Abstract
Target-matched treatment with PI3K/AKT/mTOR pathway inhibitors in patients with diverse advanced cancers with PIK3CA mutations have shown promise. Tumors from patients with colorectal cancer (CRC) were analyzed for PIK3CA, KRAS and BRAF mutations. PIK3CA mutated tumors were treated, whenever feasible, with agents targeting the PI3K/AKT/mTOR pathway. Of 194 patients analyzed, 31 (16%) had PIK3CA mutations, and 189 (97%) were assessed for KRAS mutations. Patients with PIK3CA mutations had a higher prevalence of simultaneous KRAS mutations than patients with wild-type (wt) PIK3CA (71%, 22/31 vs. 43%, 68/158; p=0.006). Of 31 patients with PIK3CA mutations, 17 (55%) were treated with protocols containing PI3K/AKT/mTOR pathway inhibitors (median age, 57; median number of prior therapies, 4; mTORC1 inhibitors [11], PI3K inhibitors [5] or an AKT inhibitor [1]). None (0/17) had a partial or complete response (PR/CR) and only 1 (6%, 95% CI 0.01–0.27) had stable disease (SD)≥6 months, which was not significantly different from a SD≥6 month/PR/CR rate of 16% (11/67; 95% CI 0.09–0.27) in CRC patients without PIK3CA mutations treated with PI3K/AKT/mTOR pathway inhibitors (p=0.44). Median progression-free survival was 1.9 months (95% CI 1.5–2.3). In conclusion, our data provide preliminary evidence that in heavily pretreated patients with PIK3CA-mutant advanced CRC, protocols incorporating PI3K/AKT/mTOR inhibitors have minimal activity. PIK3CA mutations are associated with simultaneous KRAS mutations, possibly accounting for therapeutic resistance.
Keywords: PIK3CA mutation, PI3K/AKT/mTOR, Colorectal cancer, Target-matched therapy, KRAS mutation, Phase I trial
INTRODUCTION
The PIK3CA gene encodes the 110α subunit of phosphatidylinositol 3-kinase (PI3K) and is commonly mutated in a myriad of human cancers. (1) PIK3CA mutations activate the PI3K/AKT/mammalian target of rapamycin (mTOR) pathway, which leads to carcinogenesis and tumor progression. (2–4) Preclinical and early clinical data suggest that PIK3CA mutations can render tumors sensitive to PI3K/AKT/mTOR pathway inhibition, whereas simultaneous KRAS mutations can drive therapeutic resistance. (3, 5–9) Many of the latest advances in cancer medicine have occurred when tumor-specific molecular abnormalities were matched with appropriately selected targeted therapies. (10–12) Examples in solid tumors include treatment with KIT inhibitors in gastrointestinal stromal tumors with KIT mutations(13), EGFR inhibitors in non-small cell lung cancer harboring EGFR mutations(14) and BRAF inhibitors in melanoma with BRAF mutations. (15, 16) It is plausible that matching patients with colorectal cancer harboring PIK3CA mutations with therapies targeting the PI3K/AKT/mTOR pathway may lead to improved therapeutic benefit, as has been suggested in breast and gynecological cancers. (7, 8) PIK3CA mutations occur in approximately 17% of colorectal cancers; however, there are limited data on the outcomes of matched targeting of the PI3K/AKT/mTOR pathway in these patients. (17–20) We investigated patients with colorectal cancer referred to the Clinical Center for Targeted Therapy at MD Anderson Cancer Center (MD Anderson) for the presence of PIK3CA mutations and analyzed their treatment outcomes.
METHODS
Patients
Patients with advanced colorectal cancer refractory to standard therapies referred for early clinical trials with targeted therapeutic agents to the Clinical Center for Targeted Therapy at MD Anderson were eligible for analysis providing they had adequate tissue available for mutation analysis. The registration of patients in the database, pathology assessment, and mutation analysis were performed at MD Anderson. All treatments and analyses were performed in accordance with MD Anderson IRB guidelines.
Tissue Samples and Mutation Analyses
PIK3CA and KRAS mutations were investigated in archival formalin-fixed, paraffin-embedded tissue blocks or material from fine needle aspiration biopsy obtained from diagnostic and/or therapeutic procedures. All histologies were centrally reviewed at MD Anderson. PIK3CA and KRAS mutation testing was done at the Clinical Laboratory Improvement Amendment (CLIA)–certified Molecular Diagnostic Laboratory within the Division of Pathology and Laboratory Medicine at MD Anderson. DNA was extracted from micro-dissected, paraffin-embedded tumor sections and further studied using a polymerase chain reaction–based DNA sequencing method for PIK3CA mutations in codons c532 to c554 of exon 9 (helical domain) and c1011 to c1062 of exon 20 (kinase domain), which included the mutation hotspot region of the PIK3CA proto-oncogene by Sanger sequencing after amplification of 276– and 198–base pair amplicons, respectively, using primers designed by the MD Anderson Molecular Diagnostic Laboratory. After January 2011, the assay used was mass spectrometric detection (Sequenom MassARRAY) to screen for the mutational hot spots in exon 1 (Q60K, R88Q, E110K and K111N), exon 4 (N345K), exon 6 (S405S), exon 7 (E418K, C420R, E453K), exon 9 (P539R, E542 [base 1 and 2], E545 [all 3 bases] and Q546 [base 1 and 2]), exon 18 (F909L) and exon 20 (Y1021 [base 1 and 2], T1025 [base 1], M1043I, M1043V, A1046V, H1047Y, H1047R, H1047L, G1049R). The mutations identified during the initial screening were confirmed by Sanger sequencing assay. The lower limit of detection is approximately 10%. Additionally, whenever possible, mutation analyses for KRAS, NRAS codons 12, 13, and 61 mutations of exons 2–3 and BRAF mutations in exon 15 were carried out using PCR-based DNA sequencing mutation, as previously described. (21)
Treatment and Evaluation
Consecutive patients with underlying PIK3CA mutations were offered, whenever possible, a clinical trial, which included an inhibitor of the PI3K/AKT/mTOR pathway. Treatment continued until disease progression or the occurrence of unacceptable toxicity. Treatment was carried out according to the requisites in the treatment protocols selected. Assessments, including history, physical examination, and laboratory evaluations, were performed as specified in each protocol, typically before the initiation of therapy, weekly during the first cycle, and then, at a minimum, at the beginning of each new treatment cycle. Efficacy was assessed from computed tomography (CT) scans and/or magnetic resonance imaging (MRI) at baseline before treatment initiation and then every 2 cycles (6–8 weeks). All radiographs were read in the Department of Radiology at MD Anderson and reviewed in the Department of Investigational Cancer Therapeutics tumor measurement clinic. Responses were categorized per Response Evaluation Criteria in Solid Tumors (RECIST) 1.0.(22) In brief, complete response (CR) was defined as the disappearance of all measurable and non-measurable disease; partial response (PR) was defined as at least a 30% decrease in the sum of the longest diameter of measurable target lesions; progressive disease (PD) was defined as at least a 20% increase in the sum of the longest diameter of measurable target lesions, or unequivocal progression of a non-target lesion, or the appearance of a new lesion; and stable disease (SD) was defined as neither sufficient shrinkage to qualify for PR nor sufficient increase to qualify for PD.
Statistical analysis
Two-way contingency tables were employed to summarize the relationship between two categorical variables. Fisher’s exact test was used to assess the association among categorical variables and PIK3CA mutation status. Progression-free survival (PFS), estimated by the Kaplan-Meier method, was defined as the time interval from the start of therapy to the first observation of disease progression or death, whichever occurred first. Patients alive and without disease progression were censored at the last follow-up date. All statistical analyses were carried out using SPSS 19 computer software (SPSS Chicago, IL).
RESULTS
Patients characteristics
A total of 194 patients with advanced colorectal cancer were screened for the presence of PIK3CA mutations starting January 2009. Their median age was 58 years (range, 25 to 81 years); 140 (72%) were White, 28 (14%) African American, 19 (10%) Hispanic, and 7 (4%) Asian. Detailed patient characteristics are listed in Table 1.
TABLE 1.
BASELINE PATIENT CHARACTERISTICS
| Variable | Number | PIK3CA mutation (%) | Wild-type PIK3CA (%) | P value |
|---|---|---|---|---|
| All | 194 | 31 (100) | 163 (100) | Not applicable |
| Median age | 58 range, 25–81 |
56 range, 36–71 |
59 range, 25–81 |
0.21 |
| Ethnicity | ||||
| White | 140 | 21(68) | 119 (73) | 0.58 |
| African-American | 28 | 6 (19) | 22 (13.5) | |
| Hispanic | 19 | 2 (6.5) | 17 (10.5) | |
| Asian | 7 | 2 (6.5) | 5 (3) | |
| Tumor site | ||||
| Colon | 156 | 28 (90) | 128 (79) | 0.15 |
| Rectum | 38 | 3 (10) | 35 (21) | |
| KRAS (n=189)* | ||||
| KRAS mutation | 84 | 22 (71) | 68 (43) | 0.006 |
| KRAS wild-type | 91 | 9 (29) | 90 (57) |
Only patients tested for KRAS mutations are included in the analysis
PIK3CA mutations
PIK3CA mutations were detected in 31 (16%) of the 194 patients, with 22 mutations in the kinase domain in exon 9, and 8 mutations in the helical domain in exon 20. The most frequent point mutation was E545K (1633G>A) in 11 (35%) patients, followed by E542K (1624G>A) in 8 patients (26%), and H1047L (3140A>T) in 3 (10%) patients (Table 2). No association between PIK3CA mutation and age or ethnicity or tumor site was identified (Table 1).
TABLE 2.
TYPES OF PIK3CA MUTATIONS IDENTIFIED
| Mutation type | N (%) |
|---|---|
| PIK3CA mutations | 31 |
| N345K | 1 (<5) |
| E542K | 8 (26) |
| E545A | 1 (<5) |
| E545K | 11 (35) |
| Q546P | 1 (<5) |
| Q546R | 1 (<5) |
| R1023Q | 1 (<5) |
| M1043V | 1 (<5) |
| H1047R | 2 (6) |
| H1047L | 3 (10) |
| G1049R | 1 (<5) |
KRAS and BRAF mutations
Of the 194 patients, 189 patients were tested for KRAS mutations and 90 (47%) had a mutation. Among patients with KRAS mutations the most prevalent ones were G12D (35G>A) in 30% (27/90), G12V (35G>T) in 20% (18/90), G13D (38G>A) in 11% (10/90) and G12A (35G>C) in 11% (10/90) of patients. Patients with PIK3CA mutations were more likely to have simultaneous KRAS mutations compared to patients with wild-type (wt) PIK3CA (71%, 22/31 vs. 43%, 68/158; p=0.006). KRAS mutations compared to wt KRAS were associated with PIK3CA mutations in exon 9 (18%, 16/90 vs. 6%, 6/99; p=0.01), but not in exon 20 (6%. 5/90 vs. 3%, 3/99; p=0.48) (Table 3).
TABLE 3.
COEXISTENCE OF PIK3CA AND KRAS MUTATIONS
| Variable | Number | KRAS mutation (%) | Wild-type KRAS (%) | P value |
|---|---|---|---|---|
| Tested | 189 | 90 (100) | 99 (100) | Not applicable |
| PIK3CA mutations (any) | 31 | 22 (24) | 9 (9) | 0.006 |
| Wild-type PIK3CA | 158 | 68 (76) | 90 (91) | |
| PIK3CA exon 9 mutations | 21 | 16(18) | 6 (6) | 0.01 |
| Wild-type or exon 20 PIK3CA mutations | 168 | 74 (82) | 93 (94) | |
| PIK3CA exon 20 mutations | 8 | 5 (6) | 3 (3) | 0.48 |
| Wild-type or exon 9 PIK3CA mutations | 181 | 85 (94) | 96 (97) |
Of the 194 patients, 167 patients were tested for BRAF mutations and 11 (7%) had mutated BRAF (10 had a V600E mutation and 1 patient had a D594G mutation). All patients with a BRAF mutation were negative for KRAS mutations. Of these 11 patients with BRAF mutations, 2 (18%) had co-existent PIK3CA mutations. There was no difference in the incidence of BRAF mutations among those with or without PIK3CA mutations (7%, 2/27 vs. 6%, 9/140; p=0.69)
Response rate to PI3K/AKT/mTOR-directed therapies
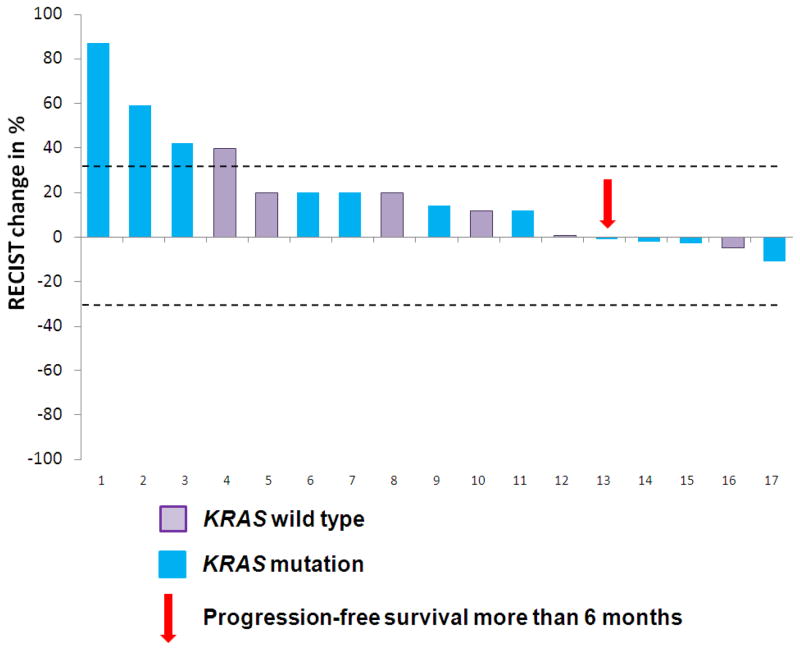
Of the 31 patients with PIK3CA mutations, 17 (55%) were treated in clinical trials including a PI3K/AKT/mTOR pathway inhibitor. These patients had been refractory to a median of 4 prior therapies (range 2– 7). Most patients (11/17, 65%) received mTORC1 inhibitor (rapalog)-based therapy; 5/17 (29%) received PI3K inhibitor-based therapy, and 1 (6%) received an AKT inhibitor-based therapy (Supplementary Table S1, online only). Most patients (13/17, 76%) received PI3K/AKT/mTOR inhibitors at 100% of the maximum tolerated dose/recommended phase II dose/FDA-approved dose (range, 30% to 100%). Of the 11 patients treated with mTORC1 inhibitors, 10 (91%) received the maximum tolerated dose/recommended phase II dose/FDA-approved dose. In contrast, only 1 (20%) of 5 patients treated with PI3K inhibitors was treated with the maximum tolerated dose (Supplementary Table S1, online only). Of the 17 patients, none achieved partial or complete responses (PR/CR) and only 1 (6%, 95% CI 0.01–0.27) patient had stable disease for more than 6 months (SD ≥6) (Figure 1). This was similar to the SD ≥6/PR/CR rate of 16% (11/67; 95% CI 0.09–0.27) in colorectal cancer patients without PIK3CA mutations treated on the same protocols targeting the PI3K/AKT/mTOR pathway (p=0.44).
Figure 1.
Waterfall plot describing responses per RECIST in 17 patients with metastatic colorectal cancer and PIK3CA mutations treated with PI3K/AKT/mTOR pathway inhibitors
We also reviewed 14 of 31 patients (45%) with PIK3CA mutations who were not treated with PI3K/AKT/mTOR inhibitors. Of these 14 patients, 5 were not treated due to ineligibility or patient/doctor preference and 9 received other experimental therapies, often because PIK3CA status was not available at the time of decision making. None (0%, 95% CI 0.00–0.29) of these 9 patients attained a PR or SD ≥6, which was similar to SD ≥6/PR/CR rate of 6% (1/17, 95% CI 0.01–0.27) in the remaining 17 patients treated with PI3K/AKT/mTOR inhibitors (p=1.00).
Progression-free survival on PI3K/AKT/mTOR therapies
The median PFS for these 17 patients with a PIK3CA mutation on target-matched therapy with PI3K/AKT/mTOR inhibitors was 1.9 months (95% CI; 1.5–2.3). There was no difference in median PFS in 11 patients with KRAS mutations vs. 6 patients with wt KRAS (1.8 months; 95%CI, 1.4–2.2 vs. 1.9 months; 95%CI, 1.1–2.7; p=0.59). The same 17 patients had a median PFS of 3.1 months (95% CI; 0.4–5.8) on their last FDA-approved therapy prior to referral to the Clinical Center for Targeted Therapy (p= 0.1) (Figure 2). Patients (n=67) without PIK3CA mutations treated with the same therapies with PI3K/AKT/mTOR inhibitors had a similar median PFS (2.3 months; 95%CI 2.1–2.6) as did patients with PIK3CA mutations treated with PI3K/AKT/mTOR inhibitors (1.9 months; 95% CI; 1.5–2.3; p=0.44, Figure 2).
Figure 2.
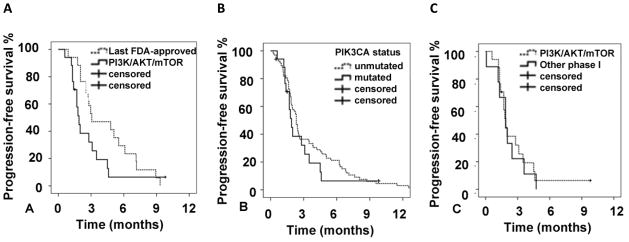
A. Progression-free survival (PFS) in 17 patients with metastatic colorectal cancer and PIK3CA mutations treated with PI3K/AKT/mTOR pathway inhibitors compared to median PFS in the same patients treated with the last FDA-approved therapy for metastatic disease (median PFS of 1.9 months vs. 3.1 months; p=0.1). B. PFS in patients with PIK3CA mutations who received PI3K/AKT/mTOR-directed therapy (n=17) compared to patients without PIK3CA mutations (n=67) treated with same therapies (median PFS of 1.9 months vs. 2.3 months, p=0.44) C. PFS in patients with PIK3CA mutations who received PI3K/AKT/mTOR directed therapy (n=17) compared to patients (n=9) treated with other phase 1 therapies (median PFS of 1.9 months vs. 1.9 months; p=0.77).
In addition, we reviewed PFS in 9 patients with colorectal cancer and PIK3CA mutations who were treated with experimental therapies other than PI3K/AKT/mTOR inhibitors. The median PFS for those 9 patients was 1.9 months (95%CI 1.0–2.7), which was similar to the median PFS of 1.9 months (95%CI 1.5–2.3) in 17 patients with PIK3CA mutations treated with matched therapy (p=0.77, Figure 2).
DISCUSSION
We identified PIK3CA mutations in 16% of the 194 patients with colorectal cancer referred to the Clinical Center for Targeted Therapy. The most common types of PIK3CA mutations associated with colorectal cancer in our patients were E545K and E542K mutations in exon 9. These results are similar to those in published studies. (18, 19) Earlier studies suggested that H1047R mutations may be more sensitive to PI3K/AKT/mTOR pathway inhibition, but in our study, only 2 patients (6%) had this particular mutation and that can potentially account for their lack of therapeutic response. (8)
We found a strong association between KRAS mutations and PIK3CA mutations in our patients that supports previously published data. (18, 19, 23–25) We found that KRAS mutations were more frequently associated with exon 9 than exon 20 PIK3CA mutations. A similar association was noted by De Roock et al. in a study of 743 patients with metastatic colorectal cancer, but a larger study of 1,170 patients with predominantly localized colorectal cancer did not find a similar association. (18, 19)
The paradigm for matching targeted therapy based on specific molecular defects identified in the cancer cell was established by the remarkable success of imatinib in chronic myeloid leukemia. (26) This model has been successfully replicated in solid tumors such as BRAF V600E mutated melanoma and non-small cell lung cancer with ALK rearrangement. (15, 16, 27) In the context of targeting PIK3CA mutated cancers with PI3K/AKT/mTOR inhibitors, we have observed encouraging responses in heavily pretreated patients with breast and gynecological malignancies and PIK3CA mutations treated with PI3K/AKT/mTOR pathway inhibitors. (9, 28, 29) In the current study, 17 heavily pretreated patients with colorectal cancer and PIK3CA mutations treated with PI3K/AKT/mTOR inhibitors did not achieve better outcomes compared to patients with colorectal cancer without PIK3CA mutations treated with identical therapies (SD ≥6/PR/CR rate 6% vs.16%, p=0.44) and their outcomes were strikingly similar response rates of 4–11% previously reported from early-phase clinical trials in an unselected population, including a 1.4% response rate and 9% rate of SD at 6 months for unselected colorectal cancer patients from our center. (30–33) In addition, the median PFS in patients with PIK3CA mutations treated with PI3K/AKT/mTOR inhibitors demonstrated a trend toward inferiority to the median PFS achieved by the same patients on their last FDA-approved therapy (1.9 months vs. 3.1 months; p=0.1). Our observations are in line with the experience from a major European cancer center reporting lack of therapeutic benefit with therapeutic matching (including PIK3CA) in colorectal cancer. (20) Reasons why these patients do not derive similar benefits from PI3K/AKT/mTOR targeted therapies as patients do with PIK3CA mutations and breast or gynecological cancers remains unknown. Preclinical models demonstrated that simultaneous mutations in the MAPK pathway (RAS, RAF or MEK) can negate the effect of mTOR inhibitors in patients with PIK3CA mutations. (5, 6) Therefore, it is plausible that this strong association between PIK3CA and KRAS mutations in colorectal cancer can contribute to therapeutic resistance since, in our series, 71% of patients with colorectal cancer and PIK3CA mutations had simultaneous KRAS mutations. In contrast, in patients with breast and gynecological malignancies with PIK3CA mutations, only 23% had co-existing KRAS mutations, which were often in different exons than in colorectal cancer. (9) Furthermore, Shimizu et al. analyzed phase I therapies targeting PI3K and MAPK pathways in diverse advanced cancers and demonstrated superior outcomes with combined PI3K and MAPK inhibition in patients with cancers having alterations in both pathways. (20, 34)
The majority of patients in our study were treated with rapalogs and it is possible that the outcomes with these inhibitors may be inferior due to feedback activation of AKT, as has been previously described in vitro and in patients. (35, 36) Other biomarkers besides activating mutations in PIK3CA may be required to select for inhibitors of the PI3K pathway, including loss of the tumor suppressor PTEN or gene amplification of members the insulin growth factor family. Ongoing clinical efforts in PIK3CA-mutant colorectal cancer are utilizing AKT or PI3K inhibitors and excluding concurrent KRAS mutations in an effort to isolate a responsive population.
Our study has several important limitations. First, there were only a small number of patients with PIK3CA mutations who were treated with PI3K/AKT/mTOR inhibitors; the majority of patients (65%) received mTORC1-targeting agents. Second, the analysis was retrospective. Third, patients received drugs that impacted different parts of the pathway (mTORC1 inhibitors, 11 patients; PI3K inhibitors, 5 patients; AKT inhibitor, 1 patient), though the latter might also imply that conclusions are not confined to one type of drug. Fourth, doses were variable. For instance, only 76% of patients received a maximum tolerated dose/recommended phase II dose/FDA-approved dose of a PI3K/AKT/mTOR inhibitor. Of interest, 91% of patients treated with mTORC1 inhibitors received a maximum tolerated dose/recommended phase II dose/FDA-approved dose compared to only 20% of patients treated with a PI3K inhibitor, which precludes firm conclusions about the latter class of drugs. (32, 37) Fifth, none of the patients in our analysis received combined PI3K and MAPK targeted therapy.
In conclusion, our data, though preliminary, suggest that targeted matched therapy with PI3K/AKT/mTOR pathway inhibitors (in particular mTORC1 inhibitors) in patients with metastatic colorectal cancer harboring PIK3CA mutations may be associated with minimal activity. The lack of benefit can conceivably be explained, in part, by the high prevalence of co-existing KRAS mutations driving therapeutic resistance.
Supplementary Material
Acknowledgments
Funding: This work was supported in part by NIH grant U01 CA062461 (R. Kurzrock). Molecular testing was supported in part by the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy.
We thank Ms. Joann Aaron for scientific review and editing of this article.
Footnotes
Conflicts of interest/ Disclosure: Filip Janku has research support from Novartis, Roche, Biocartis, Transgenomic, Trovagene. Razelle Kurzrock has research support from GlaxoSmithKline, Novartis, Merck, and Bayer. Scott Kopetz is a consultant for Roche, Sanofi, Amgen, Bristol-Meyers Squibb, Bayer.
References
- 1.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 2.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102(3):802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelmann JC, Rahmann S, Wolf M, Schultz J, Fritzilas E, Kneitz S, et al. Modelling cross-hybridization on phylogenetic DNA microarrays increases the detection power of closely related species. Mol Ecol Resour. 2009;9(1):83–93. doi: 10.1111/j.1755-0998.2008.02199.x. [DOI] [PubMed] [Google Scholar]
- 4.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3(10):1221–4. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 5.Ihle NT, Lemos R, Jr, Wipf P, Yacoub A, Mitchell C, Siwak D, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res. 2009;69(1):143–50. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120(8):2858–66. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10(3):558–65. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–84. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30(8):777–82. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer--is it becoming a reality? Nat Rev Clin Oncol. 2010;7(7):401–14. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 11.Janku F, Garrido-Laguna I, Petruzelka LB, Stewart DJ, Kurzrock R. Novel therapeutic targets in non-small cell lung cancer. J Thorac Oncol. 2011;6(9):1601–12. doi: 10.1097/JTO.0b013e31822944b3. [DOI] [PubMed] [Google Scholar]
- 12.Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18(22):6373–83. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347(7):472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 14.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 15.Falchook GS, Long GV, Kurzrock R, Kim KB, Arkenau TH, Brown MP, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379(9829):1893–901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 19.Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–68. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dienstmann R, Serpico D, Rodon J, Saura C, Macarulla T, Elez E, et al. Molecular profiling of patients with colorectal cancer and matched targeted therapy in phase I clinical trials. Mol Cancer Ther. 2012;11(9):2062–71. doi: 10.1158/1535-7163.MCT-12-0290. [DOI] [PubMed] [Google Scholar]
- 21.Zuo Z, Chen SS, Chandra PK, Galbincea JM, Soape M, Doan S, et al. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod Pathol. 2009;22(8):1023–31. doi: 10.1038/modpathol.2009.59. [DOI] [PubMed] [Google Scholar]
- 22.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 23.Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27(9):1477–84. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrido-Laguna I, Hong DS, Janku F, Nguyen LM, Falchook GS, Fu S, et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One. 2012;7(5):e38033. doi: 10.1371/journal.pone.0038033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihle NT, Powis G. Inhibitors of phosphatidylinositol-3-kinase in cancer therapy. Mol Aspects Med. 2010;31(2):135–44. doi: 10.1016/j.mam.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348(11):994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 27.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moulder S, Moroney J, Helgason T, Wheler J, Booser D, Albarracin C, et al. Responses to liposomal Doxorubicin, bevacizumab, and temsirolimus in metaplastic carcinoma of the breast: biologic rationale and implications for stem-cell research in breast cancer. J Clin Oncol. 2011;29(19):e572–5. doi: 10.1200/JCO.2010.34.0604. [DOI] [PubMed] [Google Scholar]
- 29.Moroney JW, Schlumbrecht MP, Helgason T, Coleman RL, Moulder S, Naing A, et al. A phase I trial of liposomal doxorubicin, bevacizumab, and temsirolimus in patients with advanced gynecologic and breast malignancies. Clin Cancer Res. 2011;17(21):6840–6. doi: 10.1158/1078-0432.CCR-11-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horstmann E, McCabe MS, Grochow L, Yamamoto S, Rubinstein L, Budd T, et al. Risks and benefits of phase 1 oncology trials, 1991 through 2002. N Engl J Med. 2005;352(9):895–904. doi: 10.1056/NEJMsa042220. [DOI] [PubMed] [Google Scholar]
- 31.Kurzrock R, Benjamin RS. Risks and benefits of phase 1 oncology trials, revisited. N Engl J Med. 2005;352(9):930–2. doi: 10.1056/NEJMe058007. [DOI] [PubMed] [Google Scholar]
- 32.Jain RK, Lee JJ, Hong D, Markman M, Gong J, Naing A, et al. Phase I oncology studies: evidence that in the era of targeted therapies patients on lower doses do not fare worse. Clin Cancer Res. 2010;16(4):1289–97. doi: 10.1158/1078-0432.CCR-09-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong DS, Patel JC, Wheler J, Naing A, Garrido-Laguna I, Falchook G, et al. Outcomes in 144 patients with colorectal cancer treated in a phase I clinic: the MD Anderson Cancer Center experience. Clin Colorectal Cancer. 2012;11(4):297–303. doi: 10.1016/j.clcc.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu T, Tolcher AW, Papadopoulos KP, Beeram M, Rasco DW, Smith LS, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res. 2012;18(8):2316–25. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 35.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66(3):1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1(3):248–59. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta S, Hunsberger S, Boerner SA, Rubinstein L, Royds R, Ivy P, et al. Meta-analysis of the Relationship Between Dose and Benefit in Phase I Targeted Agent Trials. J Natl Cancer Inst. 2012;104(24):1860–6. doi: 10.1093/jnci/djs439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.