Abstract
Huntington's disease (HD) is caused by cytosine-adenine-guanine (CAG) repeat expansions in the huntingtin (Htt) gene. Although early energy metabolic alterations in HD are likely to contribute to later neurodegenerative processes, the cellular and molecular mechanisms responsible for these metabolic alterations are not well characterized. Using the BACHD mice that express the full-length mutant huntingtin (mHtt) protein with 97 glutamine repeats, we first demonstrated localized in vivo changes in brain glucose use reminiscent of what is observed in premanifest HD carriers. Using biochemical, molecular, and functional analyses on different primary cell culture models from BACHD mice, we observed that mHtt does not directly affect metabolic activity in a cell autonomous manner. However, coculture of neurons with astrocytes from wild-type or BACHD mice identified mutant astrocytes as a source of adverse non-cell autonomous effects on neuron energy metabolism possibly by increasing oxidative stress. These results suggest that astrocyte-to-neuron signaling is involved in early energy metabolic alterations in HD.
Keywords: astrocyte, glucose metabolism, Huntington's disease, oxidative stress
Introduction
Huntington's disease (HD) is an autosomal dominant neurodegenerative disease caused by a cytosine-adenine-guanine (CAG) repeat expansion in the first exon of the HTT gene that encodes huntingtin (Htt). HD is characterized by involuntary abnormal movements and postures (chorea, dyskinesia, and dystonia) associated with psychiatric disturbances and cognitive deficits.1 The most striking neuropathological hallmark of this disorder is atrophy of the striatum2 with preferential loss of GABAergic medium-size spiny neurons. Other regions such as cortex, hypothalamus, and hippocampus also undergo degeneration in the course of the disease.
The proposed mechanisms by which mutant Htt (mHtt) is neurotoxic include transcriptional modulation, protein aggregation, excitotoxicity, and mitochondrial dysfunction.3 Several lines of evidence support the notion that defective energy metabolism significantly contributes to the pathogenesis of HD.4 In HD patients, there is strong evidence for reduced glucose consumption in the brain, especially in basal ganglia5 and even in presymptomatic mutation carriers.6 At the cellular level, mitochondrial abnormalities are a major cause of energy deficiency triggered by mHtt.7 Studies of striatal samples from late-stage HD patients revealed reduced activity for almost all components of the oxidative phosphorylation pathway.8 These results point to the existence of cell autonomous mechanisms by which mHtt compromises mitochondria bioenergetics and dynamics in medium-size spiny neurons. There is also some evidence that mHtt can impair cellular bioenergetics by affecting extra-mitochondrial pathways.9, 10 In addition, expression of mHtt in non-neuronal cells such as astrocytes may also exacerbate HD neuropathology.11, 12, 13, 14, 15 Astrocytes regulate energy metabolism by providing neuronal mitochondria with energy substrates derived from glycolysis.16 Mutant Htt is expressed in astrocytes in HD patients,13 but very little is known regarding its contribution to brain energy metabolism dysregulation. A positron emission tomography study performed in HD patients reported a selective reduction in striatal glucose consumption (CMRglu) without any change in oxygen consumption (CMRO2), suggesting a selective defect in glycolytic flux rather than a reduced cerebral oxidative phosphorylation in HD17 and therefore raising the possibility that astrocytes may be involved in those metabolic defects.
To better understand whether astrocyte–neuron interactions contribute to brain energy deficits in HD, we performed in vivo measurement of glucose uptake in a mouse model of HD expressing the full-length human mutant Htt (BACHD mice) and in vitro characterization of the metabolic profile of BACHD neurons and astrocytes. Our results strongly suggest that HD astrocytes are the source of adverse non-cell autonomous effects on neuron energy metabolism.
Materials and methods
Animals
BACHD founder mice expressing expanded human Htt with 97 mixed CAA-CAG repeats were kindly provided by Dr William Yang (University of California, Los Angeles, CA, USA). These mice were bred with FVB/NJ mice (Taconic, Bomholt, Denmark) to generate male and female BACHD and wild-type (WT) littermates. To determine mice genotype, extraction of genomic DNA was performed using the REDExtract-N-Amp tissue PCR kit (Sigma-Aldrich, Lyon, France). A PCR was performed using the following primers HTT5: 5′-gagccatgattgtgctatcg-3′, HTT3: 5′-agctacgctgctcacagaaa-3′. All animal experimental procedures were fully compliant with the French regulation (Code Rural R214/87 to R214/130), the recommendations of the EEC (86/609/EEC) for care and use of laboratory animals, and conformed to the ethical guidelines of the French National Charter on the ethic of animal experimentation. The animal facility is accredited by the French authorities (Veterinary Inspectors) under the number B9-032-02.
In Vivo [14C]-2-Deoxyglucose Uptake
We measured [14C]-2-Deoxyglucose (2-DG) uptake in six BACHD (64.3±6.1 weeks old, ranging from 56 to 68) and six control (63.4±7.6 weeks old, ranging from 54 to 69) male mice. Experiments were performed in conscious, lightly restrained animals that were previously habituated to the constraint. Animals were fasted for 12 hours before the experiment but had free access to water. On the day of the experiment, mice were anesthetized with isoflurane (2% in O2) and two catheters were inserted into the femoral artery and vein, respectively. All whiskers except the two caudal of row C (C1C2) of both whiskerpads were clipped. Mu metal pieces (1.5 mm long, 0.2 mm diameter) were fastened onto the right C1C2 whiskers with cyanoacrylic glue. Body temperature was maintained at 37 °C. Mice were allowed to recover from anesthesia for 1 hour and were placed into the Lausanne whisker stimulator as previously described.18 The stimulation consisted of magnetic field bursts that were delivered at 50 Hz, during 46 ms with 90-ms interval. Mice were intravenously injected with [14C]-2DG (16.5 μCi/100 g body weight; Perkin Elmer, Boston, MA, USA) 5 minutes after the onset of the stimulation. Glycemia was measured using an automatic blood glucose meter (Onetouch Ultra lifescan, Johnson and Jonhson, New Brunswick, NJ, USA). Mice were stimulated for a period of 50 minutes and then were euthanized by injection of a lethal dose of sodium pentobarbital. The brains were rapidly removed and immediately frozen at −40 °C in isopentane. They were then embedded in a custom-made mixture of M1 embedding matrix (Shandon, Pittsburgh, PA, USA) and Fast Green (Sigma-Aldrich) and cut into 20 μm-thick coronal sections with a CM3050S cryostat (Leica, Rueil-Malmaison, France). Every second section was photographed using a digital camera (high in-plane resolution of 30 × 30 μm2), to serve as a reference for postmortem image three-dimensional (3D) reconstruction. Every second section was mounted on Superfrost glass slides, rapidly heat-dried, and placed against autoradiographic film (Kodak Biomax MR) for 10 days together with radioactive [14C] standards (146C, American Radiochemical Company, St Louis, MO, USA). The same sections were then processed for Nissl staining, to obtain additional information about brain anatomy.
Postmortem Image Registration, Three-Dimensional Reconstruction and Analysis
To be reconstructed in 3D, autoradiographic and Nissl-stained sections were digitized using a high-resolution flatbed scanner (ImageScanner III; GE Healthcare Europe, Orsay, France) at 600 d.p.i. (pixel size 42 × 42 μm2) and 1,200 d.p.i. (pixel size 21 × 21 μm2) in-plane resolution, respectively. Image processing was performed using our in-house software BrainVISA/Anatomist (http://brainvisa.info/). First, blockface photographs were reconstructed in 3D as previously described19 to create a spatially coherent photographic volume of the whole brain (resolution of 0.024 × 0.024 × 0.04 mm3). Each anatomic volume was reconstructed in 3D (resolution of 0.021 × 0.021 × 0.04 mm3) using the corresponding photographic volume as geometrical reference for co-registration. Then, each autoradiographic volume was reconstructed in 3D (resolution of 0.042 × 0.042 × 0.04 mm3) using the corresponding anatomic volume as geometrical reference for co-registration. Autoradiographic images were converted from gray levels to normalized activity (nCi/g) using the [14C] standards. The BrainRAT module of BrainVISA was used to perform the sequential rigid and affine co-registrations as well as the image conversion. Regional differences in cerebral glucose uptake between BACHD transgenic mice and WT mice were then assessed with SPM5 software as previously described,19 the statistical significance being set up at P<0.01 (uncorrected for multiple comparisons) for the whole brain exploration and at P<0.05 for whisker stimulation experiments, and the minimum cluster size being set at 1,000 contiguous voxels (∼0.07 mm3).
Cell Culture
Primary astrocytes
Highly purified astrocytes cultures were prepared from 1- or 2-day-old pups from FVB/NJ or BACHD mice of either sex as previously described.20 Briefly, forebrains were removed aseptically from the skulls, the meninges were carefully excised under a dissecting microscope and the neocortex was dissected free of brainstem, thalamus, striatum, and hippocampus. The cells were dissociated by passage through needles of decreasing gauge size three times with a 5-ml syringe. The cells were seeded on six-well plates in Dulbecco's Modified Eagle's medium (DMEM Life Technologies, Saint Aubin, France) containing 5.5 mM glucose and supplemented with 10% fetal bovine serum, 2 mM L-glutamine (Gibco, Life Technologies) and antibiotics (Penicillin-Streptomycin, Gibco, Life Technologies) in a final volume of 2 ml/well and incubated at 37 °C in an atmosphere containing 5% CO2/95% air. The culture medium was renewed 3 to 4 days after seeding and subsequently three times per week. Cultures were used between 19 to 21 DIV.
Neuron–Astrocyte Cell-Insert Coculture
Astrocytes cultures were prepared as described above. After 10 days in vitro, and 1 week before neuronal culture preparation, astrocytes were treated with trypsin and seeded on cell culture inserts (35-3102, Falcon) with 1 μm pores. Primary culture of mixed cortical and striatal neurons was made from embryonic day 15.5 (E15.5) mouse embryos. Timed pregnant FVB/NJ mice or BACHD mice were killed with a lethal dose of pentobarbital and embryos quickly removed and dissected on cooled Hank's balanced sodium salts without Ca2+ and Mg2+. Cortices and striatum were isolated and incubated for 15 min at 37 °C in 0.3 mg/ml DNase I (Sigma-Aldrich). Tissues were mechanically dissociated with P1000 and P200 pipettes. Cells were finally concentrated by centrifugation (20 °C, 10 minutes, 1,000 g) and resuspended in serum-free Neurobasal medium supplemented with 2% B27 supplement (Invitrogen, Carlsbad, CA, USA), 1% antibiotic–antifungal mixture (Invitrogen) and 0.5 mM L-glutamine (Sigma-Aldrich). Cells were plated at a density of 2 × 105 cells/cm2 in 12- or 24-well plates coated with 50 μg/ml 30 to 70 kDa poly-D-lysine (Sigma-Aldrich). Twenty-four hours after seeding, neuron cultures were treated with 10 μM Cytosine β-D-arabinofuranoside hydrochloride (Sigma-Aldrich) for 24 hours. Astrocytes inserts were placed onto neurons 2 days after neurons seeding. Cultures were used for experiment 13 days after neuron seeding.
In Vitro [14C]-2-Deoxyglucose Uptake
Culture medium was removed and cells were preincubated for 2 hours for astrocytes or 30 minutes for neuron–astrocyte cell-insert coculture in 2 ml of serum-free DMEM (Gibco) supplemented with 5 mM glucose, 44 mM NaHCO3, and antibiotics for astrocytes and with 5 mM glucose, 44 mM NaHCO3, and L-glutamine 0.25% for neurons. Medium was then replaced by 2 ml of DMEM containing [14C]-2DG (final concentration 33 nM) for 20 min. Some astrocytes wells were also treated with glutamate (200 μM) during the same period. The reaction was terminated by aspiration of the medium and cells were rinsed three times with 2 ml of ice-cold phosphate-buffered saline (PBS). Astrocytes were then lysed by addition of 2 ml of 10 mM NaOH containing 0.1% Triton X-100. Neuron–astrocyte cocultures were lysed by addition of 750 μL of the same buffer. A 300 μL aliquot was assessed for [14C] using a liquid scintillation cocktail (Insta-Gel Plus, Perkin Elmer, Courtaboeuf, France) and counted with a liquid scintillation analyzer (Tri-Carb B2910TR, Perkin Elmer). The protein content was measured in 10 μL of the remaining lysate using bicinchoninic acid (BCA kit, Fischer Scientific, Illkirch, France).
Reverse Transcriptase–Quantitative Polymerase Chain Reaction
Mouse glucose metabolism quantitative polymerase chain reaction array on neuron–astrocyte cell-insert coculture
Total neuronal RNA was extracted using TRIzol (Invitrogen) according to21 and genomic DNA was eliminated by two successive DNAse treatments (DNAse RQ1 and DNAse GE, Qiagen, Courtaboeuf, France). cDNA was synthetized from 0.6 μg RNA using RT2 first strand kit (Qiagen, 330401) following manufacturer's instructions. cDNA was mixed with RT2 SYBER Green ROX qPCR Mastermix (Qiagen, 330520) and dispensed in Mouse glucose metabolism qPCR array plate (Qiagen, PAMM-006ZA). For each sample (i.e., plate), we checked that the three quality controls (absence of contamination with genomic DNA, efficiency of reverse transcriptase (RT) and quantitative polymerase chain reactions (qPCRs)) were fulfilled, according to the manufacturer's instructions. The abundance of the gene of interest was normalized to the abundance of the mean of housekeeping genes Actin beta, Gapdh, Hsp90ab, Gusb, and B2m using the ΔCt method. Expression of all housekeeping genes was not different between the four groups. Data were analyzed using MEV software (http://www.tm4.org/mev/).
Custom-Made qPCR Array Plate on 12-Month-Old BACHD Mouse Striata
Mouse brains were rapidly collected and each striatum was dissected out on ice from 1 mm coronal slices and stored in RNA later (Sigma-Aldrich) until further processing. Total RNA was isolated from striatal samples using TRIzol, purified on RNA clean-up columns and residual DNA was digested by on-column treatment with DNAse (Macherey-Nagel, Bethlehem, PA, USA). cDNA was synthesized from 0.5 μg RNA using the RT2 PCR Array First Strand kit from SABiosciences (Qiagen) following manufacturer's instructions. cDNA was mixed with the RT2 Real-Time SYBR Green PCR mix (SABiosciences, Qiagen) and dispensed in a custom-made qPCR array plate containing specific primers for a gene of interest in each well. The abundance of selected genes of interest (mouse Htt, Darpp32, Drd1a, Drd2, penk1, gfap, GS) was normalized to the mean abundance of the housekeeping genes 18s, Gapdh, Hprt1, and Gusb using the ΔCt method. Expression of all housekeeping genes was not different between the two groups.
Reverse transcriptase–quantitative polymerase chain reaction of individual genes
Total mRNA was extracted from BACHD mouse striata or cell culture samples with TRIzol and treated with DNAse RQ1. cDNA was synthesized from 400 ng of total RNA and random oligo primers using the SuperScriptVilocDNA synthesis kit (Invitrogen). Two nanograms of cDNA were used in triplicate to perform real-time PCR using TaqPlatinium enzyme (SYBR Green PCR kit, Invitrogen) and the Master cycle eprealplex (Eppendorf, Le Pecq, France) system. The following primers were used: mouse and human Htt 5′-CCGCTGCACCGACCAAAGAA-3′ (forward) and 5′-ATTTCTGAGAGACTGTGCCA-3′ (reverse); mouse glut3 5′-TGCTGACTGCCCTCTGGTCCTTATG-3′ (forward) and 5′-CGTCTGCCAAAGCGGTTGACA-3′ (reverse); mouse and rat cyclophiline A 5′-ATGGCAAATGCTGGACCAAA-3′ (forward) and 5′-GCCTTCTTTCACCTTCCCAAA-3′ (reverse); mouse pgc1α 5′-CCGCACACACCGCAAT-3′ (forward and 5′- CATCCCTCTTGAGCCTTTCGT-3′ (reverse). A dissociation step was added at the end of the PCR to confirm the amplification reaction of a single product. No-template and no-reverse transcriptase controls were included in each assay and they do not produce detectable signals during 40 cycles of amplification. mRNA levels were normalized to cyclophiline A level.
Histochemistry (COX)
Following an overdose of sodium pentobarbital, 12-month-old mice (six WT and six BACHD) were perfused transcardially with 4% paraformaldehyde in 0.1 M phosphate buffer (PB). Brains were post-fixed by incubation in the same solution overnight and cryoprotected by incubation in a 30% sucrose solution during 48 hours. Floating coronal brain section (40 μm) were cut on a freezing microtome, collected serially, and stored at −20 °C in a storing solution containing ethylene glycol, glycerol, and 0.1 M PB until analysis. Sections were incubated for 2 to 3 hours at 37 °C protected from light in a solution containing 4% sucrose, 0.005% 3,3′-diaminobenzidine tetrahydrochloride dihydrate and 0.003% cytochrome C in 0.1 M PB. Intensity and surface staining were measured by using a computer-based image analysis system (MCID Analysis, St Catharines, Ontario, Canada).
Immunohistochemistry (8-Hydroxydeoxyguanosine)
Half-brains of 12-month-old mice were post-fixed with 4% paraformaldehyde in 0.1 M PB (pH 7.4) and then cryoprotected in 30% sucrose overnight at 4 °C. Free-floating sections were cut using a cryostat. For immunohistochemistry, a modified avidin-biotin-peroxidase technique was employed. Briefly, the sections were pretreated with 3% H2O2 in 0.1 M sodium PBS for 30 minutes. The sections were rinsed in PBS twice for 5 minutes each. The sections were incubated sequentially in (a) 5% bovine serum albumin (BSA)/5% normal goat serum/5% non-fat dry milk/0.2% Triton X-100 (Sigma-Aldrich) for 30 minutes, (b) goat anti-8-OH-dG (anti-8-hydroxydeoxyguanosine) antibody (Millipore, Billerica, MA, USA; diluted at 1:100 in PBS/0.5% BSA) for 18 hours, (c) biotinylated anti-goat IgG (Vector Laboratories, Burlingame, CA, USA; 1:200, diluted in PBS/0.5% BSA) for 1 hour, and (d) avidin-biotin-peroxidase complex (Vector Laboratories; 1:200 in PBS) for 1 hour. The immunoreaction was visualized using 3,3′-diaminobenzidine tetrahydrochloride dihydrate as the chromogen. All incubations and rinses were performed at room temperature with agitation using an orbital shaker. The sections were mounted onto slides, dehydrated, cleared in xylene, and coverslipped.
Western Blots
Cell cultures (astrocytes or neurons) were scraped and homogenized on ice in 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% SDS, with protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Western blots were performed using ECL detection as previously described22 using antibodies against actin (1:2,000, Sigma-Aldrich) and Htt (2b4, 1:1,000, Chemicon). For carbonyl groups detection, the OxyBlot Protein Oxidation Detection kit (Millipore) was used according to manufacturer's instructions on 10 μg of homogenized neuronal cells coming from neuron/astrocyte cell insert coculture samples (DIV13). Samples were loaded onto 12% polyacrylamide gels. Primary antibody dilution was 1:150 in T-TBS BSA 1% and the secondary antibody dilution was 1:300 in T-TBS BSA 1%.
Förster Resonance Energy Transfer Glucose Measurement
HEK293 cells were co-transfected at 60% confluence with a plasmid coding for the glucose sensor FLII12Pglu700μδ623 and with pARIS-htt, a synthetic cDNA coding for a tagged full-length version of human Htt expressing either 23 or 100Q24 using Lipofectamine 2000 and studied after 48 hours. Cells were imaged at room temperature (22 to 25 °C) in a 95% air/5% CO2-gassed solution of the following composition (in mM): 112 NaCl, 1.25 CaCl2, 1.25 MgSO4, 5 KCl, 5 glucose, 10 HEPES, 24 NaHCO3, pH 7.4, using an upright Olympus FV1000 Confocal Microscope equipped with a × 20 water immersion objective (N.A. 1.0) and a 440 nm solid-state laser.
Live Imaging of Astrocyte–Neuron Insert-Cell Cocultures
After 13DIV, astrocyte–neuron insert-cell coculture medium was replaced by serum-free DMEM (Gibco) supplemented with 5 mM glucose, 44 mM NaHCO3, and 0.25% L-glutamine and cells were incubated for 30 minutes at 37 °C. Astrocyte inserts were then removed and cells were incubated for 30 minutes at 37 °C in an atmosphere containing 5% CO2/95% air, in the same medium containing both calcein-AM (2 μM, Invitrogen, 485/530 nm) to assess cell viability, and Rhodamine 123 (0.5 μM, Sigma-Aldrich) to assess mitochondrial membrane potential, or in medium containing both 0.5 μM Rhodamine 123 and carboxy-2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA; 25 μM, Image it live green reactive oxygen species detection kit, Molecular Probes, 495/529 nm) to assess the production of oxidative species in neurons. Some wells were treated with butyl hydroperoxyde (100 μM, Molecular probes, Life Technologies) for 1 hour or 1 hour 30 minutes as positive control of DCF staining. Medium was replaced by a control medium and cells were observed using a Zeiss microscope (Axio Observer Z1, Marly le roi, France) in an atmosphere at 37 °C containing 5% CO2/95% air (chamber PeCon, incubator XL-S1). Pictures were taken with a Hamamatsu camera (C10600, Orca) in four random fields per well. Intensities of calcein-AM and Rhodamine staining and positive DCF cell number were quantified by using Image J software (http://rsb.info.nih.gov/ij/) and ‘ROI manager' plug-in.
Statistical Analysis
Data are expressed as mean±s.e.m., n indicates the number of mice for in vivo experiments and the number of independent pups or embryos for in vitro experiments. In vitro data were repeated at least in three independent cultures. Statistical analyses were performed using paired and unpaired Student's t-test for two groups comparison and one-way or two-way analysis of variance followed by post hoc Dunnett's or Scheffe's test for more than two groups comparison (Statistica, Statsoft, Maisons-Alfort, France).
Results
Neuropathological Status of BACHD Mice
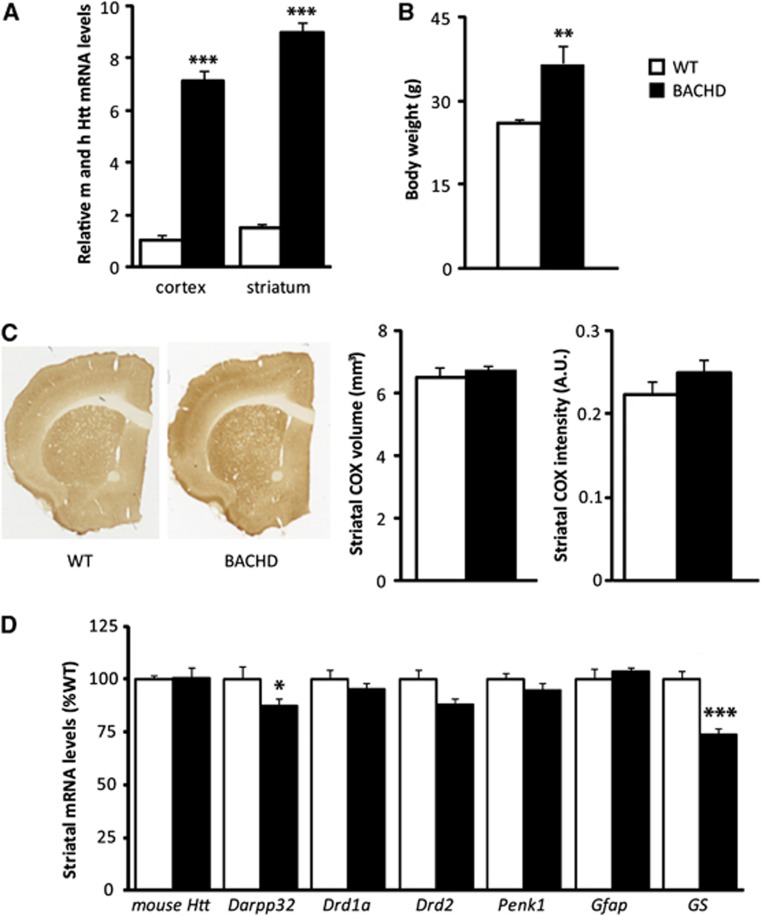
Reverse transcriptase–quantitative polymerase chain reaction was performed on cortical and striatal samples of BACHD and WT to assess mRNA relative quantity of either mouse htt alone or total htt (mouse+human). The level of endogenous htt was unchanged, whereas the level of total htt was significantly higher in the cortex ( × 7) and in the striatum ( × 6) of BACHD mice compared with WT mice (Figures 1A and 1D). The body weight of BACHD mice was significantly higher than WT animals (+35%, Figure 1B) as previously reported.25, 26 Using Cytochrome Oxidase (COX)-stained sections (Figure 1C), we found no evidence for striatal atrophy in 12-month-old BACHD mice. Intensity of COX staining was not different between WT and BACHD mice indicating no major mitochondrial deficits at that age. Using RT–qPCR, we found a significant decrease in darpp32 and GS in BACHD versus WT mice and no significant change for mouse drd1a, drd2, penk1, and gfap transcripts (Figure 1D). These results show that 12-month-old BACHD mice display a mild HD phenotype with no neuronal death and no major striatal dysfunction, modeling the early stage of the disease.
Figure 1.
Characterization of BACHD mice. (A) Levels of total huntingtin (mouse, m and human, h Htt) mRNA in cortical and striatal tissues of 12-month-old BACHD mice were assessed using quantitative reverse transcription polymerase chain reaction (RT–qPCR). Expression level of total Htt was significantly higher in the cortex ( × 7) (WT n=5, BACHD n=6; t-test P=4.0 × 10−7) and in the striatum ( × 6) of BACHD mice (n=6) compared with WT (n=6) (t-test P=1.1 × 10−8). (B) The body weight was significantly higher in BACHD mice (n=6) than in WT animals (n=6) (t-test P=0.01). (C) No significant atrophy of the caudate putamen as measured on COX-stained sections (WT n=6, BACHD n=6; t-test for volume P=0.52 and for intensity P=0.17) and (D) no major transcriptional defects were observed in BACHD mice at 12 months of age except for Darpp32 and GS (WT n=6, BACHD n=6, Darpp32, t-test P=0.021 and GS, t-test P=0.00079). All data represent means±s.e.m. *P<0.05, **P<0.01, and ***P<0.001 compared with WT. a.u., arbitrary unit; WT, wild-type.
Localized Cerebral Metabolic Alterations in BACHD Mice in vivo
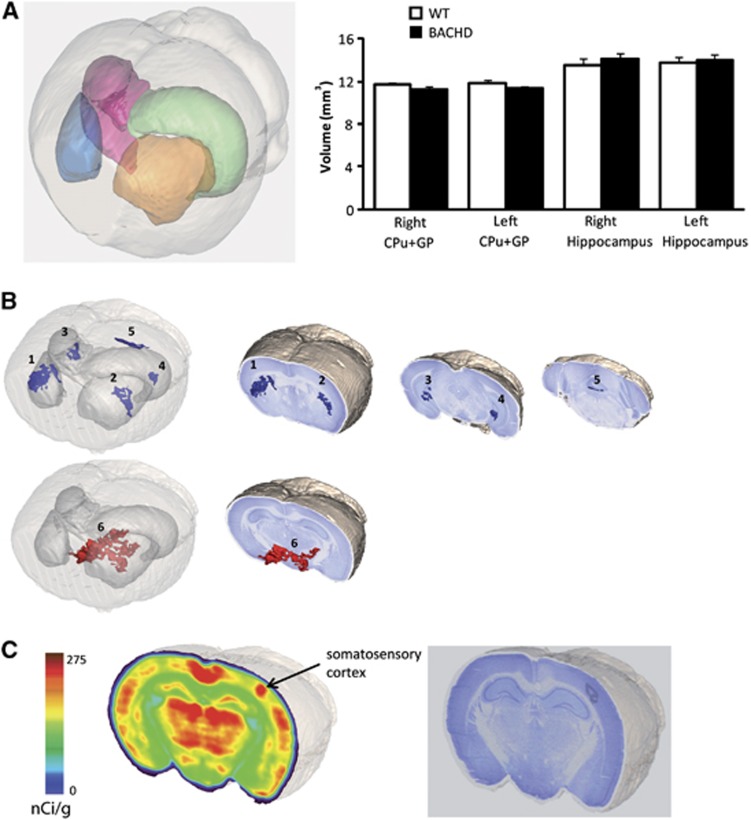
We performed an in vivo 3D autoradiographic measurement of glucose uptake including a voxel-wise statistical analysis to map the cerebral metabolic differences between WT and BACHD mice. Such an approach allows for an unbiased identification of statistical significance at the voxel level between two groups of animals without operator-dependent delineation of regions of interest19 as it is widely employed for human studies using Statistical Parametric Mapping. Thanks to the 3D reconstruction of the whole brain, we calculated the volume of both left and right striatum and pallidum, and hippocampus (Figure 2A). We found no significant differences between BACHD and WT mice, thus confirming our results using COX staining. We found that BACHD mice display very restricted and localized decreases in glucose uptake in the dorsal and lateral part of the caudate putamen (Figure 2B regions 1 and 2), in the posterior part of the hippocampus (dentate gyrus, Figure 2B regions 3 and 4) and in the cerebellum (second lobule, Figure 2B, region 5, Table 1). The dorsal part of the hypothalamus and a fraction of the zona incerta (Figure 2B, region 6) were the only brain regions where the glucose uptake was higher in BACHD mice. These results indicate that BACHD mice expressing the full-length human mHtt display localized metabolic changes reminiscent to what is observed in premanifest HD patients.27 Finally, we found that the metabolic increase due to unilateral whisker stimulation observed in the contralateral somatosensory cortex was higher in BACHD compared with WT mice (Figure 2C, Table 1). We next sought to determine the respective contribution of neurons and astrocytes to these functional changes.
Figure 2.
Alterations of brain energy metabolism in BACHD mice. In vivo glucose uptake was measured using three-dimensional-reconstructed autoradiography and voxel-wise statistical analysis to map the cerebral metabolic differences between 15-month-old WT and BACHD mice. (A) No atrophy was observed in BACHD mice as the volumes of both left and right striatum, and pallidum (CPu+GP), and of the hippocampus were not significantly different between BACHD (n=6) and WT mice (n=6) (t-test, right GPu+GP P=0.16 and left GPu+GP P=0.11; right hippocampus P=0.5 and left hippocampus P=0.77). (B) BACHD mice display localized decreases in glucose uptake in the dorsal and lateral part of the caudate putamen (regions 1 and 2), in the posterior part of the hippocampus (dentate gyrus, regions 3 and 4), and in the cerebellum (second lobule, region 5); and an increase in the dorsal part of the hypothalamus and a fraction of the zona incerta (region 6). (C) The metabolic increase induced by whisker stimulation observed in the corresponding somatosensory cortex was higher in BACHD mice compared with WT. CPu, caudate putamen; GP, globus pallidus; WT, wild-type.
Table 1. Glucose uptake in 15-month-old BACHD and WT mice.
| Region (cluster number) | WT | BACHD | % Change | Statistics (P-value) |
|---|---|---|---|---|
| Caudate putamen (right-1) | 241.0±3.0 | 223.9±2.2 | −7.1 | 0.00094 |
| Caudate putamen (left-2) | 234.8±2.7 | 213.7±3.5 | −9.0 | 0.00074 |
| Dentate gyrus (right-3) | 197.2±1.5 | 182.7±2.1 | −7.4 | 0.00021 |
| Dentate gyrus (left-4) | 196.9±2.0 | 181.3±2.5 | −8.0 | 0.00062 |
| Cerebellum (5) | 179.5±2.7 | 158.6±3.1 | −11.6 | 0.00046 |
| Hypothalamus+zona incerta (6) | 212.1±5.0 | 232.4±1.3 | +9.6 | 7.8 × 10−6 |
| Activated barrel cortex | 231.8±4.5 | 252.6±3.0 | +9.0 | 0.003 |
Regional 2-DG ([14C]-2-Deoxyglucose) uptake values, mean±s.e.m. (nCi/g), percentage difference, and associated P-values measured between 15-month-old BACHD and WT (wild-type) mice, using autoradiography three-dimensional reconstruction and voxel-wise statistical analysis (see Materials and Methods).
Mutant Htt Affects Neuronal Metabolism by a Non-Cell Autonomous Mechanism
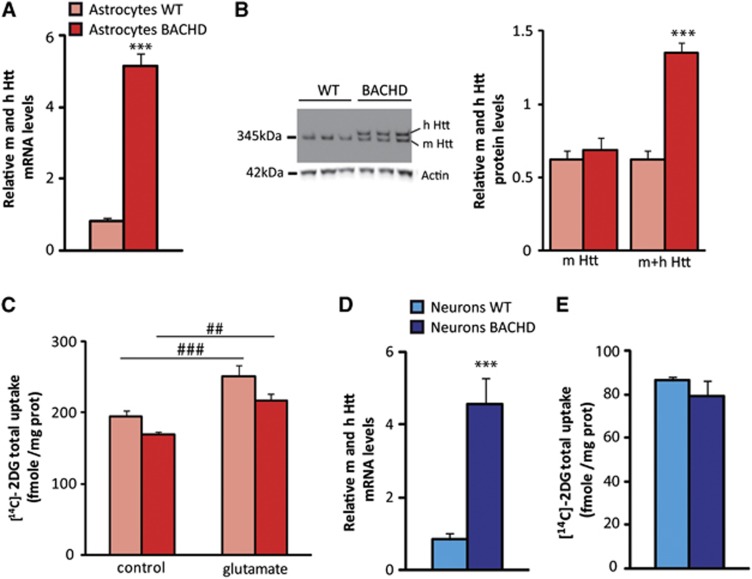
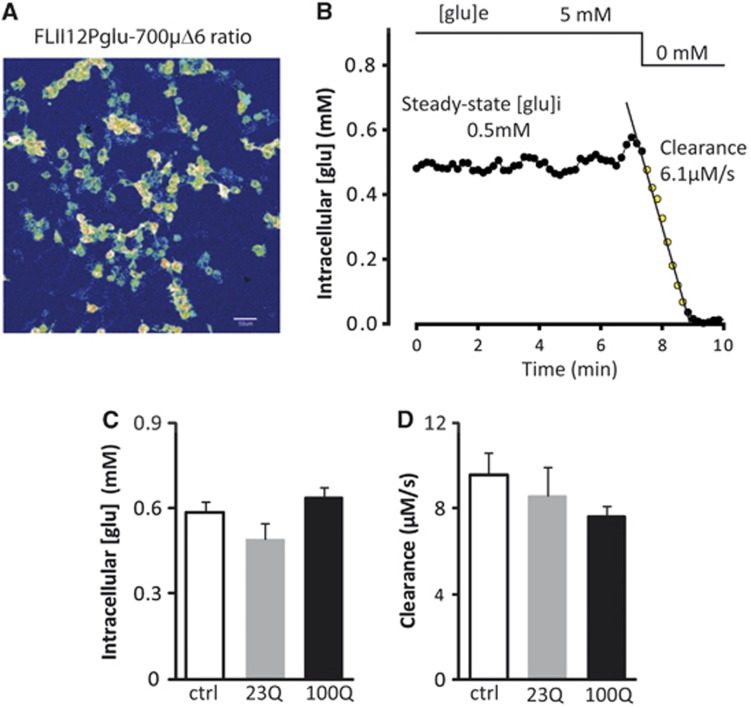
Using RT–qPCR and immunoblotting, we first confirmed that BACHD astrocytes express both human Htt mRNA and protein (Figures 3A and 3B). We then measured and compared glucose uptake in both resting and glutamate-stimulated conditions in those primary astrocytes. Even if a global effect of astrocytes genotype was observed by two-way analysis of variance, glucose uptake in resting and glutamate-stimulated conditions was moderately diminished (−14% in both conditions) but not significantly different between BACHD and WT astrocytes using a post hoc test. The metabolic response induced by glutamate was maintained (+29%) in both conditions (Figure 3C). We next evaluated the metabolic consequences of mHtt expression into neurons. We measured 2DG uptake in a neuron–astrocyte coculture model at 13DIV in which neurons represent the principal cell population as characterized by a very low level of gfap and a high level of fox-3 (NeuN) transcripts (data not shown). Those neuronal cells express mHtt at the mRNA level (Figure 3D) and as for primary astrocytes, we did not find any metabolic changes compared with WT neurons (Figure 3E). To further confirm the absence of a direct metabolic effect of mHtt, glucose homeostasis was evaluated using a Förster Resonance Energy Transfer glucose nanosensor23 that allows the monitoring of glucose concentration in real-time, in single cells, and its response to glucose withdrawal (Figures 4A and 4B). HEK293 cells were co-transfected with a synthetic cDNA encoding a tagged full-length version of human Htt with either 23Q or 100Q.24 As shown in Figure 4C, neither 23Q nor 100Q affected the intracellular concentration of glucose in the steady-state. The steady-state glucose level is determined by the balance between glucose transport and glucose consumption, so that in principle, a modulation of both processes in the same direction and degree may cancel out regarding glucose levels. However, such possibility was discarded by the observation that 23Q and 100Q did not affect the rate of glucose clearance after extracellular glucose withdrawal (see Figure 4D). Altogether, these results mean that mHtt does not alter glucose metabolism in a cell autonomous manner. We next tested the hypothesis that cell–cell interactions were required to explain the metabolic deficit.
Figure 3.
Mutant Htt does not directly affect energy metabolism in both neurons and astrocytes in vitro. Human mutant Htt is ubiquitously expressed in the brain of BACHD mice. (A, D) BACHD astrocytes and neurons express significantly higher levels ( × 5) of Htt mRNA compared with WT as measured by quantitative reverse transcription polymerase chain reaction (RT–qPCR) (astrocytes WT n=12, BACHD n=12, t-test P=1.9 × 10−13; neurons WT n=11, BACHD n=8, t-test P=1.18 × 10−8). (B) The protein level of Htt (human mutated (h) and mouse (m) form) is significantly higher in primary culture of astrocytes prepared from BACHD (n=5) compared with WT mice (n=7) as observed by immunoblotting (mouse Htt WT versus mouse Htt BACHD, t-test P=0.56; mouse+human Htt WT versus mouse+human Htt BACHD, t-test P=1 × 10−4). (C) Even if a global effect of astrocytes genotype was observed by two-way analysis of variance (ANOVA, astrocytes genotype) F(1,60)=8.27, P=0.006, glucose uptake in resting and glutamate-stimulated conditions was not significantly different between BACHD (n=16) and WT astrocytes (n=16; Scheffé post hoc test: WT control versus BACHD control P=0.3, WT glutamate versus BACHD glutamate P=0.1). The metabolic response induced by glutamate was maintained (+29%) in both conditions (two-way ANOVA (glutamate treatment) F(1,60)=25.14; P=1 × 10−5; Scheffé post hoc test: WT control versus WT glutamate P=0.0044, BACHD control versus BACHD glutamate P=0.019). (E) Glucose uptake was not significantly altered in BACHD neurons (n=8) compared with WT (n=7; t-test, P=0.33). All data represent means±s.e.m. #P<0.05 and ##P<0.005 compared with control; ***P<0.001 compared with WT. 2-DG, [14C]-2-Deoxyglucose; Htt, huntingtin; WT, wild-type.
Figure 4.
Mutant Htt does not modify the glycolytic rate in single cells as measured by FRET. (A) FLII12Pglu-700δμ6, a FRET glucose nanosensor was expressed in HEK293 cells. (B) Typical example of real-time measurement of intracellular glucose concentrations and of the rate of glucose clearance after extracellular glucose withdrawal in a single cell. (C, D) Expression of a synthetic cDNA encoding a tagged full-length version of human Htt with either 23Q or 100Q did not affect the intracellular concentration of glucose in the steady-state (one-way analysis of variance (ANOVA), F(2,75)=3.081; P=0.052) and the rate of glucose clearance during glucose deprivation (one-way ANOVA, F(2,74)=0.915; P=0.405). Control: n=25 cells, 23Q: n=25 cells, and 100Q: n=28 cells. All data represent means±s.e.m. FRET, Förster Resonance Energy Transfer; Htt, huntingtin.
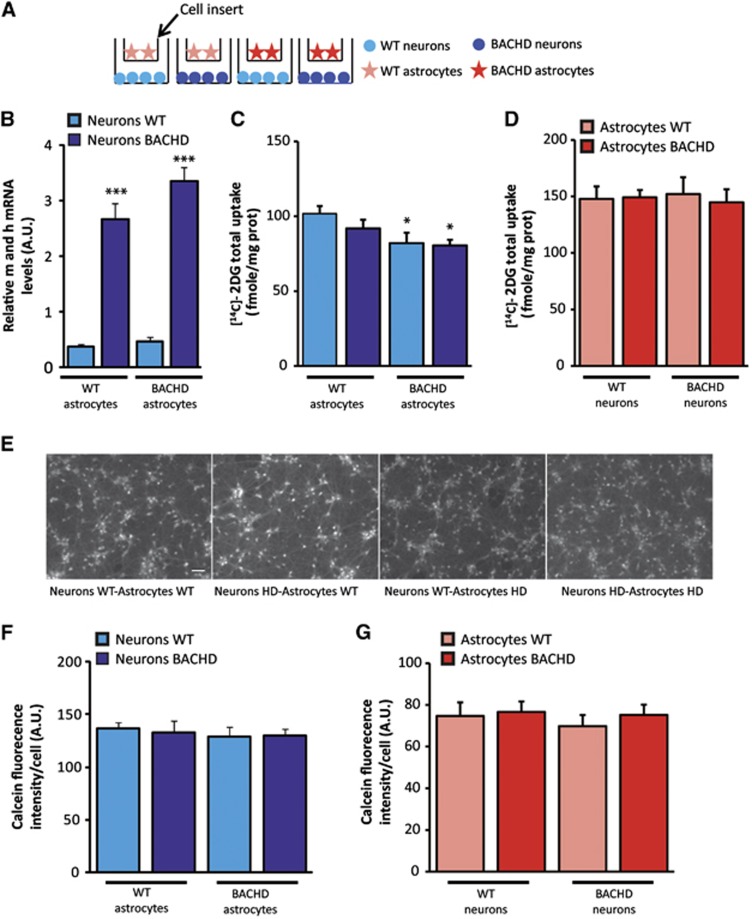
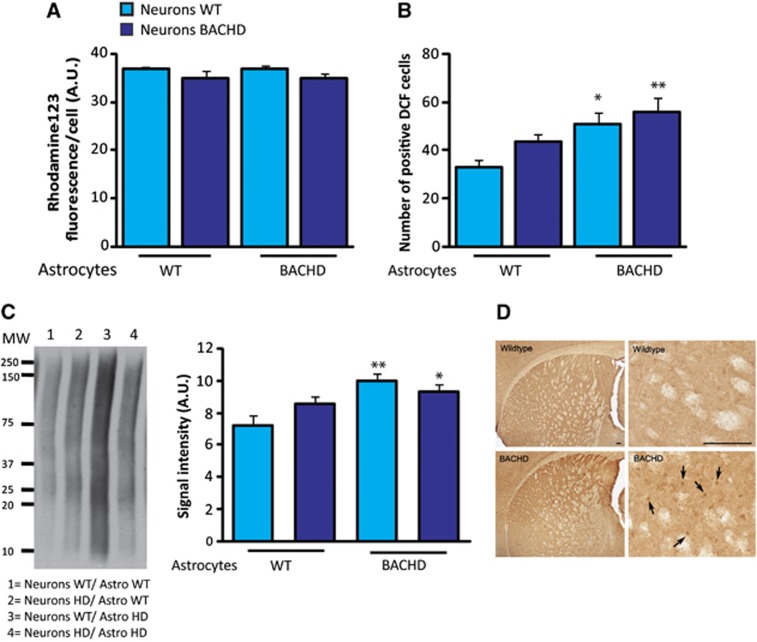
We then developed a neuron–astrocyte cell-insert coculture model system in which astrocytes are seeded on cell culture inserts and can be placed on neurons so that different genotype can be mixed and metabolism can be measured in both cell types independently (Figure 5A). We first verified that the level of mHtt transcripts in neurons was not significantly different depending on the genotype of the astrocytes (Figure 5B). We found that BACHD as well as WT neurons have a reduced glucose uptake only when cultured with BACHD astrocytes whereas neurons genotype does not affect their metabolism (Figure 5C). The uptake of glucose was unchanged in astrocytes in all four conditions (Figure 5D), thus confirming the data obtained in primary astrocytes (Figure 3C). Such functional effect was not the consequence of a major loss of neuronal or astrocytic viability as confirmed by a similar calcein-AM staining performed on live cells in all four conditions (Figures 5E–G). These coculture experiments therefore identified mutant astrocytes as a source of adverse non-cell autonomous effects on neuron energy metabolism via a diffusible factor. To determine whether the glial-derived diffusible factor or its absence affects neuronal metabolic activity via a transcriptional effect, we next examined the expression of 84 key genes involved in the regulation and enzymatic pathways of glucose metabolism using the mouse Glucose Metabolism PCR Array. Expression of all genes involved in glycolysis, TCA cycle, pentose phosphate pathways, gluconeogenesis, and glycogen metabolism was not altered (Figure 6) in all conditions. Using RT–qPCR, we also found that the expression levels of pgc1α and glut3 transcripts were not different in all four conditions (data not shown). These results suggest that mHtt does not directly or indirectly modify the transcriptional activity of the major genes involved in the regulation of neuronal glucose metabolism. We next assessed mitochondrial function in neurons using Rhodamine 123 staining in living neurons. This dye redistributes according to the negative membrane potential across the mitochondrial inner membrane so that a loss of potential will result in the loss of the fluorescence intensity. We did not observe any significant difference in fluorescence intensity/cell between all conditions suggesting that the mitochondrial membrane potential was not affected by the expression of mHtt into neurons (Figure 7A). We next sought to determine whether oxidative stress contributes to the metabolic demise of neurons as excessive oxidative damage is occurring in cells and tissues from HD patients. To address this question, we pursued a series of assays designed to measure oxidative protein damage and oxidative DNA damage. We first used the cell-permeant H2DCFDA as an indicator for reactive oxygen species (ROS) production. We observed a significant increase in ROS production in BACHD neurons and WT neurons cultured with BACHD astrocytes (Figure 7B). Mutant Htt expression into neurons has no effect on oxidative stress. We then performed immunoblot analysis to measure carbonyl adduct formation in neurons and found an increased expression of carbonyl residues of neuronal proteins in the two conditions with HD astrocytes (Figure 7C). Interestingly, we observed an increase in the immunostaining of 8-OH-dG, an indicator of oxidative stress in HD patients,28 in particular, in the dorsal part of the striatum of BACHD mice (Figure 7D), a region where we found in vivo 2DG uptake impairment.
Figure 5.
Astrocytic mHtt is a source of adverse non-cell autonomous effects on neuron energy metabolism via a diffusible factor. (A) Schematic representation of the neuron–astrocyte cell-insert coculture model, astrocytes are seeded on cell culture inserts and are placed on neurons. (B) BACHD neurons express fivefold more mHtt than WT neurons (two-way analysis of variance (ANOVA, neuron genotype), F(1,20)=172.21; P=1 × 10−6). Astrocytes genotype does not modify neuronal mHtt transcript levels (n=6 per condition) (two-way ANOVA (astrocyte genotype), F(1,20)=3.94; P=0.061). (C) BACHD as well as WT neurons have a reduced glucose use only when cultured with BACHD astrocytes (two-way ANOVA (neuron genotype), F(1,45)=0.95; P=0.33; (astrocyte genotype), F(1,45)=6.90; P=0.012) whereas the uptake of glucose was unchanged in the astrocytes in all four conditions (D) (two-way ANOVA (neuron genotype), F(1,45)=5 × 10−5; P=0.99; (astrocyte genotype), F(1,45)=0.07; P=0.79), (neurons WT/astrocytes WT n=15, neurons HD/astrocytes WT n=9, neurons WT/astrocytes HD n=14, neurons HD/astrocytes HD n=11). Such functional effect was not the consequence of a major loss of neuronal or astrocytic viability as confirmed by a similar calcein-AM staining performed on live cells in all four conditions (E–G) (neurons WT/astrocytes WT n=5, neurons HD/astrocytes WT n=5, neurons WT/astrocytes HD n=7, neurons HD/astrocytes HD n=8, scale bar, 100 μm; two-way ANOVA, for neurons: (neuron genotype) F(1,21)=1.25; P=0.28 and (astrocyte genotype) F(1,21)=0.16; P=0.69; for astrocytes: (neuron genotype) F(1,21)=0.057; P=0.94 and (astrocyte genotype) F(1,21)=0.89; P=0.36). All data represent means±s.e.m. *P<0.05 and ***P<0.001 compared with respective control. a.u., arbitrary unit; Htt, huntingtin; WT, wild-type.
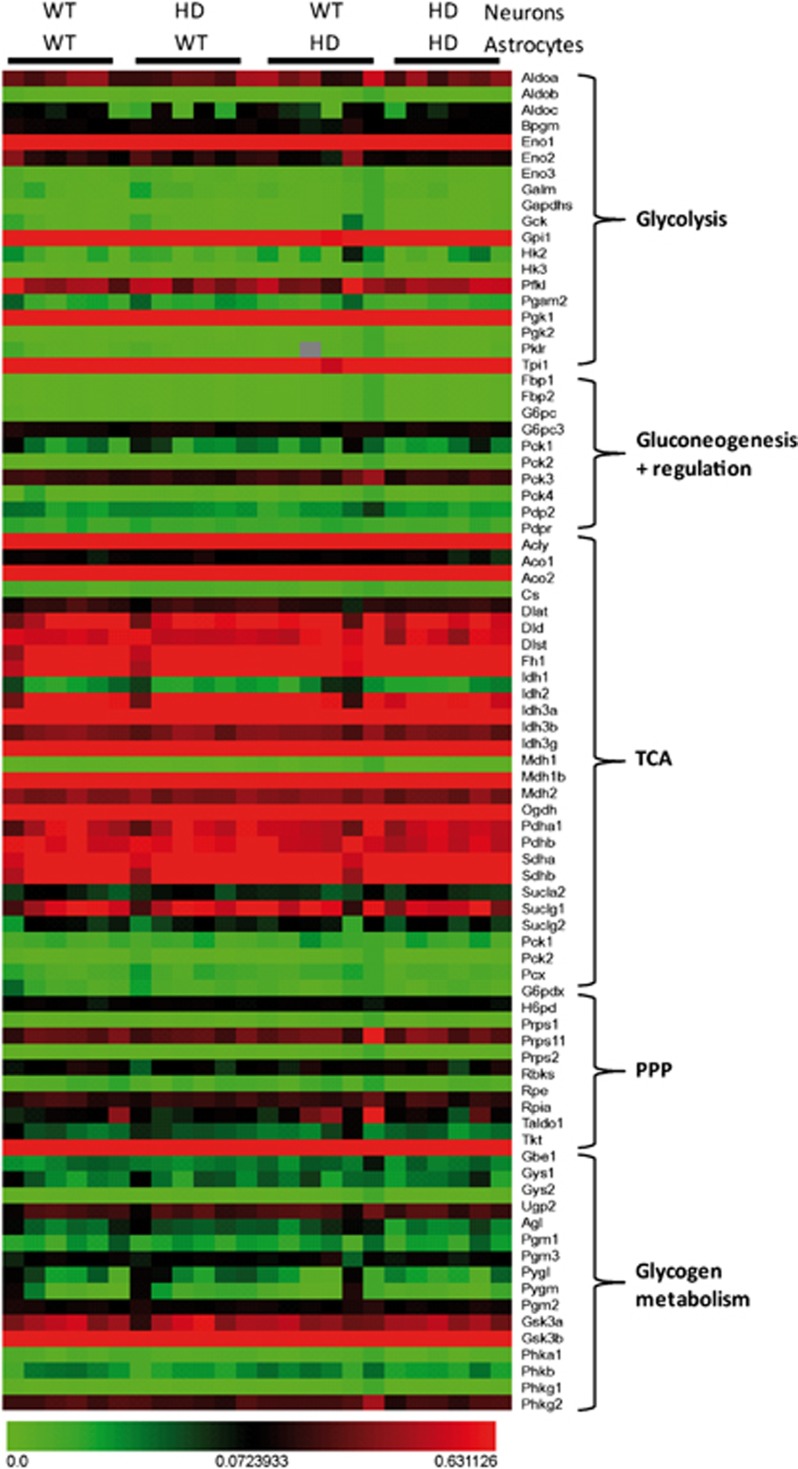
Figure 6.
Mutant Htt expressed in neurons or astrocytes does not modify the transcriptional activity of the major genes involved in the regulation of neuronal glucose metabolism. The expression of 84 key genes involved in the regulation and enzymatic pathways of glucose metabolism was assessed using the mouse Glucose Metabolism PCR Array. Genes expression levels involved in glycolysis, TCA cycle, pentose phosphate pathways (PPP), gluconeogenesis, and glycogen metabolism are unchanged between the four different conditions (n=6 per condition). HD, Huntington's disease; Htt, huntingtin; TCA, tricarboxylic acid; WT, wild-type.
Figure 7.
Astrocytic mHtt increases oxidative stress in neurons. (A) The mitochondrial membrane potential is not affected by the expression of mHtt in neurons as assessed using Rhodamine 123 staining in living neurons (neurons WT/astrocytes WT n=5, neurons HD/astrocytes WT n=5, neurons WT/astrocytes HD n=7, neurons HD/astrocytes HD n=8; (two-way analysis of variance (ANOVA, neuron genotype), F(1,21)=1.10; P=0.31; (astrocyte genotype), F(1,21)=1.80; P=0.19). (B) A significant increase in ROS production was observed in BACHD neurons and WT neurons cultured with BACHD astrocytes using 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), a cell-permeant indicator for ROS (two-way ANOVA (astrocyte genotype), F(1,28)=11.75; P=0.0019) whereas neurons genotype did not affect oxidative stress (two-way ANOVA (neuron genotype), F(1,28)=2.92; P=0.098) (neurons WT/astrocytes WT n=7, neurons HD/astrocytes WT n=7, neurons WT/astrocytes HD n=8, neurons HD/astrocytes HD n=8). (C) Immunoblot of carbonyl adduct formation shows an increased expression of carbonyl residues of neuronal proteins only when neurons are cultured in the presence of HD astrocytes (n=4 per condition, two-way ANOVA (neuron genotype), F(1,12)=0.36; P=0.57; (astrocyte genotype), F(1,12)=11.84; P=0.0049). Immunostaining of 8-hydroxydeoxyguanosine (8-OH-dG) is increased in the dorsal striatum of 12-month-old BACHD mice (arrow) compared with WT mice (D) (n=3 per genotype). All data represent means±s.e.m. *P<0.05 and **P<0.01 compared with WT/WT condition. a.u., arbitrary unit; DCF, 2',7'-dichlorofluorescein; HD, Huntington's disease; ROS, reactive oxygen species; WT, wild-type.
Discussion
Numerous studies have demonstrated that deficits in measures of energy metabolism become manifest in presymptomatic and symptomatic HD brain (see references in Mochel and Haller (2011)4). The shared downstream consequences of the mutant CAG tract and 3-nitropropionic acid (a succinate dehydrogenase inhibitor) as well as the direct association of Htt with mitochondria have implied that the polyglutamine tract in Htt may directly affect the mitochondrial metabolic function. The mechanisms identified until now include impaired oxidative phosphorylation, abnormal mitochondrial calcium handling, abnormal mitochondria trafficking, and deregulation of key factors of mitochondrial biogenesis, such as the transcriptional coactivator PGC-1α.7 In addition to these well-documented effects, we herein provide evidence that astrocytes expressing mutant Htt may be a supplementary source of adverse metabolic events affecting neurons.
Early Metabolic Alterations Of Energy Metabolism In Vivo in BACHD Mice
Using the same experimental paradigm that is used in HD patients to measure brain energy metabolism with [18F]-FDG-positron emission tomography, we found that BACHD mice expressing expanded human Htt with 97 CAA-CAG repeats, do present localized alterations of brain glucose use at 15 months of age. In contrast to others studies,25, 26 we did not find any significant striatal atrophy at that age. We used two different approaches, the first one based on the Cavalieri method to estimate striatal volume after COX staining (Figure 1C) and the second, thanks to the 3D brain reconstruction of Nissl-stained sections allowing segmentation of the entire striatum and pallidum (Figure 2A). Using magnetic resonance imaging, it was shown that BACHD brain volumes were identical to control mice until 65 weeks (Janet Dubinsky, personal communication). Therefore, the metabolic changes we observed cannot be attributed to secondary consequences of major neuronal death in our BACHD colony. In HD patients also, changes in brain metabolism appear to precede significant volumetric changes.6 Very few studies have examined in vivo brain glucose use in mouse models of HD. At variance to what we presently observed in BACHD mice, recently published 2-DG maps from 6-week-old R6/2 mice expressing high levels of Htt exon1 bearing 110 to 150 CAG repeats, demonstrated more widespread (striatum, primary motor cortex, and prefrontal cortex) and more pronounced decreases (up to 30%) in glucose uptake compared with WT mice.29
In HD patients, the more recent positron emission tomography studies have shown reductions in striatal and anterior cingulate metabolic activity, associated with relative metabolic increases in the ventrolateral/ventral posterolateral thalamus, cerebellar vermis, and in the primary motor and visual regions of the cerebral cortex.6 In addition to the expected decrease in the striatum, we found moderate metabolic defects also in the hippocampus and the cerebellum. There is now evidence for hippocampal dysfunction in HD accumulating in recent years because this may contribute to specific cognitive and affective symptoms of the disease. In fact, in multiple transgenic lines of HD mice, adult hippocampal neurogenesis and synaptic plasticity is disrupted30 and emotional as well as cognitive deficits have been recently observed also in adult BACHD mice.31 Concerning the cerebellum, it has been recently shown in R6/2 and HdhQ200 knock-in mouse models of HD a loss of viability and function of cerebellar Purkinje cells that may contribute to motor dysfunction in the course of the disease32 but no information is yet available in BACHD mice. We also observed a higher glucose uptake in the hypothalamus, a region that is also under focus in HD because of the well described neuroendocrine disturbances and also sleep and circadian cycle perturbations that occur early in HD. In this context it is worth noting that a recent study has shown that in BACHD mice, the expression of the mutant Htt in the hypothalamus causes peripheral metabolic abnormalities33 suggestive for a local dysfunction of the hypothalamic circuits. Finally, our intriguing observation of an increased metabolic response to whisker simulation in BACHD mice is supported by functional magnetic resonance imaging data also showing hyper-activations in brain areas of HD patients subjected to various tasks.34
We next sought to investigate the cellular mechanism responsible for these metabolic defects using an in vitro approach and found that mHtt does not induce direct metabolic alteration into neurons but that the mechanisms are driven by mHtt expressed into astrocytes.
Cellular Basis for the Altered Bioenergetics in BACHD
Using different in vitro cell culture models and methodological approaches, we did not find any evidence for a direct functional effect of mHtt on glucose metabolism. Accordingly, mutant Htt did not modify the expression of many genes involved in the regulation and enzymatic pathways of glucose metabolism including PGC1α in BACHD neurons. Indeed, BACHD mice do not display transcriptional alterations of other mRNA enriched in the striatum, such as dopamine receptors D1 and D2 as reported recently.26 Even if mRNA expression was not changed, we cannot rule out that the enzymatic activity may be altered by changes in substrate concentrations.
In addition to neurons, it is now well-accepted that the expression of mHtt in glial cells can contribute to the pathogenesis of HD.11, 12, 13, 14, 15 Our data suggest that BACHD astrocytes are the source of adverse non-cell autonomous effects on neuron energy metabolism via a diffusible factor released by astrocytes. As BACHD astrocytes did not compromise neuronal energy metabolism by altering the transcription of the major genes involved in the metabolic pathways, what mechanisms could be involved?
The first hypothesis is that BACHD astrocytes may fail to provide adequate metabolic support to neurons, in particular lactate. Lactate, released by astrocytes, has been shown to sustain glutamatergic neurotransmission and synaptic plasticity.35, 36 Although we did not measure extracellular lactate and the expression of the monocarboxylate transporters responsible for lactate fluxes across cells, this hypothesis deserves further examination particularly in the context of increased oxidative stress (see below).
The second hypothesis is related to chronic inflammation, which involves glia and is known to accelerate disease progression in HD. Indeed, R6/2 astrocytes exhibit high IκB kinase (IKK) activity that causes prolongation of NF-κB activation, thus upregulating proinflammatory factors during inflammation and producing a more-damaging effect on primary R6/2 neurons.37 Although we did not directly test this hypothesis, we performed immunostaining of microglia using Iba-1 antibody in the brain of 14-month old BACHD and did not find activated microglia, a surrogate marker of neuroinflammation (data not shown), indicating no significant inflammation in these mice.
The third hypothesis is that BACHD astrocytes have a reduced antioxidant potential leading to an increased oxidative stress in neurons. We indeed provided evidence that oxidative stress is increased in BACHD mice and is induced in vitro in neurons when cocultured with BACHD astrocytes. Oxidative stress is mediated by increases in ROS including superoxide (O2−), hydrogen peroxide (H2O2), hydroxyl radical (OH−), and reactive nitrogen species such as peroxynitrite (ONOO−). Mitochondria dysfunction may be a source of ROS overproduction in neurons but nicotinamide adenine dinucleotide phosphate oxidase could also contribute.38 ROS impairs cellular function by degrading proteins, lipids, and nucleic acids. Evidence of enhanced oxidative stress in HD brains includes an accumulation of lipofuscin, 8-OH-dG, and carbonyl formation.39 However, although oxidative damage is well described in HD, little is known about the specific protein targets of this damage. The major carbonylated proteins in both striatum and cortex from human HD samples are those involved in glucose metabolism and mitochondrial energy pathways in accordance with the impaired energy metabolism described in HD.40 Of interest, it was very recently observed that a defective shuttling of ascorbic acid from astrocytes to neurons could be responsible for the early metabolic failure occurring in HD by shifting the neuronal metabolic substrate preferences between lactate and glucose.41
In conclusion, our data suggest that in vivo functional imaging is useful to obtain pattern of brain metabolism before HD symptoms onset and to identify regions at risk during the course of the disease. We propose that astrocyte-to-neuron signaling is involved in the early energy metabolic alterations that occur in HD.
Acknowledgments
We are grateful to Julien Mitja, Marion Chaigneau, Diane Houitte, Martine Guillermier, Charlène Joséphine, Ashu Johri, and Abhishek Chandra for expert technical assistance during in vivo autoradiographic experiments and immunohistochemical stainings. We thank William Yang for the BACHD mice and Frédéric Saudou for sharing the cDNAs pARIS huntingtin Q23 and Q100. We thank Michelle Gray for critical reading of the manuscript.
The authors declare no conflict of interests.
Footnotes
This work was supported by Fondation de la Recherche Médicale to L.B., ECOS-Sud program C10S04 to L.F.B. and G.B., Commissariat à l'Energie Atomique et aux Energies Alternatives (CEA), and Centre National de la Recherche Scientifique (CNRS).
References
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011;121:493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini A, Leenders KL, Spiegel R, Meier D, Vontobel P, WeigellWeber M, et al. Striatal glucose metabolism and dopamine D-2 receptor binding in asymptomatic gene carriers and patients with Huntington's disease. Brain. 1996;119:2085–2095. doi: 10.1093/brain/119.6.2085. [DOI] [PubMed] [Google Scholar]
- Feigin A, Tang C, Ma Y, Mattis P, Zgaljardic D, Guttman M, et al. Thalamic metabolism and symptom onset in preclinical Huntington's disease. Brain. 2007;130:2858–2867. doi: 10.1093/brain/awm217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano M, Galvan L, Deglon N, Brouillet E. Mitochondria in Huntington's disease. Biochim Biophys Acta. 2010;1802:52–61. doi: 10.1016/j.bbadis.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Browne SE, Beal MF. The energetics of Huntington's disease. Neurochem Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- Guidetti P, Charles V, Chen EY, Reddy PH, Kordower JH, Whetsell WO, Jr, et al. Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp Neurol. 2001;169:340–350. doi: 10.1006/exnr.2000.7626. [DOI] [PubMed] [Google Scholar]
- Lee JM, Ivanova EV, Seong IS, Cashorali T, Kohane I, Gusella JF, et al. Unbiased gene expression analysis implicates the huntingtin polyglutamine tract in extra-mitochondrial energy metabolism. PLoS Genet. 2007;3:e135. doi: 10.1371/journal.pgen.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171:1001–1012. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford J, Shin JY, Roberts M, Wang CE, Li XJ, Li S. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc Natl Acad Sci USA. 2009;106:22480–22485. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faideau M, Kim JH, Cormier K, Gilmore R, Welch M, Auregan G, et al. In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington's disease subjects. Hum Mol Genet. 2010;19:3053–3067. doi: 10.1093/hmg/ddq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza M, Leoni V, Karasinska JM, Petricca L, Fan JJ, Carroll J, et al. Cholesterol defect is marked across multiple rodent models of Huntington's disease and is manifest in astrocytes. J Neurosci. 2010;30:10844–10850. doi: 10.1523/JNEUROSCI.0917-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford J, Shin JY, Roberts M, Wang CE, Sheng GQ, Li SH, et al. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J Biol Chem. 2010;285:10653–10661. doi: 10.1074/jbc.M109.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14:724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Powers WJ, Videen TO, Markham J, McGee-Minnich L, Antenor-Dorsey JV, Hershey T, et al. Selective defect of in vivo glycolysis in early Huntington's disease striatum. Proc Natl Acad Sci USA. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutsinos-Porche B, Bonvento G, Tanaka K, Steiner P, Welker E, Chatton JY, et al. Glial glutamate transporters mediate a functional metabolic crosstalk between neurons and astrocytes in the mouse developing cortex. Neuron. 2003;37:275–286. doi: 10.1016/s0896-6273(02)01170-4. [DOI] [PubMed] [Google Scholar]
- Dubois A, Herard AS, Delatour B, Hantraye P, Bonvento G, Dhenain M, et al. Detection by voxel-wise statistical analysis of significant changes in regional cerebral glucose uptake in an APP/PS1 transgenic mouse model of Alzheimer's disease. Neuroimage. 2010;51:586–598. doi: 10.1016/j.neuroimage.2010.02.074. [DOI] [PubMed] [Google Scholar]
- Vega C, Pellerin L, Dantzer R, Magistretti PJ. Long-term modulation of glucose utilization by IL-1 alpha and TNF-alpha in astrocytes: Na+ pump activity as a potential target via distinct signaling mechanisms. Glia. 2002;39:10–18. doi: 10.1002/glia.10080. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Escartin C, Won SJ, Malgorn C, Auregan G, Berman AE, Chen PC, et al. Nuclear factor erythroid 2-related factor 2 facilitates neuronal glutathione synthesis by upregulating neuronal excitatory amino acid transporter 3 expression. J Neurosci. 2011;31:7392–7401. doi: 10.1523/JNEUROSCI.6577-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanaga H, Chaudhuri B, Frommer WB. GLUT1 and GLUT9 as major contributors to glucose influx in HepG2 cells identified by a high sensitivity intramolecular FRET glucose sensor. Biochim Biophys Acta. 2008;1778:1091–1099. doi: 10.1016/j.bbamem.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo R, Molina-Calavita M, Poizat G, Keryer G, Humbert S, Saudou F. pARIS-htt: an optimised expression platform to study huntingtin reveals functional domains required for vesicular trafficking. Mol Brain. 2010;3:17. doi: 10.1186/1756-6606-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouladi MA, Stanek LM, Xie YY, Franciosi S, Southwell AL, Deng Y, et al. Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum Mol Genet. 2012;21:2219–2232. doi: 10.1093/hmg/dds037. [DOI] [PubMed] [Google Scholar]
- Tang CC, Feigin A, Ma Y, Habeck C, Paulsen JS, Leenders KL, et al. Metabolic network as a progression biomarker of premanifest Huntington's disease. J Clin Invest. 2013;123:4076–4088. doi: 10.1172/JCI69411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JD, Matson WR, Juhl AR, Leavitt BR, Paulsen JS, PREDICT-HD Investigators and Coordinators of the Huntington Study Group 8OHdG as a marker for Huntington disease progression. Neurobiol Dis. 2012;46:625–634. doi: 10.1016/j.nbd.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda-Prado E, Popp S, Khan U, Stefanov D, Rodriguez J, Menalled LB, et al. R6/2 Huntington's disease mice develop early and progressive abnormal brain metabolism and seizures. J Neurosci. 2012;32:6456–6467. doi: 10.1523/JNEUROSCI.0388-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransome MI, Renoir T, Hannan AJ. Hippocampal neurogenesis, cognitive deficits and affective disorder in Huntington's disease. Neural Plast. 2012;2012:874387. doi: 10.1155/2012/874387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abada YS, Schreiber R, Ellenbroek B. Motor, emotional and cognitive deficits in adult BACHD mice: a model for Huntington's disease. Behav Brain Res. 2013;238:243–251. doi: 10.1016/j.bbr.2012.10.039. [DOI] [PubMed] [Google Scholar]
- Dougherty SE, Reeves JL, Lesort M, Detloff PJ, Cowell RM. Purkinje cell dysfunction and loss in a knock-in mouse model of Huntington disease. Exp Neurol. 2013;240:96–102. doi: 10.1016/j.expneurol.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hult S, Soylu R, Bjorklund T, Belgardt BF, Mauer J, Bruning JC, et al. Mutant huntingtin causes metabolic imbalance by disruption of hypothalamic neurocircuits. Cell Metab. 2011;13:428–439. doi: 10.1016/j.cmet.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Bohanna I, Georgiou-Karistianis N, Hannan AJ, Egan GF. Magnetic resonance imaging as an approach towards identifying neuropathological biomarkers for Huntington's disease. Brain Res Rev. 2008;58:209–225. doi: 10.1016/j.brainresrev.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci USA. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Stern SA, Bozdagi O, Huntley GW, Walker RH, Magistretti PJ, et al. Astrocyte-neuron lactate transport is required for long-term memory formation. Cell. 2011;144:810–823. doi: 10.1016/j.cell.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao HY, Chen YC, Chen HM, Tu PH, Chern Y. A critical role of astrocyte-mediated nuclear factor-kappaB-dependent inflammation in Huntington's disease. Hum Mol Genet. 2013;22:1826–1842. doi: 10.1093/hmg/ddt036. [DOI] [PubMed] [Google Scholar]
- Valencia A, Sapp E, Kimm JS, McClory H, Reeves PB, Alexander J, et al. Elevated NADPH oxidase activity contributes to oxidative stress and cell death in Huntington's disease. Hum Mol Genet. 2013;22:1112–1131. doi: 10.1093/hmg/dds516. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Browne SE, Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Sign. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- Sorolla MA, Rodriguez-Colman MJ, Tamarit J, Ortega Z, Lucas JJ, Ferrer I, et al. Protein oxidation in Huntington disease affects energy production and vitamin B6 metabolism. Free Radic Biol Med. 2010;49:612–621. doi: 10.1016/j.freeradbiomed.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Acuna AI, Esparza M, Kramm C, Beltran FA, Parra AV, Cepeda C, et al. A failure in energy metabolism and antioxidant uptake precede symptoms of Huntington's disease in mice. Nat Commun. 2013;4:2917. doi: 10.1038/ncomms3917. [DOI] [PMC free article] [PubMed] [Google Scholar]