Abstract
Background:
For the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections and decolonization of MRSA carriers, the use of mupirocin a topical antibiotic is increasing day by day.
Aim:
The present study was carried out to determine the prevalence rate of high-level and low-level mupirocin resistant MRSA isolates among patients admitted to a tertiary care hospital.
Materials and Methods:
This is a prospective study carried out on MRSA isolated from the various clinical specimens from outpatient and inpatient departments during period of one year. A total of 82 MRSA isolates were recovered from 6468 different clinical specimens. Mupirocin resistant MRSA was detected by two different methods: Epsilometer test (E-test) and agar dilution method. D-shaped zone test (D-zone test) was also performed for determination of inducible clindamycin resistance in MRSA isolates.
Results:
Out of 82 non-duplicate MRSA isolates mupirocin resistance were found in 15 (18.3%) isolates by both E-test and agar dilution method. Of these 15 mupirocin resistant, 8 (53.3%) isolates were high-level resistant (MuH) and 7 (46.7%) isolates were low-level resistant (MuL). Four isolates were D-zone test positive showing simultaneous inducible clindamycin resistance among mupirocin resistant MRSA isolates.
Conclusion:
Higher prevalence of both high-level and low-level of mupirocin resistant MRSA was observed in patient from the population. It is advisable to perform routine test to detect MRSA colonization among health care workers and nasal decolonization to prevent spread of MRSA infections among hospitalized patients.
Keywords: High-level, MRSA, MuH, MuL, Mupirocin resistance, Low-level
Introduction
Staphylococcus aureus infections are one of the most common and serious hospital-acquired infections seen in developing countries.[1,2,3] Various studies have shown an increased prevalence of staphylococcus infections which may be attributed to its carriage in anterior nares and hands of health care workers and patients.[4,5] Along with that drug resistance seen in cases of Staphylococcus aureus infections is a great concern for the clinicians to prevent spread of infections. Methicillin an important drug of penicillin group was commonly used for these infections before the emergence of methicillin-resistant Staphylococcus aureus (MRSA) strains.[6] The important risk factors for development of MRSA are irrational use of antibiotic, prolong duration of hospital stay, nasal and hand carriage in health care staff.[7,8]
Vancomycin and linezolid are commonly used antibiotics for MRSA infections whereas mupirocin a topical antibiotic is used for treatment of skin and soft tissue infections as well as decolonization of carriers.[9,10] It inhibits the protein synthesis by binding specifically to isoleucyl-tRNA synthetase enzyme.[11] The increased pressure of MRSA infections among patients and its carriage in health care staff has led to indiscriminate use of mupirocin which has resulted in emergence of its resistance.[10] Phenotypically mupirocin resistant strains are grouped into low-level (MuL) resistance and high-level (MuH) resistance phenotypes.[11] The minimum inhibitory concentrations (MIC) are <4 μg/ml, 8-256 μg/ml and >512 μg/ml for mupirocin sensitive, low-level resistance and high-level resistance respectively.[12] Although there is no guideline for detection of mupirocin resistance, traditionally disc diffusion method using 5 μg and 200 μg mupirocin disc, agar dilution method and broth micro dilution method have been used.[13]
Various studies have suggested that treatment of infections with low-level resistant strains is still possible with normal dosage schedule of 0.2% mupirocin ointment.[14] Whereas, high-level resistant strains are frequently associated with failure of decolonization as well as treatment of skin and soft tissues infections.[15,16,17] Thus the present study was aimed to determine the prevalence of high-level mupirocin resistance strains, low-level mupirocin resistance strains and mupirocin sensitive phenotypes.
Material and Methods
MRSA isolates recovered from clinical specimens such as pus, blood, urine, tracheal aspirate, wounds swab, surgical pus and synovial fluid from patients who attended various outpatient departments or admitted to various wards of Mayo Institute of Medical Sciences, Barabanki, UP, India during one year period from May 2013 to April 2014 were included in the study. Informed consent was obtained from each patient and a detailed clinical history and demographic profile was taken and recorded from the patients whose culture was positive for MRSA.
Isolation and identification of MRSA
Clinical specimens obtained in the microbiology laboratory were processed as per routine microbiological procedures for isolation and identification of Staphylococcus aureus. All the clinical specimens excluding urine were inoculated on 5% sheep blood agar and MacConkey agar media, whereas urine samples were inoculated on Cysteine-lactose electrolyte deficient (CLED) agar media and incubated at 37°C aerobically. The growth was identified as Staphylococcus aureus by using conventional biochemical methods according to standard microbiological techniques.
Detection of MRSA
MRSA was detected by using cefoxitin (30 μg) disc as per Clinical and Laboratory Standards Institute (CLSI) guidelines.[18] A lawn culture was made on Mueller-Hinton agar (MHA) with 4% NaCl from suspension of turbidity equivalent to 0.5 MacFarland Standards from overnight growth in nutrient agar and incubated for 24 hours aerobically at 35°C. After incubation the plates were examined for zone of inhibition. Zone of inhibition ≥22 mm was considered as sensitive and such isolates were excluded from the study, whereas ≤21 was considered as resistant and reported as a MRSA and included in the study.
Antimicrobial susceptibility testing of MRSA
Antimicrobial susceptibility testing was done according to the CLSI guidelines by Kirby-Bauer disc diffusion method for the following antibiotics: Cefuroxime (30 μg), cephalothin (30 μg), clindamycin (2 μg), cotrimoxazole (1.25/23.75 μg), erythromycin (15 μg), oxacillin (1 μg), penicillin (10 units), chloramphenicol (30 μg), linezolid (30 μg), vancomycin (30 μg).[18] Four to five morphological identical colonies picked from overnight growth in nutrient agar, were inoculated into 5 ml of peptone water and incubated at 37°C until turbid and compared with 0.5 McFarland Standards. After standardization of turbidity, using a sterile cotton swab a lawn culture was done on the surface of MHA. Antibiotic discs were applied by pressing gently using a sterile forceps on the surface of media and placed at least 20 mm apart from each other.
Epsilometer test (E-test) for determination of minimum inhibitory concentration for mupirocin
E-test was performed by Kirby Bauer disc-diffusion method as per CLSI guidelines by using HiComb mupirocin strip.[18] Lawn culture was made on the surface of MHA medium. HiComb strip with mupirocin antibiotic ranges from 0.1-240 μg/ml was applied perfectly by gently pressing using a sterile forceps. The plates were then incubated aerobically at 35°C for 24 hours. After incubation plates were examined for the minimum inhibitory concentration (MIC). Isolates with MICs > 512 μg/ml were considered as MuH, those with MICs 8-256 μg/ml were considered as MuL and with <4 μg/ml were considered as mupirocin sensitive.
Agar dilution method for determination of MIC for mupirocin
Agar dilution method was performed by doubling dilution of mupirocin incorporated in MHA plate. A suspension of turbidity equivalent to 0.5 MacFarland Standards was prepared from overnight growth of Staphylococcus aureus on nutrient agar. The surface of each MHA plate was inoculated with 1 μl of suspension and plate was incubated at 37°C aerobically for 24 hours. Plates were examined for growth and compared with positive growth control plate without the antibiotic agent, more than one colony or light film of growth was considered as mupirocin resistant. By this method, more than one bacteria were tested per plate. MIC values were same as used for E-test.
D-shaped zone test (D-test) for determination of inducible clindamycin resistance
D-test was performed for determination of inducible clindamycin resistance in MRSA isolates. While performing antimicrobial susceptibility testing, erythromycin disk was placed in close proximity (20 mm) to clindamycin disk. After 16-18 hours of incubation the plates were observed for flattening of the zone of inhibition adjacent to the erythromycin disk (referred to as a D-zone) which indicates inducible clindamycin resistance. These isolates were reported as resistant to clindamycin. Hazy growth within the zone of inhibition around clindamycin was also considered as clindamycin resistance, even if no D-zone is apparent.[18]
Statistical analysis
All the data were recorded in Microsoft excel sheet. Data were analyzed statistically by using Chi-square test to calculate significant levels. P values <0.05 were considered statistically significant.
Results
A total of 361 Staphylococcus aureus were obtained from 6468 different clinical samples. Among 361 Staphylococcus aureus, 82 isolates were MRSA. MRSA isolates were obtained in highest number from pus (72%) followed by sputum (8.5%), urine (7.3%) and blood (6.1%). Distribution of Staphylococcus aureus and MRSA on the basis of source of specimen is listed in Table 1.
Table 1.
Distribution of Staphylococcus aureus and MRSA on the basis of source
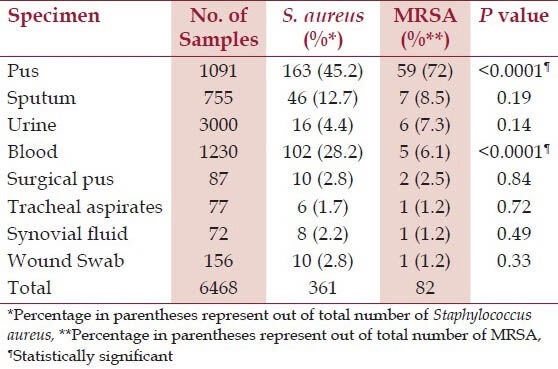
Out of 82 MRSA isolates, mupirocin resistance were seen in 15 (18.3%) isolates by both E-test and agar dilution method. Of these 15 mupirocin resistant MRSA, 8 (53.3%) isolates were MuH and 7 (46.7%) isolates were MuL [Table 2]. Four isolates were D-zone test positive showing simultaneous inducible clindamycin resistance among mupirocin resistant MRSA isolates.
Table 2.
Mupirocin resistant Staphylococcus aureus among MRSA isolates (n = 82)
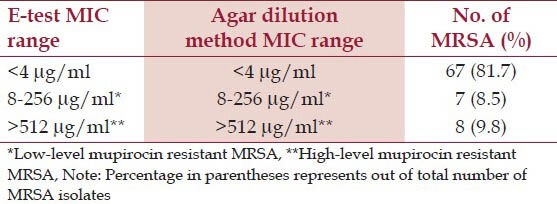
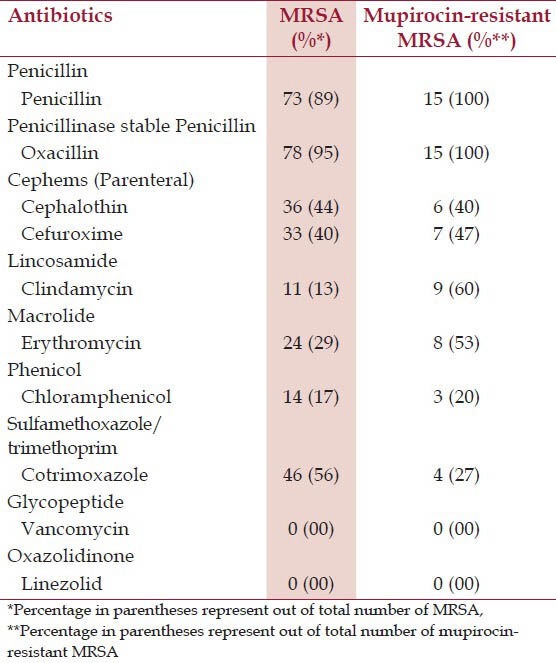
Table 3 shows the antimicrobial susceptibility profile of MRSA isolates. In this study, it is observed that MRSA isolates were resistant to 88.4%, 55.9%, 44.2%, and 40.3% of penicillin, cotrimoxazole, cephalothin, and cefuroxime respectively. Vancomycin and linezolid were uniformily sensitive to all MRSA isolates.
Table 3.
Antimicrobial resistance pattern of MRSA and Mupirocin-resistant MRSA isolates (n = 82)
Discussion
MRSA is one of the leading cause of infections among health care staff and hospitalized patients. It also causes community associated infections. It causes a wide range of infections such as abscesses, impetigo, cellulitis, deep seated pyogenic lesions, meningitis, septicaemia, and pneumonia.[19] Prevalence of MRSA infections may increase because of improper hand hygiene and handling of MRSA carrier patients. Mupirocin is a commonly used antibiotic for decolonization of MRSA in carriers and for treatment of skin and soft tissue infections caused by MRSA.[9,10] Emergence of mupirocin resistance due to its irrational use for treatment of skin and soft tissue infections is further worsening the problem of MRSA infections. Studies also suggest that mupirocin resistance may be transferred from the commensal flora of skin to MRSA during mupirocin therapy.[20] In this study, a total of 82 (22.7%) MRSA isolates were obtained from 361 Staphylococcus aureus. The prevalence is quite low as compared to several other studies conducted in this region.[1,21] This may be attributed to lesser exposure to antibiotics due to low-level of healthcare facilities in this region in past years.
Among 82 MRSA isolates, 15 (18.3%) isolates were mupirocin resistant which is high as compared to other studies conducted in this region.[22,23] The reason for higher prevalence of mupirocin resistance may be an increased use of mupirocin ointment for skin and soft tissue infections. Eight (9.7%) MuH and 7 (8.5%) MuL MRSA isolates determined by two different MIC methods were comparable. Whereas MuL isolates may be treated with normal dosage schedule of mupirocin ointment, but MuH has been found to be associated with treatment and decolonization failure.[14,15,16,17]
In this study 4 (4.9%) mupirocin resistant MRSA isolates were showing D-test positive indicating inducible clindamycin resistance. This is low in comparison to several studies conducted in different regions.[24,25,26] Antibiotic susceptibility pattern of MRSA and mupirocin-resistant MRSA isolates showed no significant association of methicillin resistance and mupirocin resistance with resistance to other antibiotics in this study.
Conclusion
The study has demonstrated a higher prevalence of both MuH and MuL in MRSA isolates which is a serious challenge to the clinicians to deal with increasing problem of MRSA transmission in the hospital. This may be due to an increased prevalence of MRSA infections in the health care setup and over the counter sale of drugs. Mupirocin resistance along with MRSA resistance may enhance the spread of infections. Thus it is advisable to routinely perform nasal decolonization of health care workers to prevent spread of infections among hospitalized patients and to detect mupirocin resistance in MRSA strains isolated from the carriers so that alternative decolonization methods may be used.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Joshi S, Ray P, Manchanda V, Bajaj J, Chitnis DS, Gautam V, et al. Methicillin-resistant Staphylococcus aureus (MRSA) in India: Prevalence and susceptibility pattern. Indian J Med Res. 2013;137:363–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Dubey D, Rath S, Sahu MC, Pattnaik L, Debata NK, Padhy RN. Surveillance of infection status of drug resistant Staphylococcus aureus in an Indian teaching hospital. Asian Pac J Trop Dis. 2013;3:133–42. [Google Scholar]
- 3.Kaur H, Purwar S, Saini A, Kaur H, Karadesai SG, Kholkute SD, Roy S. Status of Methicillin-resistant Staphylococcus aureus Infections and Evaluation of PVL Producing Strains in Belgaum, South India. JKIMSU. 2012;1:43–51. [Google Scholar]
- 4.Sivaraman K, Venkataraman N, Cole AM. Staphylococcus aureus Nasal Carriage and its Contributing Factors. Future Microbiol. 2009;4:999–1008. doi: 10.2217/fmb.09.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–62. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 6.Rayner C, Munckhof WJ. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern Med J. 2005;35:S3–16. doi: 10.1111/j.1444-0903.2005.00976.x. [DOI] [PubMed] [Google Scholar]
- 7.Klutymans J, van Balkum A, Verbrughi H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms and associated risk. Clin Microbiol Rev. 1997;10:505–20. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graffunder EM, Venezia RA. Risk factors associated with nosocomial methicillin-resistant Staphylococcus aureus (MRSA) infection including previous use of antimicrobials. J Antimicrob Chemother. 2002;49:999–1005. doi: 10.1093/jac/dkf009. [DOI] [PubMed] [Google Scholar]
- 9.Rodvold KA, McConeghy KW. Methicillin-resistant Staphylococcus aureus therapy: Past, present, and future. Clin Infect Dis. 2014;58:S20–7. doi: 10.1093/cid/cit614. [DOI] [PubMed] [Google Scholar]
- 10.Hogue JS, Buttke P, Braun LE, Fairchok MP. Mupirocin resistance related to increasing mupirocin use in clinical isolates of methicillin-resistant Staphylococcus aureus in a pediatric population. J Clin Microbiol. 2010;48:2599–600. doi: 10.1128/JCM.02118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagisawa T, Lee JT, Wu HC, Kawakami M. Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J Biol Chem. 1994;269:24304–9. [PubMed] [Google Scholar]
- 12.de Oliveira NE, Cardozo AP, Marques De A, dos Santos KR, Giambiagi-deMarval M. Interpretive criteria to differentiate low- and high-level mupirocin resistance in Staphylococcus aureus. J Med Microbiol. 2007;56:937–9. doi: 10.1099/jmm.0.46965-0. [DOI] [PubMed] [Google Scholar]
- 13.Patel JB, Gorwitz RJ, Jernigan JB. Mupirocin resistance. Clin Infect Dis. 2009;49:935–41. doi: 10.1086/605495. [DOI] [PubMed] [Google Scholar]
- 14.Hudson IR. The efficacy of intranasal mupirocin in the prevention of staphylococcal infections: A review of recent experience. J Hosp Infect. 1994;27:81–98. doi: 10.1016/0195-6701(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 15.Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus: Does mupirocin remain effective? Infect Control Hosp Epidemiol. 2003;24:342–6. doi: 10.1086/502218. [DOI] [PubMed] [Google Scholar]
- 16.Simor AE, Phillips E, McGeer A, Konvalinka A, Loeb M, Devlin HR, et al. Randomized controlled trial of chlorhexidine gluconate for washing, intranasal mupirocin, and rifampin and doxycycline versus no treatment for the eradication of methicillin-resistant Staphylococcus aureus colonization. Clin Infect Dis. 2007;44:178–85. doi: 10.1086/510392. [DOI] [PubMed] [Google Scholar]
- 17.Robicsek A, Beaumont JL, Thomson RB, Jr, Govindarajan G, Peterson LR. Topical therapy for methicillin-resistant Staphylococcus aureus colonization: Impact on infection risk. Infect Control Hosp Epidemiol. 2009;30:623–32. doi: 10.1086/597550. [DOI] [PubMed] [Google Scholar]
- 18.CLSI. CLSI document M100-S23. Vol. 33. Wayne: Clinical and Laboratory Standards Institute; 2013. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement; pp. 74–85. [Google Scholar]
- 19.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:S350–9. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurdle JG, O Neill AJ, Mody L, Chopra I, Bradley SF. In vivo transfer of high-level mupirocin resistance from Staphylococcus epidermidis to methicillin-resistant Staphylococcus aureus associated with failure of mupirocin prophylaxis. J Antimicrob Chemother. 2005;56:1166–8. doi: 10.1093/jac/dki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora S, Devi P, Arora U, Devi B. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in a tertiary care hospital in northern India. J Lab Physicians. 2010;2:78–81. doi: 10.4103/0974-2727.72154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh AK, Venkatesh V, Singh M. Mupirocin resistance in clinical isolates of Staphylococcus aureus in a tertiary care hospital set up in north India. Int J Med Res Health Sci. 2013;2:840–7. [Google Scholar]
- 23.Gadepalli R, Dhawan B, Mohanty S, Kapil A, Das BK, Chaudhry R, et al. Mupirocin resistance in Staphylococcus aureus in an Indian hospital. Diagn Microbiol Infect Dis. 2007;58:125–7. doi: 10.1016/j.diagmicrobio.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Gadepalli R, Dhawan B, Mohanty S, Kapil A, Das BK, Chaudhry R. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus. Indian J Med Res. 2006;123:571–3. [PubMed] [Google Scholar]
- 25.Gupta V, Datta P, Rani H, Chander J. Inducible clindamycin resistance in Staphylococcus aureus: A Study from North India. J Postgrad Med. 2009;55:176–9. doi: 10.4103/0022-3859.57393. [DOI] [PubMed] [Google Scholar]
- 26.Prabhu K, Rao S, Rao V. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. J Lab Physicians. 2011;3:25–7. doi: 10.4103/0974-2727.78558. [DOI] [PMC free article] [PubMed] [Google Scholar]


