Abstract
Fungal pneumonia is increasingly common, particularly in highly immunosuppressed patients, such as solid organ or hematopoietic stem cell transplant recipients, and the diagnosis is evolving. While standard techniques such as microscopy and culture remain the mainstay of diagnosis, relatively recent advances in serologic and molecular testing are important additions to the field. This chapter will review the laboratory tools used to diagnose fungal respiratory disease.
Keywords: Fungal diagnosis, laboratory methods, fungal tests
Introduction
The term “fungus” is used to encompass a very broad range of organisms and the number capable of causing respiratory infection is extensive, varying from yeasts to complex, highly resistant moulds. Fungi are ubiquitous in the environment, typically found in soil and decaying vegetation, thus exposure of the respiratory system to these organisms is common and, accordingly, many fungal infections present with pulmonary involvement. A few fungi are highly virulent and can cause disease even in hosts with intact immunity, however, the virulence of most fungal organisms is low and exposure frequently results in transient colonization which is cleared by the intact immune system. The advent of highly immunosuppressed hosts states, such as with organ transplantation and cancer chemotherapy, has resulted in an increasing incidence of invasive fungal infections over the last several decades.1 In fact, many fungal organisms previously not thought to be pathogenic have been found to cause disease, most commonly as opportunistic infections in immunosuppressed populations.
The diagnosis of fungal disease is evolving. As outlined by the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) definitions for invasive fungal disease, laboratory information must be interpreted with caution and requires consideration of the virulence of the specific fungal pathogen recovered as well as the host’s immunocompetence and clinical presentation.2 In this chapter we will review the laboratory tools used in the diagnosis of pulmonary fungal infection, beginning with a discussion of traditional methods including culture and microscopy and ending with molecular tools and other emerging technologies.
Microbiological Diagnostics
Direct Microscopy
Direct microscopy of clinical specimens can be an initial, cost-effective test, providing prompt recognition of many invasive fungal infections.3 Staining for fungal elements utilizes relatively inexpensive materials, does not require highly specialized equipment, and results can often be available within several hours of specimen collection. The most common direct microscopic procedure relies on the use of 10% to 20% potassium hydroxide (KOH) which degrades the proteinaceous components of specimens while leaving the fungal cell wall intact allowing their visualization. The visibility of fungi within clinical specimens can be further enhanced by the addition of Calcofluor white, a fluorophore, which binds to the chitin in fungal cell walls4 or lactophenol cotton blue, which stains the outer cell wall. Other stains are frequently used in direct microscopy, such as the India ink wet mount which is useful for visualization of encapsulated fungi, particularly Cryptococcus neoformans. Although a negative direct examination cannot rule out fungal disease, visualization of fungal elements in specimens can often secure initial information helpful in the selection of empiric antifungal therapy.5 Direct microscopy also has utility in the surgical management of some invasive fungal infections. For example, the use of real-time KOH examinations performed during surgical debridement of fungal lesions is gaining popularity as a means of preserving non-infected tissues, particularly for mucormycosis in which infected tissues often appear grossly normal.
Table 1 presents a partial list of fungi capable of causing pulmonary infection, sorted into broad categories based on their morphologic characteristics when visualized in specimens by direct microscopy. As noted, while direct microscopy typically only provides information regarding the gross morphology of the organism present (e.g., yeast or mould), it can occasionally provide sufficient information for identification of the etiologic pathogen (e.g., the thermally dimorphic fungi). In some cases it has been the sole mechanism for diagnosis, such as with Pneumocystis jiroveci which cannot be cultured in routine microbiology laboratories.
Table 1.
Fungi associated with pulmonary infections and their characteristic morphology in clinical specimens by direct microscopic examination
| Fungal Group and Representative Pathogens | Microscopic Morphology in Clinical Specimens |
|---|---|
| YEASTS | |
|
| |
| Cryptococcus neoformans | Spherical budding yeasts of variable size, 2–15 μm in diameter. Capsule may be present or absent. No hyphae or pseudohyphae. |
| Trichosporon | Hyaline arthroconidia, blastoconidia, and pseudohyphae, 2–4 by 8 μm. |
|
| |
| MOULDS | |
|
| |
| Mucormycotinaa | Broad, thin-walled, pausi-septate hyphae, 6–25 μm wide with non-parallel sides and random branches. |
| Absidia | |
| Cunninghamella | |
| Mucor | |
| Rhizomucor | |
| Rhizopus | |
| Saksenaea | |
|
| |
| Hyaline Hyphomycetes | |
| Aspergillus | Hyaline, septate, dichotomously branching hyphae of uniform width (3–6 μm). |
| Acremonium | |
| Fusarium | Aspergillus may produce conidial heads in specimens from cavitary pulmonary lesions. Adventitious conidiation may be visualized on histopathologic examination of some of the non-Aspergillus hyphomycetes. |
| Paecilomyces | |
| Phialemonium | |
| Scedosporium | |
| Scopulariopsis | |
| Trichoderma | |
|
| |
| Dematiaceous Hyphomycetes | |
| Alternaria | Pigmented (brown, tan, or black), septate hyphae, 2–6 μm wide. |
| Bipolaris | |
| Curvularia | |
| Cladophialophora | |
| Dactylaria | |
| Exophiala | |
| Phialophora | |
| Ramichloridium | |
| Wangiella | |
|
| |
| OTHER | |
|
| |
| Dimorphic (geographically restricted) Fungi | |
| Blastomyces dermatitidis | Large (8–15 μm diameter) thick-walled budding yeast cells. The junction between mother and daughter cells is typically broad-based. Cells may appear multinucleate. |
| Coccidioides immitis | Spherical, thick-walled spherules, 20–30 μm diameter. Mature spherules contain small, 2–5 μm diameter endospores. Released endospores may be mistaken for yeast. Arthroconidia and hyphae may form in cavitary lesions. |
| Histoplasma capsulatum | Small (2–4 μm diameter), intracellular, budding yeasts. |
| Paracoccidioides brasiliensis | Large (2–30 μm diameter), multiple-budding yeasts ; 12 or more narrow neck buds of variable size may arise from the mother cell. |
| Penicillium marneffei | Oval, intracellular yeast cells bisected with a septum (fission yeast). |
| Sporothrix schenckii | Elongated or “cigar-shaped” yeast cells of varying size (rare). Tissue reaction forms asteroid bodies. |
| Pneumocystis jiroveci | Cysts are round, collapsed, or crescent shaped. Trophozoites seen on staining with giemsa or immunofluorescent stains. |
Moulds of the subphylum Mucormycotina, previously referred to as Zygomycetes
Historically, the diagnosis of P. jiroveci pneumonia (PCP) has been comprised of direct visual examination of the specimen after some type of staining method, including but not limited to KOH, Grocott-Gomori methenamine silver (GMS), and newer immunofluourescent antibody stains. The MeriFluor-Pneumocystis Direct Flourescent Antibody (DFA) ® test (Meridian Bioscience, Inc., Cincinatti, OH) is an FDA-approved immunofluorescent stain for the diagnosis of PCP. When compared with more traditional microscopic staining methods such GMS or Calcofluor white, the MeriFluor DFA performs reasonably well with a sensitivity of 91% but with a positive predictive value of only 82%, in contrast with 96% with GMS or Calcofluor white.6 In addition, it is comparatively labor intensive thus hampering its widespread utility. More recently, a variety of nucleic acid amplification assays (discussed below) have been developed for detection of P. jiroveci in respiratory samples and are now increasingly used as the primary method of diagnosing PCP in many laboratories.
Culture
Culture remains one of the key methods for diagnosing fungal infection. As it is more sensitive than direct microscopy, culture should be performed on all acceptable clinical specimens collected in the evaluation of suspected fungal disease, regardless of whether organisms are visualized on direct microscopy. The handling of clinical specimens is essential to the successful recovery of a fungal organism. In general, for respiratory samples, the most purulent and/or bloody portions of the specimen are inoculated directly onto specialized fungal culture media containing antibiotics to inhibit overgrowth with bacteria. Centrifugation of respiratory samples is not recommended as it has not been shown to increase the isolation of fungi and is more likely to result in bacterial overgrowth. In addition, the frequently used process of grinding tissue samples for bacterial culture can destroy fungal organisms, particularly the zygomycetous moulds.
Despite the use of specialized media and appropriate handling of specimens, fungi are difficult to cultivate and frequently do not grow at all. In one study, up to 70% of tissue samples with invasive septate hyphae visualized on histopathology, did not grow a mould in culture.7 In another, approximately one third of zygomycetous moulds visualized on histopathology failed to grow.8
If a fungus is recovered in culture, identification of the organism can also be difficult and sometimes inconclusive. Definitive identification of fungi relies in part on the macro- and microscopic morphologic characteristics of the organism; however, while most moulds do not produce reproductive structures in vivo, others fail to do so even in vitro, making identification by microscopic taxonomy impossible.9 Molecular sequencing is an emerging technique under investigation to assist in identifying fungi to the species level (discussed below).10
Susceptibility Testing
Recovery and identification of the infecting fungal pathogen in culture also provides the opportunity for antimicrobial susceptibility testing and standardized procedures for testing both yeasts11 and moulds12 have been established. Clinical susceptibility breakpoints are only available for Candida species as correlation between in vitro antifungal susceptibility test results and clinical outcomes for moulds is limited. However, there is much work on-going in this area. Epidemiological cutoff values (ECVs) are now established for Aspergillus and the triazoles, which aids in discriminating wild-type strains from those with acquired resistance mutations.13 For several other moulds, intrinsic patterns of antifungal activity have been established, historically based on clinical response to treatment. For example, Fusarium has variable response to amphotericin B and the extended-spectrum triazoles, but little to no response to the echinocandins. For Scedosporium, the activity of antifungal drugs varies according to species: S. apiospermum is primarily resistant to amphotericin B whereas S. prolificans is resistant to treatment with all antifungal agents. Availability of a standardized susceptibility testing method for moulds has allowed for expansion of the understanding of fungal response to therapy by establishing wild type minimal inhibitory concentration distributions for many of the less common moulds. As more information is obtained, susceptibility testing will be increasingly used to guide the selection of therapy, particularly for pathogens with unpredictable antifungal activity profiles and to rule out resistance in the setting of prior antifungal exposure and/or clinical failure.
Histopathologic Diagnostics
Microscopy
As with direct microscopy of clinical specimens, visualization of fungal elements within tissue samples can provide definitive evidence of invasive infection. Identification of fungi in tissue sections, however, can be difficult owing to limited biopsy tissue specimen size with few organisms present, or as per Figure 1, only the presence of mycelial elements rendering common organism types indistinguishable from one another. Unfortunately, most in situ manifestations of fungi are not pathognomonic. In addition, the technique of preserving tissue excludes the possibility of recovering the pathogen in culture. Thus, when fungal infection is suspected, care must be made to specifically request fungal cultures of the fresh tissue specimen.
Figure 1.
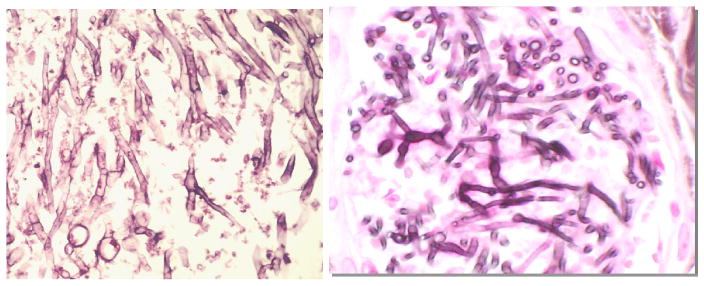
Histopathology of Moulds.
Aspergillus in lung tissue (left) and Fusarium in skin (right) cannot be differentiated by histopathologic morphology. (Photos: BD Alexander)
Immunohistochemistry
Immunohistochemistry is a means of using antibodies directed at fungal cell antigens to identify organisms visualized in tissue. Advantages of immunohistochemistry include relatively rapid results, preservation of tissue morphology, and the ability to use Formalin Fixed Paraffin Embedded (FFPE) tissue. Unfortunately, the application of immunohistochemistry is hindered by the fact that available antibodies have a high degree of cross-reactivity between taxonomic lines. They have also been unable to identify fungi to species level. 14–15 At this time, there are currently no commercially approved immunohistochemical tests for identification of fungi in tissue. Further, antibodies for many moulds, including non-Aspergillus hyphomycetes and zygomycetous moulds, are not commercially available.5
Immunologic and Biochemical Diagnostics
Serum antibody detection has been used predominantly for the diagnosis of dimorphic fungal infections. Techniques used for antibody detection, include, but are not limited to complement fixation, tube precipitins, and immunoenzyme assays. Variants of these assays may employ immunodiffusion (ID) in agar. Tube precipitins antibodies indicate IgM activity, whereas complement-fixing antibodies represent IgG. In general, tube precipitins antibodies are detectable early in illness and, at times during recurrence, but wane quickly while complement-fixing antibodies are usually detectable after 2–3 weeks of illness and frequently only with more severe disease.16 The complement-fixing antibody titer has prognostic implications, with high titers portending severe or disseminated disease and decreases in titer with recovery from the infection. Testing for antibodies can be complicated in the immunosuppressed patient, depending on the etiology of the immunosuppression and the element of the immune system that is predominantly affected. In the immune compromised host, significant antibody increases simply come too late, if at all, and tests for IgM often remain negative.
The use of direct fungal antigen detection in blood and other body fluids has been used for several decades, most commonly in the diagnosis of histoplasmosis and cryptococcosis, although new tests for detecting galactomannan and (1→3)-β-D-glucan have provided alternative techniques for diagnosis of invasive aspergillosis and other invasive fungal diseases. These tests allow for a more rapid diagnosis of disease in comparison to histopathologic and microbiologic culture methods. The application and performance of specific serologic methods follows.
Serologic Tests for Endemic Fungi
The use of serologic tests for the diagnosis of endemic fungal infections has been limited to a large extent by the high degree of test cross reactivity between the different pathogens. For example, cross-reactivity of Coccidioides with the commonly used Histoplasma urinary antigen enzyme immunoassay has been well described.17 Further, performance of the tests depends to a large degree on the extent and site of infection. The Histoplasma urine antigen test has been found to be positive in 92% of patients with disseminated disease18 but only in 80% with acute diffuse pulmonary disease, 34% in subacute pulmonary disease, and 14% in chronic pulmonary disease.19 Hage et al20 recently found antigen detection in BAL fluid may be complementary to other conventional diagnostic methods in identification of histoplasmosis. In cases of pulmonary histoplasmosis with or without disseminated disease diagnosed by histopathologic or microbiologic data, 94% of patients had Histoplasma antigen detected in BAL fluid. In combining BAL antigen detection and BAL cytopathology, the sensitivity was 97% for diagnosis of histoplasmosis. Evaluation of Blastomyces antigen detection revealed the antigen was detectable in BAL in 80% of patients with confirmed blastomycosis. However, combination of Blastomyces BAL antigen detection and BAL cytopathology did not result in an increased sensitivity over BAL antigen detection alone. For Coccidioides, an immunodiffusion test was positive in 53% of immunosuppressed patients compared with 73% of non-immunosuppressed patients, and the complement fixation test had positive results in 67% and 75%, respectively.21 Another study reported seropositivity in approximately one-half of patients with Coccidioides pneumonia.22–23 Alternatively, antigen detection may also prove useful for the early diagnosis of coccidioidomycosis. Durkin et al24 recently evaluated a new Coccidioides urine antigen detection test, the Coccidioides EIA test (MiraVista Diagnostics, Indianapolis, IN), and detected antigenuria in 71% of patients with coccidioidomycosis. Importantly, the test did exhibit some residual cross reactivity in patients with histoplasmosis and the majority of patients studied were immunosuppressed (79%) with severe forms of the disease (46% had disseminated disease). Thus, utility of the test in nonimmunocompromised patients with primary pulmonary coccidioidomycosis remains unclear.
The sensitivity and specificity of serologic tests for paracoccidioidomycosis has been historically compromised by the use of complex mixtures of undefined antigens. The use of combinations of purified, well-characterized antigens for the detection of serum antibodies appears to improve diagnostic performance. Fernandes et al25 evaluated two recombinant antigens derived from Paracoccidioides brasiliensis and found a sensitivity of 96% with a specificity of 100% when the combination of antigens was used.
Cryptococcocal Antigen Test
The detection of the capsular polysaccharide antigen in serum and cerebrospinal fluid (CSF) plays a significant role in the diagnosis of cryptococcosis, both in pulmonary and disseminated disease. The antigen test detects both C. neoformans and C. gattii. The sensitivity and specificity of serum antigen testing for the diagnosis of cryptococcal meningoencephalitis approaches that of testing CSF, however the test has a lower diagnostic yield for isolated pulmonary cryptococcosis.26 Additionally, the serum cryptococcal antigen test is more sensitive in patients with disseminated disease and in immunocompromised hosts likely due to greater fungal burden in these settings. Routine testing of BAL fluid for the cryptococcal antigen has been evaluated and found to have a sensitivity of 71% with a positive predictive value of 59%, and is thus not recommended.27 The serum cryptococcal capsular polysaccharide antigen has been shown to be cross-reactive with Trichosporon, another fungal organism known to cause both pulmonary and disseminated disease.28–29
Asperigllus Galactomannan Antigen Test
Galactomannan is a polysaccharide cell-wall component of Aspergillus that can be measured in serum and other body fluids to evaluate for evidence of invasive aspergillosis. Approval by the U.S. Food and Drug Administration in 2003 for testing serum to aid in the diagnosis of invasive aspergillosis was based on data in hematopoietic stem cell transplant (HSCT) and leukemia patients, with a reported sensitivity of 81% and a specificity of 89%.30 But as demonstrated in a meta-analysis by Pfeiffer et al31, the serum test has variable results in other populations. A compilation of 27 studies from 1995 to 2005, the meta-analysis found the sensitivity of the assay to be 70% and specificity 92% in patients with hematologic malignancy. In HSCT recipients, sensitivity was 82% and specificity 86%. This is in stark contrast to data from the solid organ transplant population in which sensitivity was only 22% and specificity 84%.
In an attempt to improve the diagnostic yield for invasive pulmonary aspergillosis, attention has been aimed at directly testing BAL fluid for the presence of galactomannan. The utility of using BAL galactomannan detection in identifying invasive pulmonary aspergillosis is complicated by the inability to differentiate between Aspergillus colonization, pulmonary aspergilloma, and invasive pulmonary aspergillosis. This distinction is clinically important in that the management is very different for these conditions. In pulmonary aspergilloma without invasive disease, Park et al32 found 32 of 36 BAL samples in patients were positive for galactomannan. The sensitivity of the BAL galactomannan assay for the identification of Aspergillus associated with pulmonary aspergilloma was 92% whereas the sensitivity of the serum galactomannan assay was 38%. Patients with pulmonary aspergilloma and positive serum galactomannan were significantly more likely to have hemoptysis, perhaps reflecting locally invasive disease allowing for leakage of galactomannan into the bloodstream.
As with serum, performance of the test in BAL seems to also vary by patient population. In a study by Park et al33 evaluating the utility of galactomannan BAL testing in a population with various underlying medical conditions, sensitivity was 64% and specificity 89%. Two recent studies in patients with hematologic malignancies or HSCT found the BAL galactomannan assay to have a sensitivity of 57–73% and specificity of 89–95%.34–35 In a study of intensive care unit (ICU) patients, the test had a sensitivity of 88% and specificity of 87%.36 Although the US Food and Drug Administration (FDA) approved positive test cutoff threshold for galactomannan in BAL is an index of 0.5, use of a higher cutoff value improved utility of the BAL galactomannan assay in HSCT or hematologic malignancy patients with radiographic evidence of invasive pulmonary aspergillosis (sensitivity 100%, specificity 100%). 37–38,39 Among lung transplant patients, three recent studies have shown improved sensitivities using a positive cutoff index value of ≥1 (sensitivity 82–100% and specificity of 81–96%).40–41
Galactomannan results can be affected by a number of factors. The use of β-lactam antibiotics, including piperacillin-tazobactam42–43 and amoxicillin-clavulanic acid44 can result in false positive galactomannan test results in serum. Although it is unclear the effect of intravenous administration of Plasma-Lyte (Baxter, Deerfield, IL) on serum galactomannan results, the use of Plama-Lyte as the solution for collection of a BAL sample and respiratory colonization with other moulds including the common saprophyte, Penicillium, has been shown to result in a false positive test in BAL.45 On the other hand, false negative galactomannan results may be the result of mould-active antifungal medications. In patients receiving antifungal therapy, the sensitivity of serum galactomannan increased from 31% to 85% when the positive test cutoff value was decreased from 1.5 to 0.5, suggesting patients receiving mould active antifungals may have lower circulating levels of antigen in the blood, thus affecting the test results.46
(1→3)-β-D-Glucan Test
The detection of (1→3)-β-D-glucan (BG), a component of the fungal cell wall, in serum is another useful tool in diagnosing invasive fungal infections. (1→3)-β-D-glucan is detected in patients with Aspergillus and Candida and can also be useful in the identification of infection with Pneumocystis, Fusarium, and Trichosporon, among others. 47–48 Based on a multicenter study of patients with IFI and healthy controls49, the BG assay (Fungitell®; Associates of Cape Cod, Falmouth, MA) was approved by the FDA for the qualitative detection of BG in the serum of patients with symptoms of or medical conditions predisposing to IFI and as an aid in the diagnosis of deep-seated mycoses and fungemia.
In the evaluation of patients with invasive disease secondary to Aspergillus, Candida, Fusarium, and Trichosporon, the BG test had a sensitivity of 70%, negative predictive value of 75–100%, specificity of 87–90%, and positive predictive value of 79–84%.50–51 In one study, specificity increased to 96% with ≥2 sequential positive tests and the BG test was positive a median of 10 days before the clinical diagnosis of invasive fungal infection.18 Pazos et al52 found improved specificity and positive predictive value, both to 100%, without affecting the sensitivity or negative predictive value when the BG test was used in conjunction with the galactomannan assay to serially monitor cancer patients at high risk for invasive aspergillosis. On the other hand, a recent study evaluating the utility of serially monitoring lung transplant recipients with the test suggests marginal performance in this population. The majority (92%) of lung transplant patients not diagnosed with an IFI had at least one BG level ≥60 pg/mL and 90% had at least one BG level ≥80 pg/mL. Although the negative predictive value of the BG test was high, the low positive predictive value limited its utility as a screening tool for early diagnosis of IFI in the lung transplant population.53 Further, (1→3)-β-D-glucan is ubiquitous in the environment and, as with the galactomannan test, multiple sources of false positive test results exist. For example, poor specimen handling, hemodialysis, surgical packing, bacteremia, receipt of blood products, and intravenous therapy with a beta-lactam/inhibitor combination may cause false-positive test results.52,54,55,56
Molecular Diagnostics
Nucleic acid-based assays have potential to complement and enhance conventional methods used in diagnostic mycology and their use is gaining popularity; some large referral clinical laboratories are now routinely employing in-house developed molecular assays for fungal diseases. However, widespread use of molecular tools to diagnose fungal disease is currently limited by a lack of standardization in testing methods. The fungal cell wall is hard to penetrate making extraction of intact fungal DNA difficult. Further complicating the issue is that many commercially produced reagents and blood collection tubes have been found to be contaminated with fungal DNA, resulting in false positive tests.57–58. Other drawbacks for clinical laboratories include high equipment costs, labor-intensive procedures, and the need for molecularly trained technologists.59
The majority of molecular tests developed to date for the diagnosis of fungal disease are polymerase chain reaction (PCR)-based assays focused primarily on the detection of Candida, Aspergillus, or Pneumocystis jirovecci from clinical samples. With that said, development of an assay for detection of fungal nucleic acid circulating in blood during invasive aspergillosis has been problematic. A meta-analysis inclusive of sixteen studies published from 2000–2008 (>10,000 samples from 1618 patients) reporting on the performance of various PCR assays on blood components for the diagnosis of invasive aspergillosis revealed an overall sensitivity of 88% and specificity of 75% based on the results of a single positive test.60 Requiring two consecutive positive test results led to decreased sensitivity (75%) with a modest improvement in specificity (87%). The authors concluded a single PCR-negative result was sufficient to exclude a diagnosis of proven or probable invasive aspergillosis, but 2 positive tests were required to confirm the diagnosis. Recent evaluation of a commercially prepared Aspergillus serum-based PCR assay (Myconostica, Ltd., Manchester, United Kingdom) found a sensitivity of 60–70% and specificity of 91–100%.61 The diagnosis of invasive pulmonary aspergillosis via PCR-based testing of respiratory specimens has also been studied. Distinguishing between Aspergillus colonization and invasive aspergillosis has been a major limitation. 62 Whether quantitative PCR results can address this issue is still being sorted out. Based on a meta-analysis of the literature examining Aspergillus PCR testing of BAL fluid, although quite heterogeneous among the various studies, pooled sensitivity was 79% and specificity 94%.63 Overall, the lack of standardization in molecular methods continues to be a major outstanding problem in the diagnosis of invasive aspergillosis.
For diagnosis of PCP, a commercially prepared PCR assay for Pnuemocystis jirovecci (Myconostica, Ltd., Manchester, United Kingdom) designed for testing respiratory specimens was compared with the FDA-approved MeriFluor-Pneumocystis DFA test and found a sensitivity of 93%, specificity of 91%, and positive and negative predictive values of 59% and 99%, respectively.64 Many clinical laboratories are moving towards the use of molecular based assays for the diagnosis of PCP.
Another application for molecular technology includes identification of fungi recovered in culture. As previously mentioned, identification of moulds by conventional methods relies heavily on in vitro sporulation which can be a slow, arduous process. Molecular sequencing is an emerging platform under investigation as a method for identifying fungi recovered in culture.65 While the genomes for the majority of fungi have not yet been fully sequenced, certain fungal gene regions contain species-specific sequences that permit the identification of hundreds of pathogenic fungi to species level. While open access databases such as GenBank currently contain information for a few reference mould strains, the amount of information in these databases is rapidly expanding.
Similarly, the use of sequencing technology to identify fungi in tissue samples has also been evaluated. A recent study used FFPE tissue from histopathologically proven cases of mucormycosis and recovered DNA by PCR from 22 of 27 tissue specimens, including from 10 of 12 culture confirmed cases for which the identification was concordant to the genus level in 9 samples.66 Rickerts et al67, using an extraction procedure that included boiling and freezing, were able to successfully identify the infecting mould from FFPE tissue in 21 cases of culture proven Aspergillus fumigatus and various zygomycetous moulds. In another study68, PCR was found to be superior to culture in evaluating 27 tissue specimens with histopathologically confirmed cases of mould infection (26 of 27 samples vs. 17 of 27 samples, respectively; p = 0.006).
Finally, another promising molecular technology for use with tissue specimens is Peptide Nucleic Acid Fluorescent in situ Hybridization (PNA FISH), a fluorescent DNA probe. In a study using culture proven fungal infections, PNA FISH was found to have a slightly lower sensitivity than conventional staining methods (95% vs. 98% respectively) but with a higher positive predictive value (97% vs. 86% respectively) in distinguishing between Aspergillus, zygomycetous moulds, and Candida.69 However, to enhance permeability of the fungal cell wall, the molecular probes utilized are typically limited in length to 20–40 base pairs, which limits identification of moulds to species level.70 For example, 250–300 base pairs are needed to reliably differentiate members of the subphylum Mucormycotina.
New and Emerging Diagnostics
Proteomics
The use of proteomic “fingerprints” in the rapid diagnosis of fungal infections is in early stages of investigation and is not in clinical use at this time. However, early studies are intriguing. Using plasma and BAL from neutropenic rabbit models of Pseudomonas and Aspergillus pneumonia, Gonzales and colleagues demonstrated unique protein profiles that are shared in early infection and diverge at later stages of infection. Haptoglobin, apolipoprotein A1, transthyretin, and C-reactive protein are differentially expressed in these infections suggesting possible future diagnostics contributions.71
Genomics
While exogenous immune suppression is perhaps the greatest risk factor for acquiring many invasive fungal infections, polymorphic variation in key host immune effector genes likely contributes to disease susceptibility as well. Detection of host polymorphisms that contribute to fungal disease susceptibility and response is being evaluated. Identification of single nucleotide polymorphisms (SNPs) that may predispose hosts to invasive aspergillosis and which may be useful adjunctive diagnostic information in the future are being delineated.72,73,74,75
Conclusion
Fungal pneumonia has become increasingly common in the age of highly immunosuppressed patients such as with HIV/AIDS or following transplantation. Diagnosis has been limited by the standard methods of direct microscopy, culture, and histopathologic evaluation. Newer techniques are evolving, aiding in the earlier diagnosis and subsequent appropriate treatment of these infections. Molecular methods including PCR coupled with sequencing offer the best promise as replacements for culture but standardization of such methods are needed.
Acknowledgments
This project was supported by an United States Public Health Service grant from the National Institutes of Health, K24 AI 072522 (BDA).
References
- 1.Marr KA, Carter RA, Crippa F, et al. Epidemiology and Outcome of Mould Infections in Hematopoietic Stem Cell Transplant Recipients. Clin Infect Dis. 2002;34:909–17. doi: 10.1086/339202. [DOI] [PubMed] [Google Scholar]
- 2.De Pauw B, Walsh TJ, Donnelly JP, et al. Revised Definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections/cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–21. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murrary PR, Witebsky FG. The Clinician and the Microbiology Lab. In: Mandell JL, Bennet JE, Dolin R, editors. Principles and Practice of Infectious Diseases. 7. Philadelphia: Churchill Livingstone; 2009. pp. 233–65. [Google Scholar]
- 4.Harrington BJ, Hageage GJ. Calcofluor White; A Review of its Uses and Applications in Clinical Mycology and Parasitology. Lab Med. 2003;34:361–7. [Google Scholar]
- 5.Bennet JE. Introduction to Mycoses. In: Mandell JL, Bennet JE, Dolin R, editors. Principles and Practice of Infectious Diseases. 7. Philadelphia: Churchill Livingstone; 2009. pp. 3221–4. [Google Scholar]
- 6.Procop GW, Haddad S, Quinn J, et al. Detection of Pneumocystis jiroveci in Respiratory Specimens by Four Staining Methods. J Clin Microbiol. 2004;42:3333–5. doi: 10.1128/JCM.42.7.3333-3335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarrand JJ, Lichterfeld M, Warraich I, et al. Diagnosis of Invasive Septate Mold Infections: A Correlation of Microbiological Culture and Histologic or Cytologic Examination. Am J Clin Pathol. 2003;119:854–8. doi: 10.1309/EXBV-YAUP-ENBM-285Y. [DOI] [PubMed] [Google Scholar]
- 8.Kontoyiannis DP, Wessel VC, Bodey GP, et al. Zygomycosis in the 1990’s in a Tertiary-Care Cancer Center. Clin Infect Dis. 2000;30:851–6. doi: 10.1086/313803. [DOI] [PubMed] [Google Scholar]
- 9.Alexander BD, Pfaller MA. Contemporary Tools for the Diagnosis and Management of Invasive Mycoses. Clin Infect Dis. 2006;43:S15–27. [Google Scholar]
- 10.Balagee SA, Kano R, Baddley JW, et al. Molecular Identification of Aspergillus Species Collected for the Transplant-Associate Infection Surveillance Network. J Clin Microbiol. 2009;47:3138–41. doi: 10.1128/JCM.01070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI) CLSI Document M27-A3. 3. [Accessed July 25, 2011]. Reference Method for Broth Dilution Antifungal SusceptibilityTesting of Filamentous Fungi; Approved Standard. Available at: http://www.clsi.org.
- 12.Clinical and Laboratory Standards Institute (CLSI) CLSI Document M38-A2. 2. [Accessed July 25, 2011]. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard. Available at: http://www.clsi.org.
- 13.Pfaller MA, Diekema DJ, Ghannoum MA, et al. Wild-Type MIC Distribution and Epidemiological Cutoff Values for Aspergillus fumigatus and Three Triazoles as Determined by the Clinical and Laboratory Standards Institute Broth Microdilution Methods. J Clin Microbiol. 2009;47:3142–6. doi: 10.1128/JCM.00940-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed JA, Hemann BA, Alexander JL, et al. Immunomycology: Rapid and Specific Immunocytochemical Identificant of Fungi in Formalin-fixed, Paraffin-embedded Material. J Histochem Cytochem. 1993;41:1217–21. doi: 10.1177/41.8.8331285. [DOI] [PubMed] [Google Scholar]
- 15.Verweij PE, Smedts F, Poot T, et al. Immunoperoxidase staining for identification of Aspergillus species in routinely processed tissue sections. J Clin Pathol. 1996;49:798–801. doi: 10.1136/jcp.49.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borchers AT, Gershwin ME. The immune response in Coccidioidomycosis. Autoimmun Rev. 2010;10:94–102. doi: 10.1016/j.autrev.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Wheat J, Wheat H, Connolly P, et al. Cross-Reactivity in Histoplasma capsulatum variety capsulatum Antigen Assays of Urine Samples from Patients with Endemic Mycoses. Clin Infect Dis. 1997;24:1169–71. doi: 10.1086/513647. [DOI] [PubMed] [Google Scholar]
- 18.Williams B, Fojtasek M, Connolly-Stringfield P, Wheat J. Diagnosis of histoplasmosis by antigen detection during an outbreak in Indianapolis, Ind. Arch Pathol Lab Med. 1994;118:1205–8. [PubMed] [Google Scholar]
- 19.Wheat LJ, Conces D, Allen SD, et al. Pulmonary histoplasmosis syndromes: recognition, diagnosis, and management. Semin Respir Crit Care Med. 2004;25:129–44. doi: 10.1055/s-2004-824898. [DOI] [PubMed] [Google Scholar]
- 20.Hage CA, Davis TE, Fuller D, et al. Diagnosis of Histoplasmosis by Antigen Detection in BAL Fluid. Chest. 2010;137:623–28. doi: 10.1378/chest.09-1702. [DOI] [PubMed] [Google Scholar]
- 21.Blair JE, Coakley B, Santelli AC, et al. Serologic testing for symptomatic coccidioidomycosis in immunocompetent and immunosuppressed hosts. Mycopathol. 2006;162:317–24. doi: 10.1007/s11046-006-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarosi GA, Lawrence JP, Smith DK, et al. Rapid diagnostic evaluation of bronchial washings in patients with suspected coccidioidomycosis. Semin Respir Infect. 2001;16:238–41. doi: 10.1053/srin.2001.29323. [DOI] [PubMed] [Google Scholar]
- 23.DiTommaso JP, Ampel NM, Sobonya RE, et al. Bronchoscopic diagnosis of pulmonary coccidioidomycosis: Comparison of cytology, culture, and transbronchial biopsy. Diagn microbial Infect Dis. 1994;18:83–7. doi: 10.1016/0732-8893(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 24.Durkin M, Connolly P, Kuberski T, et al. Diagnosis of Coccidioidomycosis with Use of the Coccidioides Antigen Enzyme Immunoassay. Clin Infect Dis. 2008;47:e69–73. doi: 10.1086/592073. [DOI] [PubMed] [Google Scholar]
- 25.Fernandes VC, Coitinho JB, Veloso JMR, et al. Combined use of Paracoccidioides brasiliensis recombinant rPb27 and rPB40 antigens in an enzyme-linked immunosorbent assay for immunodiagnosis of paracoccidioidomycosis. J Immunol Methods. 2011;367:78–84. doi: 10.1016/j.jim.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Singh N, Alexander BD, Lortholary O, et al. Pulmonary Crytococcosis in Solid Organ Transplant Recipients: Clinical Relevance of Serum Cryptococcal Antigen. Clin Infect Dis. 2008;46:e12–8. doi: 10.1086/524738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kralovic SM, Rhodes JC. Utility of Routine Testing of Bronchoalveolar Lavage Fluid for Cryptococcal Antigen. J Clin Microbiol. 1998;36:3088–9. doi: 10.1128/jcm.36.10.3088-3089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyman CA, Devi SJN, Nathanson J, et al. Detection and Quantitation of the Glucuronoxylomannan-Like Polysaccharide Antigen from Clinical and Nonclinical Isolates of Trichosporon beigelii and Implications for Pathogenicity. J Clin Microbiol. 1995;33:126–30. doi: 10.1128/jcm.33.1.126-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fekkar A, Brun S, D’Ussel M, et al. Serum Cross-Reactivity with Aspergillus Galactomannan and Cryptococcal Antigen during Fatal Disseminated Trichosporon dermatis Infection. Clin Infect Dis. 2009;49:1457–8. doi: 10.1086/644499. [DOI] [PubMed] [Google Scholar]
- 30.Platelia aspergillus EIA [package insert] Redmond, WA: Bio-Rad Laboratories; 2003. [Google Scholar]
- 31.Pfeiffer CD, Fine JP, Safdar N. Diagnosis of Invasive Aspergillosis Using a Galactomannan Assay: A Meta-Analysis. Clin Infect Dis. 2006;42:1417–27. doi: 10.1086/503427. [DOI] [PubMed] [Google Scholar]
- 32.Park SY, Lee SO, Choi SH, et al. Serum and Bronchoalveolar Lavage Fluid Galactomannan Assays in Patients with Pulmonary Aspergilloma. Clin Infect Dis. 2011;52:e149–52. doi: 10.1093/cid/cir027. [DOI] [PubMed] [Google Scholar]
- 33.Park SY, Lee SO, Choi SH, et al. Aspergillus galactomannan antigen assay in bronchoalveolar lavage fluid for diagnosis of invasive pulmonary aspergillosis. J Infect. 2010;61:492–8. doi: 10.1016/j.jinf.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Bergeron A, Belle A, Sulahian A, et al. Contribution of Galactomannan Antigen Detection in BAL to the Diagnosis of Invasive Pulmonary Aspergillosis in Patients With Hematologic Malignancies. Chest. 2010;137:410–5. doi: 10.1378/chest.09-0701. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen MH, Leather H, Clancy CJ, et al. Galactomannan Testing in Bronchoalveolar Lavage Fluid Facilitates the Diagnosis of Invasive Pulmonary Aspergillosis in Patients with Hematologic Malignancies and Stem Cell Transplant Recipients. Biol Blood Marrow Transplant. 2011 doi: 10.1016/j.bbmt.2010.11.013. In print. [DOI] [PubMed] [Google Scholar]
- 36.Meersseman W, Lagrou K, Maertens J, et al. Galactomannan in Bronchoalveolar Lavage Fluid: A Tool for Diagnosing Aspergillosis in Intensive Care Unit Patients. Am J Respir Crit Care Med. 2008;177:27–34. doi: 10.1164/rccm.200704-606OC. [DOI] [PubMed] [Google Scholar]
- 37.Maertens J, Maertens V, Theunissen K, et al. Bronchoalveolar Lavage Fluid Galactomannan for the Diagnosis of Invasive Pulmonary Aspergillosis in Patients with Hematologic Diseases. Clin Infect Dis. 2009;49:1688–93. doi: 10.1086/647935. [DOI] [PubMed] [Google Scholar]
- 38.Musher B, Fredricks D, Leisenring W, et al. Aspergillus Galactomannan Enzyme Immunoassay and Quantitative PCR for Diagnosis of Invasive Aspergillosis with Bronchoalveolar Lavage Fluid. J Clin Microbiol. 2004;42:5517–22. doi: 10.1128/JCM.42.12.5517-5522.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker MJ, Lugtenburg EJ, Corneliessen JJ, et al. Galactomannan detection in computerized tomography-based broncho-alveolar lavage fluid and serum in heamatological patients at risk for invasive pulmonary aspergillosis. Br J Haematol. 2003;121:448–57. doi: 10.1046/j.1365-2141.2003.04308.x. [DOI] [PubMed] [Google Scholar]
- 40.Pasqualotto AC, Xavier MO, Sánchez LB, et al. Diagnosis of Invasive Aspergillosis in Lung Transplant recipients by Detection of Galactomannan in the Bronchoalveolar Lavage Fluid. Transplantation. 2010;90:306–11. doi: 10.1097/TP.0b013e3181e49bc1. [DOI] [PubMed] [Google Scholar]
- 41.Husain S, Clancy CJ, Nguyen MJ, et al. Performance Characteristics of the Platelia Aspergillus Enzyme Immunoassay for Detection of Aspergillus Galactomannan Antigen in Bronchoalveolar Lavage Fluid. Clin Vacc Immuno. 2008;15:1760–3. doi: 10.1128/CVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sulahian A, Touratier S, Ribaud P. False-positive test for Aspergillus antigenemia related to concomitant administration of piperacillin and tazobactam. N Engl J Med. 2003;349:2366–7. doi: 10.1056/NEJM200312113492424. [DOI] [PubMed] [Google Scholar]
- 43.Adam O, Auperin A, Wilquin F, et al. Treatment with pipieracillin-tazobactam and false-positive Aspergillus galactomannan antigen test results for patients receiving with hematological malignancies. Clin Infect Dis. 2004;38:917–20. doi: 10.1086/383148. [DOI] [PubMed] [Google Scholar]
- 44.Mattei D, Rapezzi D, Mordini N, et al. False-positive Aspergillus galactomannan enzyme-linked immunosorbent assay results in vivo during amoxicillin-clavulanic acid treatment. J Clin Microbiol. 2004;42:5362–3. doi: 10.1128/JCM.42.11.5362-5363.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hage CA, Reynolds JM, Durkin M, et al. Plasmalyte as a Cause of False-Positive Results for Aspergillus Galactomannan in Bronchoalveolar Lavage Fluid. J Clin Microbiol. 2007;45:676–7. doi: 10.1128/JCM.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marr KA, Laverdiere M, Gugel A. Antifungal therapy decreases sensitivity of the Aspergillus galactomannan enzyme immunoassay. Clin Infect Dis. 2005;40:1762–9. doi: 10.1086/429921. [DOI] [PubMed] [Google Scholar]
- 47.Desmet S, Van Wijngaerden E, Maertens J, et al. Serum (1→3)-β-D-glucan as a tool for diagnosis of Pneymocystis jirovecii pneumonia in patients with human immunodeficiency virus infection or hematologic malignancy. J Clin Microbiol. 2009;47:3871–4. doi: 10.1128/JCM.01756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida M, Obayashi T, Iwama A, et al. Detection of plasma (1→3)- β-D-flucan in patients with Fusarium, Trichosporon, Saccharomyces, and Acremonium fungaemias. J Med Vet Mycol. 1997;35:371–4. [PubMed] [Google Scholar]
- 49.Ostrovsky-Zeichner L, Alexander BD, Kett DH, et al. Multicenter Clinical Evaluation of the (1→3) β-D-Glucan Assay as an Aid to Diagnosis of Fungal Infections in Humans. Clin Infect Dis. 2005;41:654–9. doi: 10.1086/432470. [DOI] [PubMed] [Google Scholar]
- 50.Odabasi Z, Mattiuzzi G, Estey E, et al. β-D-Glucan as a Diagnostic Adjunct for Invasive Fungal Infections: Validation, Cutoff Development, and Performance in Patients with Acute Myelogenous Leukemia and Myelodysplastic Syndrome. Clin Infect Dis. 2004;39:199–205. doi: 10.1086/421944. [DOI] [PubMed] [Google Scholar]
- 51.Ostrovsky-Zeichner L, Alexander BD, Kett DH, et al. Multicenter Clinical Evaluation of the (1→3)- β-D-Glucan Assay as an Aid to the Diagnosis of Fungal Infections in Humans. Clin Infect Dis. 2005;41:654–9. doi: 10.1086/432470. [DOI] [PubMed] [Google Scholar]
- 52.Pasos C, Pontón J, Del Palacio A. Contribution of (1→3)- β-D-glucan chromogenic assay to diagnosis and therapeutic monitoring of invasive aspergillosis in neutropenic adult patients: a comparison with serial screening for circulating galactomannan. J Clin Microbiol. 2005;43:299–305. doi: 10.1128/JCM.43.1.299-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander BD, Smith PB, Davis RD, et al. The (1,3) β-D-Glucan Test as an Aid to Early Diagnosis of Invasive Fungal Infections following Lung Transplantation. J Clin Microbiol. 2010;48:4083–8. doi: 10.1128/JCM.01183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pickering JW, Sant HW, Bowles CA, et al. Evaluation of a (1→3)- β-D-Glucan Assay for Diagnosis of Invasive Fungal Infections. J Clin Microbiol. 2005;43:5957–62. doi: 10.1128/JCM.43.12.5957-5962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mennink-Kersten MA, Warris A, Verweij PE, et al. 1,3-β-D-Glucan in Patients Receiving Intravenous Amoxicillin-Clavulanic Acid. N Engl J Med. 2006;354:2834–5. doi: 10.1056/NEJMc053340. [DOI] [PubMed] [Google Scholar]
- 56.Nakao A, Yasui M, Kawagoe T, et al. False-positive endotoxemia derives from gauze glucan after hepatectomy for hepatocellular carcinoma with cirrhosis. Hepatogastroenterology. 1997;44:1413–8. [PubMed] [Google Scholar]
- 57.Harrison E, Stalhberger T, Whelan R, et al. Aspergillus DNA contamination in blood collection tubes. Diagn Microbiol Infect Dis. 2010;67:392–4. doi: 10.1016/j.diagmicrobio.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer J, Francesconi A, Kasai M, et al. Sources of false positive Aspergillus DNA by PCR from normal human blood. 41st Interscience Conference on Antimicrobial Agents and Chemotherapy; Chicago, IL. Dec 16–19, 2001; p. abstract no. J-844. [Google Scholar]
- 59.Donnelly JP. Polymerase Chain Reaction for Diagnosing Invasive Aspergillosis: Getting Closer but Still a Ways to Go. Clin Infect Dis. 2006;42:487–9. doi: 10.1086/499818. [DOI] [PubMed] [Google Scholar]
- 60.Mengoli C, Cruciani M, Barnes RA, et al. Use of PCR for the diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect Dis. 2009;9:89–96. doi: 10.1016/S1473-3099(09)70019-2. [DOI] [PubMed] [Google Scholar]
- 61.White PL, Perry MD, Moody A, et al. Evaluation of Analytical and Preliminary Clinical Performance of Myconostica MycAssay Aspergillus When Testing Serum Specimens for Diagnosis and Invasive Aspergillosis. J Clin Microbiol. 2011;49:2169–74. doi: 10.1128/JCM.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayette MP, Vaira D, Susin F, et al. Detection of Aspergillus species DNA by PCR in Bronchoalveolar lavage fluid. J Clin Microbiol. 2001;39:2338–40. doi: 10.1128/JCM.39.6.2338-2340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuon FF. A systematic literature review on the diagnosis of invasive aspergillosis using polymerase chain reaction (PCR) from Bronchoalveolar lavage clinical samples. Rev Iberoam Micol. 2007;24:89–94. [PubMed] [Google Scholar]
- 64.Hauser PM, Bille J, Lass-Flörl, et al. Multicenter, Prospective Clinical Evaluation of Respiratory Samples from Subjects at Risk for Pneumocystis jirovecci Infection by Use of a Commercial Real-Time PCR Assay. J Clin Microbiol. 2011;49:1872–8. doi: 10.1128/JCM.02390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balajee SA, Kano R, Baddley JW, et al. Molecular Identification of Aspergillus Species Collected for the Transplant-Associated Infection Surveillance network. J Clin Microbiol. 2009;47:3138–41. doi: 10.1128/JCM.01070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hammond SP, Bialek R, Milner DA, et al. Molecular Methods to Improve Diagnosis and Identification of Mucormycosis. J Clin Microbiol. 2011;49:2151–3. doi: 10.1128/JCM.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rickers V, Just-Nübling G, Knorad F, et al. Diagnosis of invasive aspergillosis and mucormycosis in immunocompromised patients by seminesterd PCR assay of tissue samples. Eur J Clin Microbiol Infect Dis. 2006;25:8–13. doi: 10.1007/s10096-005-0078-7. [DOI] [PubMed] [Google Scholar]
- 68.Rickers V, Mousset S, Lambrecht E, et al. Comparison of Histopathological Analysis, Culture, and Polymerase Chain Reaction Assays to Detect Invasive Mold Infections from Biopsy Specimens. Clin Infect Dis. 2007;44:1078–83. doi: 10.1086/512812. [DOI] [PubMed] [Google Scholar]
- 69.Hayden RT, Qian X, Procop GW, et al. In situ hybridization for the identification of filamentous fungi in tissue section. Diagn Mol Pathol. 2002;11:119–26. doi: 10.1097/00019606-200206000-00009. [DOI] [PubMed] [Google Scholar]
- 70.Hayden RT, Isotalo PA, Parrett T, et al. In situ hybridization for the differentiation of Aspergillus, Fusarium, and Pseudallescheria species in tissue section. Diagn Mol Pathol. 2003;12:21–6. doi: 10.1097/00019606-200303000-00003. [DOI] [PubMed] [Google Scholar]
- 71.Gonzales DA, De Torre C, Wang H, et al. Protein expression profiles distinguish between experimental invasive pulmonary aspergillosis and Pseudomonas pneumonia. Proteomics. 2010;10:4270–80. doi: 10.1002/pmic.200900768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sainz J, Salas-Alvarado I, López-Fernández E, et al. TNFR1 mRNA expression level and TNFR1 gene polymorphisms are predictive markers for susceptibility to develop invasive pulmonary aspergillosis. Int J Immunopathol Pharmacol. 2010;23:423–36. doi: 10.1177/039463201002300205. [DOI] [PubMed] [Google Scholar]
- 73.Loeffler J, Ok M, Morton OC, et al. Genetic polymorphisms in the cytokine and chemokine system: their possible importance in allogenic stem cell transplantation. Curr Top Microbiol Immunol. 2010;341:83–96. doi: 10.1007/82_2010_22. [DOI] [PubMed] [Google Scholar]
- 74.Zaas AK, Liao G, Chien JW, et al. Plasminogen alleles influence susceptibility to invasive aspergillosis. PloS Genet. 2008;4:e1000101. doi: 10.1371/journal.pgen.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bochud PY, Chien JW, Marr KA, et al. Toll-like Receptor 4 Polymorphisms and Aspergillosis in Stem-Cell Transplantation. New Engl J Med. 2008;359:1766–77. doi: 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]


