorporating Mo-MuLV Env into the Gag particles.
Acknowledgments
We are grateful to Mathilda Sjöberg and Maarit Suomalainen for critical reading of the manuscript and Kjell Hultenby for help with EM.
This work was supported by Swedish Research Council grants B5107-20006266/2000 and K2002-99XG-13279-04A to H.G.
REFERENCES
- 1.Arthur, L. O., J. W. Bess, Jr., R. C. Sowder II, R. E. Benveniste, D. L. Mann, J. C. Chermann, and L. E. Henderson. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 258:1935-1938. [DOI] [PubMed]
- 2.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134-144. [DOI] [PubMed] [Google Scholar]
- 3.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, R. H., R. J. Kuhn, N. H. Olson, M. G. Rossmann, H. K. Choi, T. J. Smith, and T. S. Baker. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dettenhofer, M., and X. F. Yu. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esser, M. T., D. R. Graham, L. V. Coren, C. M. Trubey, J. W. Bess, Jr., L. O. Arthur, D. E. Ott, and J. D. Lifson. 2001. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J. Virol. 75:6173-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Follenzi, A., G. Sabatino, A. Lombardo, C. Boccaccio, and L. Naldini. 2002. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum. Gene Ther. 13:243-260. [DOI] [PubMed] [Google Scholar]
- 10.Forster, M. J., B. Mulloy, and M. V. Nermut. 2000. Molecular modelling study of HIV p17gag (MA) protein shell utilising data from electron microscopy and X-ray crystallography. J. Mol. Biol. 298:841-857. [DOI] [PubMed] [Google Scholar]
- 11.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Gelderblom, H. R., P. G. Bauer, M. Özel, S. Höglund, M. Niedrig, H. Renz, B. Morath, P. Lundquist, Å. Nilsson, J. Mattow, C. Grund, and G. Pauli. 1992. Morphogenesis and morphology of human immunodeficiency virus, p. 33-54. In R. C. Aloia and C. C. Curtain (ed.), Advances in membrane fluidity. Wiley-Liss, Inc., New York, N.Y.
- 15.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 16.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125-133. [DOI] [PubMed] [Google Scholar]
- 17.Goila-Gaur, R., D. G. Demirov, J. M. Orenstein, A. Ono, and E. O. Freed. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 77:6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammarstedt, M., K. Wallengren, K. W. Pedersen, N. Roos, and H. Garoff. 2000. Minimal exclusion of plasma membrane proteins during retroviral envelope formation. Proc. Natl. Acad. Sci. USA 97:7527-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriksson, P., and V. Bosch. 1998. Inhibition of cellular glycoprotein incorporation into human immunodeficiency virus-like particles by coexpression of additional cellular interaction partner. Virology 251:16-21. [DOI] [PubMed] [Google Scholar]
- 20.Holm, K., K. Weclewicz, R. Hewson, and M. Suomalainen. 2003. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55gag associates with membrane domains that are largely resistant to Brij 98 but sensitive to Triton X-100. J. Virol. 77:4805-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 22.Kirchhausen, T. 2000. Three ways to make a vesicle. Nat. Rev. Mol. Cell. Biol. 1:187-198. [DOI] [PubMed] [Google Scholar]
- 23.Lallos, L. B., S. Laal, J. A. Hoxie, S. Zolla-Pazner, and J. C. Bandres. 1999. Exclusion of HIV coreceptors CXCR4, CCR5, and CCR3 from the HIV envelope. AIDS Res. Hum. Retrovir. 15:895-897. [DOI] [PubMed] [Google Scholar]
- 24.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 25.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 28.Medalia, O., I. Weber, A. S. Frangakis, D. Nicastro, G. Gerisch, and W. Baumeister. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298:1209-1213. [DOI] [PubMed] [Google Scholar]
- 29.Nermut, M. V., D. J. Hockley, P. Bron, D. Thomas, W. H. Zhang, and I. M. Jones. 1998. Further evidence for hexagonal organization of HIV gag protein in prebudding assemblies and immature virus-like particles. J. Struct. Biol. 123:143-149. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott, D. E. 1997. Cellular proteins in HIV virions. Rev. Med. Virol. 7:167-180. [DOI] [PubMed] [Google Scholar]
- 33.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder II, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 34.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retrovir. 11:1003-1006. [DOI] [PubMed] [Google Scholar]
- 35.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 36.Schmid, S. L. 1997. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu. Rev. Biochem. 66:511-548. [DOI] [PubMed] [Google Scholar]
- 37.Sjoberg, E. M., M. Suomalainen, and H. Garoff. 1994. A significantly improved Semliki Forest virus expression system based on translation enhancer segments from the viral capsid gene. Bio/Technology 12:1127-1131. [DOI] [PubMed] [Google Scholar]
- 38.Suomalainen, M., K. Hultenby, and H. Garoff. 1996. Targeting of Moloney murine leukemia virus gag precursor to the site of virus budding. J. Cell Biol. 135:1841-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 40.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 41.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 42.Zhao, H., M. Ekstrom, and H. Garoff. 1998. The M1 and NP proteins of influenza A virus form homo- but not heterooligomeric complexes when coexpressed in BHK-21 cells. J. Gen. Virol. 79:2435-2446. [DOI] [PubMed] [Google Scholar]