Abstract
A series of degrasyn-like symmetrical compounds have been designed, synthesized, and screened against B cell malignancy (multiple myeloma, mantle cell lymphoma) cell lines. The lead compounds T5165804 and CP2005 showed higher nanomolar potency against these tumor cells in comparison to degrasyn and inhibited Usp9x activity in vitro and in intact cells. These observations suggest that this new class of compounds holds promise as cancer therapeutic agents
1. Introduction
Protein ubiquitination is altered in several diseases, including cancer.1 The ubiquitination process is controlled by the opposing action of E3 ligases, which mediate Ub transfer2 and deubiquitinases (DUBs) that catalyze its removal.3 This process is therefore capable of controlling the destruction, activity and localization of several target proteins and has emerged as a novel focus for targeted therapy in cancer. Usp9x is a DUB whose activity and expression are highly elevated in several tumors (i.e. lymphoma, myeloma, breast, colon, cervical) and knockdown or gene silencing studies demonstrate that Usp9x is essential for tumor cell survival and response to conventional cancer therapy.4–11 This suggests that Usp9x inhibitors may have a wide anti-tumor activity. In our previous study, we reported library syntheses and SAR studies of tyrphostin-type compounds and demonstrated that those with tumor cytotoxic activity also possessed ubiquitin-modulatory activity in B-cell derived tumors.12 Here we report a bio-informatics based approach to improve on the cell-based activity of the degrasyn (WP1130) template. In this study we designed and tested several modified degrasyn-like symmetrical molecules (Figure 1) for Usp9x inhibitory activity and cytotoxicity in B-cell lines representative of multiple myeloma and mantle cell lymphoma. These novel structures have improved anti-tumor and Usp9x inhibitory activity when compared to our previously described DUB inhibitory compound (WP1130).
Figure 1.
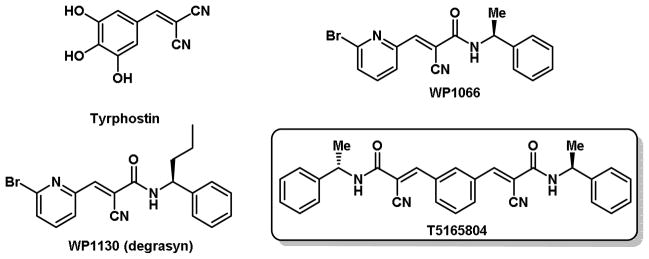
Tyrphostin, degrasyn and degrasyn like compounds
2. Modeling
In order to locate new starting point for our synthetic efforts, we chose to conduct in-silico searches followed by the purchase of a limited set of compounds. Compounds identified from that set as legitimate hits were followed up by full synthetic efforts to explore the SAR around the active compounds. This method was efficient because the bulk of resources were directed towards active compounds, but this was somewhat countered by the reduced novelty of the compounds. To balance both aspects, we chose to conduct both a structural similarity search that would undoubtedly lead to structures with the same core and a pharmacophoric fingerprint followed by a similarity search to identify compounds near, but not too close to the parent compound. The pharmacophore fingerprint is a representation of the features and the distances between them and is calculated from the 3D structure and conformations of the molecule. Molecules with similar fingerprints are expected to have similar activities.
As expected, most of the compounds resulting from the structural similarity search contained the same structural core as the WP1130, though the attached group varied considerably. This is largely due to the considerable number of degrasyn contained in the vendor databases. Only a handful of compounds from the combined pharmacophoric fingerprint followed by similarity had the identical central core, but there were slight variations of the core, such as a cyclized version that would not have been obvious. Other compounds were not obvious when observed from a strict structural viewpoint. However, upon closer examinations of these compounds, it is clear that they make “chemical sense” in that they have similar elements to the parent structure in terms of H-bond donors, acceptors, and hydrophobic regions. Not surprisingly, these are the same features that are incorporated into the pharmacophore fingerprints. In addition, they also have a certain structure similarity to the original compound, yet would not be obvious changes to the parent compound. Structures obtained from the structural similarity search were screened against a multiple myeloma (MM1.S) cell line for anti-tumor and ubiquitin-modulatory activity (Figure 2). The lead compound T5165804 inhibited at the same level as WP1130 (degrasyn) at 10 μM and was later confirm to be more active when an IC50 was determined. The most obvious difference between T5165804 and degrasyn is that T5165804 is symmetrical and has two cores which could be a possible explanation for its increased activity against these tumor cells. A focused library was synthesized and screened on the basis of the core structure.
Figure 2.
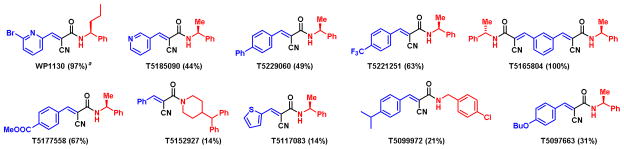
Anti-tumor activity of degrasyn-like commercial compounds against MM-1 cell lines. aFor all the compounds %inhibition determined at 10 μM.
3. Chemistry
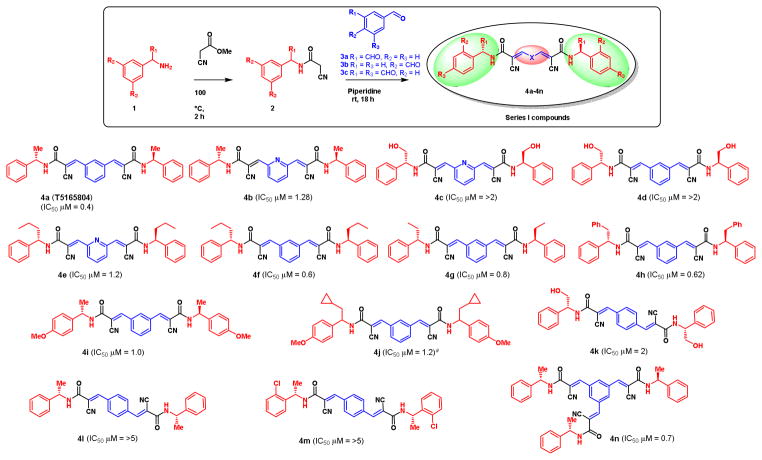
The first series of compounds was designed on the basis of the structure of T5165804 (Figure 1) as illustrated in Figure 3. First, the corresponding arylamine 1 was coupled with cyanoacetic methyl ester to form the cyano amide intermediate 2 quantitatively. Knoevenagel condensation of 2 with the corresponding dicarboxaldehyde 3 gave the symmetrical degrasyn like compounds 4 (Figure 3). In a similar way, Knoevenagel condensation with 1,3,5-benzenetricarboxaldehyde 3c resulted a trimer-like compound 4n. Various α-alkyl substituted amines have been used to make a library of degrasyn like symmetrical compounds (compounds 4a–4p, Figure 3). We synthesized 14 symmetrical degrasyn like compounds including one trimer compound for series I library (4a–4n, Figure 3).
Figure 3.
Anti-tumor activity of series I compounds against MM-1 cell lines. aRacemic compound.
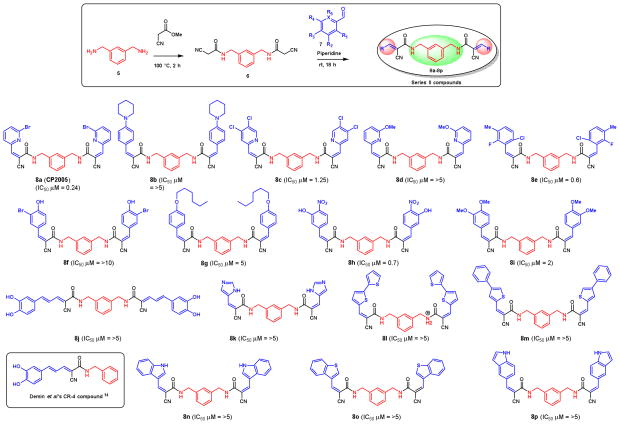
The second series of compounds were designed on the basis of the inside-out structure of series I compounds as shown in Figure 4. Coupling of m-xylylenediamine 5 with cyanoacetic methyl ester to form the bis cyano amide intermediate 6, then, Knoevenagel condensation with various aldehydes 7 produced a library of 16 symmetrical compounds for series II library (8a–8p, Figure 4).
Figure 4.
Anti-tumor activity of series II compounds against MM-1 cell lines.
4. Biology activity
The newly synthesized compounds were tested for their ability to inhibit abnormal cell proliferation. In those experiments, multiple myeloma cells (MM1.S) were chosen as a model for the primary screen. In a typical experiment, cell lines were incubated with a range of compound concentrations for 72 h to determine the concentration required to inhibit cell growth based on an MTT assay or induce apoptosis detected by PARP cleavage. Those with activity greater than WP1130 were screened and compared for their Usp9x inhibitory activity against recombinantly expressed Usp9x catalytic domain (AA 1553-1960) and activity against Usp9x in intact cells using a HA-tagged Ub-vinylsulfone covalent substrate labeling assay as previously described.10,13
Nine out of possible seventy-five compounds designated from modeling were purchased and screened in an MM1.S cellular assay to determine cell proliferation at the 10 μM level and the results are shown in Figure 2. Compounds T5221251 and T5177558 showed more than 60% inhibition whereas compound T5229060 showed only 49% inhibition (Figure 2). The compound T5165804 (100% inhibition) was the only compound which had comparable activity to WP1130 in the assay (97% inhibition). A complete inhibition curve was carried out for T5165804 and full IC50 was calculated to be 400 nM (see supporting information). The most obvious difference between T5165804 and degrasyn is that T5165804 is symmetrical and presents two cores, which could account for the increased activity.
In compound series I, we tested different rigid structures at the central X position (Figure 3). The compounds containing substituted groups at 1,3-positions of benzene ring were most effective against MM1.S cells (4a–4j, Figure 3); most of the compounds eliminated MM1.S cell growth at submicromolar doses. In compounds where the substitution groups are at 1,4-positions of benzene ring, the biological activity was significantly reduced by at least 5-fold. This indicates that three carbon distances on the central X group are important for the activity. The activity of the compounds in which the meta-substituted X group was changed to pyridine was significantly reduced by at least 2–3 times (4b, 4c and 4e, Figure 3). The activity of hydrophobic substitutions at R1 with Me, Et, n-propyl, and cyclopropyl, even with benzyl group, was tolerated within 2-fold differences. The S-configuration of the R1 group is important for activity (data not shown). Surprisingly, the exchange of the hydrophobic R1 group with methylene alcohol abolishes biological activity. It is very interesting to note that the trimer 4n has the same potency as the dimer structure 4g, suggesting that the increase of potency of dimer structure, compared with that of the monomer degrasyn series, is not caused by the larger number of core units. A different mechanism may underlie their differences in biological activity.
In compound series II, we tested the inside-out structure of series I. The structures and biological activity of these compounds are shown in Figure 4. Compounds containing phenyl or pyridine groups at each side were most effective against MM1.S cells in the high nanomolar range (8a–8p). Substitution with other heterocyclic rings such as imidazole, thiophene, indole, and benzothiophene, abolishes biological activity completely (8k–8p). Interestingly, CR-4 which was developed by Demin et al. has been shown to be an inhibitor of all cell lines at nanomolar concentrations,14 however, neither CR-4 nor the dimer of CR-4 (8j) showed activity against MM1.S cell line. Among all the compounds in Series II, the slightly modified degrasyn (WP1130) or WP1066 dimer-like structure 8a (also called CP2005) showed the most potency at 240 nM against MM1.S cell line. On the other hand, bulky substitution at the R3 position, such as piperidine, and n-hexyloxy were not tolerable for the activity (8b and 8g). Changing the R2 group to chloro- (8c), bromo- (8f), and methoxy- (8i) resulted in less effect on activity against MM1.S cell line.
The most active compounds toward MM1.S cell line in both series I and series II compounds were T5165804 at 400 nM and 8a at 240 nM. We have further tested T5165804 (4a) and 8a, along with WP1130 (degrasyn) in other cancer cell lines including CML (K562), melanoma (A375), and mantle cell lymphoma (Mino) and the results were shown in Table 1. Cell lines were incubated with a range of compound concentrations for 72 h to determine the IC50 value. MTT assays were used to conduct this analysis. Interestingly the 4a and 8a were 2–6-fold more active than WP1130 in all cell lines.
Table 1.
Anti-tumor activity of T5165804 (4a), CP2005 (8a), and WP1130 against various cell lines.
| Compound | IC5O(μM) | |||
|---|---|---|---|---|
| K562 | A375 | MM-1 | Mino | |
| WP1130 | 2.4 | 1.7 | 1.2 | 0.8 |
| T5165804 (4a) | 0.6 | 0.8 | 0.4 | 0.4 |
| CP2005 (8a) | 0.4 | 0.6 | 0.24 | 0.2 |
WP1066 analogs have been identified as JAK2 and downstream Stat3 and c-myc inhibitors.18 To evaluate whether the CP2005 competes with the ATP kinase binding site, it was screened for affinity against a T7-bacteriophage library displaying 228 human kinases (Ambit Biosciences)15 (supplemental material). However, CP2005 did not compete with the ATP binding site and showed no direct inhibition against known kinase targets, such as Bcr-Abl, c-kit, or JAK2. This indicates that the mechanism of action is different from that of ATP binding kinase inhibitors and is in agreement with the finding that WP1066 and similar analogs block the STAT pathway by inducing its ubiquitination and sequestration of the kinase in the aggresome.13,16 WP1066 has also been reported to induce a rapid and specific JAK2 degradation through a proteolytic mechanism in some cell types.17
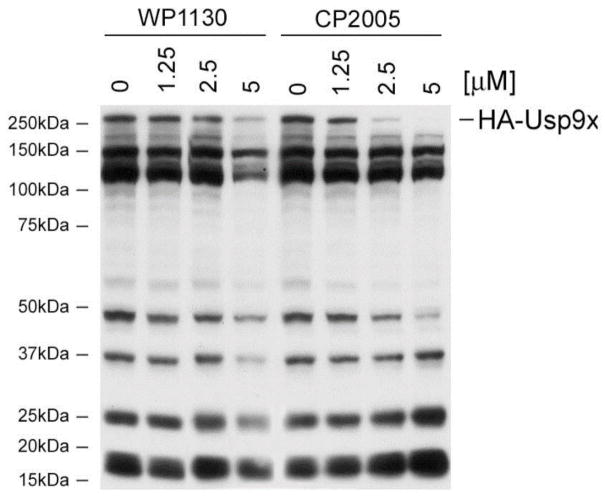
We have previously demonstrated that WP1130 directly inhibits Usp9x, a deubiquitinase (DUB) that regulates the stability of Mcl-1, an anti-apoptotic protein expressed in many tumors, including hematologic malignancies,10 thus, we assessed the direct inhibitory activity of WP1130 and CP2005 in an enzyme assay utilizing recombinant enzyme composed of the catalytic domain of Usp9x (aa1553-1960). The recombinant catalytic domain retains enzymatic activity against ubiquitin–AMC and was used to screen and compare Usp9x inhibition by these compounds. Usp9x DUB inhibitory activity was monitored by measuring a change in isopeptide bond cleavage-mediated substrate fluorescence as a function of time in the presence or absence of various WP1130 and CP2005 concentrations (Figure 5).13 The results demonstrate that inhibition of Usp9x by either compounds (WP1130 and CP2005), with IC50 calculations reporting greater Usp9x inhibition by CP2005 vs. WP1130 (IC50 2.5 vs 5.9 μM, respectively). To confirm DUB inhibition in cells, Z138 mantle cell lymphoma cells were treated with the indicated concentration (Figure 6) of WP1130 or CP2005 for 4 h before assessing DUB activity by HA-Ub labeling and HA immunoblotting as previously described.13 CP2005 demonstrated greater Usp9x inhibitory activity in Z138 cells when compared to WP1130 (Figure 6). The results also suggest greater DUB inhibitory specificity by CP2005 as Usp9x activity was dose-dependently reduced with limited inhibition of other DUBs in these cells. WP1130 inhibited a greater number of DUBs, as previously described.
Figure 5.
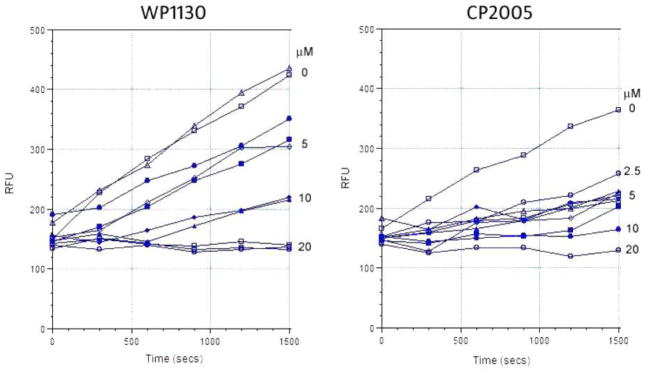
Effects of WP1130 and CP2005 (8a) on Usp9xCD activity. Usp9xCD activity was assessed in the presence or absence of the indicated concentration of each compound. The rate of substrate (Ub-AMC) cleavage was measured over time by monitoring fluorescence. Compounds were incubated with enzyme for 30 min at 37 °C before the addition of substrate
Figure 6.
Effect of WP1130 and CP2005 (8a) on DUB activity in Z138 cells. The activity of Usp9x (HA-Usp9x) is depicted.
5. Conclusion
We synthesized two series of dimer-like symmetrical degrasyn compounds. These compounds were evaluated for their ability to inhibit tumor cell growth. Among all the analogs T5165804 and CP2005 had higher nanomolar potency against various cell lines when compared with degrasyn. In addition, CP2005 was shown to inhibit Usp9x enzymatic activity in vitro and in intact cells. Further chemical modification, mechanism of action studies and pharmacological evaluation are underway.
6. Experimental
6.1. Modeling
Similarity Searching
The similarity searching was completed on a 4-processer SGI Tezro using the Sybyl Modeling Suite v6.92 from Tripos, Inc.19 A database of several million compounds from 36 vendors was converted from a 2D SDF file format to a 2D/3D searchable database with the Unity package in Sybyl. A pharmacophore fingerprint for each molecule was calculated using the default options of the Tuplets module. These structures in their various forms were the object of the computational searches. The structure of WP1130 with propyl replaced by methyl was drawn in and converted to a 3D structure. The structurally search consisted of a shell script written to sequentially search each of the databases using the dbsimilar command and the best 25 structures having >60% similarity with the parent structure were kept. The list of 273 hits resulting from this search was read into a molecular spreadsheet and the hierarchical clustering method in Selector was used to obtain the diversity clusters based on Atom pairs and 2D Fingerprint metrics. Fifty compounds were selected randomly from these clusters. Another search was conducted by a similar shell script and specifying molecular fingerprint with >30% similarity as measured by a stochastic cosine coefficient distance. This led to 894 hits that were processed to find 63 compounds with structural similarity >35%. Out of the combined 113 compounds from both searches, only 75 were found to be available. Nine compounds were received from the vendor Ambinter and no further compounds were ordered.
6.2. Synthesis of degrasyn dimer libraries
6.2.1 General methods
All chemicals and solvents were obtained from Sigma-Aldrich (Milwaukee, WI) of Fisher Scientific (Pittsburg, PA) and used without further purification. Analytical HPLC was performed on a Varian Prostar system, with a Varian Microsorb-MW C18 column (250 × 4.6 mm; 5μ) using the following solvent system A = H2O/0.l% TFA and B = acetonitrile/0.1% TFA. Varian Prepstar preparative system equipped with a Prep Microsorb-MW C18 column (250 × 41.4 mm; 6μ; 60 Å) was used for preparative HPLC with the same solvent systems. Program A: Gradient: 0–5 min. 30% B. 5–35 min. 95% B. 30–35 min. 95% B. Program B: Gradient: 0–5 min. 10% B. 5–35 min. 95% B. 30–40 min. 95% B. Low resolution Mass spectra (ionspray, a variation of electrospray, unless otherwise noted) were acquired on an Applied Biosystems Q-trap 2000 LC-MS-MS. High resolution Mass spectra (ion spray) were acquired on an Agilent Time of Flight Accurate Mass 6220A system with Agilent LC1200 HPLC at the front end. UV was measured on Perkin Elmer Lambda 25 UV/Vis spectrometer. IR was measured on Perkin Elmer Spectra One FT-IR spectrometer. 1H-NMR and 13C-NMR spectra were recorded on a Bruker Biospin spectrometer with a B-ACS 60 autosampler: 600.13 MHz (1H-NMR and 150.92 MHz for 13C-NMR.), IBM-Bruker Avance 500 (500 MHz for 1H NMR and 125.76 MHz for 13C NMR), and IBM-Bruker Avance 300 (300 MHz for 1H NMR and 75.48 MHz for 13C NMR). Chemical shifts (δ) are determined relative to d4-methanol (referenced to 3.34 ppm (δ) for 1H-NMR and 49.86 ppm for 13C-NMR) or CDCl3 (referenced to 7.27 ppm (δ) for 1H NMR and 77.0 ppm for 13C NMR) or DMSO-d6 (referenced to 2.49 ppm (δ) for 1H NMR and 39.5 ppm for 13C NMR). Proton-proton coupling constants (J) are given in Hertz and spectral splitting patterns are designated as singlet (s), doublet (d), triplet (t), quadruplet (q), multiplet or overlapped (m), and broad (br). Flash chromatography was performed using Merck silica gel 60 (mesh size 230–400 ASTM) or using an Isco (Lincoln, NE) combiFlash Companion or SQ16x flash chromatography system with RediSep columns (normal phase silica gel (mesh size 230–400 ASTM)) and Fisher Optima TM grade solvents. Thin-layer chromatography (TLC) was performed on Merck (Darmstadt, Germany) silica gel F-254 aluminum-backed plates with visualization under UV (254nm) and by staining with potassium permanganate or ceric ammonium molybdate.
6.3. General Procedure for Synthesizing Series I compound
In step 1, a mixture of methyl cyanoacetate (25 mmol) and the Amine 1 (25 mmol) was stirred vigorously for overnight. The resulting solid was triturated with 8 mL 95% ethanol and the product filtered. The amide 2 from the step1 reaction (5.0 mmol), aldehyde 3 (5 mmol), and piperidine (five drops) were stirred in anhydrous ethanol (10 mL). Ethanol was evaporated and solid was triturated with water and dried under high vacuum to give the desired product 4.
6.3.1 (2E,2′E)-3,3′-(1,3-phenylene)bis(2-cyano-N-((S)-1-phenylethyl)acrylamide) (4a)
The compound was purified using prep HPLC using Program B as described in above (general methods section). 1H NMR (600 MHz, CDCl3) δ 8.40 (s, 2H), 8.28 (s, 1H), 8.11 (d, J = 7.9, 2H), 7.65 (s, 1H), 7.35 (dd, J = 18.2, 7.1, 10H), 6.71 (s, 2H), 4.62 (d, J = 5.7, 4H).; 13C NMR (151 MHz, CDCl3) δ 159.5, 151.6, 136.9, 133.4, 132.9, 132.5, 130.3, 128.9, 128.0, 127.9, 116.3, 106.1, 44.7.; HRMS (C30H26N4O2) calcd. 475.2129 found 475.2131 [M+H].
6.3.2 (2E,2′E)-3,3′-(1,3-phenylene)bis(2-cyano-N-((S)-1-phenylethyl)acrylamide) (4b)
The compound was purified using preparative HPLC and Program B as described above. 1H NMR (DMSO-d6) δ 9.02 (d, 2H), 8.21 (m, 1H), 8.16 (s, 2H), 8.03 (m, 2H), 7.40 (m, 4H), 7.34 (m, 4H), 7.25 (m, 2H), 5.07 (m, 2H), 1.50 (m, 6H).; HRMS (C29H25N5O2) calcd. 476.2081 found 476.2066 [M+H].
6.3.3 (2E,2′E)-3,3′-(pyridine-2,6-diyl)bis(2-cyano-N-(2-hydroxy-1-phenylethyl)acrylamide) (4c)
1H NMR (600 MHz, CDCl3) δ 8.36 (s, 2H), 8.33 (s, 1H), 8.17 (d, J = 7.8 Hz, 1H), 8.03 (d, J = 7.8 Hz, 1H), 7.66 (t, J = 7.8 Hz, 1H), 7.38 (m, 8H), 7.31 (m, 2H), 7.22 (d, J = 7.2 Hz, 2H), 5.22 (q, J = 4.8 Hz, 2H), 3.97 (m, 4H), 2.13 (bs, 2H).; 13C NMR (150 MHz, CDCl3) δ 159.8, 151.8, 138.2, 137.1, 135.4, 132.8, 132.7, 132.3, 130.1, 129.1, 128.2, 126.7, 116.4, 106.0, 65.7, 56.5.; HRMS (C29H25N5O4) calcd. 508.1979 found 508.1991 [M+H].
6.3.4 (2E,2′E)-3,3′-(1,3-phenylene)bis(2-cyano-N-(1-phenylpropyl)acrylamide) (4g)
1H NMR (600 MHz, CDCl3) δ 8.33 (s, 2H), 8.23 (s, 1H), 8.07 (dd, J = 7.8, 1.2 Hz, 2H), 7.62 (t, J = 8.4 Hz, 1H), 7.33 (m, 10H), 6.60 (d, J = 7.8 Hz, 2H), 4.99 (q, J = 7.8 Hz, 2H), 1.94 (m, 4H), 0.95 (t, J = 7.8 Hz, 6H).; 13C NMR (150 MHz, CDCl3) δ 158.6, 151.5, 141.0, 133.4, 132.9, 132.5, 130.2, 128.9, 128.8, 126.6, 116.5, 106.2, 55.4, 29.0, 10.7.; HRMS (C32H30N4O2) calcd. 503.2442 found 503.2428 [M+H].
6.3.5 (2E,2′E)-3,3′-(1,3-phenylene)bis(2-cyano-N-((S)-1-(4-methoxyphenyl)ethyl)acrylamide) (4i)
1H NMR (600 MHz, CDCl3) δ 1.59 (d, J = 7.8 Hz, 6H), 3.81 (s, 6H), 5.20 (t, J = 7.2 Hz, 6H), 6.51 (d, J = 7.8 Hz, 2H), 6.90 (d, J = 9.0, 4H), 7.29 (d, J = 9.0 Hz, 4H), 7.63 (t, J = 7.8 Hz, 1H), 8.07 (d, J = 8.4 Hz, 2H), 8.25 (s, 1H), and 8.35 (s, 2H).; HRMS (C32H31N4O4) calcd. 535.2340 found 535.2355 [M+H].
6.3.6 (2E,2′E)-3,3′-(1,3-phenylene)bis(2-cyano-N-(cyclopropyl(phenyl)methyl)acrylamide) (4j)
1H NMR (600 MHz, CDCl3) δ 0.44 (m, 2H), 0.53 (m, 2H), 0.68 (m, 4H), 1.29 (m, 2H), 4.51 (t, J = 8.4 Hz, 2H), 6.81 (d, J = 7.8 Hz, 2H), 7.30 (t, J = 7.2 Hz, 2H), 7.36–7.41 (m, 8H), 7.64 (t, J = 7.8 Hz, 1H), 8.09 (dd, J = 7.8, 1.2 Hz, 2H), 8.27 (s, 1H), 8.35 (s, 2H).; HRMS (C38H38N4O4) calcd. 615.2966 found 615.2980 [M+H].
6.3.7 (2E,2′E)-3,3′-(1,4-phenylene)bis(2-cyano-N-(2-hydroxy-1-phenylethyl)acrylamide) (4k)
1H NMR (600 MHz, DMSO-d6) δ 8.86 (d, J = 7.8 Hz, 2H), 8.27 (s, 2H), 8.11 (s, 4H), 7.41 (d, J = 7.8 Hz, 4H), 7.36 (t, J = 7.2 Hz, 4H), 5.06 (t, J = 6.0 Hz, 2H), 5.01 (m, 2H), 3.73 (m, 4H), 1.38 (d, J = 7.0 Hz, 3H).; 13C NMR (150 MHz, CDCl3) δ 162.3, 149.5, 140.8, 135.5, 130.9, 128.7, 127.6, 127.5, 116.6, 109.1, 64.6, 56.9.; HRMS (C30H26N4O4) calcd. 507.2027 found 507.2041 [M+H].
6.3.8 (2E,2′E)-3,3′-(1,4-phenylene)bis(2-cyano-N-((S)-1-phenylethyl)acrylamide) (4l)
1H NMR (DMSO-d6) δ 8.98 (m, 2H), 8.23 (s, 2H), 8.09 (s, 4H), 7.41 (m, 4H), 7.36 (m, 4H), 7.26 (m, 2H), 5.08 (m, 2H), and 1.50 (m, 6H).; 13C NMR 160.32, 148.90, 143.85, 134.96, 130.37, 128.31, 126.86, 126.14, 116.02, 108.68, 49.25, 21.83.; HRMS (C30H26N4O2) calcd. 475.2129 found 475.2150 [M+H].
6.3.9 (2E,2′E)-3,3′-(1,4-phenylene)bis(N-(2-chlorobenzyl)-2-cyanoacrylamide) (4m)
1H NMR (DMSO-d6) δ 9.14 (m, 2H), 8.31 (s, 2H), 8.13 (s, 4H), 7.48 (m, 2H), 7.41 (m, 2H), 7.36 (m, 2H), 7.33 (m, 2H), 4.53 (m, 4H).; 13C NMR (150 MHz, CDCl3) δ 160.26, 149.54, 135.46, 135.00, 130.52, 129.17, 128.85, 128.80, 127.23, 115.99, 107.99, 41.13.; HRMS (C30H24Cl2N4O2) calcd. 543.1349 found 543.1383 [M+H].
6.3.10 (2Z,2′Z)-3,3′-(5-((E)-2-cyano-3-oxo-3-(((S)-1-phenylpropyl)amino)prop-1-en-1-yl)-1,3-phenylene) bis(2-cyano-N-((S)-1-phenylpropyl)acrylamide) (4n)
1H NMR (CDCl3) δ 8.42 (s, 3H), 8.39 (s, 3H), 7.29–7.40 (m, 15H), 6.68 (d, J = 8.0 Hz, 3H), 4.99–5.03 (m, 3H), 1.92–2.02 (m, 6H), and 0.98 (t, J = 7.3 Hz, 9H).; 13C NMR (150 MHz, CDCl3) δ 158.5, 150.1, 141.0, 134.2, 134.0, 129.0, 128.0, 126.8, 116.0, 108.1, 56.6, 29.1, 10.8.; HRMS (C42H36N6O3) calcd. 673.2922 found 673.2941 [M+H].
6.4. General Procedure for Synthesizing Series II Compound
m-Xylylene diamine 5 (14 mL, 100 mM) and 22 mL (220 mM) of methyl cyanoacetate were stirred for 2 h at room temperature and then heated at 60 °C for 3 h. After the exothermic reaction subsided, a solid was formed. Trituration with ethyl acetate and filtering yielded 23 g of 6 as a white solid (85% yield). 1H NMR δ (DMSO-d6) 8.755 (m, 2H), 7.329 (m, 1H), 7.204 (m, 3H), 4.31 (s, 4H), 3.713 (s, 4H), 3.215 (s, 4H).; 13C NMR (150 MHz, CDCl3) δ 162.7, 139.2, 128.9, 126.9, 126.6, 116.6, 43.1, 25.8. The amide 6 from the step1 reaction (5.0 mmol), aldehyde 7 (5 mmol), and piperidine (five drops) were added and stirred in anhydrous ethanol (10 mL). Ethanol was evaporated and solid was triturated with water and dried under high vacuum to give the desired product 8.
6.4.1 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(3-(6-bromopyridin-2-yl)-2-cyanoacrylamide) (8a)
N,N′-(1,3-phenylenebis(methylene))bis(2-cyanoacetamide) (2.7 g, 1 mM), 6-bromo-2-pyridine carboxaldehyde (3.8 g, 2 mM), and 300 μL of piperidine in 50 mL of acetonitrile were heated at 60 °C for 16 h. Cooling and filtering yielded 4 g of a white solid (67% yield). 1H NMR (500 MHz, DMSO) δ 9.13 (s, 2H), 8.11 (s, 2H), 7.92 (t, J = 7.7, 2H), 7.86 (d, J = 7.2, 2H), 7.79 (d, J = 7.8, 2H), 7.32 (d, J = 7.4, 1H), 7.29 (s, 1H), 7.23 (d, J = 7.6, 2H), 4.44 (d, J = 5.9, 4H).; 13C NMR (126 MHz, DMSO) δ 161.2, 151.4, 147.1, 141.6, 141.1, 139.3, 131.0, 128.9, 127.0, 126.7, 126.5, 115.4, 110.7, 43.6.; HRMS (C26H18Br2N6O2) calcd. 604.9931 found 604.9947 [M+H].
6.4.2 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(4-(piperidin-1-yl)phenyl)acrylamide) (8b)
1H NMR (500 MHz, DMSO) δ 8.72 (s, 2H), 7.98 (s, 2H), 7.82 (d, J = 9.0, 4H), 7.29 (s, 1H), 7.25 (s, 1H), 7.19 (d, J = 7.5, 2H), 6.99 (d, J = 9.1, 4H), 4.40 (s, 4H), 3.46 (s, 7H), 1.57 (s, 13H).; 13C NMR (126 MHz, DMSO) δ 162.65, 153.6, 150.7, 139.8, 133.2, 128.7, 126.6, 126.4, 120.1, 118.4, 113.8, 98.0, 47.9, 43.4, 25.4, 24.4.; HRMS (C38H40N6O2) calcd. 613.3286 found 613.3297 [M+H].
6.4.3 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(3,4-dichlorophenyl)acrylamide) (8c)
1H NMR (500 MHz, DMSO) δ 9.10 (t, J = 5.9, 2H), 8.17 (s, 2H), 8.10 (d, J = 2.0, 2H), 7.88 (d, J = 2.0, 1H), 7.86 (d, J = 2.0, 1H), 7.81 (s, 1H), 7.79 (s, 1H), 7.35 – 7.29 (m, 1H), 7.27 – 7.17 (m, 3H), 4.43 (d, J = 5.9, 4H).; 13C NMR (126 MHz, DMSO) δ 161.0, 148.6, 139.3, 135.1, 132.8, 132.4, 132.0, 131.8, 130.0, 128.8, 126.5, 126.2, 116.2, 108.6, 43.5.; HRMS (C25H16Cl4N6O2) calcd. 585.0162 found 585.0191 [M+H].
6.4.4 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(6-methoxypyridin-2-yl)acrylamide) (8d)
1H NMR (500 MHz, DMSO) δ 9.03 (t, J = 6.0, 2H), 8.06 (s, 2H), 7.82 (dd, J = 8.4, 7.2, 2H), 7.34 (d, J = 6.9, 2H), 7.31 (d, J = 7.4, 1H), 7.27 (s, 1H), 7.23 (d, J = 7.7, 2H), 6.99 (d, J = 8.1, 2H), 4.44 (d, J = 5.9, 4H), 3.97 (s, 6H).; 13C NMR (126 MHz, DMSO) δ 163.4, 161.6, 148.9, 147.7, 140.6, 139.4, 128.8, 126.4, 122.4, 116.5, 115.3, 108.7, 54.6, 43.6.; HRMS (C28H24N6O4) calcd. 509.1932 found 509.1921 [M+H].
6.4.5 (2E,2′E)-N,N′-(1,3-phenylenebis(methylene))bis(3-(6-chloro-2-fluoro-3-methylphenyl)-2-cyanoacrylamide) (8e)
1H NMR (DMSO-d6) δ 9.287 (t, 2H), 8.15 (s, 2H), 7.49 (m, 2H), 7.40 (m, 2H), 7.31 (mt, 1H), 7.25 (s, 1H), 7.25 (m, 2H), 4.45 (m, 4H), and 2.27 (s, 6H).; 13C NMR (150 MHz, CDCl3) δ 159.4, 143.2, 138.7, 134.3, 128.4, 126.5, 126.3, 125.2, 124.6, 119.7, 115.5, 114.4, 43.5, 13.8.; HRMS (C30H22Cl2F2N4O2) calcd. 579.1161 found 597.1155 [M+H].
6.4.6 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(3-(3-bromo-4-hydroxyphenyl)-2-cyanoacrylamide) (8f)
1H NMR (500 MHz, DMSO) δ 8.81 (t, J = 5.9, 2H), 8.16 (d, J = 2.1, 2H), 8.00 (s, 2H), 7.77 (dd, J = 8.7, 2.1, 2H), 7.34 – 7.27 (m, 1H), 7.25 (s, 1H), 7.20 (d, J = 7.5, 2H), 6.95 (d, J = 8.6, 2H), 4.40 (d, J = 5.8, 4H).; 13C NMR (126 MHz, DMSO) δ 162.2, 161.5, 149.7, 139.6, 135.6, 132.6, 128.7, 126.7, 126.5, 126.4, 122.7, 117.8, 111.9, 100.5, 44.3, 43.5.; HRMS (C28H20Br2N4O4) calcd.634.9924 found 634.9928 [M+H].
6.4.7 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(4-(hexyloxy)phenyl)acrylamide) (8g)
1H NMR (500 MHz, DMSO) δ 8.92 (t, J = 5.9, 2H), 8.10 (s, 2H), 7.91 (d, J = 9.0, 4H), 7.35 – 7.28 (m, 1H), 7.25 (s, 1H), 7.21 (d, J = 7.7, 2H), 7.07 (d, J = 8.9, 4H), 4.41 (d, J = 5.9, 4H), 4.06 (t, J = 6.5, 4H), 1.85 – 1.64 (m, 4H), 1.42 (s, 4H), 1.37 – 1.18 (m, 8H), 0.88 (t, J = 7.1, 6H).; 13C NMR (126 MHz, DMSO) δ 162.6, 161.9, 150.7, 139.6, 133.0, 128.7, 126.4, 126.3, 124.6, 117.4, 115.6, 102.7, 68.4, 43.4, 31.4, 28.9, 25.5, 22.5, 14.3.; HRMS (C40H46N4O4) calcd. 646.3592 found 647.3599 [M+H].
6.4.8 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(3-hydroxy-4-nitrophenyl)acrylamide) (8h)
1H NMR (DMSO-d6) δ 9.17 (m, 2H), 8.16 (s, 2H), 7.96 (m, 2H), 7.60 (s, 2H), 7.42 (m, 2H), 7.33 (m, 1H), 7.30 (s, 1H), 7.20 (m,2H), 4.37 (m, 4H).; 13C NMR (150 MHz, CDCl3) δ160.5, 148.7, 138.8, 138.7, 137.4, 126.1, 125.8, 120.2, 115.5, 109.3, 43.1.; HRMS (C28H20N6O8) calcd. 569.1415 found 569.1413 [M+H].
6.4.9 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(3,4-dimethoxyphenyl)acrylamide) (8i)
1H NMR (500 MHz, DMSO) δ 9.28 (s, 2H), 8.13 (s, 2H), 7.48 (d, J = 8.2, 2H), 7.39 (d, J = 8.3, 2H), 7.35 (s, 1H), 7.30 (s, 1H), 7.24 (d, J = 7.6, 2H), 4.44 (d, J = 5.9, 5H), 3.79 (s, 6H);2.27 (s, 3H) 2.26 (s, 3H).; 13C NMR (150 MHz, CDCl3) δ 161.4, 152.5, 150.6, 148.6, 139.1, 128.2, 125.9, 124.4, 117.0, 112.2, 111.7, 102.3, 55.7, 55.4, 43.0.; HRMS (C32H30N4O6) calcd. 567.2238 found 567.2236 [M+H].
6.4.10 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(1H-imidazol-5-yl)acrylamide) (8k)
1H NMR (500 MHz, DMSO) δ 8.79 (s, 2H), 8.08 (s, 2H), 7.96 (d, J = 9.4, 4H), 7.30 (s, 1H), 7.25 (s, 1H), 7.20 (s, 2H), 4.40 (d, J = 5.9, 4H).; 13C NMR (126 MHz, DMSO) δ 162.2, 143.6, 139.7, 138.8, 128.7, 126.8, 126.4, 117.3, 100.5, 43.5.; HRMS (C28H22N8O2) calcd. 427.1625 found 427.1640 [M+H].
6.4.11 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(3-([2,2′-bithiophen]-5-yl)-2-cyanoacrylamide) (8l)
1H NMR (500 MHz, DMSO) δ 8.92 (t, J = 5.9, 2H), 8.36 (s, 2H), 7.80 (d, J = 4.2, 2H), 7.68 (dd, J = 5.1, 1.0, 2H), 7.52 (dd, J = 3.6, 1.0, 2H), 7.46 (d, J = 4.0, 2H), 7.34 – 7.27 (m, 1H), 7.27 – 7.18 (m, 3H), 7.16 (dd, J = 5.0, 3.7, 2H), 4.41 (d, J = 5.9, 4H).; 13C NMR (126 MHz, DMSO) δ 161.4, 145.0, 143.8, 140.3, 139.5, 135.5, 134.5, 129.4, 128.7, 128.7, 127.1, 126.4, 126.3, 125.2, 117.1, 100.9, 43.5.; HRMS (C32H22N4O2S4) calcd. 623.0698 found 623.0700 [M+H].
6.4.12 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(4-phenylthiophen-2-yl)acrylamide) (8m)
1H NMR (500 MHz, DMSO) δ 8.99 (t, J = 5.9, 2H), 8.44 (s, 2H), 8.35 (s, 2H), 8.26 (d, J = 1.2, 2H), 7.71 (d, J = 7.2, 4H), 7.45 (t, J = 7.7, 4H), 7.30 (ddd, J = 44.1, 22.9, 7.5, 7H), 4.43 (d, J = 5.9, 4H).; 13C NMR (126 MHz, DMSO) δ 161.3, 144.2, 142.5, 139.5, 136.9, 136.6, 134.1, 130.0, 129.6, 128.8, 128.4, 126.5, 126.5, 116.9, 102.3, 43.6.; HRMS (C32H26N4O2S2) calcd. 611.1570 found 611.1562 [M+H].
6.4.13 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(1H-indol-3-yl)acrylamide)MS 525.6 (M+H) (8n)
1H NMR (500 MHz, DMSO) δ 8.86 (t, J = 5.9, 2H), 8.51 (s,1, 2H), 8.44 (s,1, 2H), 7.89 (d, J = 7.8, 2H), 7.64 – 7.15 (m, 11H), 4.44 (d, J = 5.9, 4H).; 13C NMR (126 MHz, DMSO) δ 162.7, 142.8, 139.4, 136.5, 130.5, 128.2, 127.0, 126.2, 126.4, 123.7, 122.0, 119.1, 118.8, 113.2, 110.0, 97.6, 43.4.; HRMS (C32H24N6O2) calcd. 525.2034 found 525.2051 [M+H], 547.1859 [M+Na].
6.4.14 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(3-(benzo[b]thiophen-3-yl)-2-cyanoacrylamide) (8o)
1H NMR (500 MHz, DMSO) δ 9.23 (t, J = 5.9, 2H), 8.77 (s, 2H), 8.45 (s, 2H), 8.09 (d, J = 7.5, 4H), 7.49 (dd, J = 13.7, 7.0, 4H), 7.38 – 7.31 (m, 2H), 7.27 (d, J = 7.5, 2H), 4.49 (d, J = 5.8, 4H).; 13C NMR (126 MHz, DMSO) δ 161.7, 140.4, 139.5, 139.3, 138.1, 133.3, 128.9, 128.2, 126.6, 126.1, 125.7, 123.6, 122.0, 117.3, 106.9, 43.5.; HRMS (C32H22N6O2S2) calcd. 559.1257 found 559.1251 [M+H], 581.1069 [M+Na].
6.4.15 (2Z,2′Z)-N,N′-(1,3-phenylenebis(methylene))bis(2-cyano-3-(1H-indol-5-yl)acrylamide) (8p)
1H NMR (500 MHz, DMSO) δ 11.59 (s, 2H), 8.90 (t, J = 5.9, 2H), 8.27 (s, 2H), 8.22 (s, 2H), 7.81 (d, J = 8.7, 2H), 7.70 – 7.42 (m, 4H), 7.37 – 7.26 (m, 2H), 7.23 (d, J = 7.9, 2H), 6.59 (s, 2H), 4.44 (d, J = 5.9, 4H).; 13C NMR (126 MHz, DMSO) δ 162.3, 153.2, 139.7, 138.6, 128.8, 128.3, 128.1, 126.8, 126.5, 125.4, 123.5, 123.4, 118.0, 112.8, 103.2, 101.3, 43.6.; HRMS (C32H24N6O2) calcd. 525.2034 found 525.2037 [M+H], 547.1856 [M+Na].
7. Biological Activity
7.1. Screening for Anti-Tumor Activity
All compounds are tested for purity (HPLC, NMR) and the calculated molecular weight is used to make up a 10 mM stock solution of each compound (in 100% dimethylsulfoxide or 50% dimethylsulfoxide:50% polyethylene glycol 300). Compounds are diluted into cell culture media consisting of RPMI1640 with 10% fetal bovine serum to a final starting concentration of 10 μM. [Highest final DMSO content = 0.1%.]. Tumor cells (20,000 for non-adherent cells; 5,000 for adherent cells) are plated into individual wells of a 96-well culture plate in 0.1 mL of culture media. Diluted compounds are added to individual wells containing pre-plated cells (final volume 0.2 mL) and incubated at 37 °C for 24 to 72 h. Cells received vehicle alone as a control.
7.2. MTT (viability) assay
MTT reagent (20 μL of 5 mg/mL stock solution, Sigma) is added to the cells and the plates are incubated at 37 °C for another 2 h. Cells are lysed by adding 100 μL of lysis buffer (20% SDS in 50% N,N-dimethylformamide (Sigma) adjusted to pH 4.7 by 80% acetic acid and 1M HCl such that the final concentration of acetic acid is 2.5% and HCl is 2.5%) into each well and incubated for 6 h. The OD570 of each sample is determined by using a SPECTRA MAX M2 plate reader (Molecular Devices). The OD in control and treated wells is used as an estimate of the effect of compounds on cell growth and survival.
7.3. Cell Lines
B cell malignancies MM1.S (multiple myeloma), mantle cell lymphoma (Z138, Mino), and CML K562 (cell line derived from a patient with CML erythroid blast crisis), were grown in RPMI 1640 containing 10% heat-inactivated fetal bovine serum and 2 mM glutamine. A375 melanoma cells were grown and maintained in the same medium.
7.4. Expression and Purification of the Recombinant Usp9x Catalytic Domain
(Usp9xCD) DNA corresponding to amino acids 1553-1960 of human Usp9x was synthesized using codons optimized for protein expression in E. coli (Genscript). The DNA was cloned into a ULP1-protease cleavable N-terminal His6-Smt3-fusion tag expression vector, derived from pET-28. Protein expression was induced at OD600 2.0, in TB media with Kanamycin overnight at 16 C. Cells were harvested and flash frozen before use. Purification involved a Ni-NTA affinity column, followed by protease-cleavage to remove the affinity tag, passage through a second Ni-NTA column to remove the protease and fusion-tag, and then a final S-200 column equilibrated with 100 mM KCl, 20 mM HEPES, pH 7.4, 2 mM DTT. Protein was concentrated to roughly 20–40 mg/mL before aliquots were flash-frozen. All steps were performed in the presence of reducing agents, either BME, DTT, or TCEP.
7.5. Usp9x catalytic domain activity assay
Purified recombinant enzyme in buffer containing 2 mM DTT was buffer exchanged into 25 mM Tris-HCl, 50 mM NaCl and 1 mg/mL BSA using a spin column. Three hundred nM of Usp9xCD was incubated with the indicated final concentration of inhibitor for 30 min at 37 °C before the addition of 1.5 μM Ub-AMC (BostonBiochem) in a final reaction volume of 25 μL. Fluorescence was monitored (Ex 380 nm, Em 460 nm) in a 384 well plate and read over time in a Molecular Devices SPECTRA MAX M2 plate reader (heated to 37 °C). IC50 values were estimated by integrating the slope of each reaction using GraphPad 6.
7.6. Assessment of DUB activity in intact cells
DUB activity was measured in control and treated cells as previously described. In brief, cells were lysed by sonication in DUB assay buffer (50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 250 mM Sucrose, 1 mM PMSF, 1x Roche proteinase inhibitory cocktail) and 20 μg of protein from the supernatant fraction (after a 14,000 × g spin) were incubated with 200 nM HA-Ub vinyl-sulfone (Boston Biochem) in a final volume of 20 μL. After 90 min at 37 °C, reactions were stopped with the addition of 5 X-sample buffers. DUB activity was detected by SDS-PAGE resolution of the protein followed by nitrocellulose membrane transfer and immunoblotting with anti-HA.
Supplementary Material
Acknowledgments
The authors thank Cancer Center support grant CA016672 for the support of the translational chemistry core Facility and NMR facility at M. D. Anderson Cancer Center. These studies were also partially supported by a grant from the Leukemia Lymphoma Society (to N.J.D.).
References and notes
- 1.Kessler BM. Curr Opin Chem Biol. 2013;17:59–65. doi: 10.1016/j.cbpa.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Micel LN, Tentler JJ, Smith PG, Eckhardt GS. J Clin Oncol. 2013;31:1231–1238. doi: 10.1200/JCO.2012.44.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]; (b) Zhang J-J, Ng K-M, Lok C-N, Sun RW-Y, Che C-M. Chem Commun. 2013;49:5153–5155. doi: 10.1039/c3cc41766b. [DOI] [PubMed] [Google Scholar]
- 4.Harris DR, Mims A, Bunz F. Cancer Biol Ther. 2012;13:1319–1324. doi: 10.4161/cbt.21792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogarty MD. Cell Res. 2010;20:391–393. doi: 10.1038/cr.2010.37. [DOI] [PubMed] [Google Scholar]
- 6.Opferman JT, Green DR. Cancer Cell. 2010;17:117–119. doi: 10.1016/j.ccr.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Peddaboina C, Jupiter D, Fletcher S, Yap JL, Rai A, Tobin RP, Jiang W, Rascoe P, Rogers MK, Smythe WR, Cao X. BMC Cancer. 2012;12:541. doi: 10.1186/1471-2407-12-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolen U, Kobzeva V, Gasparjan N, Ovaa H, Winberg G, Kisseljov F, Masucci MG. Mol Carcinog. 2006;45:260–269. doi: 10.1002/mc.20177. [DOI] [PubMed] [Google Scholar]
- 9.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O’Rourke K, Bazan F, Eastham-Anderson J, Yue P, Dornan D, Huang DC, Dixit VM. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 10.Sun H, Kapuria V, Peterson LF, Fang D, Bornmann WG, Bartholomeusz G, Talpaz M, Donato NJ. Blood. 2011;117:3151–3162. doi: 10.1182/blood-2010-03-276477. [DOI] [PubMed] [Google Scholar]
- 11.Trivigno D, Essmann F, Huber SM, Rudner J. Neoplasia. 2012;14:893–904. doi: 10.1593/neo.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng Z, Pal A, Han D, Shimei W, Maxwell D, Sun D, Levitzki A, Talpaz M, Donato NJ, Bornmann W. Bioorg Med Chem. 2011;19:7194–7204. doi: 10.1016/j.bmc.2011.09.057. [DOI] [PubMed] [Google Scholar]
- 13.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Cancer Res. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 14.Demin P, Rounova O, Grunberger T, Cimpean L, Sharfe N, Roifman CM. Bioorg Med Chem. 2004;12:3019–3026. doi: 10.1016/j.bmc.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 15.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lélias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 16.Kapuria V, Levitzki A, Bornmann WG, Maxwell D, Priebe W, Sorenson RJ, Showalter HD, Talpaz M, Donato NJ. Cell Signal. 2011;23:2076–2085. doi: 10.1016/j.cellsig.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Verstovsek S, Manshouri T, Quintas-Cardama A, Harris D, Cortes J, Giles FJ, Kantarjian H, Priebe W, Estrov Z. Clin Cancer Res. 2008;14:788–796. doi: 10.1158/1078-0432.CCR-07-0524. [DOI] [PubMed] [Google Scholar]
- 18.Kong1 L–Y, Abou-Ghazall MK, Weil J, Chakraborty A, Sun W, Qiao W, Fuller GN, Fokt I, Grimm EA, Schmittling RJ, Archer GE, Jr, Sampson JH, Priebe W, Heimberger AB. Clin Cancer Res. 2008;14:5759–5768. doi: 10.1158/1078-0432.CCR-08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SYBYL. Version 6.9. St. Louis, MO, USA: Tripos Inc; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.