Abstract
In the title compound, C26H19NO, the plane of the aromatic heterocycle makes a dihedral angle of 75.22 (4)° with that of the attached phenyl ring. In the crystal, molecules are connected by C—H⋯O interactions, generating R 2 2(12) dimers. These dimers are further connected by C—H⋯π interactions, linking the molecules into chains running along the a-axis direction.
Keywords: crystal structure
Related literature
For background to acridines, see: Kumar et al. (2012 ▶). For the biological activity of acridine derivatives, see: Pigatto et al. (2011 ▶); Das et al. (2011 ▶); Kumar et al. (2012 ▶); Prommier et al. (2006 ▶) Denny et al. (1982 ▶); Baguley & Ferguson (1998 ▶). For the synthesis of acridines, see: Tomar et al. (2010 ▶). For related structures, see: Buckleton & Waters (1984 ▶); Chantrapromma et al. (2010 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶).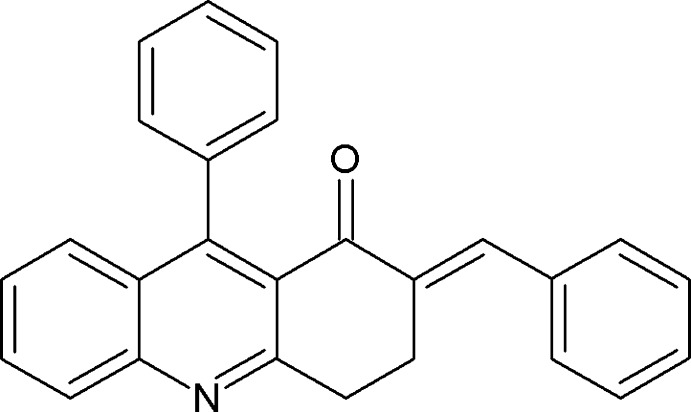
Experimental
Crystal data
C26H19NO
M r = 361.42
Monoclinic,

a = 9.2222 (3) Å
b = 10.7555 (4) Å
c = 19.4962 (5) Å
β = 95.503 (2)°
V = 1924.90 (11) Å3
Z = 4
Mo Kα radiation
μ = 0.08 mm−1
T = 293 K
0.20 × 0.20 × 0.20 mm
Data collection
Bruker SMART APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.662, T max = 0.746
18382 measured reflections
4776 independent reflections
3205 reflections with I > 2σ(I)
R int = 0.029
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.131
S = 1.00
4776 reflections
254 parameters
H-atom parameters constrained
Δρmax = 0.23 e Å−3
Δρmin = −0.16 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814015943/bt6954sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814015943/bt6954Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814015943/bt6954Isup3.cml
CCDC reference: 1012814
Additional supporting information: crystallographic information; 3D view; checkCIF report
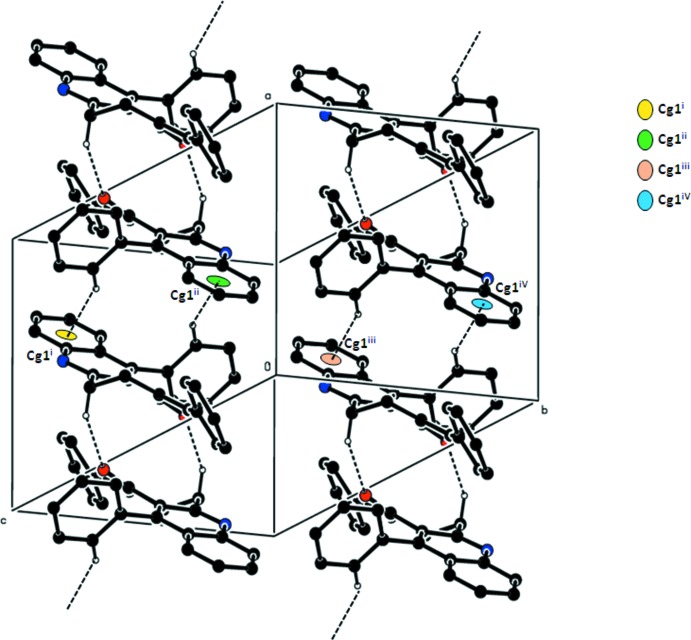
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C10—H10B⋯O1i | 0.97 | 2.58 | 3.2700 (18) | 128 |
| C26—H26⋯Cg1ii | 0.93 | 2.71 | 3.577 (18) | 156 |
Symmetry codes: (i) 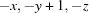 ; (ii)
; (ii)  .
.
Acknowledgments
The authors thank the TBI X-ray facility, CAS in Crystallography and Biophysics, University of Madras, India, for the data collection. TV and DV also thank the UGC (SAP–CAS) for the facilities to the department.
supplementary crystallographic information
S1. Comment
Acridine is structurally related to anthracene with one of the central CH group replaced by nitrogen. Amsacrine which is an acridine derivative is clinically used for the treatment of cancer (Denny et al., 1982; Baguley & Ferguson, 1998). The strong activity of acridine derivatives is due to their ability to intercalate into DNA base pairs and leading to cell cycle arrest and apoptosis (Prommier et al., 2006).
The phenyl (C21—C26) and benzyl (C14—C20) rings deviate from the plane of the acridine system by 72.48 (6) ° and 49.24 (6) °, respectively. The crystal packing is stabilized by intermolecular C—H···O (C10—H10B···O1) interactions generating a R22(12) ring motif (Bernstein et al., 1995). These dimers are further connected by C—H···π (C26—H26···Cg1) interactions generating chains running along the a-axis.
S2. Experimental
A 1:2 molar mixture of 9-phenyl-3,4-dihydroacridin-1(2H)-one was treated with aromatic aldehydes in the presence of NaOH and allowed to stir at room temperature for 5–7 h. After completion of the reaction as inferred by the TLC, the mixture was poured into 200 g of crushed ice and neutralized with dil HCl. The precipitate thus formed after adding into crushed ice was filtered off and the residue subjected to column chromatography using petroleum ether: ethyl acetate mixture (3:1) v/v as eluent and compound obtained as a pale yellow solid.
S3. Refinement
All H atoms were located in a difference map. Nevertheless, they were positioned geometrically (C—H = 0.93–0.98 Å) and allowed to ride on their parent atoms, with Uiso(H) =1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound with the atom numbering scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are presented as small spheres of arbitrary radius.
Fig. 2.
The cystal packing of the title compound showing the intermolecular C—H···O and C—H···π interactions chain running along aaxis, where Cg1 is the centroid of ring atoms C1—C6. Symmetry codes: (i) X,1/2-Y,1/2+Z; (ii) 1-X,-1/2+Y,1/2-Z; (iii) X,3/2-Y,1/2+Z and (iv) 1-X,1/2+Y,1/2-Z.
Crystal data
| C26H19NO | F(000) = 760 |
| Mr = 361.42 | Dx = 1.247 Mg m−3Dm = 1.25 Mg m−3Dm measured by not measured |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4776 reflections |
| a = 9.2222 (3) Å | θ = 2.1–28.3° |
| b = 10.7555 (4) Å | µ = 0.08 mm−1 |
| c = 19.4962 (5) Å | T = 293 K |
| β = 95.503 (2)° | Block, white |
| V = 1924.90 (11) Å3 | 0.20 × 0.20 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII CCD diffractometer | 4776 independent reflections |
| Radiation source: fine-focus sealed tube | 3205 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.029 |
| ω and φ scans | θmax = 28.3°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −12→10 |
| Tmin = 0.662, Tmax = 0.746 | k = −13→14 |
| 18382 measured reflections | l = −25→25 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.044 | H-atom parameters constrained |
| wR(F2) = 0.131 | w = 1/[σ2(Fo2) + (0.0587P)2 + 0.3326P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max = 0.013 |
| 4776 reflections | Δρmax = 0.23 e Å−3 |
| 254 parameters | Δρmin = −0.16 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0041 (8) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.47972 (17) | 0.49247 (14) | 0.17309 (8) | 0.0542 (4) | |
| H1 | 0.4841 | 0.4062 | 0.1757 | 0.065* | |
| C2 | 0.5587 (2) | 0.56214 (17) | 0.22142 (9) | 0.0711 (5) | |
| H2 | 0.6162 | 0.5233 | 0.2569 | 0.085* | |
| C3 | 0.5539 (2) | 0.69204 (17) | 0.21798 (10) | 0.0811 (6) | |
| H3 | 0.6089 | 0.7388 | 0.2511 | 0.097* | |
| C4 | 0.4701 (2) | 0.75058 (15) | 0.16689 (9) | 0.0677 (5) | |
| H4 | 0.4674 | 0.8370 | 0.1655 | 0.081* | |
| C5 | 0.38702 (16) | 0.68116 (12) | 0.11582 (7) | 0.0457 (3) | |
| C6 | 0.39092 (15) | 0.54970 (12) | 0.11883 (7) | 0.0416 (3) | |
| C7 | 0.30846 (14) | 0.48187 (11) | 0.06585 (6) | 0.0376 (3) | |
| C8 | 0.23412 (14) | 0.54820 (11) | 0.01290 (6) | 0.0381 (3) | |
| C9 | 0.23927 (14) | 0.68112 (11) | 0.01406 (7) | 0.0399 (3) | |
| C10 | 0.15959 (17) | 0.75179 (12) | −0.04403 (7) | 0.0484 (3) | |
| H10A | 0.1996 | 0.8350 | −0.0456 | 0.058* | |
| H10B | 0.0577 | 0.7590 | −0.0361 | 0.058* | |
| C11 | 0.17193 (18) | 0.68716 (12) | −0.11266 (7) | 0.0502 (4) | |
| H11A | 0.1159 | 0.7328 | −0.1490 | 0.060* | |
| H11B | 0.2730 | 0.6870 | −0.1227 | 0.060* | |
| C12 | 0.11714 (15) | 0.55543 (12) | −0.11151 (7) | 0.0434 (3) | |
| C13 | 0.14319 (15) | 0.48612 (11) | −0.04483 (6) | 0.0399 (3) | |
| C14 | 0.04150 (17) | 0.49483 (12) | −0.16301 (7) | 0.0475 (3) | |
| H14 | 0.0134 | 0.4146 | −0.1523 | 0.057* | |
| C15 | −0.00427 (18) | 0.53472 (12) | −0.23384 (7) | 0.0488 (3) | |
| C16 | 0.0762 (2) | 0.61459 (14) | −0.27197 (8) | 0.0565 (4) | |
| H16 | 0.1647 | 0.6460 | −0.2525 | 0.068* | |
| C17 | 0.0255 (2) | 0.64745 (16) | −0.33848 (8) | 0.0688 (5) | |
| H17 | 0.0809 | 0.6998 | −0.3635 | 0.083* | |
| C18 | −0.1053 (3) | 0.60374 (18) | −0.36787 (9) | 0.0773 (6) | |
| H18 | −0.1398 | 0.6280 | −0.4122 | 0.093* | |
| C19 | −0.1857 (2) | 0.52368 (18) | −0.33150 (9) | 0.0739 (5) | |
| H19 | −0.2745 | 0.4935 | −0.3515 | 0.089* | |
| C20 | −0.1349 (2) | 0.48783 (15) | −0.26524 (8) | 0.0601 (4) | |
| H20 | −0.1886 | 0.4319 | −0.2415 | 0.072* | |
| C21 | 0.31014 (14) | 0.34287 (11) | 0.06809 (6) | 0.0398 (3) | |
| C22 | 0.23121 (16) | 0.27801 (12) | 0.11307 (7) | 0.0486 (3) | |
| H22 | 0.1776 | 0.3208 | 0.1435 | 0.058* | |
| C23 | 0.23196 (19) | 0.14904 (14) | 0.11288 (9) | 0.0607 (4) | |
| H23 | 0.1770 | 0.1056 | 0.1425 | 0.073* | |
| C24 | 0.3134 (2) | 0.08513 (14) | 0.06919 (10) | 0.0710 (5) | |
| H24 | 0.3138 | −0.0013 | 0.0691 | 0.085* | |
| C25 | 0.3942 (2) | 0.14968 (16) | 0.02578 (10) | 0.0766 (5) | |
| H25 | 0.4509 | 0.1068 | −0.0033 | 0.092* | |
| C26 | 0.39204 (19) | 0.27804 (14) | 0.02481 (9) | 0.0604 (4) | |
| H26 | 0.4463 | 0.3209 | −0.0053 | 0.072* | |
| N1 | 0.31024 (13) | 0.74545 (10) | 0.06376 (6) | 0.0459 (3) | |
| O1 | 0.09155 (12) | 0.38323 (8) | −0.03732 (5) | 0.0517 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0594 (9) | 0.0455 (8) | 0.0552 (8) | 0.0054 (7) | −0.0065 (7) | −0.0014 (6) |
| C2 | 0.0810 (12) | 0.0635 (10) | 0.0627 (10) | 0.0080 (9) | −0.0252 (9) | −0.0038 (8) |
| C3 | 0.1013 (15) | 0.0616 (11) | 0.0722 (11) | 0.0025 (10) | −0.0351 (11) | −0.0180 (9) |
| C4 | 0.0883 (13) | 0.0435 (8) | 0.0671 (10) | 0.0003 (8) | −0.0153 (9) | −0.0135 (7) |
| C5 | 0.0519 (8) | 0.0371 (7) | 0.0476 (7) | 0.0009 (6) | 0.0017 (6) | −0.0063 (6) |
| C6 | 0.0442 (7) | 0.0374 (7) | 0.0431 (7) | 0.0019 (6) | 0.0029 (6) | −0.0019 (5) |
| C7 | 0.0401 (7) | 0.0310 (6) | 0.0421 (6) | 0.0006 (5) | 0.0059 (5) | 0.0004 (5) |
| C8 | 0.0424 (7) | 0.0301 (6) | 0.0419 (7) | −0.0008 (5) | 0.0045 (5) | 0.0012 (5) |
| C9 | 0.0441 (7) | 0.0298 (6) | 0.0460 (7) | −0.0005 (5) | 0.0054 (6) | 0.0008 (5) |
| C10 | 0.0619 (9) | 0.0281 (6) | 0.0537 (8) | −0.0006 (6) | −0.0023 (7) | 0.0052 (6) |
| C11 | 0.0655 (9) | 0.0372 (7) | 0.0470 (7) | −0.0054 (6) | 0.0013 (7) | 0.0077 (6) |
| C12 | 0.0527 (8) | 0.0340 (6) | 0.0434 (7) | 0.0007 (6) | 0.0039 (6) | 0.0030 (5) |
| C13 | 0.0467 (7) | 0.0299 (6) | 0.0428 (7) | −0.0002 (5) | 0.0028 (6) | 0.0022 (5) |
| C14 | 0.0638 (9) | 0.0346 (7) | 0.0437 (7) | 0.0006 (6) | 0.0037 (6) | 0.0023 (5) |
| C15 | 0.0669 (9) | 0.0376 (7) | 0.0415 (7) | 0.0072 (7) | 0.0042 (7) | −0.0024 (6) |
| C16 | 0.0744 (11) | 0.0466 (8) | 0.0493 (8) | 0.0060 (7) | 0.0093 (7) | 0.0015 (7) |
| C17 | 0.1057 (15) | 0.0537 (9) | 0.0488 (9) | 0.0118 (10) | 0.0160 (10) | 0.0080 (7) |
| C18 | 0.1155 (17) | 0.0686 (12) | 0.0457 (9) | 0.0241 (11) | −0.0039 (10) | 0.0050 (8) |
| C19 | 0.0860 (13) | 0.0770 (12) | 0.0544 (10) | 0.0099 (10) | −0.0154 (9) | −0.0101 (9) |
| C20 | 0.0771 (11) | 0.0542 (9) | 0.0477 (8) | −0.0002 (8) | 0.0000 (8) | −0.0060 (7) |
| C21 | 0.0437 (7) | 0.0315 (6) | 0.0429 (7) | 0.0038 (5) | −0.0031 (6) | 0.0013 (5) |
| C22 | 0.0569 (9) | 0.0395 (7) | 0.0485 (8) | 0.0022 (6) | 0.0000 (6) | 0.0072 (6) |
| C23 | 0.0715 (11) | 0.0423 (8) | 0.0648 (10) | −0.0094 (7) | −0.0124 (8) | 0.0168 (7) |
| C24 | 0.0966 (14) | 0.0291 (7) | 0.0817 (12) | 0.0063 (8) | −0.0204 (11) | 0.0012 (8) |
| C25 | 0.1012 (15) | 0.0443 (9) | 0.0846 (13) | 0.0234 (9) | 0.0111 (11) | −0.0074 (9) |
| C26 | 0.0708 (10) | 0.0422 (8) | 0.0705 (10) | 0.0103 (7) | 0.0191 (8) | 0.0008 (7) |
| N1 | 0.0549 (7) | 0.0322 (5) | 0.0499 (6) | 0.0004 (5) | 0.0015 (5) | −0.0035 (5) |
| O1 | 0.0673 (7) | 0.0343 (5) | 0.0514 (6) | −0.0106 (4) | −0.0052 (5) | 0.0058 (4) |
Geometric parameters (Å, º)
| C1—C2 | 1.359 (2) | C13—O1 | 1.2191 (15) |
| C1—C6 | 1.4151 (19) | C14—C15 | 1.4689 (18) |
| C1—H1 | 0.9300 | C14—H14 | 0.9300 |
| C2—C3 | 1.399 (2) | C15—C20 | 1.393 (2) |
| C2—H2 | 0.9300 | C15—C16 | 1.395 (2) |
| C3—C4 | 1.356 (2) | C16—C17 | 1.381 (2) |
| C3—H3 | 0.9300 | C16—H16 | 0.9300 |
| C4—C5 | 1.411 (2) | C17—C18 | 1.369 (3) |
| C4—H4 | 0.9300 | C17—H17 | 0.9300 |
| C5—N1 | 1.3682 (17) | C18—C19 | 1.377 (3) |
| C5—C6 | 1.4154 (18) | C18—H18 | 0.9300 |
| C6—C7 | 1.4236 (17) | C19—C20 | 1.386 (2) |
| C7—C8 | 1.3818 (17) | C19—H19 | 0.9300 |
| C7—C21 | 1.4957 (17) | C20—H20 | 0.9300 |
| C8—C9 | 1.4304 (17) | C21—C26 | 1.3750 (19) |
| C8—C13 | 1.4949 (17) | C21—C22 | 1.3813 (19) |
| C9—N1 | 1.3132 (16) | C22—C23 | 1.387 (2) |
| C9—C10 | 1.4970 (18) | C22—H22 | 0.9300 |
| C10—C11 | 1.522 (2) | C23—C24 | 1.373 (3) |
| C10—H10A | 0.9700 | C23—H23 | 0.9300 |
| C10—H10B | 0.9700 | C24—C25 | 1.369 (3) |
| C11—C12 | 1.5051 (18) | C24—H24 | 0.9300 |
| C11—H11A | 0.9700 | C25—C26 | 1.381 (2) |
| C11—H11B | 0.9700 | C25—H25 | 0.9300 |
| C12—C14 | 1.3361 (18) | C26—H26 | 0.9300 |
| C12—C13 | 1.4978 (17) | ||
| C2—C1—C6 | 120.76 (14) | O1—C13—C12 | 121.60 (12) |
| C2—C1—H1 | 119.6 | C8—C13—C12 | 117.57 (11) |
| C6—C1—H1 | 119.6 | C12—C14—C15 | 130.26 (13) |
| C1—C2—C3 | 120.33 (15) | C12—C14—H14 | 114.9 |
| C1—C2—H2 | 119.8 | C15—C14—H14 | 114.9 |
| C3—C2—H2 | 119.8 | C20—C15—C16 | 118.07 (14) |
| C4—C3—C2 | 120.80 (15) | C20—C15—C14 | 117.78 (14) |
| C4—C3—H3 | 119.6 | C16—C15—C14 | 124.14 (14) |
| C2—C3—H3 | 119.6 | C17—C16—C15 | 120.55 (17) |
| C3—C4—C5 | 120.37 (15) | C17—C16—H16 | 119.7 |
| C3—C4—H4 | 119.8 | C15—C16—H16 | 119.7 |
| C5—C4—H4 | 119.8 | C18—C17—C16 | 120.70 (18) |
| N1—C5—C4 | 117.62 (13) | C18—C17—H17 | 119.7 |
| N1—C5—C6 | 123.01 (12) | C16—C17—H17 | 119.7 |
| C4—C5—C6 | 119.32 (13) | C17—C18—C19 | 119.74 (16) |
| C1—C6—C5 | 118.43 (12) | C17—C18—H18 | 120.1 |
| C1—C6—C7 | 123.37 (12) | C19—C18—H18 | 120.1 |
| C5—C6—C7 | 118.18 (12) | C18—C19—C20 | 120.23 (18) |
| C8—C7—C6 | 118.02 (11) | C18—C19—H19 | 119.9 |
| C8—C7—C21 | 122.74 (11) | C20—C19—H19 | 119.9 |
| C6—C7—C21 | 119.20 (11) | C19—C20—C15 | 120.66 (17) |
| C7—C8—C9 | 119.39 (11) | C19—C20—H20 | 119.7 |
| C7—C8—C13 | 122.29 (11) | C15—C20—H20 | 119.7 |
| C9—C8—C13 | 118.27 (11) | C26—C21—C22 | 119.17 (13) |
| N1—C9—C8 | 123.48 (12) | C26—C21—C7 | 119.59 (12) |
| N1—C9—C10 | 117.68 (11) | C22—C21—C7 | 121.24 (12) |
| C8—C9—C10 | 118.84 (11) | C21—C22—C23 | 120.02 (14) |
| C9—C10—C11 | 111.14 (11) | C21—C22—H22 | 120.0 |
| C9—C10—H10A | 109.4 | C23—C22—H22 | 120.0 |
| C11—C10—H10A | 109.4 | C24—C23—C22 | 120.35 (16) |
| C9—C10—H10B | 109.4 | C24—C23—H23 | 119.8 |
| C11—C10—H10B | 109.4 | C22—C23—H23 | 119.8 |
| H10A—C10—H10B | 108.0 | C25—C24—C23 | 119.48 (15) |
| C12—C11—C10 | 111.30 (11) | C25—C24—H24 | 120.3 |
| C12—C11—H11A | 109.4 | C23—C24—H24 | 120.3 |
| C10—C11—H11A | 109.4 | C24—C25—C26 | 120.52 (17) |
| C12—C11—H11B | 109.4 | C24—C25—H25 | 119.7 |
| C10—C11—H11B | 109.4 | C26—C25—H25 | 119.7 |
| H11A—C11—H11B | 108.0 | C21—C26—C25 | 120.43 (16) |
| C14—C12—C13 | 115.98 (12) | C21—C26—H26 | 119.8 |
| C14—C12—C11 | 126.86 (12) | C25—C26—H26 | 119.8 |
| C13—C12—C11 | 117.09 (11) | C9—N1—C5 | 117.84 (11) |
| O1—C13—C8 | 120.83 (11) | ||
| C6—C1—C2—C3 | −0.3 (3) | C14—C12—C13—O1 | −3.7 (2) |
| C1—C2—C3—C4 | 0.4 (3) | C11—C12—C13—O1 | 173.38 (13) |
| C2—C3—C4—C5 | −0.5 (3) | C14—C12—C13—C8 | 176.83 (12) |
| C3—C4—C5—N1 | −176.88 (17) | C11—C12—C13—C8 | −6.04 (18) |
| C3—C4—C5—C6 | 0.6 (3) | C13—C12—C14—C15 | 179.30 (14) |
| C2—C1—C6—C5 | 0.4 (2) | C11—C12—C14—C15 | 2.5 (3) |
| C2—C1—C6—C7 | 178.48 (15) | C12—C14—C15—C20 | −148.09 (16) |
| N1—C5—C6—C1 | 176.83 (13) | C12—C14—C15—C16 | 33.2 (2) |
| C4—C5—C6—C1 | −0.5 (2) | C20—C15—C16—C17 | 1.1 (2) |
| N1—C5—C6—C7 | −1.4 (2) | C14—C15—C16—C17 | 179.87 (14) |
| C4—C5—C6—C7 | −178.70 (14) | C15—C16—C17—C18 | 0.9 (3) |
| C1—C6—C7—C8 | −175.42 (13) | C16—C17—C18—C19 | −1.6 (3) |
| C5—C6—C7—C8 | 2.67 (18) | C17—C18—C19—C20 | 0.3 (3) |
| C1—C6—C7—C21 | 2.24 (19) | C18—C19—C20—C15 | 1.8 (3) |
| C5—C6—C7—C21 | −179.67 (12) | C16—C15—C20—C19 | −2.5 (2) |
| C6—C7—C8—C9 | −1.66 (18) | C14—C15—C20—C19 | 178.72 (14) |
| C21—C7—C8—C9 | −179.24 (12) | C8—C7—C21—C26 | 74.02 (18) |
| C6—C7—C8—C13 | −179.36 (11) | C6—C7—C21—C26 | −103.53 (15) |
| C21—C7—C8—C13 | 3.06 (19) | C8—C7—C21—C22 | −106.21 (15) |
| C7—C8—C9—N1 | −0.88 (19) | C6—C7—C21—C22 | 76.24 (16) |
| C13—C8—C9—N1 | 176.92 (12) | C26—C21—C22—C23 | −1.8 (2) |
| C7—C8—C9—C10 | 179.33 (12) | C7—C21—C22—C23 | 178.44 (13) |
| C13—C8—C9—C10 | −2.87 (18) | C21—C22—C23—C24 | 1.4 (2) |
| N1—C9—C10—C11 | 141.67 (13) | C22—C23—C24—C25 | 0.0 (3) |
| C8—C9—C10—C11 | −38.53 (17) | C23—C24—C25—C26 | −1.2 (3) |
| C9—C10—C11—C12 | 56.53 (17) | C22—C21—C26—C25 | 0.7 (2) |
| C10—C11—C12—C14 | 142.07 (15) | C7—C21—C26—C25 | −179.55 (15) |
| C10—C11—C12—C13 | −34.70 (18) | C24—C25—C26—C21 | 0.8 (3) |
| C7—C8—C13—O1 | 24.48 (19) | C8—C9—N1—C5 | 2.25 (19) |
| C9—C8—C13—O1 | −153.24 (13) | C10—C9—N1—C5 | −177.96 (12) |
| C7—C8—C13—C12 | −156.08 (12) | C4—C5—N1—C9 | 176.28 (13) |
| C9—C8—C13—C12 | 26.19 (17) | C6—C5—N1—C9 | −1.1 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C10—H10B···O1i | 0.97 | 2.58 | 3.2700 (18) | 128 |
| C26—H26···Cg1ii | 0.93 | 2.71 | 3.577 (18) | 156 |
Symmetry codes: (i) −x, −y+1, −z; (ii) −x+1, −y+1, −z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: BT6954).
References
- Baguley, B. C. & Ferguson, L. R. (1998). Biochim. Biophys. Acta, 1400, 213–222. [DOI] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2008). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Buckleton, J. S. & Waters, T. N. (1984). Acta Cryst. C40, 1587–1589.
- Chantrapromma, S., Boonnak, N., Razak, I. A. & Fun, H.-K. (2010). Acta Cryst. E66, o81–o82. [DOI] [PMC free article] [PubMed]
- Das, S., Kundu, S. & Suresh, K. G. (2011). DNA Cell Biol. 30, 525–535. [DOI] [PubMed]
- Denny, W. A., Cain B. F., Atwell, G. J., Hansch, C. & Panthananickal, A. (1982). J. Med. Chem. 25, 276–315. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kumar, R., Kaur, M. & Kumari, M. (2012). Acta Pol. Pharm. 69, 3–9. [PubMed]
- Pigatto, M. C., Lima, M. C. A., Galdino, S. L., Pitta, I. R., Vessecchi, R., Assis, M. D., Santos, J. S., Costa, T. C. T. D. & Lopes, P. N. (2011). Eur. J. Med. Chem. 1, 4245–4251. [DOI] [PubMed]
- Prommier, Y. & Goldwasser, F., Chabner, B. A. & Longo, D. L. (2006). Editors. Cancer Chemother. Biother. pp. 451–475.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tomar, V., Bhattacharjee, G., Uddin, K., Rajakumar, S., Srivastava, K. & Puri, S. K. (2010). Eur. J. Med. Chem. 45, 745–751. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536814015943/bt6954sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814015943/bt6954Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814015943/bt6954Isup3.cml
CCDC reference: 1012814
Additional supporting information: crystallographic information; 3D view; checkCIF report