Abstract
The genetic architecture of human diseases governs the success of genetic mapping and the future of personalized medicine. Although numerous studies have queried the genetic basis of common disease, contradictory hypotheses have been advocated about features of genetic architecture (e.g., the contribution of rare vs. common variants). We developed an integrated simulation framework, calibrated to empirical data, to enable systematic evaluation of such hypotheses. For type 2 diabetes (T2D), two simple parameters – (a) the target size for causal mutation and (b) the coupling between selection and phenotypic effect – define a broad space of architectures. While extreme models are excluded, many models remain consistent with epidemiology, linkage, and genome-wide association studies for T2D, including those where rare variants explain little (<25%) or most (>80%) of heritability. Ongoing sequencing and genotyping studies will further constrain architecture, but very large samples (e.g., >250K unselected individuals) will be required to localize most of the heritability underlying traits like T2D.
INTRODUCTION
The genetic architecture of human diseases – that is, the number, frequencies, and effect sizes of causal alleles – has profound implications for the future of genetic research and its impact on clinical medicine. Targeting of diagnosis and therapeutics based on individual genome sequence will be more tractable for diseases caused by rare mutations of large effect than for diseases where many genes and variants together contribute.1–4 Similarly, the efficiency and power of research study designs5–8 and analytical methods9,10 depend critically on the underlying distribution of causal allele frequencies and effect sizes.
Complex disease architecture can be examined via several methods: epidemiological studies of twin and sibling concordance11–14, family-based linkage scans15–17, genome-wide association studies (GWAS)7, including ‘polygene’ analyses combining data from large numbers of common variants18,19, and (more recently) genome sequencing in phenotyped individuals.20–25 Each individual study design, however, provides a limited glimpse into the full architecture of a given trait, and to date only ~5–20% of heritability for most common diseases has been explained (most due to loci identified in GWAS).26,27
There has been much focus on this so-called “missing heritability” of disease.28 Some have argued that the unexplained heritability lies in a large number of common, individually weak alleles.18,19,29,30 Conversely, the numerous rare variants revealed by exome sequencing studies31–35 have been interpreted as evidence that rare alleles explain the majority of heritability; it has been proposed that hundreds of rare monogenic sub-phenotypes exist for each common disease,36–38 and that GWAS results may be due to ‘synthetic’ associations caused by rare variants on common disease-associated haplotypes39. Others have suggested epistasis, epigenetics, or parent-of-origin-specific effects.27,40,41
In order to systematically evaluate these and other hypotheses, it is necessary to compare the predictions of each model to empirical data from not just one but all available genetic studies in a unified framework. Here, we asked: which models are consistent with the cumulative results of studies already performed, and which models can be excluded? Are models where common variants predominate plausible despite the large number of rare alleles segregating in human populations? Are rare variant models compatible with the generally negative findings of family-based linkage studies, and the numerous disease loci found in GWAS?
To address these questions, we developed a population genetic framework to directly simulate, in large populations, a wide space of genetic architectures. Focusing on the test case of type 2 diabetes (T2D), we quantitatively evaluate each hypothesis about genetic architecture by simulating genetic studies as they were conducted for T2D, and asking whether simulated results are consistent with empirical observation.
RESULTS
Simple models of complex diseases
The genetic architecture of any trait has – by necessity – been shaped by population genetic forces. Mutations at some (but not all) genomic loci have the potential to alter disease risk (we refer to these as the disease ‘target’). Genetic drift and gene flow, influenced by demographic history and population migration, cause fluctuations in allele frequencies independent of phenotype. Finally, natural selection results in directional changes in the frequencies of alleles that influence evolutionary ‘fitness’, which is itself a composite of many traits (including, potentially, the disease of interest).
Analytical or simulation-based models have yielded insight into the qualitative dependencies of genetic architecture (usually at single loci) on subsets of these parameters.10,42–47 For example, explosive population growth following a bottleneck can allow even deleterious disease alleles to reach common population frequency.45 Conversely, strong selection against disease47, or high mutation rates coupled with mild selection46, could, in principle, enable rare alleles to explain most of heritability.
To quantitatively investigate the extent to which such models are consistent with emerging data from association studies and population-based sequencing, we performed simulations that enabled granular predictions of genome-wide genetic architecture and study results. Although the number of disease model parameters is potentially without bound, we sought to generate the simplest possible models considering only mutations (of additive effect), genetic drift, and purifying selection. If such simple models produce predictions inconsistent with empirical data, this does not imply that more complex models could not be consistent. However, if a simple model is consistent, then we can conclude that its features are indeed plausible given current data.
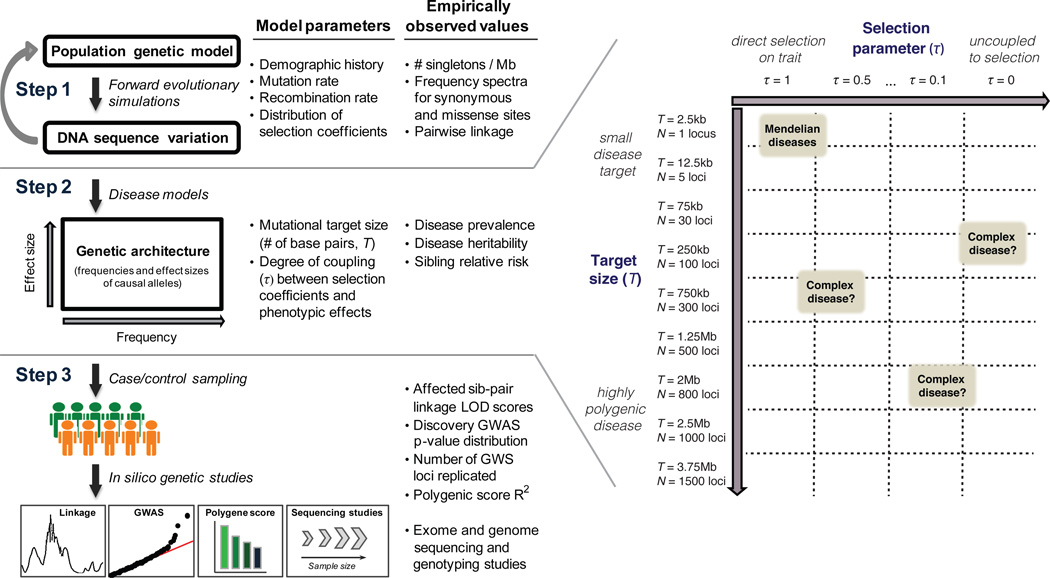
Based on these considerations, we developed a three-stage framework: (1) forward evolutionary simulation to generate multi-locus DNA sequence variation at large scale (hundreds of thousands of individuals) that is consistent with empirical sequencing data, (2) mapping of genotype to phenotype under a range of disease models, and (3) in silico prediction of genetic study results under each model (Fig. 1). For simplicity, our analysis focuses on Northern European populations.
Figure.1. Framework for specification and evaluation of disease models.
Stage 1: Simulation of DNA variation at population-scale
Three main processes determine the spectrum of DNA variation: (a) mutation and recombination, (b) demographic history, and (c) natural selection on segregating alleles. To fit values for these parameters, we generated forward simulations of thousands of unlinked loci resembling protein-coding genes in 500,000 individuals using the software ForSim (Supplementary Fig. 1).48
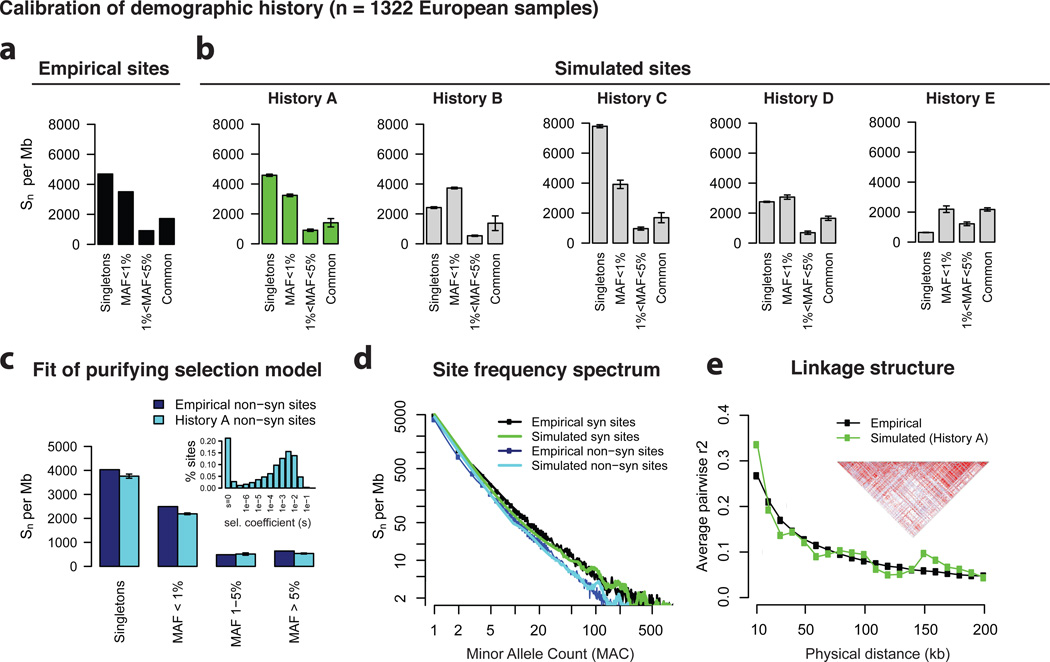
We initially parameterized simulations using previously published demographic histories9,49,50, and then tested a grid over parameter space (Supplementary Table 1) to find the best fit to empirical data; for comparison, we also tested a naïve history of constant population size (N=10K). Specifically, we asked whether the simulated site frequency spectrum (SFS) under each history matched the empirical SFS of synonymous sites (assumed evolutionary neutral) observed in 1,322 European exomes sequenced by the GoT2D Consortium (Fig. 2a–b, Supplementary Figs. 2–4). The best match to empirical data was produced by a hybrid model with features of two published histories (‘History A’, parameters in Fig. 2); this model recapitulated the number and frequency distribution of both rare and common synonymous sites (Fig. 2b), as well as empirically observed patterns of linkage disequilibrium between common variants (Fig. 2e, Supplementary Fig. 5).
Figure.2. Patterns of genetic variation: forward simulated vs. empirically observed.
a) Number of singleton, rare (MAF<1%), intermediate frequency (1%<MAF<5%), and common (MAF>5%) synonymous sites per Mb of mutational target in empirical data from GoT2D Consortium, n=1322 European samples. b) Number of simulated neutrally evolving sites per Mb under different human demographic histories: A = history chosen in this study (µ=2e-8, Na=8.1K, Nb=2K, te=370 generations, re=1.3%, Ne=228K), B = Gravel et al (µ=2.4e-8, Na=7.3K->14.4K, Nb=1.8K->1.0K, te=920 generations, re=0.4%, Ne=35.9K), C = Kryukov et al (µ=1.8e-8, Na=8.1K, Nb=7.9K, te=370 generations, re=1.3%, Ne=900K), D = Schaffner et al (µ=1.5e-8, Na=12.5K, Nb=7.7K->540, te=350 generations, re=0.7%, Ne=100K), E = Fixed 10K population (Na=Nb=Ne=10K). c) Number of non-synonymous (under purifying selection) sites per Mb in empirical data (dark blue) and in forward simulated data (light blue) using chosen demographic history and distribution of selection coefficients (inset). d) Full site frequency spectrum (n = 1322 samples) of simulated synonymous (green) and non-synonymous (light blue) sites compared to those in empirical data (black, dark blue). e) Average pairwise LD (measured by r2) as a function of physical distance between frequency-matched common (MAF > 5%) in simulated (green) and empirical (black) data. Linkage structure at a representative 200kb forward simulated locus, as generated in Haploview (inset).
We next fit the distribution of purifying selection on protein-coding mutations by performing forward simulations under History A while applying per-variant selection coefficients drawn from a range of gamma distributions (as in previous reports9,51, we assume ~20% of non-synonymous sites are neutrally evolving). The best-fit gamma distribution produced a SFS closely matching that of non-synonymous sites in empirical data (Fig. 2c–d, Supplementary Figs. 6–7). We assume that all disease loci are under the same distribution of purifying selection (of strength comparable to selection against protein-coding changes). This simplifying assumption is likely reasonable for at least a portion of conserved non-coding regulatory elements52, but future work will consider selection distributions matched to different classes of biologically functional loci.
As Fig. 2 demonstrates, a few simple evolutionary parameters can produce sequence variation consistent with results of a large-scale exome sequencing study (parameters available as a ForSim configuration file). We next used these simulations to explore the relationship between allele frequency and deleteriousness. There has been much recent debate, based on abundant rare variation observed in sequencing studies8,34, about what fraction of deleterious variation is rare, and conversely, what fraction of rare variation is deleterious. Under our simulations, we find that while >90% of deleterious (s>0.0005) non-synonymous variants are indeed rare (MAF<0.1%; Supplementary Fig. 8), fewer than 45% of all rare non-synonymous variants are deleterious (consistent also with empirical estimates using prediction tools such as PolyPhen34). Thus, most rare variants are simply of recent age (Supplementary Figs. 9–10), and it would be inappropriate to infer functional consequence based on frequency alone.
Stage 2: Specification of disease models
Under an additive liability threshold model, the relationship between genotype and phenotype is controlled by (a) the number of disease variants each individual carries; (b) causal variants’ effects on disease (these may or may not be related to variant selection coefficients); (c) the magnitude of non-genetic (e.g., environmental) influences; and (d) the liability threshold above which disease ensues. By modulating these levers, it is possible to model a principled distribution of causal variant frequencies and effect sizes rather than specify them ad hoc.
To map genotype to phenotype for a specific disease, we focused on type 2 diabetes (T2D). The prevalence of T2D (~8%53) determines the liability threshold, and the heritability (~45%, estimated from family studies54) determines the magnitude of genetic (compared to environmental) effects (Supplementary Fig. 11, Supplementary Note). We confirmed that the T2D heritability specified under each disease model could be recovered via phenotypic regression and analysis of variance in full-sibling pairs sampled from simulated populations (Supplementary Fig. 12).
The number of disease variants carried by an individual is determined by the mutational target size (T), or the sum total of nucleotides that, if mutated, would influence risk of disease. In the current study, we assume that causal mutations occur only at sites under evolutionary constraint similar to that at non-synonymous changes under purifying selection; thus only protein-coding loci and some conserved non-coding regions55,56 (collectively spanning ~10%, or ~300Mb, of the human genome52,57) contribute to disease. We simulated models with T ranging from 75kb-3.75Mb, corresponding to 0.02%–1.2% of constrained genome sequence. To model linkage between variants at structurally contiguous regions, we grouped the disease target into ‘loci’ (N=30, 100, 300, 500, 800, or 1500 causal loci in each model). Each locus contains 2.4kb of functional target (under selection) flanked by neutrally evolving regions (Supplementary Fig. 1).
While purifying selection against lethal Mendelian diseases is direct and evident, the relationship between selection and post-reproductive common diseases is less clear. We therefore model apparent selection, where fitness is a composite of many traits, with a range of possible mappings between a variant’s effect on fitness (measured by the selection coefficient, s) and its effect on a particular disease (g). We model this mapping with a single parameter (τ), which quantifies this ‘coupling’: g = sτ(Eyre-Walker47). We performed simulations with τ = 0, 0.1, 0.2, 0.3, 0.4, 0.5, and 1. Where τ=1 (‘tightly coupled’), variants with large effects on fitness have large effects on disease. Where τ=0 (‘uncoupled’), there is no relationship between the selection coefficients of causal mutations and their impact on the disease of interest (Supplementary Fig. 13).
Genetic architecture resulting under each disease model
We first verified that models in the two-dimensional parameter space (of T and τ) produce architectures with qualitative features consistent with analytical expectation.
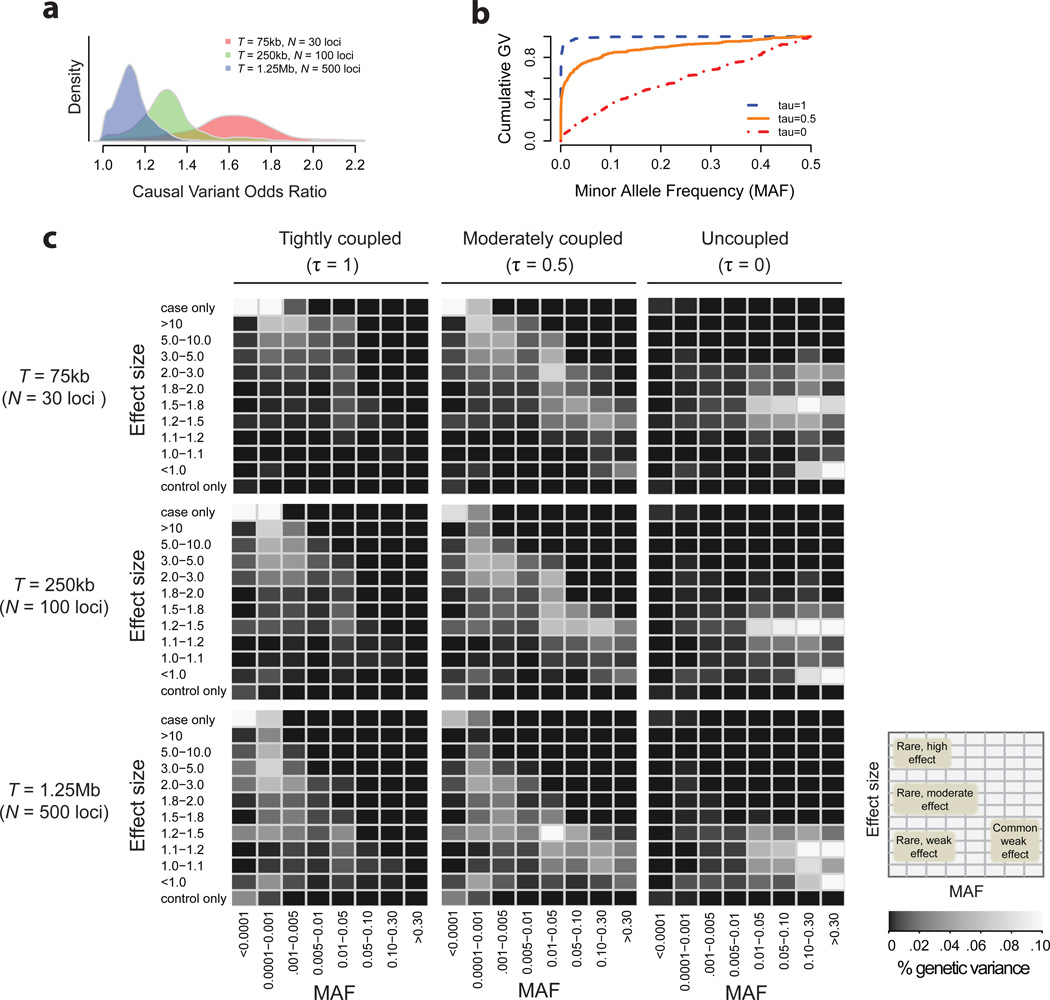
We first asked: how do rare and common variant effect sizes compare under different models? Under tightly coupled (τ=1) models, rare variants (those under strong purifying selection) have much larger effects than common variants, while under uncoupled (τ=0) models, rare and common alleles have comparable phenotypic effects (Supplementary Fig. 13–14). In contrast, the target size does not impact the relative effect sizes of rare and common variants; rather, increases in target size reduce causal variant effects across the entire frequency spectrum. This occurs because T2D prevalence and heritability are fixed, so a larger number of causal variants must be counteracted by smaller per-variant effects (Fig. 3a; Supplementary Table 2). Notably, under all models, the high prevalence and modest heritability of T2D constrain common (MAF>5%) variants to odds ratios <2, even at relatively small target sizes (e.g. T=75kb).
Figure.3. Sensitivity of genetic architecture to parameters of disease models.
a) Density of odds ratios (as measured in a sampled cohort of 10K individuals) for common (MAF > 2%) causal variants under disease models with varying target sizes; for all three models shown here there is no coupling to selection (τ = 0). b) Cumulative portion of population genetic variance explained by causal variants as a function of their minor allele frequency under disease models with different degrees of coupling to selection; for all three models shown here target size (T) is fixed at 500 functional loci. c) Heat maps showing distribution of population genetic variance in the two-dimensional minor allele frequency (x-axis) and effect size (y-axis) space of causal variants; models shown are for τ = 1.0, 0.5, and 0 and T = 75kb, 250kb, and 1.25Mb (N = 30, 100, and 500 causal loci).
Next, we asked: how is disease heritability partitioned by allele frequency across models? The contribution of each causal variant to heritability (population genetic variance) is: Va = = 2 * (g2) * (1 – f) * f58, where is the variant’s additive effect and f is its frequency. Under tightly coupled (τ=1) models, where is very large for some rare alleles (often private to cases), the rare class (MAF<1%) collectively explains >90% of heritability. Conversely, under uncoupled models (τ=0), common (MAF>5%) alleles with modest effects (OR<1.2) explain ~95% of heritability (Fig. 3b–c, Supplementary Fig. 15). These relationships hold regardless of target size.
Finally, we examined the distribution of variant effects within each individual (rather than population-wide) to evaluate the potential of individualized risk prediction. Under tightly coupled models, patients with T2D have only few (1–5) high-effect risk alleles that are rarely seen among unaffected individuals. Conversely, under weakly coupled models with similar target size, each patient has hundreds of risk alleles with similar individual and cumulative effect (Supplementary Fig. 16–17); moreover, most of these are also commonly observed among controls. Thus, for a given target size, genetic risk prediction will be far more informative (diagnostic for some patients, assuming effects at rare alleles can be discovered and accurately quantified) if there is strong coupling to selection. This confirms the widely-discussed intuition that, under rare variant models of common disease, sequencing studies may greatly enhance clinical prediction.
In summary, simple disease models with only two free parameters (target size, coupling to selection) generate diverse genetic architectures (Fig. 3c) with qualitative features consistent with prior expectation.
Stage 3: Simulation of genetic study results
We next addressed our main question: which models produce genetic study results compatible with observed data in genetic studies of T2D?
To define the set of genetic studies to simulate, we collated results from published studies of T2D in European populations. These data included: (a) epidemiological estimates of sibling relative risk (~1.8–3.454,59,60); (b) meta-analysis of linkage scans in ~4,200 affected sibling pairs (ASPs) with T2D (max LOD score 2.261); (c) discovery GWAS in 4,549 cases and 5,579 controls (DIAGRAMv162; two genome-wide significant loci with p<5e-08); (d) replication of the top (p<0.0001) signals from the discovery GWAS in an effective sample size of ~55K (~16 genome-wide significant loci62); (e) larger-scale meta-analysis in 12,171 cases and 56,862 controls (DIAGRAMv3), followed by genotyping of top (p<0.005) signals on the Metabochip array in 34K cases and 115K controls63,64 (39 genome-wide significant loci; Supplementary Table 3); and (f) ‘polygene score’ logistic regression18 using thousands of common marker effects learned in discovery GWAS (together, these explain 2.0–2.5% of test sample variance, measured by Nagelkerke’s R2).19
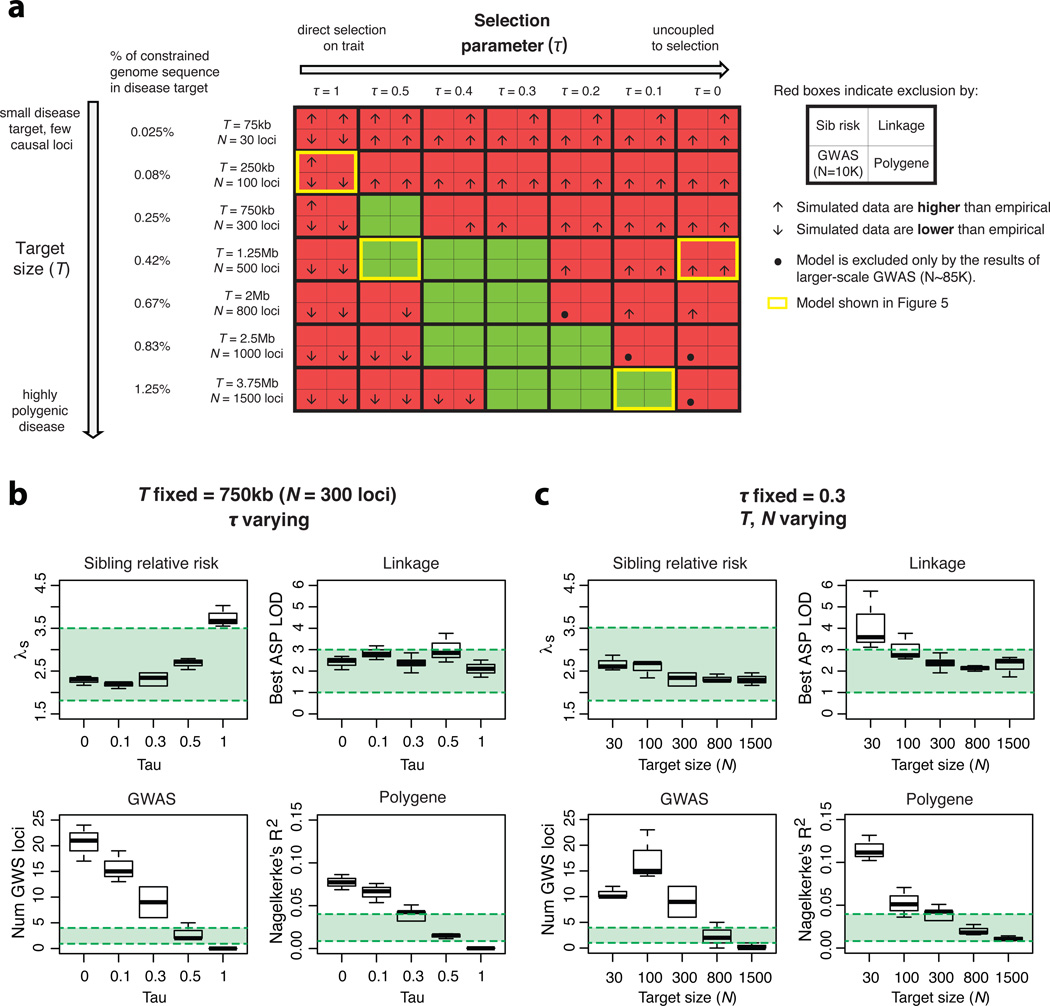
We simulated over 50 distinct disease models spanning a range of target sizes and selection parameters (Fig. 4a). Under each model, we performed the above genetic studies, matching assay type and effective sample size (rather than total sample size, due to cohort structure; Supplementary Table 3) to empirical studies. Each simulated study was analyzed without knowledge of which variants were causal (as would be the case in an actual study). As expected, the results of each study depend heavily on the genetic architecture (Fig. 4b–c, Supplementary Fig. 18–19).
Figure.4. Genetic study results for type 2 diabetes under different disease models.
a) Space of disease models tested, each varying in target size (vertical axis) and selection coupling (horizontal axis). All models have fixed prevalence (8%) and heritability (45%), matching values observed for T2D. Each model produces results that are either inconsistent (red) or consistent (green) with empirical data for T2D. Inside red models, arrows indicate whether simulated results were too high or too low relative to empirical data (see Supp. Figure 17 for further detail). Dots in GWAS boxes indicate that the model is excluded by an excess of findings in large-scale (N~85K) GWAS (though results in 10K samples are consistent). b–c) Sensitivity of study results under models with N fixed at 300 loci and τ varying (b) or τ fixed at 0.3 and N varying (c). In each box, simulated data are shown (clockwise) for sibling relative risk, best genome-wide LOD score in an affected sibling pair (ASP) study of 4200 ASPs, number of genome-wide significant (p-value < 5*10^-8) loci detected in a GWAS of ~10K samples, and the Nagelkerke’s R2 value in a polygene score logistic regression in 5K samples, using common variants with a discovery p-value < 0.01 (PT = 0.01). Green zones are centered (vertically) on empirically observed values for T2D, and represent the simulated values deemed consistent with empirical data (see Methods).
We first evaluated ‘tightly coupled’ models (τ=1) for consistency with empirical T2D data (Fig. 5a–b). These models produce relatively high (λs=4.2) sibling risk due to rare, high effect mutations shared by ASPs. Linkage peaks (LOD>3.0) in simulated ASP studies are rarely observed, however, and only at small target sizes: 90% of replicates under models with T<75kb (N<30 loci) yield a linkage peak, whereas fewer than 20% do when T>250kb (N=100 loci). Thus, while empirical linkage data (where no LOD>2.2 was observed) can exclude oligogenic ‘tightly coupled’ hypotheses, they cannot rule out models with larger T. GWAS results, however, are sufficient to exclude all ‘tightly coupled’ models, regardless of target size: under complete coupling, too few causal variants are common enough to reach genome-wide significance even after large-scale follow-up (4–5 loci when T=250kb, compared to 39 in empirical data). Under tightly coupled models, polygene score regression is less successful than empirically observed (R2<0.5%, compared to ~2% for T2D; Fig. 4; Supplementary Fig. 18). A mixed linear modeling approach using common SNPs65 also recovers a much smaller fraction (<10%) of T2D heritability than has been empirically reported64 (Supplementary Fig. 19).
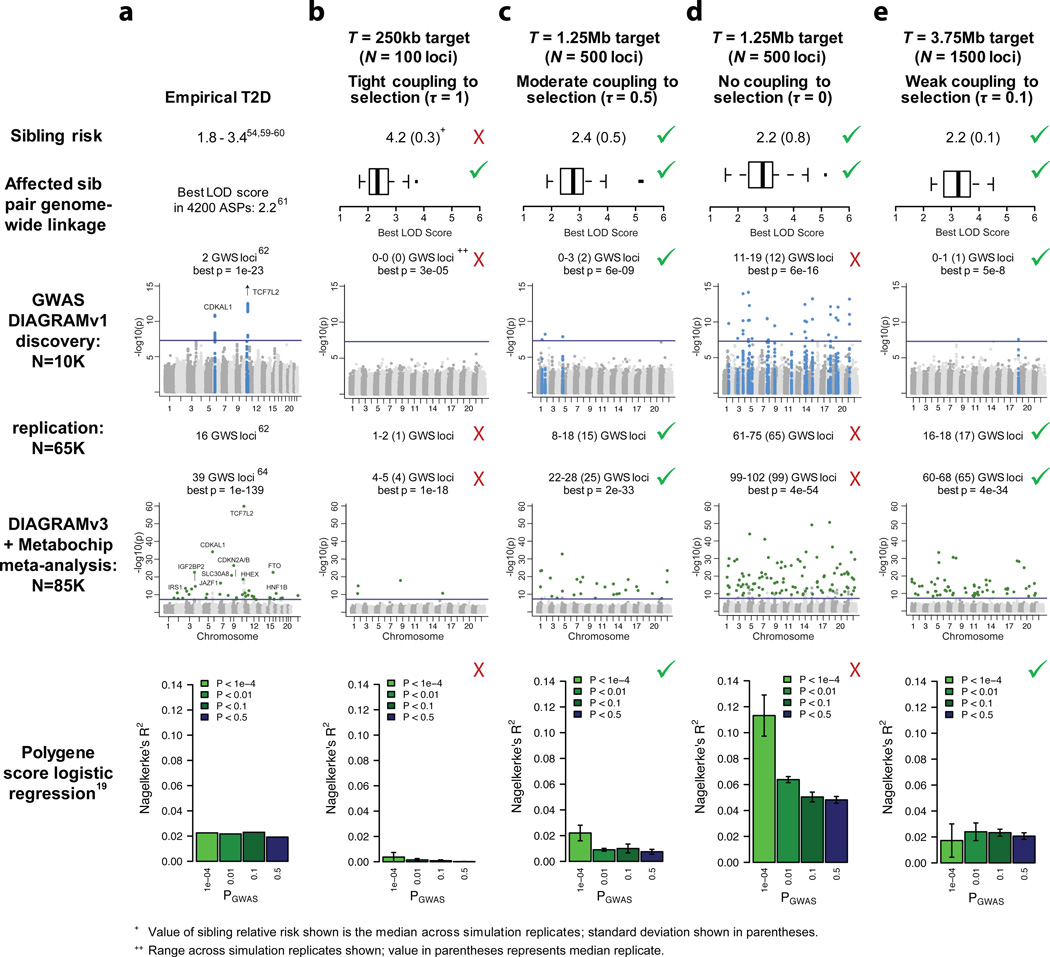
Figure.5. Simulated study results under representative disease models and comparison to T2D empirical data.
At left (a) are empirical genetic study results for type 2 diabetes (black outline, see Methods for detail). At right are simulated genetic study results for four different disease models. b) T = 250kb (N = 100 loci), τ =1 (tight coupling to selection); an ‘extreme’ rare variant model. c) T = 1.25Mb (N = 500 loci), τ =0.5 (moderate coupling to selection); an intermediate model. d) T = 1.25Mb (N = 500 loci), τ =0 (no coupling to selection); a ‘common polygenic’ model. e) T = 3.75Mb (N = 1500 loci), τ =0.1 (weak coupling to selection); a highly polygenic hybrid model. Red crosses indicate inconsistency with empirical data for T2D; green checks indicate consistency with empirical data. ‘GWS loci’ refers to the number of unique loci at which a variant is associated to disease at genome-wide significance levels (p<5e-8).
Next, we evaluated ‘uncoupled’ (τ=0) hypotheses. These models produce modest risk to sibs (λs≈2) and lack positive linkage results (for T>250kb), consistent with observed data. However, across a wide range of uncoupled models (up to T=3.75Mb, or N=1500 loci), an excess of GWAS findings is observed. An example of such a model (τ=0, T=1.25Mb, or N=500 loci) is shown in Fig. 5d; 11–19 GWAS loci are found in discovery (as compared to 2 in empirical data), 61–71 loci after replication (16 empirically), and 99–102 loci in the large-scale GWAS followed by Metabochip genotyping (39 empirically). Under this uncoupled model, polygene score regression also explains a larger proportion of phenotypic variance than observed for T2D (R2>10% at p<1e-4, compared to ~2% in empirical data; Fig. 4; Supplementary Fig. 18).
While these extreme models of genetic architecture are inconsistent with empirical data, a broad continuum of intermediate models remains consistent (Fig. 4a). This class of consistent models includes those with moderate coupling and smaller target sizes, as well as those with weak coupling and larger target sizes. Two examples are shown in Fig. 5c (‘moderate’; τ=0.5, T=1.25Mb, or N=500 loci) and Fig. 5e (‘weakly coupled’; τ=0.1, T=3.75Mb, or N=1500 loci). Predicted outcomes under both models are consistent with empirical data. However, these architectures have quite distinct properties: under the ‘moderate’ model, rare (MAF<5%) alleles explain ~80% of heritability, while under the ‘weakly coupled’ model, rare variants explain <25% of heritability (Supplementary Fig. 15).
Prediction of results from future studies
Ongoing studies are now using (a) exome and whole-genome sequencing and (b) genotyping via an exome array to study rare and intermediate frequency variants in modest (thousands) and large (tens of thousands) samples, respectively. In coming years, it is predicted that sequencing will be performed in hundreds of thousands or even millions of people. To what extent will these ongoing and future studies further constrain T2D genetic architecture?
We simulated high-coverage, whole-genome sequencing of 3K and 10K individuals (sample sizes similar to those of studies being performed by the Go-T2D and T2D-GENES Projects, respectively), as well as a study in which a large proportion of rare coding variants are genotyped in 20K cases and 35K controls (also similar to ongoing studies for T2D). In each study, we simulated single variant association as well as gene-based association (Methods). To project studies that might be done in coming years, such as in the UK Biobank, we simulated complete genome sequencing of an unselected population cohort of 250K individuals (20K cases, 230K controls).
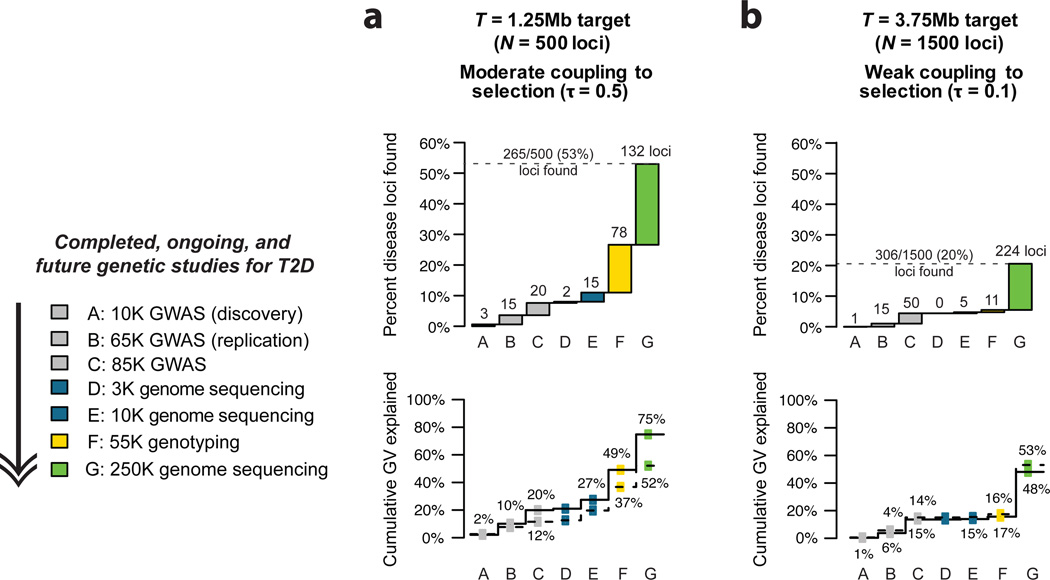
We then asked: at what point will disease models that are currently consistent with all available data diverge in future studies? As examples, we focused on the two consistent models depicted in Fig. 5c (‘moderate’) and Fig. 5e (‘weakly coupled’). For both models, whole-genome sequencing in 3K individuals discovers few signals not previously detected by GWAS. In 10K samples, the models diverge slightly: ~15 novel loci (representing ~6% of heritability) are predicted under the ‘moderate’ model, whereas ~5 loci (representing <1% of heritability) are predicted under the ‘weakly coupled’ model. The most significant constraint, however, is predicted to come from large exome array studies: ~80 novel loci under the ‘moderate’ model (bringing cumulative heritability explained to ~50%), but only ~10 loci under the ‘weakly coupled’ model (and ~15% of heritability explained). Thus, at least one of these models will likely be inconsistent with the results of studies already planned for T2D.
As sample size is expanded to 250K unselected individuals, these models diverge further. In both cases, substantial discovery is predicted, but the total fraction of heritability explained, as well as the frequency distribution of identified causal variants, differs. Under the ‘moderate’ model, over half (~265 out of 500) of all disease loci would be discovered, and would collectively explain ~75% of T2D heritability. At a majority of loci, the most disease-associated variant would be rare (MAF<2%). Under the ‘weakly coupled’ model, a much larger fraction of disease loci would remain undetected (due to the individually small effect sizes of very many causal variants), and a smaller proportion of total heritability (~48%) would be explained. However, the most associated variant at virtually all these loci would be common (MAF>2%), and thus likely discoverable by GWAS of comparable sample size, without need for complete sequencing.
Thus, ongoing sequencing and genotyping studies (and the extent to which they are successful) will likely place substantial bounds on T2D genetic architecture. However, enumerating the full set of causal loci contributing to inherited risk of disease will be extremely challenging even in the limit of very large samples.
DISCUSSION
We developed a hypothesis testing framework, calibrated to empirical data, in which precisely defined disease models produce falsifiable predictions66 about the results of genetic studies. Application of this framework to T2D excludes a subset of extreme architectures inconsistent with linkage, GWAS, or polygene results, but also identifies a range of consistent models with widely varying features. Importantly, all simulated global and locus-level architectures (genotype and phenotype) under these consistent models are freely available for use in the development and evaluation of study designs or novel analytical methods.
The current study has many limitations. Although only two model parameters were sufficient to generate diverse architectures, more parameters could be included. For example, causal variants were simulated only at regions under purifying selection (alternate models where neutrally evolving alleles have effects on disease are explored in Supplemental Fig. 20). Positive selection was not simulated, and derived alleles were only modeled as increasing disease risk (though interestingly, this does not preclude the occurrence of significantly associated markers of protective effect, which may have implications for interpreting the causal direction of effect from GWAS associations; Supplementary Fig. 21).
Additionally, locus structure in our study was uniform; heterogeneity in phenotypic contributions across loci arose only from stochastic sequence variation (Supplementary Fig. 22). Adding skew in the distribution of length, overall phenotypic contribution, and coupling to selection across disease loci could produce more varied models. Finally, non-additive inheritance, epistasis, or gene-environment interactions were not modeled. In future work, if the outcomes of many genetic studies (such as those directly simulated here) in human populations could be accurately predicted using analytical solutions, an inferential approach could enable efficient traversal across disease models defined by many more variable parameters.
Nonetheless, in the current study, specifying simple parameters enabled us to systematically evaluate and characterize in depth a broad space of easily-understood disease models. Although an infinite number of more complex models exist, a single simple model which produces results consistent with empirical data is alone sufficient to conclude that its properties remain currently plausible. Having found plausible models with widely varied genetic architectures, our results have a number of implications.
First, many specific hypotheses about genetic architecture cannot be adjudicated using single pieces of empirical data. For example, our results suggest that ‘synthetic associations’39,67,68, while rarely observed in simulations under consistent models, cannot be excluded based on the absence of linkage findings alone (Supplementary Fig. 23). Linkage data, in fact, do not place substantial bounds on global T2D architecture at all; only oligogenic models in which variants at a single gene have very large effects (Supplementary Fig. 24) can be excluded because empirical studies were under-powered69 to differentiate other models. Similarly, observation of numerous rare alleles in sequencing studies is not, in itself, evidence to support Mendelian models of common disease.
Second, our work shows that even broad conclusions about the validity of the ‘common disease common variant’ (CDCV) and ‘common disease rare variant’ (CDRV) hypotheses45,46,70 are premature – and further, that the answer may long remain elusive. For T2D, empirical data firmly exclude extreme models such as those where rare variants are entirely responsible for disease, but even with only two free parameters it is possible to generate models that are consistent with all available data, and yet have nearly opposite properties with respect to rare vs. common variant contributions. Multiple recent studies29,30,71,72 have demonstrated that a large fraction (~50% on average71) of common disease heritability is tagged by common markers, but our study suggests these data may still be consistent with a significant causal role for rare variants (Supplementary Fig. 19); studies directly assaying such variation will provide further discriminatory power.
Finally, our simulations indicate that hundreds of thousands of individuals will be required to discover most of the genes underlying complex diseases like T2D, and that even then a substantial fraction of heritability (and causal loci) may remain undiscovered. This is not meant as nihilistic – already much has been learned about the genetic basis of T2D, and our study suggests that in coming years a great deal more will be discovered, including further constraints on models of genetic architecture. However, the challenge of localizing disease heritability may simply be the expected outcome for a population genetic process which results in many causal alleles, strong and weak, that are both common and rare.
ONLINE METHODS
Forward simulation of population-scale data
Large populations were forward simulated according to a wide range of demographic histories and selection coefficient distributions (see S. Tables 1–2) using the publicly available software package ForSim. We varied demographic history parameters including the mutation rate (µ), recombination rate (R), ancestral population size (Na), bottleneck size (Nb), duration (te) and rate (re) of exponential growth, and modern effective population size (Ne). Selection coefficient distributions were modeled as gamma distributions, which a shape and scale parameter; a grid search was performed around values previously published by Kryukov et al. Best-fit parameters were determined by repeatedly sampling individuals (n=63, n=243, and n=1322) from simulated populations, and comparing the average sample SFS to the observed SFS in empirical data (n=63 CEU and n=243 EUR in 1000G exome data, n=1300 Europeans in T2D-GO exome data). Frequency spectra were compared by normalizing the mutation target to 1Mb and correcting for imperfect sensitivity in empirical data (see Supplementary Note).
Simulated disease locus structure
Simulated disease loci were modeled as protein-coding loci (exons with causal variation, alternating with neutrally evolving introns). 100kb of neutrally evolving target was simulated flanking each gene to facilitate downstream genetic studies requiring markers in a large window around causal loci.
Modeling of complex disease phenotype
Simulated genotypes were mapped to phenotype using an additive liability threshold model. Each variant is modeled as having additive effect g = s τ (Eyre-Walker et al). Here g is the variant’s effect on the quantitative trait underlying disease phenotype, and s is the selection coefficient under which that variant evolved. Each individual is assigned a total ‘genetic phenotype’ G by summing effects across all variants for which an individual carries the novel allele, across all disease-causing loci as: , where gij is the effect of the ith variant at the jth locus at which individual k carries a disease-causing allele. The target size is represented by N, the total number of causal loci over which genetic effects is summed. These total genetic phenotypes are then converted to z-scores, and environmental phenotypes E are randomly assigned to each individual such that the total fraction of phenotypic variance attributable to genetic risk is given by the disease heritability (h): . Given this constraint, each individual’s total phenotype P is given by: , where values of E are drawn from a normal distribution. The disease prevalence (8% for T2D) determines the threshold for P above which individuals are assigned categorical disease status. We repeat this full population-scale simulation of disease 25 times for each disease model (represented by a pairing of N and τ.
Simulation of linkage and sibling measurements
From each simulated population, we sampled 10K unrelated cases and controls. Because we simulate nuclear families with multiple offspring (mean two offspring per mating) in each generation, knowledge of sibling genotype and phenotype is available in each simulation. For each of the sampled cases and controls, we ask whether their siblings are also affected with T2D. The fraction of cases’ siblings who are affected divided by the fraction of controls’ siblings who are affected yields the sibling relative risk. To perform affected sibling pair (ASP) linkage studies, we sample 4200 (matching the size of the largest European ASP meta-analysis for T2D) sibling pairs in which both siblings are affected with T2D. SNP data provides a marker map that is significantly denser, but less polymorphic, than the microsatellite marker maps that were used in published studies. To model this, full sequence data was down-sampled across all causal loci; we included only variants with MAF>5% and pairwise LD (measured by r2) < 0.2. The software package MERLIN (http://www.sph.umich.edu/csg/abecasis/merlin/) was used to conduct non-parametric linkage analysis. The Z-scores resulting from such analyses are normally distributed; to generate LOD scores across ‘background’ non-causal loci, we randomly sampled 500 independent Z-scores from a normal distribution (representing a unique marker every ~5Mb of the human genome, similar to typical microsatellite map densities) and converted these to LOD scores using the relation: LOD = Z2/(2*ln(10)). We recorded the genome-wide (across both causal and non-causal loci) maximum LOD score in each simulated study. Simulated models yielding a sibling relative risk of 1.8–3.5 (similar to the range observed across epidemiological studies of T2D) and no genome-wide LOD score greater than 3.0 (maximum LOD score observed T2D was 2.2) were deemed consistent with empirical data for T2D.
Simulation of GWAS
We simulated discovery phase GWAS for T2D (similar to DIAGRAM v1 stage 1) by sampling 4,549 cases with T2D and 5,579 controls from simulated populations under each disease model. To simulate commercial GWAS arrays, full-sequence data across all causal loci was down-sampled; we included only variants with MAF>5% and pairwise LD (measured by r2) < 0.5. We performed standard association analysis using the software PLINK. To model markers across background non-causal loci, we randomly sampled p-values between 0 and 1 to fill a total marker set of 2M SNPs (2.2M total SNPs were imputed, for comparison, in the DIAGRAMv1 study). We used the resulting distribution of genome-wide marker p-values to generate quantile-quantile plots and Manhattan plots for comparison to empirical data. We recorded the number of unique loci at which a marker p-value was < 5e-8. To simulate replication GWAS, we genotyped all markers from the discovery phase at which p < 0.0001 in 20K cases and 35K controls (effective sample size matched to that in DIAGRAMv1 replication), and performed association testing in this larger sample. The resulting p-values were used to determine the number of unique genome-wide significant loci predicted under each disease model after replication. Finally, we simulated large-scale GWAS in an effective sample size of ~35K total individuals, similar to DIAGRAMv3; we then simulated genotyping of all independent signals with p<0.005 on a genotyping array like Metabochip in an effective sample size of ~85K. When appropriate, sample sizes were corrected to account for imputation uncertainty, and p-values were adjusted to account for genomic-control corrections performed in empirical studies. The number of loci discovered at each stage of GWAS was compared to observed data for T2D from each published study (see S. Table 3). Simulated models yielding 1–4 genome-wide significant loci in discovery (N=10K; empirically 2 loci observed for T2D), 10–30 loci in replication (N=55K; empirically 16 loci observed for T2D), and 25–65 loci in large-scale meta-analysis (N=85K; empirically 39 loci observed for T2D) were deemed consistent with empirical data.
Polygenic risk score analysis
Polygene ‘score’ analysis is a method by which to assess the aggregate predictive power of SNP alleles tested in a GWAS (Purcell et al 2009, Stahl et al 2012). Following Stahl et al, we pruned common SNPs by their linkage disequilibrium, preferentially retaining the SNPs with lower discovery p-values to obtain a set of independent, maximally associated markers. We used the p-values and effect sizes from discovery GWAS to select subsets of SNPs reaching four different PGWAS thresholds (0.001, 0.01, 0.1, and 0.5). For each SNP set, we summed the log-odds-weighted risk allele counts for each individual in an independent test sample of 2K cases and 3K controls to assign each individual a polygene risk ‘score’. We then tested these risk scores for association with case-control status using logistic regression. The predictive power of the polygene score was measured by Nagelkerke’s R2. Models yielding a Nagelkerke’s R2 between 0.01–0.04 for all PGWAS thresholds were deemed consistent with empirical data for T2D (where Nagelkerke’s R2 was ~2–2.5% for all thresholds).
Prediction of results of pending sequencing and genotyping studies
Whole genome, high coverage sequencing studies were simulated in matched case-control cohorts of 3K and 10K samples. Large-scale genotyping studies similar to ongoing studies with the exome array were simulated by sampling 20K cases and 35K controls, and assaying all sites seen >=2× in a sample of 5K controls. In each cohort, we performed single variant association testing of every assayed sequence variant across all causal loci; any locus with a variant achieving p-value < 5e-8 was deemed a novel locus if not previously found (e.g., by GWAS). We also performed rare variant burden testing in each sequenced cohort, using the sequence kernel association test (SKAT); all coding variants (both neutral, synonymous variants as well as disease-causing, non-synonymous variants) with MAF<5% were included in the burden test at each locus. Loci achieving a burden test p-value better than 1*10^-4 in sequencing studies or 1*10^-6 in the larger-scale genotyping studies were deemed significantly associated. Finally, a full sequencing study in 250K unselected individuals was simulated (20K cases and 230K controls, reflecting the population prevalence of T2D). Single variant association testing was performed in this cohort to assess the number of novel loci discovered. For all simulated studies, estimates of the heritability explained after each study were made based on a) only the odds ratios and frequencies observed in the study (dotted line in Figure 6 bottom panel) and b) the true additive effects of all segregating causal variants at the discovered loci (solid line).
Figure.6. Prediction of ongoing sequencing and large-scale genotyping studies for type 2 diabetes under different disease models consistent with empirical data.
Predictions under the two consistent disease models from Figure 5 are shown here: (a) a model with ‘moderate’ coupling to selection and a target size of T=1.25Mb (N=500 causal loci), and (b) a ‘weakly coupled’ model with a target size of T=3.75Mb (N=1500 causal loci). Top charts show cumulative fraction of disease loci discovered by each study design: A = Discovery GWAS in 10K samples, followed by B = Replication genotyping of top signals in 55K independent samples (as in Zeggini et al 2008); C = large-scale GWAS with discovery in an effective sample size of ~30K, followed by genotyping of all independent signals with p<0.005 to yield a total effective sample size of ~85K (as done via the Metabochip in Morris et al 2012); D = high coverage genome sequencing in 3K samples; E = high coverage genome sequencing in 10K samples; F = genotyping in 20K cases and 35 controls of all rare variants seen >= 2× in 5K controls (similar to ExomeChip); G = high coverage genome sequencing in 20K cases and 230K controls (a 250K unselected population cohort with T2D prevalence 8%). Labels above bars indicate predicted number of novel loci (e.g. not found in the previous studies) discovered at each step (Methods). Bottom charts show cumulative fraction of population genetic variance (heritability) explained by loci uncovered in each study. Solid line indicates true variance explained by those loci; dotted line represents fraction estimated using frequencies and odds ratios (estimated in the study) of the most associated single variants at each locus.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the Doris Duke Charitable Foundation (award number 2006087 to D.A.), the NIGMS (award R01GM078598 to S.S), and the NIMH (grant R01 MH084676 to S.S.). V.A. is also supported by NIH Training Grants T32GM007753 and T32GM008313. J.F. is supported in part by NIH Training Grant T32-GM007748-33 as well as funding from Pfizer. The GO-T2D Study is supported by grant 1RC2DK088389-01 from the NIDDK.
We gratefully acknowledge B. Lambert and K. Weiss (authors of the simulation tool ForSim) for helpful conversation, encouragement, and technical assistance. Without their software this work would not have been possible. We thank B. Voight for tremendous help in matching simulated genetic studies to those empirically conducted for T2D. We also thank E. Lander, C. Hartl, P. Fontanillas, B. Neale, M. McCarthy, M. Boehnke, M. Daly, S. Purcell, and E. Stahl for discussion and insightful critiques.
Footnotes
Full lists of members and affiliations appear in the Supplementary Note.
URLs
The ForSim software package is publicly available at the Weiss group web page: http://www.anthro.psu.edu/weiss_lab/ForSimManual.pdf
The authors provide a ForSim configuration file (containing best-fit demographic parameters) as well as sample simulated data at the following link: http://www.broadinstitute.org/mpg/archsim/.
AUTHOR CONTRIBUTIONS
V.A., J.F., S.S., and D.A. conceived and designed the simulation framework. V.A., J.F., and S.S. fit population genetic parameters to match simulated and empirical data. V.A. performed the simulation studies. V.A. and J.F. analyzed simulation data under each disease model. V.A., J.F., and D.A. wrote the manuscript. All authors edited and approved the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Collins FS, McKusick V. Implications of the Human Genome Project for Medical Science. JAMA. 2001;285:540–544. doi: 10.1001/jama.285.5.540. [DOI] [PubMed] [Google Scholar]
- 2.Jostins L, Barrett JC. Genetic risk prediction in complex disease. Human Molecular Genetics. 2011;20:R182–R188. doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thanassoulis G, Vasan R. Genetic Cardiovascular Risk Prediction - Will We Get There? Circulation. 2011;122:2323–2334. doi: 10.1161/CIRCULATIONAHA.109.909309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant RW, Moore AF, Florez JC. Genetic architecture of type 2 diabetes: recent progress and clinical implications. Diabetes Care. 2009;32:1107–1114. doi: 10.2337/dc08-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 7.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Reviews Genetics. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 8.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nature Reviews Genetics. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 9.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. PNAS. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price AL, et al. Pooled association tests for rare variants in exon-resequencing studies. American Journal of Human Genetics. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher RA. The genesis of twins. Genetics. 1919;4:489–499. doi: 10.1093/genetics/4.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neale MC, Maes HHM. Methodology for Genetic Studies of Twins and Families. Dordrecht, The Netherlands: Kluwer Academic Publishers B.V; 1992. [Google Scholar]
- 13.Martin N, Boomsma DI, Machin G. A twin-pronged attack on complex traits. Nature Genetics. 1997;17:387–392. doi: 10.1038/ng1297-387. [DOI] [PubMed] [Google Scholar]
- 14.Silventoinen K, et al. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Research. 2003;6:399–408. doi: 10.1375/136905203770326402. [DOI] [PubMed] [Google Scholar]
- 15.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. PNAS. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risch N. Linkage strategies for genetically complex traits. II. The power of affected relative pairs. American Journal of Human Genetics. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- 17.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nature Genetics. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 18.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahl Ea, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nature Genetics. 2012:1–9. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romeo S, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nature Genetics. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nature Genetics. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivas Ma, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature Genetics. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raychaudhuri S, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nature Genetics. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnefond A, et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nature Genetics. 2012;44:297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fearnhead NS, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. PNAS. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manolio Ta, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nature Reviews Genetics. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maher B. The case of the missing heritability. Nature. 2008;456 doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 29.Lee SH, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nature Genetics. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coventry A, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nature Communications. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nature Genetics. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 33.Keinan A, Clark AG. Recent Explosive Human Population Growth Has Resulted in an Excess of Rare Genetic Variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson M, et al. An Abundance of Rare Functional Variants in 202 Drug Target Genes Sequenced in 14,002 People. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupski JR, Belmont JW, Boerwinkle E, Gibbs R. a Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClellan J, King M-C. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell KJ. What is complex about complex disorders? Genome Biology. 2012;13:237. doi: 10.1186/gb-2012-13-1-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biology. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lappalainen T, Montgomery SB, Nica AC, Dermitzakis ET. Epistatic selection between coding and regulatory variation in human evolution and disease. American Journal of Human Genetics. 2011;89:459–463. doi: 10.1016/j.ajhg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. PNAS. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King CR, Rathouz PJ, Nicolae DL. An evolutionary framework for association testing in resequencing studies. PLoS Genetics. 2010;6:e1001202. doi: 10.1371/journal.pgen.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning SR, Thompson E. a Detecting rare variant associations by identity-by-descent mapping in case-control studies. Genetics. 2012;190:1521–1531. doi: 10.1534/genetics.111.136937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thornton KR, Foran AJ, Long AD. Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect. PLoS Genetics. 2013;9:e1003258. doi: 10.1371/journal.pgen.1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends in Genetics. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 46.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Human Molecular Genetics. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 47.Eyre-Walker A. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. PNAS. 2010;107:1752–1756. doi: 10.1073/pnas.0906182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert BW, Terwilliger JD, Weiss KM. ForSim: a tool for exploring the genetic architecture of complex traits with controlled truth. Bioinformatics. 2008;24:1821–1822. doi: 10.1093/bioinformatics/btn317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gravel S, et al. Demographic history and rare allele sharing among human populations. PNAS. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaffner SF, et al. Calibrating a coalescent simulation of human genome sequence variation. Genome Research. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahituv N, et al. Medical sequencing at the extremes of human body mass. American Journal of Human Genetics. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward LD, Kellis M. Evidence of Abundant Purifying Selection in Humans for Recently Acquired Regulatory Functions. Science. 2012;1675 doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowie C, Rust K, Byrd-Holt D, Gregg E. Prevalence of Diabetes and High Risk for Population in 1988 – 2006. Diabetes Care. 2010;33:562–568. doi: 10.2337/dc09-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almgren P, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54:2811–2819. doi: 10.1007/s00125-011-2267-5. [DOI] [PubMed] [Google Scholar]
- 55.Zhu Q, et al. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. American Journal of Human Genetics. 2011;88:458–468. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montgomery SB, Lappalainen T, Gutierrez-Arcelus M, Dermitzakis ET. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genetics. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park J-H, et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. PNAS. 2011;108:18026–18031. doi: 10.1073/pnas.1114759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyssenko V, et al. Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes. 2005;54:166–174. doi: 10.2337/diabetes.54.1.166. [DOI] [PubMed] [Google Scholar]
- 60.Weijnen CF, Rich SS, Meigs JB, Krolewski aS, Warram JH. Risk of diabetes in siblings of index cases with Type 2 diabetes: implications for genetic studies. Diabetic Medicine. 2002;19:41–50. doi: 10.1046/j.1464-5491.2002.00624.x. [DOI] [PubMed] [Google Scholar]
- 61.Guan W, Pluzhnikov A, Cox NJ, Boehnke M. Meta-analysis of 23 type 2 diabetes linkage studies from the International Type 2 Diabetes Linkage Analysis Consortium. Human Heredity. 2008;66:35–49. doi: 10.1159/000114164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genetics. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voight BF, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genetics. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genetics. 2012;44 doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popper KR. Conjectures and Refutations: The Growth of Scientific Knowledge. Routledge classics. 2nd. Routledge; 2002. p. 417. [Google Scholar]
- 67.Wang K, et al. Interpretation of association signals and identification of causal variants from genome-wide association studies. American Journal of Human Genetics. 2010;86:730–742. doi: 10.1016/j.ajhg.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldstein DB. The importance of synthetic associations will only be resolved empirically. PLoS Biology. 2011;9:e1001008. doi: 10.1371/journal.pbio.1001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guan W, Boehnke M, Pluzhnikov A, Cox NJ, Scott LJ. Identifying Plausible Genetic Models Based on Association and Linkage Results: Application to Type 2 Diabetes. Genetic Epidemiology. 2012;9:1–9. doi: 10.1002/gepi.21668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakravarti A. Population genetics — making sense out of sequence. Nature Reviews Genetics. 1999;21 doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 71.Zaitlen N, et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genetics. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. American Journal of Human Genetics. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.