The title compounds, (I) and (II), are dispiro-indole-pyrrolidine-benzothiophene derivatives, with (II) having a chlorine substituent on the oxoindole unit. As a result, the conformation of the two molecules differs in the angle of inclination of the indole moiety with respect to the benzothiophene ring system, with a dihedral angle of 71.59 (5) in (I) and 82.27 (7)° in (II).
Keywords: dispiro, pyrrolidine-indole, benzothiophene, crystal structure
Abstract
In the title compounds, C22H20N2O4S, (I), and C22H19ClN2O4S, (II), the pyrrolidine rings have twist conformations on the spiro–spiro C—C bonds. In (I), the five-membered ring of the oxindole moiety has an envelope conformation with the spiro C atom as the flap, while in (II) this ring is flat (r.m.s. deviation = 0.042 Å). The mean planes of the pyrrolidine rings are inclined to the mean planes of the indole units [r.m.s deviations = 0.073 and 0.069 Å for (I) and (II), respectively] and the benzothiophene ring systems (r.m.s. deviations = 0.019 and 0.034 Å for (I) and (II), respectively) by 79.57 (8) and 88.61 (7)° for (I), and by 81.99 (10) and 88.79 (10)° for (II). In both compounds, the ethoxycarbonyl group occupies an equatorial position with an extended conformation. The overall conformation of the two molecules differs in the angle of inclination of the indole unit with respect to the benzothiophene ring system, with a dihedral angle between the planes of 71.59 (5) in (I) and 82.27 (7)° in (II). In the crystal of (I), molecules are linked via pairs of N—H⋯O hydrogen bonds, forming inversion dimers enclosing R 2 2(14) loops. The dimers are linked via C—H⋯O and bifurcated C—H⋯O(O) hydrogen bonds, forming sheets lying parallel to (100). In the crystal of (II), molecules are again linked via pairs of N—H⋯O hydrogen bonds, forming inversion dimers but enclosing smaller R 2 2(8) loops. Here, the dimers are linked by C—H⋯O hydrogen bonds, forming ribbons propagating along [010].
Chemical context
The spiro-indole-pyrrolidine ring system is a frequently encountered structural motif in many biologically important and pharmacologically relevant alkaloids, such as vincrinstine, vinblastine and spirotypostatins (Cordell, 1981 ▸). Highly functionalized pyrrolidines have gained much interest in the past few years as they constitute the main structural element of many natural and synthetic pharmacologically active compounds (Waldmann, 1995 ▸). Optically active pyrrolidines have been used as intermediates, chiral ligands or auxiliaries in controlled asymmetric synthesis (Suzuki et al., 1994 ▸; Huryn et al., 1991 ▸). In view of this importance, the title compounds were synthesized and we report herein on their molecular and crystal structures.
Structural commentary
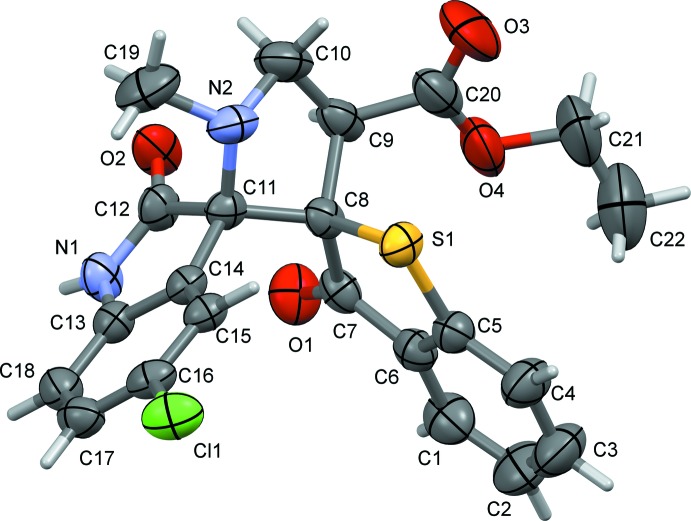
The molecular structure of molecule (I) is shown in Fig. 1 ▸. The pyrrolidine ring (N2/C8–C11) exhibits a twist conformation on bond C8—C11. The five-membered ring (N1/C11–C14) of the oxindole moiety adopts an envelope conformation with C11 as the flap atom. The C12=O2 bond length of 1.213 (1) Å confirms the presence of a keto group in the indoline moiety. The benzothiophene ring system (S1/C1–C8; r.m.s. deviation = 0.019 Å) and the mean plane of the indole ring system (N1/C11–C18; r.m.s. deviation = 0.073 Å) are inclined to one another by 71.59 (5)°, and are both almost normal to the mean plane of the pyrrolidine ring (N2/C8–C11) with dihedral angles of 88.61 (17) and 79.57 (8)°, respectively.
Figure 1.
The molecular structure of molecule (I), with the atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
The molecular structure of the compound (II) is illustrated in Fig. 2 ▸. The overall geometry of the molecule is similar to that of (II). The pyrrolidine ring (N2/C8–C11) also adopts a twist conformation on the C8—C11 bond, and the five-membered ring (N1/C11–C14) of the oxindole moiety has an r.m.s. deviation = 0.042 Å. The mean plane of the benzothiophene ring system (S1/C1–C8; r.m.s. deviation = 0.034 Å) and the mean plane of the indole ring system (N1/C11–C18; r.m.s. deviation = 0.069 Å) are inclined to one another by 82.27 (7)°, and are both almost normal to the mean plane of the pyrrolidine ring (N2/C8–C11) with dihedral angles of 88.79 (10) and 81.99 (10)°, respectively.
Figure 2.
The molecular structure of molecule (II), with the atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
Molecules (I) and (II) differ only in the presence of a chloride atom at position 5 in the oxoindole unit in (II). The conformation of the two molecules differ in the angle of inclination of the indole moiety with respect to the benzothiophene ring system, with a dihedral angle of 71.59 (5) in (I) and 82.27 (7)° in (II). This is illustrated in Fig. 3 ▸, which shows a view of the superposition of the two molecules (Mercury; Macrae et al., 2008 ▸). There is also a small difference in the orientation of the ester function, the C20—O4—C21—C22 torsion angle being 173.44 (19) in (I) and 162.3 (3)° in (II).
Figure 3.
A view of the molecular superposition of molecules (I) and (II) [red (I); blue (II); Cl atom in (II) is shown as a blue ball (Mercury; Macrae et al., 2008 ▸)].
Supramolecular features
In the crystal of (I), molecules are linked via pairs of N—H⋯O hydrogen bonds, forming inversion dimers enclosing  (14) loops (Table 1 ▸ and Fig. 4 ▸). The dimers are linked via C—H⋯O and bifurcated C—H⋯O(O) hydrogen bonds, forming sheets lying parallel to (100).
(14) loops (Table 1 ▸ and Fig. 4 ▸). The dimers are linked via C—H⋯O and bifurcated C—H⋯O(O) hydrogen bonds, forming sheets lying parallel to (100).
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
Cg is the centroid of the C1–C6 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O1i | 0.83 (2) | 2.09 (2) | 2.890 (2) | 164 (2) |
| C3—H3⋯O2ii | 0.93 | 2.56 | 3.385 (2) | 148 |
| C18—H18⋯O3iii | 0.93 | 2.56 | 3.299 (2) | 136 |
| C21—H21B⋯O2iv | 0.97 | 2.59 | 3.560 (2) | 174 |
| C2—H2⋯Cg v | 0.93 | 2.81 | 3.649 (2) | 151 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Figure 4.
The crystal packing of compound (I), viewed along the a axis. The hydrogen bonds are shown as dashed lines (see Table 1 ▸ for details; H atoms not involved in hydrogen bonding have been omitted for clarity).
In the crystal of (II), molecules are again linked via pairs of N—-H⋯O hydrogen bonds, forming inversion dimers but enclosing smaller  (8) loops (Table 2 ▸ and Fig. 5 ▸). Here the dimers are linked by C—H⋯O hydrogen bonds, forming double-stranded chains propagating along [010].
(8) loops (Table 2 ▸ and Fig. 5 ▸). Here the dimers are linked by C—H⋯O hydrogen bonds, forming double-stranded chains propagating along [010].
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
Cg is the centroid of the C1–C6 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O2i | 0.81 (2) | 2.03 (2) | 2.842 (2) | 172 (2) |
| C18—H18⋯O3ii | 0.93 | 2.57 | 3.496 (3) | 171 |
| C2—H2⋯Cg iii | 0.93 | 2.83 | 3.649 (2) | 155 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Figure 5.

A partial view along the a axis of the crystal packing of compound (II). The hydrogen bonds are shown as dashed lines (see Table 2 ▸ for details; H atoms not involved in hydrogen bonding have been omitted for clarity).
Database survey
A search of the Cambridge Structural Database (Version 5.35, last update November 2013; Allen, 2002 ▸) revealed that the title compounds are the first examples of dispiro-indole-pyrrolidine derivatives with a benzothiophene substituent on the pyrrolidine ring creating the second spiro C atom. There are a large number of indole-spiro-pyrrolidine compounds but there was only one hit for a dispiro-indole-pyrrolidine-‘cyclopentane-type’ compound, namely 4′-(p-methoxyphenyl)-1′-methyl-1H-indole-3-spiro-2′-pyrrolidine-3′-spiro-1′′-cyclopentane-2(3H),2′′-dione (refcode: ILIMUL; Govind et al., 2003 ▸). The geometry of the pyrrolidine and oxindole ring systems of the two molecules compare well with those reported for similar structures, for example, ethyl 1′′-benzyl-2′′-oxo-2′,3′,5′,6′,7′,7a′-hexahydro-1′H-dispiro[indeno[1,2-b]-quinoxaline-11,2′-pyrrolizine-3′,3′′-indoline]-1′-carboxylate monohydrate (refcode: IFOVUW; Kannan et al., 2013a ▸) and methyl 5′′-chloro-1′,1′′-dimethyl-2,2′′-dioxodispiro[indoline-3,2′-pyrrolidine-3′,3′′-indoline]-4′-carboxylate (refcode: IFOQUR; Kannan et al., 2013b ▸).
Synthesis and crystallization
The two compounds were prepared in a similar manner using isatin (1.1 mmol) for (I) and 5-chloro isatin (1.1 mmol) for (II). A mixture of (E)-ethyl 2-(3-oxobenzo[b]thiophen-2(3H)-ylidene) acetate (1.0 mmol) and the relevant isatin together with sarcosine (1.1 mmol) was refluxed in methanol (20 ml) until completion of the reaction, as evidenced by TLC analysis. After completion of the reaction, the solvent was evaporated under reduced pressure. The crude reaction mixture was dissolved in dichloromethane (2 × 50 ml) and washed with water followed by brine solution. The organic layer was separated and dried over sodium sulfate. After filtration, the solvent was evaporation under reduced pressure. The product was separated by column chromatography using hexane and ethyl acetate (9:1) as eluent to give a white solid. This was dissolved in chloroform (3 ml) and heated for 2 min. The resulting solutions were allowed to evaporate slowly at room temperature and yielded colourless block-like crystals of compounds (I) and (II).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. For both molecules (I) and (II), the NH H atoms were located in difference Fourier maps and freely refined. The C-bound H atoms were included in calculated positions and treated as riding atoms: C—H = 0.93–0.98 Å with U iso(H) = 1.5U eq(C-methyl) and = 1.2U eq(C) for other H atoms.
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C22H20N2O4S | C22H19ClN2O4S |
| M r | 408.46 | 442.90 |
| Crystal system, space group | Triclinic, P

|
Triclinic, P

|
| Temperature (K) | 293 | 293 |
| a, b, c (Å) | 8.7196 (4), 10.7874 (5), 11.3488 (5) | 10.4678 (5), 10.9074 (5), 11.5652 (5) |
| α, β, γ (°) | 82.624 (2), 82.775 (2), 79.214 (2) | 85.973 (2), 65.612 (2), 62.089 (2) |
| V (Å3) | 1034.27 (8) | 1050.26 (9) |
| Z | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.19 | 0.31 |
| Crystal size (mm) | 0.35 × 0.30 × 0.30 | 0.35 × 0.30 × 0.30 |
| Data collection | ||
| Diffractometer | Bruker Kappa APEXII CCD | Bruker AXS kappa APEX2 CCD |
| Absorption correction | Multi-scan (SADABS; Sheldrick, 1996 ▸) | Multi-scan (SADABS; Sheldrick, 1996 ▸) |
| T min, T max | 0.938, 0.946 | 0.931, 0.940 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18982, 3745, 3389 | 16876, 3788, 3178 |
| R int | 0.024 | 0.024 |
| (sin θ/λ)max (Å−1) | 0.600 | 0.600 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.032, 0.087, 1.05 | 0.036, 0.103, 1.11 |
| No. of reflections | 3745 | 3788 |
| No. of parameters | 268 | 276 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.25, −0.17 | 0.29, −0.27 |
Supplementary Material
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S1600536814015426/su2728sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814015426/su2728Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S1600536814015426/su2728IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank Dr Babu Vargheese, SAIF, IIT, Madras, India, for his help with the data collection.
supplementary crystallographic information
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Crystal data
| C22H20N2O4S | V = 1034.27 (8) Å3 |
| Mr = 408.46 | Z = 2 |
| Triclinic, P1 | F(000) = 428 |
| Hall symbol: -P 1 | Dx = 1.312 Mg m−3 |
| a = 8.7196 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.7874 (5) Å | θ = 2.4–25.0° |
| c = 11.3488 (5) Å | µ = 0.19 mm−1 |
| α = 82.624 (2)° | T = 293 K |
| β = 82.775 (2)° | Block, colourless |
| γ = 79.214 (2)° | 0.35 × 0.30 × 0.30 mm |
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Data collection
| Bruker Kappa APEXII CCD diffractometer | 3745 independent reflections |
| Radiation source: fine-focus sealed tube | 3389 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.024 |
| ω and φ scans | θmax = 25.3°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −10→10 |
| Tmin = 0.938, Tmax = 0.946 | k = −12→12 |
| 18982 measured reflections | l = −13→13 |
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.032 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.087 | w = 1/[σ2(Fo2) + (0.0417P)2 + 0.2979P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 3745 reflections | Δρmax = 0.25 e Å−3 |
| 268 parameters | Δρmin = −0.17 e Å−3 |
| 0 restraints | Extinction correction: SHELXL2013 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0099 (17) |
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.30889 (4) | 0.13663 (3) | 0.21159 (3) | 0.03693 (12) | |
| O1 | −0.05178 (12) | 0.36372 (10) | 0.34437 (10) | 0.0491 (3) | |
| O2 | 0.15077 (17) | 0.58243 (10) | 0.29000 (11) | 0.0647 (4) | |
| O3 | 0.2116 (2) | 0.32476 (19) | −0.07349 (11) | 0.1024 (6) | |
| O4 | 0.01130 (15) | 0.28291 (12) | 0.05809 (10) | 0.0580 (3) | |
| N1 | 0.18901 (15) | 0.47295 (12) | 0.47309 (12) | 0.0449 (3) | |
| H1N | 0.145 (2) | 0.5301 (17) | 0.5141 (16) | 0.054* | |
| N2 | 0.42620 (15) | 0.38340 (12) | 0.21313 (10) | 0.0452 (3) | |
| C1 | −0.11000 (18) | 0.09770 (16) | 0.37801 (14) | 0.0466 (4) | |
| H1 | −0.1955 | 0.1524 | 0.4109 | 0.056* | |
| C2 | −0.1138 (2) | −0.02952 (17) | 0.38248 (16) | 0.0569 (4) | |
| H2 | −0.2027 | −0.0613 | 0.4179 | 0.068* | |
| C3 | 0.0142 (2) | −0.11030 (16) | 0.33441 (16) | 0.0556 (4) | |
| H3 | 0.0100 | −0.1962 | 0.3379 | 0.067* | |
| C4 | 0.14759 (19) | −0.06681 (14) | 0.28153 (14) | 0.0448 (4) | |
| H4 | 0.2334 | −0.1225 | 0.2503 | 0.054* | |
| C5 | 0.15164 (16) | 0.06238 (12) | 0.27566 (11) | 0.0337 (3) | |
| C6 | 0.02306 (15) | 0.14358 (13) | 0.32381 (11) | 0.0343 (3) | |
| C7 | 0.04308 (15) | 0.27552 (13) | 0.30944 (11) | 0.0333 (3) | |
| C8 | 0.20210 (16) | 0.29422 (12) | 0.24261 (11) | 0.0326 (3) | |
| C9 | 0.1918 (2) | 0.38436 (14) | 0.12494 (12) | 0.0447 (4) | |
| H9 | 0.1131 | 0.4595 | 0.1411 | 0.054* | |
| C10 | 0.3518 (2) | 0.42508 (18) | 0.10187 (14) | 0.0581 (4) | |
| H10A | 0.4142 | 0.3851 | 0.0355 | 0.070* | |
| H10B | 0.3404 | 0.5165 | 0.0835 | 0.070* | |
| C11 | 0.30303 (16) | 0.36257 (12) | 0.30895 (12) | 0.0339 (3) | |
| C12 | 0.20227 (18) | 0.48789 (13) | 0.35267 (13) | 0.0425 (3) | |
| C13 | 0.28113 (15) | 0.36078 (13) | 0.51820 (12) | 0.0354 (3) | |
| C14 | 0.35773 (15) | 0.29409 (12) | 0.42447 (11) | 0.0314 (3) | |
| C15 | 0.46804 (16) | 0.18729 (13) | 0.44844 (13) | 0.0382 (3) | |
| H15 | 0.5232 | 0.1431 | 0.3867 | 0.046* | |
| C16 | 0.49556 (18) | 0.14672 (15) | 0.56609 (14) | 0.0480 (4) | |
| H16 | 0.5709 | 0.0755 | 0.5832 | 0.058* | |
| C17 | 0.4122 (2) | 0.21104 (16) | 0.65801 (14) | 0.0504 (4) | |
| H17 | 0.4297 | 0.1805 | 0.7365 | 0.060* | |
| C18 | 0.30339 (18) | 0.31964 (15) | 0.63592 (13) | 0.0448 (4) | |
| H18 | 0.2475 | 0.3632 | 0.6978 | 0.054* | |
| C19 | 0.5353 (2) | 0.4616 (2) | 0.23767 (18) | 0.0693 (5) | |
| H19A | 0.6191 | 0.4613 | 0.1741 | 0.104* | |
| H19B | 0.5777 | 0.4281 | 0.3116 | 0.104* | |
| H19C | 0.4810 | 0.5470 | 0.2435 | 0.104* | |
| C20 | 0.1438 (2) | 0.32731 (17) | 0.02394 (14) | 0.0562 (4) | |
| C21 | −0.0480 (3) | 0.2184 (2) | −0.02642 (17) | 0.0750 (6) | |
| H21A | 0.0326 | 0.1509 | −0.0550 | 0.090* | |
| H21B | −0.0799 | 0.2776 | −0.0944 | 0.090* | |
| C22 | −0.1847 (4) | 0.1653 (3) | 0.0376 (2) | 0.1083 (9) | |
| H22A | −0.2277 | 0.1220 | −0.0158 | 0.162* | |
| H22B | −0.2633 | 0.2329 | 0.0658 | 0.162* | |
| H22C | −0.1514 | 0.1066 | 0.1043 | 0.162* |
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0377 (2) | 0.0370 (2) | 0.0362 (2) | −0.00533 (14) | 0.00290 (14) | −0.01255 (14) |
| O1 | 0.0431 (6) | 0.0461 (6) | 0.0584 (7) | 0.0027 (5) | −0.0034 (5) | −0.0240 (5) |
| O2 | 0.0975 (10) | 0.0318 (6) | 0.0652 (8) | 0.0029 (6) | −0.0309 (7) | −0.0053 (5) |
| O3 | 0.1453 (15) | 0.1480 (16) | 0.0305 (7) | −0.0721 (13) | 0.0019 (8) | −0.0137 (8) |
| O4 | 0.0749 (8) | 0.0634 (7) | 0.0427 (6) | −0.0156 (6) | −0.0209 (6) | −0.0118 (5) |
| N1 | 0.0507 (7) | 0.0394 (7) | 0.0452 (7) | 0.0031 (6) | −0.0083 (6) | −0.0197 (6) |
| N2 | 0.0532 (7) | 0.0504 (7) | 0.0361 (6) | −0.0256 (6) | 0.0020 (5) | −0.0023 (5) |
| C1 | 0.0385 (8) | 0.0581 (10) | 0.0459 (8) | −0.0140 (7) | −0.0024 (6) | −0.0097 (7) |
| C2 | 0.0565 (10) | 0.0616 (11) | 0.0597 (10) | −0.0318 (9) | −0.0057 (8) | −0.0026 (8) |
| C3 | 0.0719 (11) | 0.0419 (9) | 0.0602 (10) | −0.0243 (8) | −0.0173 (9) | −0.0009 (7) |
| C4 | 0.0539 (9) | 0.0347 (7) | 0.0480 (8) | −0.0051 (6) | −0.0135 (7) | −0.0084 (6) |
| C5 | 0.0384 (7) | 0.0344 (7) | 0.0300 (6) | −0.0060 (5) | −0.0083 (5) | −0.0056 (5) |
| C6 | 0.0357 (7) | 0.0381 (7) | 0.0311 (7) | −0.0073 (6) | −0.0063 (5) | −0.0065 (5) |
| C7 | 0.0354 (7) | 0.0380 (7) | 0.0278 (6) | −0.0023 (6) | −0.0077 (5) | −0.0099 (5) |
| C8 | 0.0410 (7) | 0.0302 (6) | 0.0269 (6) | −0.0054 (5) | −0.0041 (5) | −0.0052 (5) |
| C9 | 0.0641 (10) | 0.0407 (8) | 0.0305 (7) | −0.0127 (7) | −0.0096 (6) | 0.0015 (6) |
| C10 | 0.0809 (12) | 0.0603 (10) | 0.0365 (8) | −0.0313 (9) | −0.0019 (8) | 0.0055 (7) |
| C11 | 0.0410 (7) | 0.0304 (7) | 0.0317 (7) | −0.0092 (5) | −0.0032 (5) | −0.0050 (5) |
| C12 | 0.0529 (9) | 0.0322 (7) | 0.0456 (8) | −0.0063 (6) | −0.0137 (7) | −0.0091 (6) |
| C13 | 0.0343 (7) | 0.0383 (7) | 0.0364 (7) | −0.0090 (6) | −0.0046 (5) | −0.0098 (6) |
| C14 | 0.0312 (6) | 0.0332 (7) | 0.0322 (7) | −0.0106 (5) | −0.0029 (5) | −0.0056 (5) |
| C15 | 0.0329 (7) | 0.0383 (7) | 0.0448 (8) | −0.0060 (6) | −0.0057 (6) | −0.0088 (6) |
| C16 | 0.0470 (8) | 0.0441 (8) | 0.0553 (9) | −0.0072 (7) | −0.0215 (7) | 0.0000 (7) |
| C17 | 0.0611 (10) | 0.0588 (10) | 0.0368 (8) | −0.0214 (8) | −0.0183 (7) | 0.0030 (7) |
| C18 | 0.0485 (8) | 0.0576 (9) | 0.0334 (7) | −0.0176 (7) | −0.0034 (6) | −0.0126 (6) |
| C19 | 0.0765 (13) | 0.0797 (13) | 0.0629 (11) | −0.0506 (11) | −0.0017 (9) | −0.0009 (10) |
| C20 | 0.0842 (13) | 0.0554 (10) | 0.0313 (8) | −0.0168 (9) | −0.0142 (8) | 0.0018 (7) |
| C21 | 0.1095 (17) | 0.0731 (13) | 0.0542 (11) | −0.0218 (12) | −0.0389 (11) | −0.0129 (9) |
| C22 | 0.134 (2) | 0.125 (2) | 0.0921 (18) | −0.0653 (19) | −0.0405 (17) | −0.0196 (16) |
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Geometric parameters (Å, º)
| S1—C5 | 1.7496 (14) | C9—C20 | 1.505 (2) |
| S1—C8 | 1.8330 (13) | C9—C10 | 1.522 (2) |
| O1—C7 | 1.2109 (16) | C9—H9 | 0.9800 |
| O2—C12 | 1.2125 (18) | C10—H10A | 0.9700 |
| O3—C20 | 1.187 (2) | C10—H10B | 0.9700 |
| O4—C20 | 1.327 (2) | C11—C14 | 1.5063 (18) |
| O4—C21 | 1.450 (2) | C11—C12 | 1.5696 (19) |
| N1—C12 | 1.348 (2) | C13—C18 | 1.378 (2) |
| N1—C13 | 1.3972 (19) | C13—C14 | 1.3904 (18) |
| N1—H1N | 0.827 (18) | C14—C15 | 1.3773 (19) |
| N2—C11 | 1.4561 (17) | C15—C16 | 1.387 (2) |
| N2—C19 | 1.457 (2) | C15—H15 | 0.9300 |
| N2—C10 | 1.473 (2) | C16—C17 | 1.380 (2) |
| C1—C2 | 1.373 (2) | C16—H16 | 0.9300 |
| C1—C6 | 1.388 (2) | C17—C18 | 1.380 (2) |
| C1—H1 | 0.9300 | C17—H17 | 0.9300 |
| C2—C3 | 1.381 (3) | C18—H18 | 0.9300 |
| C2—H2 | 0.9300 | C19—H19A | 0.9600 |
| C3—C4 | 1.375 (2) | C19—H19B | 0.9600 |
| C3—H3 | 0.9300 | C19—H19C | 0.9600 |
| C4—C5 | 1.394 (2) | C21—C22 | 1.486 (3) |
| C4—H4 | 0.9300 | C21—H21A | 0.9700 |
| C5—C6 | 1.3871 (19) | C21—H21B | 0.9700 |
| C6—C7 | 1.4523 (19) | C22—H22A | 0.9600 |
| C7—C8 | 1.5295 (18) | C22—H22B | 0.9600 |
| C8—C9 | 1.5495 (18) | C22—H22C | 0.9600 |
| C8—C11 | 1.5611 (18) | ||
| C5—S1—C8 | 92.66 (6) | N2—C11—C8 | 99.56 (10) |
| C20—O4—C21 | 117.69 (15) | C14—C11—C8 | 116.51 (10) |
| C12—N1—C13 | 112.19 (12) | N2—C11—C12 | 114.06 (11) |
| C12—N1—H1N | 122.9 (12) | C14—C11—C12 | 101.21 (10) |
| C13—N1—H1N | 124.0 (12) | C8—C11—C12 | 110.24 (11) |
| C11—N2—C19 | 115.58 (13) | O2—C12—N1 | 126.23 (14) |
| C11—N2—C10 | 107.91 (12) | O2—C12—C11 | 126.46 (14) |
| C19—N2—C10 | 115.00 (13) | N1—C12—C11 | 107.26 (12) |
| C2—C1—C6 | 119.16 (15) | C18—C13—C14 | 122.52 (13) |
| C2—C1—H1 | 120.4 | C18—C13—N1 | 127.67 (13) |
| C6—C1—H1 | 120.4 | C14—C13—N1 | 109.76 (12) |
| C1—C2—C3 | 120.05 (15) | C15—C14—C13 | 119.22 (12) |
| C1—C2—H2 | 120.0 | C15—C14—C11 | 131.91 (12) |
| C3—C2—H2 | 120.0 | C13—C14—C11 | 108.80 (11) |
| C4—C3—C2 | 121.62 (15) | C14—C15—C16 | 118.95 (14) |
| C4—C3—H3 | 119.2 | C14—C15—H15 | 120.5 |
| C2—C3—H3 | 119.2 | C16—C15—H15 | 120.5 |
| C3—C4—C5 | 118.61 (15) | C17—C16—C15 | 120.63 (14) |
| C3—C4—H4 | 120.7 | C17—C16—H16 | 119.7 |
| C5—C4—H4 | 120.7 | C15—C16—H16 | 119.7 |
| C6—C5—C4 | 119.78 (13) | C18—C17—C16 | 121.38 (14) |
| C6—C5—S1 | 114.47 (10) | C18—C17—H17 | 119.3 |
| C4—C5—S1 | 125.75 (11) | C16—C17—H17 | 119.3 |
| C5—C6—C1 | 120.77 (13) | C13—C18—C17 | 117.16 (14) |
| C5—C6—C7 | 113.66 (12) | C13—C18—H18 | 121.4 |
| C1—C6—C7 | 125.57 (13) | C17—C18—H18 | 121.4 |
| O1—C7—C6 | 126.02 (13) | N2—C19—H19A | 109.5 |
| O1—C7—C8 | 121.74 (12) | N2—C19—H19B | 109.5 |
| C6—C7—C8 | 112.24 (11) | H19A—C19—H19B | 109.5 |
| C7—C8—C9 | 114.43 (11) | N2—C19—H19C | 109.5 |
| C7—C8—C11 | 115.22 (10) | H19A—C19—H19C | 109.5 |
| C9—C8—C11 | 99.72 (10) | H19B—C19—H19C | 109.5 |
| C7—C8—S1 | 106.91 (9) | O3—C20—O4 | 124.34 (17) |
| C9—C8—S1 | 110.04 (9) | O3—C20—C9 | 125.41 (18) |
| C11—C8—S1 | 110.43 (9) | O4—C20—C9 | 110.23 (14) |
| C20—C9—C10 | 115.35 (14) | O4—C21—C22 | 107.01 (17) |
| C20—C9—C8 | 114.02 (12) | O4—C21—H21A | 110.3 |
| C10—C9—C8 | 103.71 (12) | C22—C21—H21A | 110.3 |
| C20—C9—H9 | 107.8 | O4—C21—H21B | 110.3 |
| C10—C9—H9 | 107.8 | C22—C21—H21B | 110.3 |
| C8—C9—H9 | 107.8 | H21A—C21—H21B | 108.6 |
| N2—C10—C9 | 105.51 (12) | C21—C22—H22A | 109.5 |
| N2—C10—H10A | 110.6 | C21—C22—H22B | 109.5 |
| C9—C10—H10A | 110.6 | H22A—C22—H22B | 109.5 |
| N2—C10—H10B | 110.6 | C21—C22—H22C | 109.5 |
| C9—C10—H10B | 110.6 | H22A—C22—H22C | 109.5 |
| H10A—C10—H10B | 108.8 | H22B—C22—H22C | 109.5 |
| N2—C11—C14 | 115.81 (11) | ||
| C6—C1—C2—C3 | 0.6 (2) | C9—C8—C11—N2 | −47.31 (12) |
| C1—C2—C3—C4 | 0.1 (3) | S1—C8—C11—N2 | 68.47 (11) |
| C2—C3—C4—C5 | −0.7 (2) | C7—C8—C11—C14 | 64.43 (15) |
| C3—C4—C5—C6 | 0.6 (2) | C9—C8—C11—C14 | −172.58 (11) |
| C3—C4—C5—S1 | −179.58 (11) | S1—C8—C11—C14 | −56.80 (13) |
| C8—S1—C5—C6 | −2.05 (11) | C7—C8—C11—C12 | −50.11 (14) |
| C8—S1—C5—C4 | 178.17 (12) | C9—C8—C11—C12 | 72.88 (13) |
| C4—C5—C6—C1 | 0.0 (2) | S1—C8—C11—C12 | −171.35 (9) |
| S1—C5—C6—C1 | −179.75 (11) | C13—N1—C12—O2 | −170.75 (15) |
| C4—C5—C6—C7 | −179.06 (12) | C13—N1—C12—C11 | 6.75 (16) |
| S1—C5—C6—C7 | 1.15 (15) | N2—C11—C12—O2 | 43.7 (2) |
| C2—C1—C6—C5 | −0.7 (2) | C14—C11—C12—O2 | 168.82 (15) |
| C2—C1—C6—C7 | 178.32 (14) | C8—C11—C12—O2 | −67.27 (19) |
| C5—C6—C7—O1 | −179.54 (13) | N2—C11—C12—N1 | −133.76 (13) |
| C1—C6—C7—O1 | 1.4 (2) | C14—C11—C12—N1 | −8.68 (14) |
| C5—C6—C7—C8 | 0.68 (16) | C8—C11—C12—N1 | 115.23 (13) |
| C1—C6—C7—C8 | −178.36 (13) | C12—N1—C13—C18 | 175.59 (14) |
| O1—C7—C8—C9 | −59.73 (17) | C12—N1—C13—C14 | −1.69 (17) |
| C6—C7—C8—C9 | 120.06 (12) | C18—C13—C14—C15 | −4.4 (2) |
| O1—C7—C8—C11 | 55.04 (17) | N1—C13—C14—C15 | 173.00 (12) |
| C6—C7—C8—C11 | −125.17 (12) | C18—C13—C14—C11 | 178.13 (12) |
| O1—C7—C8—S1 | 178.16 (11) | N1—C13—C14—C11 | −4.43 (15) |
| C6—C7—C8—S1 | −2.05 (12) | N2—C11—C14—C15 | −45.34 (19) |
| C5—S1—C8—C7 | 2.25 (9) | C8—C11—C14—C15 | 71.24 (18) |
| C5—S1—C8—C9 | −122.58 (10) | C12—C11—C14—C15 | −169.23 (14) |
| C5—S1—C8—C11 | 128.30 (9) | N2—C11—C14—C13 | 131.64 (12) |
| C7—C8—C9—C20 | −73.99 (17) | C8—C11—C14—C13 | −111.78 (12) |
| C11—C8—C9—C20 | 162.46 (14) | C12—C11—C14—C13 | 7.76 (13) |
| S1—C8—C9—C20 | 46.39 (16) | C13—C14—C15—C16 | 2.27 (19) |
| C7—C8—C9—C10 | 159.77 (12) | C11—C14—C15—C16 | 178.99 (13) |
| C11—C8—C9—C10 | 36.22 (14) | C14—C15—C16—C17 | 1.0 (2) |
| S1—C8—C9—C10 | −79.85 (13) | C15—C16—C17—C18 | −2.3 (2) |
| C11—N2—C10—C9 | −19.43 (17) | C14—C13—C18—C17 | 3.1 (2) |
| C19—N2—C10—C9 | −150.08 (15) | N1—C13—C18—C17 | −173.84 (14) |
| C20—C9—C10—N2 | −137.43 (14) | C16—C17—C18—C13 | 0.3 (2) |
| C8—C9—C10—N2 | −12.03 (16) | C21—O4—C20—O3 | 4.9 (3) |
| C19—N2—C11—C14 | −61.82 (18) | C21—O4—C20—C9 | −176.82 (15) |
| C10—N2—C11—C14 | 167.85 (12) | C10—C9—C20—O3 | −8.5 (3) |
| C19—N2—C11—C8 | 172.43 (14) | C8—C9—C20—O3 | −128.4 (2) |
| C10—N2—C11—C8 | 42.11 (14) | C10—C9—C20—O4 | 173.27 (14) |
| C19—N2—C11—C12 | 55.08 (18) | C8—C9—C20—O4 | 53.38 (19) |
| C10—N2—C11—C12 | −75.25 (15) | C20—O4—C21—C22 | 173.44 (19) |
| C7—C8—C11—N2 | −170.30 (11) |
(I) Ethyl (2S*,2'R*)-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C1–C6 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1i | 0.83 (2) | 2.09 (2) | 2.890 (2) | 164 (2) |
| C3—H3···O2ii | 0.93 | 2.56 | 3.385 (2) | 148 |
| C18—H18···O3iii | 0.93 | 2.56 | 3.299 (2) | 136 |
| C21—H21B···O2iv | 0.97 | 2.59 | 3.560 (2) | 174 |
| C2—H2···Cgv | 0.93 | 2.81 | 3.649 (2) | 151 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) x, y−1, z; (iii) x, y, z+1; (iv) −x, −y+1, −z; (v) −x, −y, −z+1.
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Crystal data
| C22H19ClN2O4S | V = 1050.26 (9) Å3 |
| Mr = 442.90 | Z = 2 |
| Triclinic, P1 | F(000) = 460 |
| Hall symbol: -P 1 | Dx = 1.401 Mg m−3 |
| a = 10.4678 (5) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.9074 (5) Å | θ = 2.0–25.0° |
| c = 11.5652 (5) Å | µ = 0.31 mm−1 |
| α = 85.973 (2)° | T = 293 K |
| β = 65.612 (2)° | Block, colourless |
| γ = 62.089 (2)° | 0.35 × 0.30 × 0.30 mm |
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Data collection
| Bruker AXS kappa APEX2 CCD diffractometer | Rint = 0.024 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | θmax = 25.3°, θmin = 2.1° |
| Tmin = 0.931, Tmax = 0.940 | h = −12→10 |
| 16876 measured reflections | k = −13→11 |
| 3788 independent reflections | l = −13→12 |
| 3178 reflections with I > 2σ(I) |
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: mixed |
| wR(F2) = 0.103 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.11 | w = 1/[σ2(Fo2) + (0.0458P)2 + 0.3849P] where P = (Fo2 + 2Fc2)/3 |
| 3788 reflections | (Δ/σ)max < 0.001 |
| 276 parameters | Δρmax = 0.29 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.78453 (7) | −0.07384 (7) | 0.27294 (5) | 0.06115 (18) | |
| S1 | 0.47022 (6) | 0.40379 (5) | 0.61542 (4) | 0.04426 (15) | |
| O1 | 0.24464 (18) | 0.30234 (17) | 0.93183 (14) | 0.0622 (4) | |
| O2 | 0.5511 (2) | 0.14387 (16) | 0.96174 (13) | 0.0596 (4) | |
| O3 | 0.5065 (3) | 0.6187 (2) | 0.8095 (3) | 0.1135 (8) | |
| O4 | 0.2715 (3) | 0.62165 (17) | 0.89290 (19) | 0.0794 (5) | |
| N1 | 0.5579 (2) | −0.00867 (18) | 0.83019 (16) | 0.0476 (4) | |
| H1N | 0.534 (3) | −0.054 (2) | 0.886 (2) | 0.057* | |
| N2 | 0.7301 (2) | 0.20503 (19) | 0.70036 (16) | 0.0493 (4) | |
| C1 | 0.0252 (3) | 0.4697 (2) | 0.8114 (2) | 0.0612 (6) | |
| H1 | −0.0287 | 0.4448 | 0.8874 | 0.073* | |
| C2 | −0.0533 (3) | 0.5469 (3) | 0.7396 (3) | 0.0782 (8) | |
| H2 | −0.1613 | 0.5752 | 0.7672 | 0.094* | |
| C3 | 0.0283 (3) | 0.5822 (3) | 0.6265 (3) | 0.0793 (8) | |
| H3 | −0.0262 | 0.6340 | 0.5788 | 0.095* | |
| C4 | 0.1878 (3) | 0.5431 (3) | 0.5823 (2) | 0.0629 (6) | |
| H4 | 0.2411 | 0.5676 | 0.5058 | 0.075* | |
| C5 | 0.2670 (2) | 0.4661 (2) | 0.65526 (19) | 0.0444 (4) | |
| C6 | 0.1862 (2) | 0.42945 (19) | 0.76880 (18) | 0.0443 (4) | |
| C7 | 0.2853 (2) | 0.34936 (19) | 0.83467 (17) | 0.0420 (4) | |
| C8 | 0.4562 (2) | 0.33081 (19) | 0.76494 (16) | 0.0378 (4) | |
| C9 | 0.4971 (3) | 0.4035 (2) | 0.84540 (19) | 0.0498 (5) | |
| H9 | 0.4533 | 0.3873 | 0.9344 | 0.060* | |
| C10 | 0.6786 (3) | 0.3230 (3) | 0.7914 (2) | 0.0643 (6) | |
| H10 | 0.7241 | 0.3823 | 0.7485 | 0.077* | |
| H11 | 0.7118 | 0.2898 | 0.8595 | 0.077* | |
| C11 | 0.5959 (2) | 0.17760 (19) | 0.73600 (16) | 0.0378 (4) | |
| C12 | 0.5630 (2) | 0.1056 (2) | 0.85934 (18) | 0.0448 (4) | |
| C13 | 0.6002 (2) | −0.03474 (19) | 0.69931 (17) | 0.0396 (4) | |
| C14 | 0.6242 (2) | 0.07240 (18) | 0.63836 (16) | 0.0358 (4) | |
| C15 | 0.6818 (2) | 0.0617 (2) | 0.50663 (17) | 0.0394 (4) | |
| H15 | 0.7009 | 0.1311 | 0.4639 | 0.047* | |
| C16 | 0.7104 (2) | −0.0557 (2) | 0.43996 (18) | 0.0439 (4) | |
| C17 | 0.6825 (2) | −0.1597 (2) | 0.5010 (2) | 0.0491 (5) | |
| H17 | 0.7014 | −0.2364 | 0.4533 | 0.059* | |
| C18 | 0.6265 (2) | −0.1503 (2) | 0.6330 (2) | 0.0492 (5) | |
| H18 | 0.6074 | −0.2198 | 0.6755 | 0.059* | |
| C19 | 0.8815 (3) | 0.0837 (3) | 0.6825 (3) | 0.0811 (8) | |
| H22A | 0.9067 | 0.0101 | 0.6227 | 0.122* | |
| H22B | 0.8732 | 0.0515 | 0.7634 | 0.122* | |
| H19 | 0.9647 | 0.1094 | 0.6496 | 0.122* | |
| C20 | 0.4289 (4) | 0.5589 (3) | 0.8460 (3) | 0.0688 (7) | |
| C21 | 0.1907 (6) | 0.7722 (3) | 0.8925 (5) | 0.1379 (18) | |
| H20A | 0.1867 | 0.8208 | 0.9620 | 0.165* | |
| H20B | 0.2502 | 0.7944 | 0.8122 | 0.165* | |
| C22 | 0.0345 (5) | 0.8180 (3) | 0.9074 (5) | 0.1294 (15) | |
| H21A | −0.0217 | 0.9186 | 0.9177 | 0.194* | |
| H21B | −0.0205 | 0.7869 | 0.9820 | 0.194* | |
| H21C | 0.0389 | 0.7797 | 0.8328 | 0.194* |
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0577 (3) | 0.0831 (4) | 0.0385 (3) | −0.0329 (3) | −0.0163 (2) | −0.0048 (2) |
| S1 | 0.0478 (3) | 0.0544 (3) | 0.0399 (3) | −0.0309 (2) | −0.0221 (2) | 0.0220 (2) |
| O1 | 0.0585 (9) | 0.0682 (10) | 0.0430 (8) | −0.0321 (8) | −0.0069 (7) | 0.0171 (7) |
| O2 | 0.0964 (12) | 0.0631 (9) | 0.0399 (8) | −0.0478 (9) | −0.0388 (8) | 0.0224 (7) |
| O3 | 0.158 (2) | 0.0845 (14) | 0.163 (2) | −0.0951 (16) | −0.0874 (18) | 0.0504 (14) |
| O4 | 0.1093 (16) | 0.0397 (9) | 0.0970 (14) | −0.0343 (10) | −0.0535 (12) | 0.0116 (8) |
| N1 | 0.0641 (11) | 0.0461 (9) | 0.0387 (9) | −0.0316 (8) | −0.0234 (8) | 0.0172 (7) |
| N2 | 0.0486 (9) | 0.0683 (11) | 0.0483 (9) | −0.0363 (9) | −0.0274 (8) | 0.0151 (8) |
| C1 | 0.0441 (11) | 0.0570 (13) | 0.0658 (14) | −0.0223 (10) | −0.0099 (10) | −0.0021 (11) |
| C2 | 0.0431 (12) | 0.0802 (18) | 0.101 (2) | −0.0206 (12) | −0.0317 (14) | 0.0097 (16) |
| C3 | 0.0645 (16) | 0.0875 (19) | 0.095 (2) | −0.0303 (14) | −0.0514 (16) | 0.0261 (16) |
| C4 | 0.0618 (14) | 0.0712 (15) | 0.0665 (14) | −0.0327 (12) | −0.0387 (12) | 0.0247 (12) |
| C5 | 0.0468 (11) | 0.0438 (10) | 0.0456 (11) | −0.0231 (9) | −0.0211 (9) | 0.0070 (8) |
| C6 | 0.0425 (10) | 0.0387 (10) | 0.0453 (11) | −0.0183 (8) | −0.0138 (9) | −0.0021 (8) |
| C7 | 0.0460 (10) | 0.0368 (10) | 0.0355 (10) | −0.0215 (8) | −0.0084 (8) | 0.0001 (8) |
| C8 | 0.0491 (10) | 0.0410 (10) | 0.0314 (9) | −0.0281 (8) | −0.0180 (8) | 0.0115 (7) |
| C9 | 0.0785 (14) | 0.0518 (12) | 0.0426 (11) | −0.0435 (11) | −0.0336 (10) | 0.0153 (9) |
| C10 | 0.0840 (17) | 0.0814 (16) | 0.0663 (15) | −0.0578 (14) | −0.0467 (13) | 0.0207 (12) |
| C11 | 0.0434 (10) | 0.0463 (10) | 0.0322 (9) | −0.0260 (8) | −0.0197 (8) | 0.0136 (7) |
| C12 | 0.0554 (11) | 0.0462 (11) | 0.0364 (10) | −0.0252 (9) | −0.0234 (9) | 0.0144 (8) |
| C13 | 0.0366 (9) | 0.0421 (10) | 0.0389 (10) | −0.0175 (8) | −0.0174 (8) | 0.0087 (8) |
| C14 | 0.0322 (8) | 0.0405 (9) | 0.0348 (9) | −0.0167 (7) | −0.0159 (7) | 0.0069 (7) |
| C15 | 0.0341 (9) | 0.0495 (11) | 0.0367 (9) | −0.0211 (8) | −0.0163 (8) | 0.0084 (8) |
| C16 | 0.0334 (9) | 0.0574 (12) | 0.0380 (10) | −0.0188 (9) | −0.0153 (8) | 0.0004 (8) |
| C17 | 0.0448 (11) | 0.0481 (11) | 0.0536 (12) | −0.0193 (9) | −0.0226 (9) | −0.0035 (9) |
| C18 | 0.0494 (11) | 0.0431 (11) | 0.0589 (13) | −0.0238 (9) | −0.0254 (10) | 0.0100 (9) |
| C19 | 0.0530 (14) | 0.104 (2) | 0.091 (2) | −0.0317 (14) | −0.0412 (14) | 0.0100 (16) |
| C20 | 0.116 (2) | 0.0585 (14) | 0.0698 (16) | −0.0582 (16) | −0.0565 (16) | 0.0242 (12) |
| C21 | 0.190 (5) | 0.0420 (16) | 0.224 (5) | −0.044 (2) | −0.142 (4) | 0.033 (2) |
| C22 | 0.142 (4) | 0.060 (2) | 0.162 (4) | −0.028 (2) | −0.071 (3) | 0.034 (2) |
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Geometric parameters (Å, º)
| Cl1—C16 | 1.7448 (19) | C8—C11 | 1.559 (3) |
| S1—C5 | 1.755 (2) | C9—C20 | 1.499 (3) |
| S1—C8 | 1.8308 (17) | C9—C10 | 1.519 (3) |
| O1—C7 | 1.198 (2) | C9—H9 | 0.9800 |
| O2—C12 | 1.221 (2) | C10—H10 | 0.9700 |
| O3—C20 | 1.199 (3) | C10—H11 | 0.9700 |
| O4—C20 | 1.318 (3) | C11—C14 | 1.506 (2) |
| O4—C21 | 1.454 (3) | C11—C12 | 1.568 (2) |
| N1—C12 | 1.344 (3) | C13—C18 | 1.374 (3) |
| N1—C13 | 1.399 (2) | C13—C14 | 1.392 (2) |
| N1—H1N | 0.81 (2) | C14—C15 | 1.378 (2) |
| N2—C19 | 1.453 (3) | C15—C16 | 1.384 (3) |
| N2—C10 | 1.459 (3) | C15—H15 | 0.9300 |
| N2—C11 | 1.460 (2) | C16—C17 | 1.377 (3) |
| C1—C2 | 1.376 (4) | C17—C18 | 1.382 (3) |
| C1—C6 | 1.388 (3) | C17—H17 | 0.9300 |
| C1—H1 | 0.9300 | C18—H18 | 0.9300 |
| C2—C3 | 1.379 (4) | C19—H22A | 0.9600 |
| C2—H2 | 0.9300 | C19—H22B | 0.9600 |
| C3—C4 | 1.374 (3) | C19—H19 | 0.9600 |
| C3—H3 | 0.9300 | C21—C22 | 1.402 (6) |
| C4—C5 | 1.390 (3) | C21—H20A | 0.9700 |
| C4—H4 | 0.9300 | C21—H20B | 0.9700 |
| C5—C6 | 1.388 (3) | C22—H21A | 0.9600 |
| C6—C7 | 1.460 (3) | C22—H21B | 0.9600 |
| C7—C8 | 1.543 (3) | C22—H21C | 0.9600 |
| C8—C9 | 1.558 (3) | ||
| C5—S1—C8 | 93.13 (9) | N2—C11—C8 | 100.04 (14) |
| C20—O4—C21 | 116.5 (3) | C14—C11—C8 | 119.28 (14) |
| C12—N1—C13 | 111.90 (16) | N2—C11—C12 | 114.02 (14) |
| C12—N1—H1N | 120.4 (16) | C14—C11—C12 | 101.08 (14) |
| C13—N1—H1N | 127.7 (16) | C8—C11—C12 | 109.68 (14) |
| C19—N2—C10 | 114.16 (18) | O2—C12—N1 | 126.35 (17) |
| C19—N2—C11 | 115.48 (18) | O2—C12—C11 | 125.72 (17) |
| C10—N2—C11 | 107.82 (16) | N1—C12—C11 | 107.87 (15) |
| C2—C1—C6 | 119.0 (2) | C18—C13—C14 | 122.24 (17) |
| C2—C1—H1 | 120.5 | C18—C13—N1 | 127.88 (17) |
| C6—C1—H1 | 120.5 | C14—C13—N1 | 109.75 (16) |
| C1—C2—C3 | 119.9 (2) | C15—C14—C13 | 119.77 (17) |
| C1—C2—H2 | 120.0 | C15—C14—C11 | 130.86 (16) |
| C3—C2—H2 | 120.0 | C13—C14—C11 | 109.00 (15) |
| C4—C3—C2 | 122.0 (2) | C14—C15—C16 | 117.82 (17) |
| C4—C3—H3 | 119.0 | C14—C15—H15 | 121.1 |
| C2—C3—H3 | 119.0 | C16—C15—H15 | 121.1 |
| C3—C4—C5 | 118.1 (2) | C17—C16—C15 | 122.18 (18) |
| C3—C4—H4 | 120.9 | C17—C16—Cl1 | 118.56 (15) |
| C5—C4—H4 | 120.9 | C15—C16—Cl1 | 119.27 (15) |
| C6—C5—C4 | 120.30 (19) | C16—C17—C18 | 120.25 (19) |
| C6—C5—S1 | 114.19 (14) | C16—C17—H17 | 119.9 |
| C4—C5—S1 | 125.49 (17) | C18—C17—H17 | 119.9 |
| C5—C6—C1 | 120.6 (2) | C13—C18—C17 | 117.71 (18) |
| C5—C6—C7 | 113.80 (17) | C13—C18—H18 | 121.1 |
| C1—C6—C7 | 125.61 (19) | C17—C18—H18 | 121.1 |
| O1—C7—C6 | 126.40 (18) | N2—C19—H22A | 109.5 |
| O1—C7—C8 | 121.59 (18) | N2—C19—H22B | 109.5 |
| C6—C7—C8 | 112.01 (15) | H22A—C19—H22B | 109.5 |
| C7—C8—C9 | 113.63 (15) | N2—C19—H19 | 109.5 |
| C7—C8—C11 | 115.91 (14) | H22A—C19—H19 | 109.5 |
| C9—C8—C11 | 100.13 (14) | H22B—C19—H19 | 109.5 |
| C7—C8—S1 | 106.50 (12) | O3—C20—O4 | 124.3 (3) |
| C9—C8—S1 | 110.40 (12) | O3—C20—C9 | 124.8 (3) |
| C11—C8—S1 | 110.23 (12) | O4—C20—C9 | 110.9 (2) |
| C20—C9—C10 | 114.2 (2) | C22—C21—O4 | 110.4 (3) |
| C20—C9—C8 | 114.23 (17) | C22—C21—H20A | 109.6 |
| C10—C9—C8 | 104.53 (16) | O4—C21—H20A | 109.6 |
| C20—C9—H9 | 107.9 | C22—C21—H20B | 109.6 |
| C10—C9—H9 | 107.9 | O4—C21—H20B | 109.6 |
| C8—C9—H9 | 107.9 | H20A—C21—H20B | 108.1 |
| N2—C10—C9 | 105.75 (16) | C21—C22—H21A | 109.5 |
| N2—C10—H10 | 110.6 | C21—C22—H21B | 109.5 |
| C9—C10—H10 | 110.6 | H21A—C22—H21B | 109.5 |
| N2—C10—H11 | 110.6 | C21—C22—H21C | 109.5 |
| C9—C10—H11 | 110.6 | H21A—C22—H21C | 109.5 |
| H10—C10—H11 | 108.7 | H21B—C22—H21C | 109.5 |
| N2—C11—C14 | 113.34 (15) | ||
| C6—C1—C2—C3 | 0.5 (4) | S1—C8—C11—N2 | 71.69 (14) |
| C1—C2—C3—C4 | −0.3 (5) | C7—C8—C11—C14 | 68.6 (2) |
| C2—C3—C4—C5 | −0.2 (4) | C9—C8—C11—C14 | −168.71 (15) |
| C3—C4—C5—C6 | 0.5 (3) | S1—C8—C11—C14 | −52.40 (18) |
| C3—C4—C5—S1 | 179.1 (2) | C7—C8—C11—C12 | −47.1 (2) |
| C8—S1—C5—C6 | −4.41 (16) | C9—C8—C11—C12 | 75.54 (16) |
| C8—S1—C5—C4 | 177.0 (2) | S1—C8—C11—C12 | −168.15 (12) |
| C4—C5—C6—C1 | −0.3 (3) | C13—N1—C12—O2 | −170.6 (2) |
| S1—C5—C6—C1 | −179.02 (16) | C13—N1—C12—C11 | 6.5 (2) |
| C4—C5—C6—C7 | −179.70 (19) | N2—C11—C12—O2 | 49.3 (3) |
| S1—C5—C6—C7 | 1.6 (2) | C14—C11—C12—O2 | 171.21 (19) |
| C2—C1—C6—C5 | −0.2 (3) | C8—C11—C12—O2 | −62.0 (2) |
| C2—C1—C6—C7 | 179.1 (2) | N2—C11—C12—N1 | −127.93 (18) |
| C5—C6—C7—O1 | −177.65 (19) | C14—C11—C12—N1 | −5.98 (19) |
| C1—C6—C7—O1 | 3.0 (3) | C8—C11—C12—N1 | 120.84 (17) |
| C5—C6—C7—C8 | 2.9 (2) | C12—N1—C13—C18 | 171.55 (19) |
| C1—C6—C7—C8 | −176.43 (18) | C12—N1—C13—C14 | −4.3 (2) |
| O1—C7—C8—C9 | −63.4 (2) | C18—C13—C14—C15 | −2.4 (3) |
| C6—C7—C8—C9 | 116.04 (17) | N1—C13—C14—C15 | 173.75 (15) |
| O1—C7—C8—C11 | 51.8 (2) | C18—C13—C14—C11 | −176.13 (16) |
| C6—C7—C8—C11 | −128.75 (16) | N1—C13—C14—C11 | 0.0 (2) |
| O1—C7—C8—S1 | 174.81 (16) | N2—C11—C14—C15 | −46.9 (2) |
| C6—C7—C8—S1 | −5.73 (18) | C8—C11—C14—C15 | 70.5 (2) |
| C5—S1—C8—C7 | 5.57 (13) | C12—C11—C14—C15 | −169.32 (18) |
| C5—S1—C8—C9 | −118.22 (15) | N2—C11—C14—C13 | 125.89 (16) |
| C5—S1—C8—C11 | 132.08 (13) | C8—C11—C14—C13 | −116.74 (16) |
| C7—C8—C9—C20 | −78.9 (2) | C12—C11—C14—C13 | 3.47 (18) |
| C11—C8—C9—C20 | 156.84 (19) | C13—C14—C15—C16 | 1.4 (2) |
| S1—C8—C9—C20 | 40.7 (2) | C11—C14—C15—C16 | 173.56 (17) |
| C7—C8—C9—C10 | 155.62 (16) | C14—C15—C16—C17 | 0.3 (3) |
| C11—C8—C9—C10 | 31.38 (17) | C14—C15—C16—Cl1 | −179.61 (13) |
| S1—C8—C9—C10 | −84.80 (16) | C15—C16—C17—C18 | −1.0 (3) |
| C19—N2—C10—C9 | −153.17 (19) | Cl1—C16—C17—C18 | 178.82 (14) |
| C11—N2—C10—C9 | −23.4 (2) | C14—C13—C18—C17 | 1.6 (3) |
| C20—C9—C10—N2 | −132.10 (19) | N1—C13—C18—C17 | −173.82 (18) |
| C8—C9—C10—N2 | −6.6 (2) | C16—C17—C18—C13 | 0.1 (3) |
| C19—N2—C11—C14 | −59.7 (2) | C21—O4—C20—O3 | 3.5 (4) |
| C10—N2—C11—C14 | 171.34 (16) | C21—O4—C20—C9 | −176.8 (3) |
| C19—N2—C11—C8 | 172.22 (17) | C10—C9—C20—O3 | −0.9 (4) |
| C10—N2—C11—C8 | 43.22 (18) | C8—C9—C20—O3 | −121.1 (3) |
| C19—N2—C11—C12 | 55.3 (2) | C10—C9—C20—O4 | 179.43 (19) |
| C10—N2—C11—C12 | −73.7 (2) | C8—C9—C20—O4 | 59.2 (3) |
| C7—C8—C11—N2 | −167.27 (14) | C20—O4—C21—C22 | 162.3 (3) |
| C9—C8—C11—N2 | −44.62 (16) |
(II) Ethyl (2S*,2'R*)-5''-chloro-1'-methyl-2'',3-dioxo-2,3-dihydrodispiro[1-benzothiophene-2,3'-pyrrolidine-2',3''-indoline]-4'-carboxylate . Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C1–C6 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O2i | 0.81 (2) | 2.03 (2) | 2.842 (2) | 172 (2) |
| C18—H18···O3ii | 0.93 | 2.57 | 3.496 (3) | 171 |
| C2—H2···Cgiii | 0.93 | 2.83 | 3.649 (2) | 155 |
Symmetry codes: (i) −x+1, −y, −z+2; (ii) x, y−1, z; (iii) −x, −y, −z+1.
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Bruker (2004). APEX2, SAINT and XPREP. Bruker AXS Inc., Madison, Wisconsin, USA.
- Cordell, G. (1981). In Introduction to Alkaloids: A Biogenic Approach. New York: Wiley International.
- Govind, M. M., Selvanayagam, S., Velmurugan, D., Ravikumar, K., Sridhar, G. & Raghunathan, R. (2003). Acta Cryst. E59, o1438–o1440.
- Huryn, D. M., Trost, B. M. & Fleming, I. (1991). Comp. Org. Synth. 1, 64–74.
- Kannan, P. S., Lanka, S., Thennarasu, S., Vimala, G. & SubbiahPandi, A. (2013a). Acta Cryst. E69, o854–o855. [DOI] [PMC free article] [PubMed]
- Kannan, P. S., Yuvaraj, P. S., Manivannan, K., Reddy, B. S. R. & SubbiahPandi, A. (2013b). Acta Cryst. E69, o825–o826. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Sheldrick, G. M. (1996). SADABS. University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Suzuki, H., Aoyagi, S. & Kibayashi, C. (1994). Tetrahedron Lett. 35, 6119–6122.
- Waldmann, H. (1995). Synlett. pp. 133–141.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S1600536814015426/su2728sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814015426/su2728Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S1600536814015426/su2728IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report