Abstract
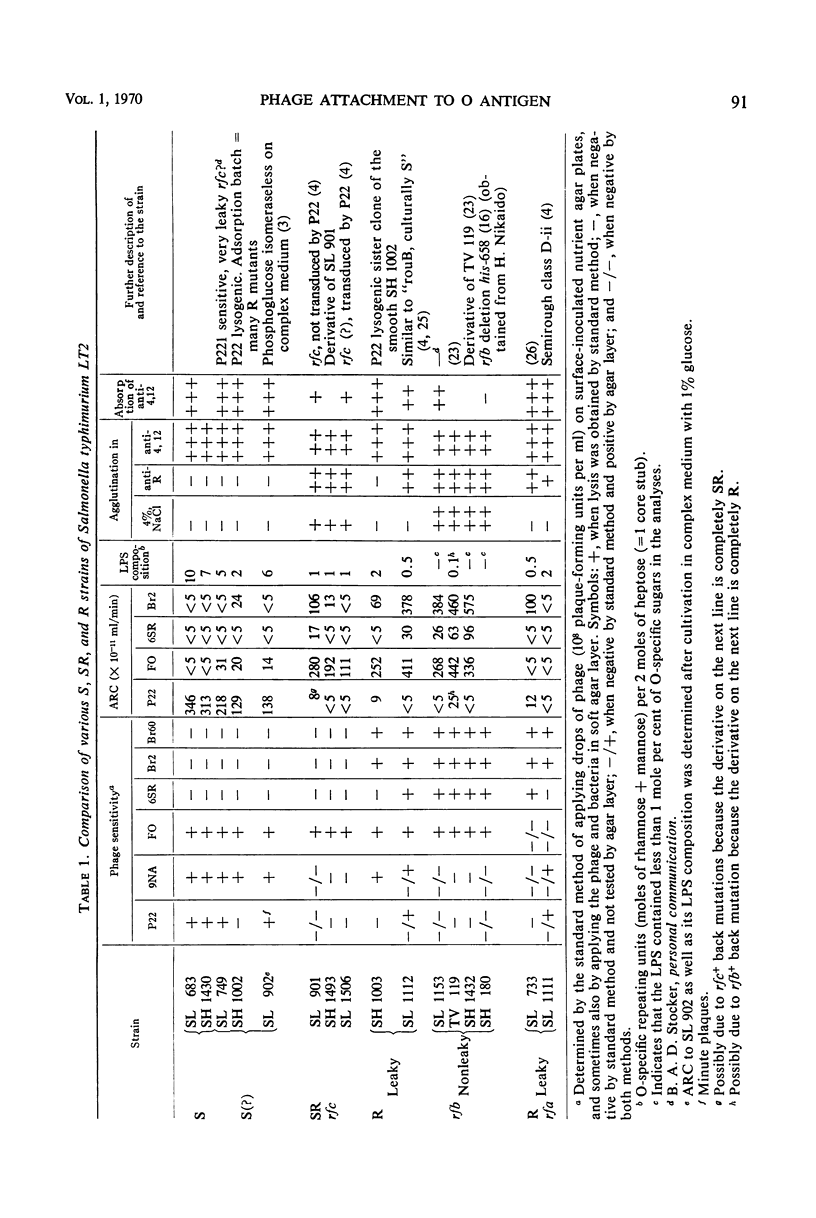
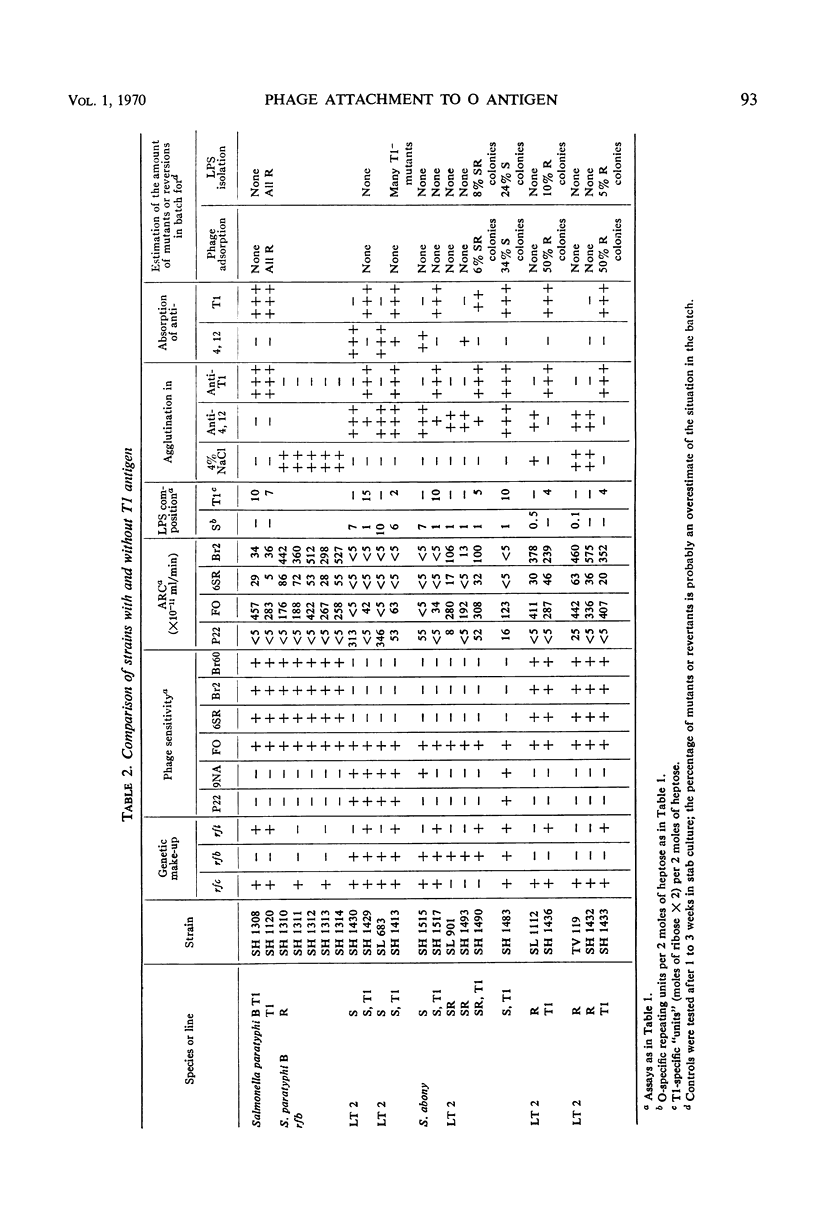
The phage adsorption ability and serological specificity of different Salmonella strains having either complete or leaky mutations in their lipopolysaccharide (LPS) synthesis were compared, together with their genotype and sugar composition, to provide a set of standards relating these parameters to LPS structure. Strains that had T1-specific side chains in their LPS, both with or without O side chains, were examined to learn more about the organization of these two side chains in the LPS and a possible competition between them. It was found that (i) adsorption of O-specific antibodies was a very sensitive test for the presence of even very small amounts of O-specific structures, (ii) that phage P22 adsorption was dependent on the presence of a nearly complete O side chain complement, and both long and numerous O side chains were required, and (iii) that the adsorption of the phages FO (Felix O-1), 6SR, and Br2, which attach to structures in the LPS core, was a sensitive indicator of any defect in O-antigen synthesis, and well developed O side chains blocked their attachment efficiently. Semirough (SR) strains with only one O-specific repeating unit per side chain adsorbed FO efficiently, whereas the access of the 6SR and Br2 phages to their receptors was blocked. Strains with T1 side chains adsorbed the FO and 6SR phages efficiently, whereas the adsorption of the Br2 phage was blocked to a large extent. The phage adsorption of four S, T1 strains (with both O and T1 side chains) showed that, as the amount of O side chain material increased, there was a reduction of the adsorption of phages in the following order: 6SR, Br2, and FO. P22 attachment appeared with the increase of O side chains. The LPS composition of these strains revealed a 10-fold reduction of the O-specific structures compared to the smooth parent strain, whereas the amount of T1-specific material was the same as in T1 strains. The short O side chains of a SR, T1 strain were, however, not reduced in number, suggesting that the apparent competition between O and T1 side chains may not be a competition for available sites in the LPS.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BECKMANN I., SUBBAIAH T. V., STOCKER B. A. ROUGH MUTANTS OF SALMONELLA TYPHIMURIUM. II. SEROLOGICAL AND CHEMICAL INVESTIGATIONS. Nature. 1964 Mar 28;201:1299–1301. doi: 10.1038/2011299a0. [DOI] [PubMed] [Google Scholar]
- FRAENKEL D., OSBORN M. J., HORECKER B. L., SMITH S. M. Metabolism and cell wall structure of a mutant of Salmonella typhimurium deficient in phosphoglucose isomerase. Biochem Biophys Res Commun. 1963 Jun 20;11:423–428. doi: 10.1016/0006-291x(63)90086-x. [DOI] [PubMed] [Google Scholar]
- Gemski P., Jr, Stocker B. A. Transduction by bacteriophage P22 in nonsmooth mutants of Salmonella typhimurium. J Bacteriol. 1967 May;93(5):1588–1597. doi: 10.1128/jb.93.5.1588-1597.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUFFMANN F. A new antigen of Salmonella paratyphi B and Salmonella typhi murium. Acta Pathol Microbiol Scand. 1956;39(4):299–304. doi: 10.1111/j.1699-0463.1956.tb03405.x. [DOI] [PubMed] [Google Scholar]
- KAY D., BRADLEY D. E. The structure of bacteriophage phi R. J Gen Microbiol. 1962 Feb;27:195–200. doi: 10.1099/00221287-27-2-195. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Holme T. Influence of O side chains on the attachment of the Felix O-1 bacteriophage to Salmonella bacteria. J Bacteriol. 1969 Aug;99(2):513–519. doi: 10.1128/jb.99.2.513-519.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. A. Studies of a receptor for felix O-1 phage in Salmonella minnesota. J Gen Microbiol. 1967 Aug;48(2):225–233. doi: 10.1099/00221287-48-2-225. [DOI] [PubMed] [Google Scholar]
- NAIDE Y., NIKAIDO H., MAEKELAE P. H., WILKINSON R. G., STOCKER B. A. SEMIROUGH STRAINS OF SALMONELLA. Proc Natl Acad Sci U S A. 1965 Jan;53:147–153. doi: 10.1073/pnas.53.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Levinthal M., Nikaido K., Nakane K. Extended deletions in the histidine-rough-B region of the Salmonella chromosome. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1825–1832. doi: 10.1073/pnas.57.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Naide Y., Mäkelä P. H. Biosynthesis of O-antigenic polysaccharides in Salmonella. Ann N Y Acad Sci. 1966 Jun 30;133(2):299–314. doi: 10.1111/j.1749-6632.1966.tb52373.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Structure of cell wall lipopolysaccharide from Salmonella typhimurium. I. Linkage between o side chains and R core. J Biol Chem. 1969 Jun 10;244(11):2835–2845. [PubMed] [Google Scholar]
- SUBBAIAH T. V., STOCKER B. A. ROUGH MUTANTS OF SALMONELLA TYPHIMURIUM. I. GENETICS. Nature. 1964 Mar 28;201:1298–1299. doi: 10.1038/2011298a0. [DOI] [PubMed] [Google Scholar]
- Sarvas M. Inheritance of Salmonella T1 antigen. Ann Med Exp Biol Fenn. 1967;45(4):447–471. [PubMed] [Google Scholar]
- Sarvas M., Lüderitz O., Westphal O. Immunochemical studies on T1,S hybrids of Salmonella paratyphi--B. Ann Med Exp Biol Fenn. 1967;45(2):117–126. [PubMed] [Google Scholar]
- Sarvas M., Mäkelä P. H. The production, by recombination, of Salmonella forms with both T-1 and O specifities. Acta Pathol Microbiol Scand. 1965;65(4):654–656. doi: 10.1111/apm.1965.65.4.654. [DOI] [PubMed] [Google Scholar]
- Shands J. W., Jr, Graham J. A., Nath K. The morphologic structure of isolated bacterial lipopolysaccharide. J Mol Biol. 1967 Apr 14;25(1):15–21. doi: 10.1016/0022-2836(67)90275-6. [DOI] [PubMed] [Google Scholar]
- Shands J. W. Localization of Somatic Antigen on Gram-Negative Bacteria by Electron Microscopy. J Bacteriol. 1965 Jul;90(1):266–270. doi: 10.1128/jb.90.1.266-270.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieu J. F., Croissant O., Dauguet C. Structure des bactériophages responsables des phénomènes de conversion chez les Salmonella. Ann Inst Pasteur (Paris) 1965 Jul;109(1):160–166. [PubMed] [Google Scholar]
- Wilkinson R. G., Stocker B. A. Genetics and cultural properties of mutants of Salmonella typhimurium lacking glucosyl or galactosyl lipopolysaccharide transferases. Nature. 1968 Mar 9;217(5132):955–957. doi: 10.1038/217955a0. [DOI] [PubMed] [Google Scholar]
- YAMAMOTO N., ANDERSON T. F. Genomic masking and recombination between serologically unrelated phages P22 and P221. Virology. 1961 Aug;14:430–439. doi: 10.1016/0042-6822(61)90334-8. [DOI] [PubMed] [Google Scholar]
- ZINDER N. D., LEDERBERG J. Genetic exchange in Salmonella. J Bacteriol. 1952 Nov;64(5):679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZINDER N. D. Lysogenization and superinfection immunity in Salmonella. Virology. 1958 Apr;5(2):291–326. doi: 10.1016/0042-6822(58)90025-4. [DOI] [PubMed] [Google Scholar]



