Abstract
β-thalassemia, one of the most common genetic diseases worldwide, is caused by mutations in the human hemoglobin beta (HBB) gene. Creation of human induced pluripotent stem cells (iPSCs) from β-thalassemia patients could offer an approach to cure this disease. Correction of the disease-causing mutations in iPSCs could restore normal function and provide a rich source of cells for transplantation. In this study, we used the latest gene-editing tool, CRISPR/Cas9 technology, combined with the piggyBac transposon to efficiently correct the HBB mutations in patient-derived iPSCs without leaving any residual footprint. No off-target effects were detected in the corrected iPSCs, and the cells retain full pluripotency and exhibit normal karyotypes. When differentiated into erythroblasts using a monolayer culture, gene-corrected iPSCs restored expression of HBB compared to the parental iPSCs line. Our study provides an effective approach to correct HBB mutations without leaving any genetic footprint in patient-derived iPSCs, thereby demonstrating a critical step toward the future application of stem cell-based gene therapy to monogenic diseases.
β-thalassemia is a genetic disorder caused by mutations in the human hemoglobin beta (HBB) gene. It is a common inherited disease extending from the Mediterranean area through the Middle East to Southeast Asia. Patients homozygous with β-thalassemia mutations have severe anemia and usually require frequent transfusions and iron chelation. So far, hematopoietic stem cell transplantation is the only cure available when histocompatible donors are available. To date, one HBE1-β-thalassemia patient treated with lentiviral delivery of a normal HBB gene into his hematopoietic stem and progenitor cells (HSPC) has required no further blood transfusions (Cavazzana-Calvo et al. 2010). However, gene therapy using viral vectors that integrate randomly into multiple sites of the host genome may potentially cause harm, as has been found in other genetic diseases (Hacein-Bey-Abina et al. 2003; Woods et al. 2006).
The ideal approach to curing a genetic disease such as β-thalassemia is to correct the mutations that cause the disease. Generation of induced pluripotent stem cells (iPSCs) from the patients’ own somatic cells could provide a rich source of cells for correction of the β-thalassemia mutation. The mutation-corrected iPSCs could be differentiated into HSPC for autologous transplantation. Such an approach would avoid the problems of immune responses to allogeneic transplantation and the possibility of insertional mutations associated with viral gene delivery.
Correcting a mutation is achievable by homologous recombination. However, the standard approach of introducing homologous DNA is inefficient (Zwaka and Thomson 2003). Efficiency can be increased by introducing DNA double-stranded breaks (DSBs) at or close to the site of the mutation. Site-specific DSBs can be achieved by introducing engineered nucleases such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), as well as the recently developed RNA-directed Cas9 nucleases (Miller et al. 2007, 2011; Cong et al. 2013; Mali et al. 2013). Corrections of the HBB mutation in β-thalassemia and sickle cell anemia using TALENs to introduce DSBs have been reported (Ma et al. 2013; Sun and Zhao 2013).
The RNA-directed Cas9 nucleases from the type II bacterial CRISPR/Cas system have been shown to be robust and versatile tools for generating DSBs in eukaryotic cells (Cong et al. 2013; Mali et al. 2013). The guide RNA (gRNA) contains a 20-nucleotide (nt) guide sequence followed by a trinucleotide (5′-NGG-3′) protospacer adjacent motif (PAM) that can direct Cas9 via Watson-Crick base-pairing to target a desired locus (Deveau et al. 2010; Deltcheva et al. 2011; Gasiunas et al. 2012; Jinek et al. 2012). To explore the potential application of the Cas9 system for gene correction in β-thalassemia, we combined this strategy together with the piggyBac system to correct the mutations in iPSCs prepared from a patient doubly heterozygous for two β-thalassemia mutations, and we achieved efficient seamless correction of both mutations in the iPSCs (Fig. 1). Additionally, we set up a new protocol for hematopoietic differentiation from iPSCs using monolayer-based culture, which enabled detection of increased HBB expression in corrected iPSCs compared to the uncorrected parental lines.
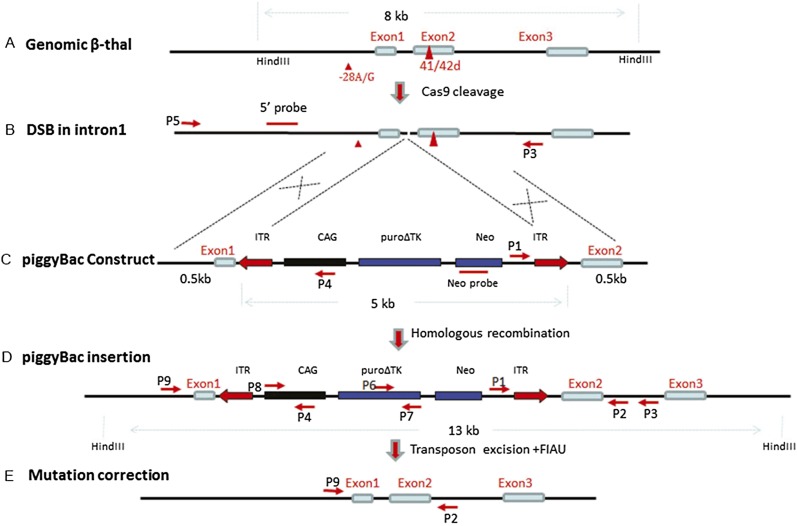
Figure 1.
Strategy for seamless correction of the β-thalassemia mutations using piggyBac and Cas9. (A) Locations of the two mutations at −28 with A/G substitution and the codon 41/42 with 4-bp deletion. (B) The DSB at intron 1 following Cas9 cleavage. (C) The targeting construct of the piggyBac transposon carrying the selectable markers, puro∆tk and Neo, flanked by 500 bp of wild-type genomic sequences. (D) Insertion of the piggyBac following homologous recombination. (E) After selection with puromycin, clones with mutation-corrected lines were identified and transiently transfected with transposase expression plasmids, followed by treatment with FIAU to eliminate piggyBac-containing clones, and the seamless mutation-corrected clones were isolated.
Results
RNA-guided DSB at the HBB gene
Cas9/gRNA, the most frequently used type II CRISPR/Cas system, has been shown to be efficient and specific in cleaving the targeted locus in cultured cells, plants, and animals (Cradick et al. 2013; Nekrasov et al. 2013; Wu et al. 2013; Yang et al. 2013). In this study, we chose the iPSCs reprogrammed from the fibroblasts of a β-thalassemia patient doubly heterozygous for the –28 (A/G) mutation of the promoter and the 4-bp (TCTT) deletion at codon 41 and 42 of exon 2, both being common mutations in the Chinese population. In order to select the proper CRISPR target sites in the human HBB gene sequence, three guide-RNA sequences were selected to target regions that are the least similar to the hemoglobin delta (HBD) gene. Each has the 20-bp guide sequences adjacent to a PAM sequence containing the canonical trinucleotide NGG to target the HBB gene, (Supplemental Fig. 1A) and was cloned into a gRNA vector. To test the ability of these gRNAs to cleave HBB, they were transfected into 293T cells together with Cas9 and pMaxGFP, which served to monitor transfection efficiency. The transfection efficiency was up to 60% based on the number of cells expressing GFP (Supplemental Fig. 1B). The DSB efficiency at the HBB induced by gRNA 1, 2, and 3 were 21%, 5.7%, and 23%, respectively, using the T7E1 assay (Supplemental Fig. 1C). gRNA1 was selected in the following studies, because it targets a site adjacent to a TTAA sequence, required by piggyBac for integration, in intron 1 located between the two mutations. To monitor the possible off-targeting introduced by Cas9-directed cleavage, six potential off-target sites similar to the gRNA1 targeted HBB site were found by BLAST search (Supplemental Table 1). These potential sites were analyzed together with the HBB gene on-target site by the T7E1 assay. Compared to the HBB locus, none of the six regions showed evidence of off-target cleavage (Supplemental Fig. 1D).
Genetic targeting of the HBB gene in the β-thalassemia iPSCs by Cas9
To correct the mutation of −28A/G and TCTT deletion at HBB in the β-thalassemia iPSCs, we constructed a targeting donor plasmid by amplifying two 500-bp segments of the genomic sequences upstream of and downstream from the TTAA sequence at intron 1 of HBB. They were then inserted into the left and right ITRs of the piggyBac transposon, respectively. The piggyBac contains the bi-functional hybrid puro∆TK gene for positive and negative selections (Chen and Bradley 2000). The patient-derived iPSCs were transfected with the donor plasmid, gRNA1, and Cas9 vectors, followed by puromycin selection. Drug-resistant colonies were picked and tested for homologous recombination by PCR amplification using two pairs of primers to screen the colonies. Each pair has one primer sequence inside the piggyBac cassette (P4 and P1) and the other at the genomic sequence outside the 5′ or 3′ homologous arm (P5 and P3) of the donor plasmid (Fig. 1B,C). The targeted clones which were positive for both primer pairs (Fig. 2A) were further confirmed to have undergone homologous recombination by Southern blot analysis (Fig. 2B). Of the 51 clones screened, 12 (23.5%) showed homologous recombination (HR).
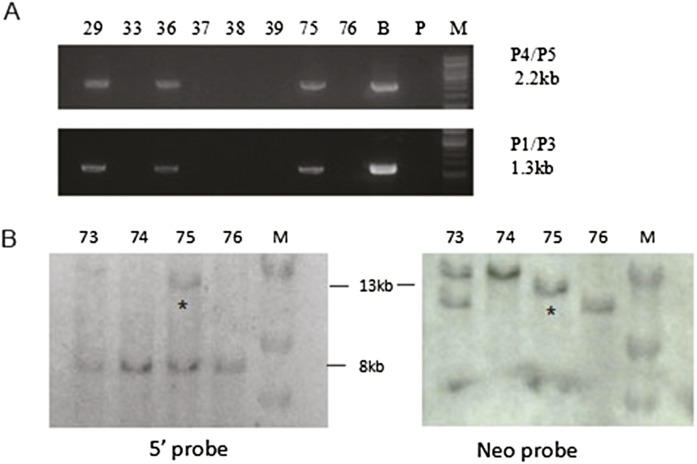
Figure 2.
Site-specific homologous recombination (HR) mediated by CRISPR/Cas9 in β-thalassemia iPSCs. (A) PCR analyses using two pairs of primers, P4/P5 and P1/P2, each pair with one in the piggyBac and the other in the HBB outside the targeting construct to detect homologous recombination. (B) Bacterial artificial chromosome (BAC) containing the piggyBac cassette inserted in the TTAA site located in intron 1 of the wild-type HBB used as a positive control; (P) parental line, (M) marker. (B) Homologous recombination confirmed by Southern blot analysis. Southern blot analysis after HindIII digestion of genomic DNA from puromycin-resistant clones using a 5′ genomic probe outside the targeting construct (left) and a Neo probe inside the piggyBac (right). Clone 75 shows the correct 13-kb band marked by an asterisk (*), detected by both the 5′ globin gene and Neo probes, indicating site-specific homologous recombination at the globin locus. The 8-kb bands are from the endogenous HBB. Other bands seen with the Neo probe are due to random integrations.
PCR-based screening for gene correction
In order to determine whether the homologous recombination corrected the β-thalassemia mutations, we designed primers that had incorporated mismatched nucleotide sequences to create restriction endonuclease cleavage sites to distinguish the wild-type from the mutated sequences. For the −28 mutation, the mutant but not the wild-type sequence would be cleaved by DdeI, and for the 4-bp deletion, the wild-type but not the mutant sequence would be cleaved by XmnI (Fig. 3A). Using this strategy, we found that of the 12 HR clones, four corrected the −28A/G mutation and five corrected the 4-bp deletion (Fig. 3B). DNA sequencing confirmed these results (Fig. 3C).
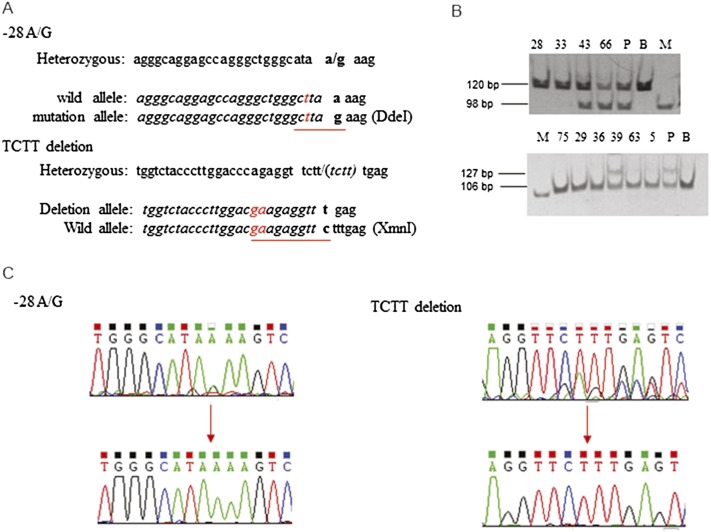
Figure 3.
Identification of gene correction in β-thalassemia iPSCs. (A) Nucleotides changed (in red) in PCR primers to generate restriction enzyme sites to distinguish the normal from the mutant alleles. For the –28 location, replacing nucleotide “a” with “t” at –31 generates a DdeI site for the mutant allele. At 41/42, replacing “cc” with “ga” generates an XmnI site for the wild-type allele. (B) PCR amplification followed by restriction enzyme digestion reveals a normal sequence for clones 28 and 33 at the –28 site and clones 75, 29, 36, 63, and 5 at the 41/42 location. (P) Parental iPSCs, (B) BAC, (M) marker. (C) Sequences of the two mutation sites showing correction of the heterozygous states (upper) to the normal sequences (lower).
Transposon excision in the corrected iPSCs
The piggyBac cassettes can be removed from mutation-corrected clones by transient expression of transposase. However, the transposase can reintegrate the released piggyBac into other chromosomal sites. Negative selection using thymidine kinase of the hybrid puroΔTK gene would allow elimination of cells that retained the piggyBac. Two corrected iPSC lines were transiently transfected by a transposase expressing plasmid, followed by negative selection with 1-(2-deoxy-2-fluoro-1-D-arabinofuranosyl)-5-iodouracil (FIAU). Resistant colonies were screened by PCR using two pairs of primers designed to amplify the CAG promoter and the puro∆TK gene in the cassette (Fig. 4A). Clones not amplified by these two primer pairs were free of PB transposon (Fig. 4B). PCR using primers flanking the insertion site of the β gene also showed that no insert remained in the genome, while primers in the HBB gene outside of the piggyBac insertion showed positive amplification of the globin band (Fig. 4B). By DNA sequencing, the seamless removal of the piggyBac transposon was confirmed by the restoration of the original intron 1 without any exogenous sequence (Fig. 4C). The excision efficiency was 12.5% (5/40 clones examined).
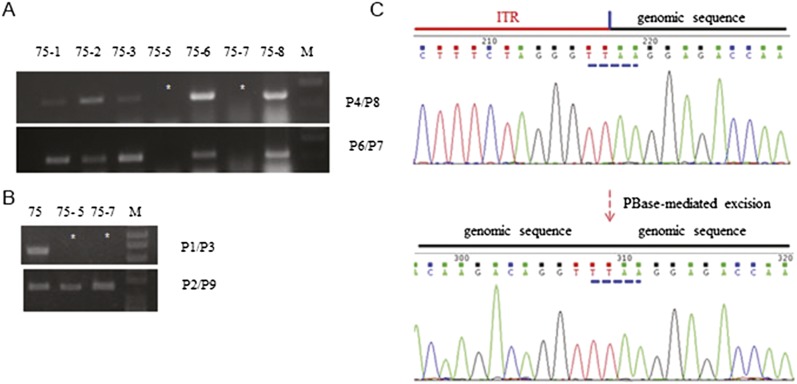
Figure 4.
Precise excision of piggyBac in mutation-corrected iPSC clones. (A) Primers P4/P8 and P6/P7 amplify the CAG promoter and puro∆TK, respectively. No amplification by both pairs indicates no piggyBac reintegration either at the targeting site or other chromosomal locations in clones 75-5 and 75-7. P1/P3 amplifies the junction between the piggyBac insert and the globin gene sequence to the 3′ genomic sequences. No amplification by these primers in these two clones indicates the removal of the piggyBac insert from HBB, while positive amplification (B) with P2/P9 further confirms PB removal. (C) Sequence analysis showing the junction between the ITR of the piggyBac and genomic sequences before transposon removal and the restoration of the normal intron 1 sequence after removal. Note the TTAA sequence of the piggyBac that is used for insertion and excision from the genome.
Characterization of the corrected β-thalassemia iPSCs
Two seamlessly corrected iPSCs, −28 mutation-corrected (59RiPSCs) and 41/42 mutation-corrected (75RiPSCs) lines, were randomly selected for subsequent analyses. They retained their pluripotency as shown by the expression of markers such as NANOG and TRA-1-60 (Supplemental Fig. 2A), maintained the normal karyotypes (Supplemental Fig. 2B), and could differentiate into cells of the three germ layers in vitro (Supplemental Fig. 2C). We also checked the corrected iPSCs for the six potential off-target sites that showed no cleavage by Cas9 in the 293T cells. Using primers which covered the regions of these sites, we did not detect any change in their sequences by Sanger sequencing as compared to sequences from the parental cells (results not shown). Hence, no off-targeting was detected at these sites in the corrected iPSCs similar to that found in the 293T cells.
Expression of HBB in erythroblast differentiation from patient-derived iPSC and gene-corrected human iPSCs
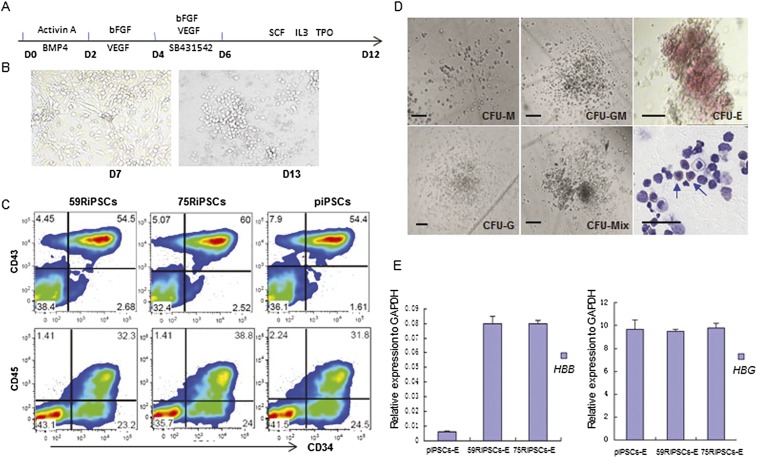
In order to evaluate whether the gene correction in patient-derived iPSCs could restore the expression of HBB, we differentiated these cell lines into the hematopoietic progenitors and erythroblasts using monolayer culture (Fig. 5A). Both corrected and parental lines changed morphology rapidly upon differentiation (Fig. 5B) and produced CD34+CD43+ or CD34+CD45+ hematopoietic progenitors efficiently (Fig. 5C). These HPCs could differentiate into various blood lineages after plating onto the semisolid culture (Fig. 5D). At day 23, the differentiated erythroblasts were picked from CFU-E for analysis of mRNA expression from HBB and hemoglobin gamma (HBG) using qRT-PCR. We showed that the transcribed expression of HBB in CFU-E colonies from two corrected iPSCs increased about 16-fold compared to the parental iPSCs (Fig. 5E). Consistent with several reports (Chang et al. 2006, 2010; Zou et al. 2011; Ma et al. 2013), we also found that erythrocyte differentiation from iPSCs preferentially expressed fetal globin (hemoglobin gamma) more than 100 times greater than adult globin (Fig. 5E) even after gene correction, indicating the need for further research on improved culture conditions to foster erythrocyte development and adult globin-switching.
Figure 5.
Hematopoietic differentiation of gene-corrected iPSCs. (A) Schematic representation of a stepwise hematopoietic differentiation strategy for iPSCs. (B) Representative morphology changes at day 7 and day 13 in hematopoietic differentiation of corrected iPSC lines. (C) Flow cytometric analysis of piPSCs (parental lines), 75RiPSCs, and 59RiPSCs lines harvested at day 14 showing the specific hematopoietic stem and progenitor markers: CD34+, CD43+, and CD45+. (D) Colony-forming assay for differentiated cells at day 10 revealed various types of hematopoietic colonies as well as nucleated cells, including erythroblasts. Blue arrows indicate erythroid cells. (E) HBB and HBG gene expression (normalized to GAPDH) were measured by quantitative RT-PCR in hematopoietic differentiation of parental lines (piPSCs-E), −28 mutation-corrected (59RiPSCs) lines, and 41/42 mutation-corrected (75RiPSCs) lines (data represent mean ± SEM, n = 3).
Discussion
β-thalassemia is one of the most common genetic diseases worldwide, with more than 200 different mutations identified (Cao and Kan 2013). Among the common ones are point mutations that affect promoter function or RNA splicing, premature chain termination due to nonsense mutation, or frame shift due to indels of one or a few nucleotides (Kan and Chang 2010). The iPSC line we studied was doubly heterozygous for a promoter mutation and a 4-bp deletion resulting in frame shift. Using Cas9 to cleave the HBB in intron 1 and a donor plasmid that contains 500 bp of the homologous arm on each side of the piggyBac transposon, homologous recombination occurred efficiently and resulted in correction of these mutations in several clones. Although no one clone was found with correction of both mutations, correction to the heterozygous state will result in the absence of clinical consequences. Since the classical work of Capecchi, Smithies, and Evans (Smithies et al. 1985; Robertson et al. 1986; Thomas and Capecchi 1986), three different systems for specific DNA cleavage to promote efficient homologous recombination have been introduced: zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and Cas9 nucleases (Urnov et al. 2005; Miller et al. 2011; Cho et al. 2013). ZFNs are difficult to design and their proprietary nature makes them expensive to acquire. TALENs can be made within a week using an established protocol, but numerous pairs have to be made to test their cutting efficiencies. In addition, their efficiency could be hampered by DNA methylation and histone acetylation in inactive chromatin (Miller et al. 2007; Mussolino et al. 2011; Sander et al. 2011; Wood et al. 2011; Sanjana et al. 2012). In contrast, the Cas9 nuclease is not limited by these obstacles and is readily available, and the guide RNA containing 20 nt of homologous sequences is easy to synthesize onto the tracrRNA (Deveau et al. 2010; Deltcheva et al. 2011; Gasiunas et al. 2012; Jinek et al. 2012; Cong et al. 2013; Hsu et al. 2013; Ma et al. 2013).
The piggyBac construct with homologous genomic sequences on each side of the selection cassette allowed positive selection of homologous recombination events that corrected the two β-thalassemia mutations. Unlike the Cre/loxP system that leaves 34 bp upon releasing the insert (Hockemeyer et al. 2011; Zou et al. 2011), negative selection with the TK gene seamlessly removed the selection cassette, leaving behind the normal HBB gene with no foreign sequences. This approach is also very efficient. The homologous recombination rate we obtained was 23%, of which four out of six clones had the −28 mutation corrected, and five out of six clones had the 4-bp deletion corrected. This strategy also could be used for correcting the sickle mutation, since the corrected −28A/G mutation is even farther upstream of the gRNA1-directed DSB than the codon 6 mutation.
In this study, we did not detect off-site nicking or targeting in the six genomic sites that bear the most homology with the gRNA-PAM sequence. Although off-targeting using Cas9 has been reported (Cradick et al. 2013; Fu et al. 2013), our judicious choice of gRNA sequences that were most dissimilar to the HBD gene might have avoided the possibility of off-targeting. As is true with cell culture work, other genomic changes cannot be ruled out without whole-genome sequencing. A modification of Cas9, called nickase, to cleave only one DNA strand may reduce the incidence of off-targeting. However, the efficiency of recombination appears to be significantly reduced (Cong et al. 2013). Recently, it has been reported that using a pair of gRNAs with nickases that cut the opposite strands of DNA at adjacent sites may increase the homologous recombination frequency and reduce off-targeting (Ran et al. 2013).
Although we succeeded in seamlessly correcting the mutation in the HBB gene, the functional analysis of the corrected HBB was hampered by the fact that erythroid precursors differentiated from iPSCs as well as ESC preferentially express embryonic and fetal, but not adult hemoglobin protein (Chang et al. 2006, 2010; Zou et al. 2011; Ma et al. 2013). Induction of a switch to adult globin has been the subject of intense investigation. Nevertheless, we could detect a 16-fold increase in the transcription of the HBB gene by qRT-PCR in erythroid cells differentiated from the mutation-corrected iPSCs over those from the parental iPSCs.
The combination of CRISPR/Cas9 and the piggyBac system achieved the site-specific correction in the HBB locus while preserving the cis-regulatory element for HBB expression without any footprint, and provides an ideal method for realizing the complete globin-switch to adult type in the future. Other hurdles such as the transplantability and the ability to produce adult globin gene expression in these gene-corrected cells still need to be overcome. Recently, a group reported globin-switching in vivo, but they infected iPSCs using lentiviral vectors carrying three transcription factors to generate hematopoietic progenitors, followed by another two transcription factors for transplantation (Doulatov et al. 2013). These progenitor cells modified by five transcription factors could undergo globin-switching in vivo, but only gave rise to short-term engraftment of myeloid and erythroid lineages. Furthermore, they did not detect any expression of HBB in vitro. Therefore, further studies are required focused on the generation of developmentally mature and transplantable HSPC.
From this study, we conclude that the combination of CRISPR/Cas9 to cleave the HBB gene and piggyBac to select for homologous recombination events has efficiently corrected two different β-thalassemia mutations and converted homozygous β-thalassemia to the heterozygous states, thus restoring HBB gene expression in erythrocytes differentiated from the corrected iPSCs. The gene-editing strategy we report here provides an important step toward therapy of monogenic disease in the future based on the genetic repair of patient-specific iPSCs.
Methods
Cell culture
293T cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). iPSCs from the β-thalassemia patient were maintained on mitomycin C (Sigma)-treated mouse embryonic fibroblast (MEF) feeder cells in DMEM/F12 GlutaMAX (Gibco) supplemented with 20% Knockout Serum Replacement (KSR), 10 ng/mL bFGF(Invitrogen), 50 μg/mL penicillin/streptomycin (Gibco), 10−4 M nonessential amino acids (Gibco), and 10−4 M β-mercaptoethanol (Sigma).
Generation of β-thalassemia iPSCs
The skin fibroblasts from an anonymous β-thalassemia patient doubly heterozygous for the −28 (A/G) mutation and for the 4-bp (TCTT) deletion at codons 41 and 42 in exon 2 were reprogrammed using temperature-sensitive Sendai viral vectors that encode four reprogramming factors (POU5F1, SOX2, KLF4, and MYC) (kindly provided by DNAVEC). The iPSCs line for this study was incubated at 38.5°C for 4 d to completely remove the Sendai viruses and thus be free of reprogramming factors. It exhibits all the pluripotent characteristics of human embryonic stem cells (Ye et al. 2013).
Surveyor assay for on/off-target analysis
1 × 105 293 T cells in 24 wells were transfected with 400 ng guide RNA and 400 ng Cas9 using Lipofectamine 2000 (Life Technologies) and harvested after 72 h for DNA preparation. The genomic region surrounding the Cas9 target site or candidate off-target sites (Supplemental Table 1) were PCR-amplified, and the PCR products purified by a gel extraction kit (Thermo). Four hundred nanograms of the purified PCR fragments were mixed with 2 μl 5× GXL buffer (Takara) and distilled water to a final volume of 10 μl and subjected to conditions that enable heteroduplex formation as previously described (Cong et al. 2013). After reannealing, the products were treated with T7 endonuclease 1 (T7E1) for 15 min at 37°C and analyzed on 10% polyacrylamide gels. The gels were stained with ethidium bromide for 20 min and examined by the Gel DOC imaging system (Bio-Rad). Quantification was calculated based on the relative band intensities.
Cas9 and PB-based donor plasmid for gene targeting
Since these mutations in the two alleles were in the HBB gene promoter and exon 2, three 20-bp guide RNAs were chosen to lie in the vicinity of the two mutations. Guide RNAs were designed according to the rule of 5′-GN20NGG-3′. The guide RNA oligonucleotides were inserted into the gRNA cloning vector (Addgene) according to the protocol provided by Mali et al. (2013) using the Gilson Assembly kit (NEB). For constructing the donor plasmid, we used the piggyBac plasmid (5′-PTK-3′, a gift from Dr. Allan Bradley) modified by inserting the CAG promoter to drive the bi-functional puro∆TK gene and the Neo gene between the two ITRs. Then, the fragments of 500-bp genomic sequences 5′ and 3′ to the TTAA site of intron 1 were amplified by primer sets of 5′ arm1 and 3′ arm1 using HBB as a template, respectively. The fragments of 5′ITR and 3′ITR were amplified by primer sets of 5′ITR and 3′ITR using the modified piggyBac plasmid as a template. Each homologous arm was then joined to one of the ITRs by PCR and cloning strategies, so that each homologous arm and the ITR would share the TTAA sequence at the junction. Finally, the 5′ homologous arm with 5′ITR, the selectable markers, and the 3′ homologous arm with 3′ITR were joined together with the vector by the standard cloning techniques, as depicted in Figure 1C.
For gene targeting, 3 × 106 iPSCs were electroporated with 3 μg each of the donor plasmid, gRNA, and Cas9 (Addgene) in 100 μl of Stem Cell Solution I (Lonza) using nucleofactor II (Lonza) set at program A023. Transfected cells were plated onto the feeders prepared from day 13 MEF of the DR4 strain (Tg [DR4], the Jackson Laboratory) and cultured in human ES medium supplemented with 10 μM of Y-27632 for 1 d. Three days after transfection, puromycin selection (1 μg/mL, InvivoGen) was started and continued for 10 d. The resistant colonies were picked and expanded for PCR screening and Southern blot verification. All primers are listed in Supplemental Table 2.
PCR detection of targeted homologous recombination, correction of mutation
Genomic PCR was performed using GXL DNA polymerase (Takara) according to the manufacturer’s instructions. To identify the integration mediated by homologous recombination, two pairs of primers were designed. Primers P1 (inside the PB cassette) and P3 (downstream from the 3′ arm) were used to amplify the 1.3-kb product. Primers P5 (HBB promoter, upstream of the 5′ arm) and P4 (in the CAG promoter) were used to amplify a 2.3-kb product. Two forward primers introduced restriction enzyme sites (DdeI in P10 and XmnI in P12) by modifying the original sequences (Fig. 3A) and were used to detect the gene correction in the targeted clones with their reverse primers (P11 and P13, respectively). All the primers are also listed in Supplemental Table 2.
Southern blot analysis
For Southern blot analysis, 10 μg of genomic DNA extracted from iPSCs was digested with HindIII (NEB) and separated on a 0.8% agarose gel by electrophoresis. The DNA was transferred onto a nylon membrane and the membrane (Perkin Elmer) hybridized at 65°C overnight with a digoxigenin-dUTP (Roche)-labeled probe. The probe was made from either a 1-kb genomic DNA fragment or neomycin gene according to the manufacturer’s instructions (Roche).
Removal of PB transposon cassette
To remove the piggyBac cassette, 3 × 106 corrected iPSCs were transfected with 10 μg hyperactive transposase vector, hyperPBase (a gift from Dr. Allan Bradley), followed by selection with FIAU (0.5 μM) for 7 d. To identify their excision, two pairs of primers, P8 and P4 (each inside of CAG), and P6 and P7 (both inside of puro∆TK), were used to detect the removal of the PB transposon cassette from the genome. PCR using primers flanking the insertion site of the HBB gene (P2 and P9) also showed that no insert remained in the genome. These primers are also listed in Supplemental Table 2.
Karyotype analysis
Chromosomal analyses of corrected iPSCs were examined and interpreted by the cytogenetics laboratory at UCSF. Metaphase spreads were prepared from cells treated with 100 ng/mL colcemid for 6 h, followed by the standard protocol for high-resolution G binding. Twenty chromosome spreads were examined for each sample.
Immunocytochemistry
Primary antibodies of pluripotent markers (NANOG and TRA-1-60) and of three germ layer markers (SMA, SOX17, and beta 3 tubulin) were used for immunocytochemistry according to the protocol previously described (Ye et al. 2013). Briefly, cells were fixed with PBS containing 4% paraformaldehyde for 10 min at room temperature. After washing with PBS, they were permeabilized and blocked with 0.1% TritonX-100, 1% BSA, and 5% FBS for 45 min. The cells were incubated with primary and secondary antibodies of appropriate dilutions according to the manufacturer’s recommendations. The stained cells were analyzed by a Nikon Eclipse TE2000-U microscope.
In vitro differentiation
To generate embryoid bodies (EBs), iPSCs were incubated with Dispase (STEMCELL Technologies) for 4 min and harvested. The clumps of cells were transferred to a 6-well plate with a low-attachment surface in DMEM supplemented with 15% FBS (Hyclone), 50 μg/mL penicillin/streptomycin (Gibco), 10−4 M nonessential amino acids (Gibco), and 10−4 M β-mercaptoethanol (Sigma). After a 1-wk culture, EBs were transferred to gelatin-coated plates and cultured with the same medium for an additional week.
Hematopoietic differentiation of human iPSCs
Human iPSCs were treated with 1 mg/mL dispase (Gibco) and replated onto the Martrigel-pretreated 12-well plate coated with mytomycin C-treated MEF and cultured in hESC medium at a density of 8 × 104 cells/well. The second day was defined as day 0. The medium was switched to STEMdiff APEL (STEMCELL Technologies) medium (AM). From day 0 to day 2, 50 ng/mL BMP4 and Activin A were added to the AM. At day 2, the cytokines in AM were changed to 50 ng/mL VEGF and 50 ng bFGF and kept for 2 d. From day 4 to day 6, 20 µM SB431542, and 50 ng/mL VEGF and bFGF were added to the AM (Wang et al. 2012). From day 6, the cells were cultured in AM supplemented with 50 μg/mL penicillin/streptomycin (Gibco), 50 ng/mL SCF, 50 ng/mL TPO, and 50 ng/mL IL-3, with a half-medium change every other day. For erythroid differentiation, the cells at day 13 were cultured in AM supplemented with 50 ng/mL hSCF, 20 ng TPO, 20 ng IL-3, and 5 units/mL EPO for 10 d, then collected and stained with Accustain Wright-Giemsa stain (Sigma-Aldrich). All the cytokines were purchased from Prospec.
Flow cytometric analysis
Cells were incubated with the indicated antibodies, or isotype antibodies were used for the flow cytometry analysis according to the protocol previously described (Ye et al. 2013; Li et al. 2014). Briefly, cells were trypsinized into single cells and suspended in PBS containing 2% FBS (Hyclone). Cells were incubated in PBS with 5% mouse serum and 0.01% NaN3 with labeled monoclonal antibodies at 4°C for 30 min, then washed and suspended in PBS with propidium iodide to stain dead cells for flow cytometric analysis on a BD LSR II cytometer. Flow cytometric data were analyzed by Flow Jo.
Colony-forming cell assay
About 1000 single cells in suspension in 0.3 mL IMDM (Gibco) with 2% FBS (Hyclone) were mixed with 3 ml MethoCult+ 4435 (STEMCELL Technologies) according to the manufacturer’s instructions. The mixtures were plated onto three low-attachment 35-mm dishes and incubated at 37°C in 5% CO2 with 100% humidity for 2 wk. The colonies were counted and classified according to morphology.
qRT-PCR
Total RNA was extracted according to the manufacturer’s instructions of the RNeasy Kit (Qiagen). Complementary DNAs were synthesized using SuperScript III reverse transcriptase (Invitrogen) following the manufacturer’s instructions. qRT-PCR was performed using SYBR Green-based PCR Master Mix (Invitrogen), and signals were analyzed with the Bio-Rad Real-time PCR system.
Acknowledgments
We thank Zhongxia Qi and Jingwei Yu for the karyotype analyses. This work was supported by NIH Grant P01-DK088760.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.173427.114.
References
- Cao A, Kan YW. 2013. The prevention of thalassemia. Cold Spring Harb Perspect Med 3: a011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, et al. . 2010. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassemia. Nature 467: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Nelson AM, Cao H, Wang L, Nakamoto B, Ware CB, Papayannopoulou T. 2006. Definitive-like erythroid from human embryonic stem cells coexpress high levels of embryonic and fetal globins with little or no adult globin. Blood 108: 1515–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Huang A, Hirata RK, Wang PR, Russell DW, Papayannopoulou T. 2010. Globin phenotype of erythroid cells derived from human induced pluripotent stem cells. Blood 115: 2553–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Bradley A. 2000. A new positive/negative selectable marker puroΔtk for use in embryonic stem cells. Genesis 28: 31–35 [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. . 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. 2013. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41: 9584–9592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64: 475–493 [DOI] [PubMed] [Google Scholar]
- Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, et al. . 2013. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 13: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109: E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. . 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302: 415–419 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. . 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shaleem O, et al. . 2013. DNA targeting specificity of the RNA-guided Cas9 nuclease. Nat Biotechnol 31: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan YW, Chang JC. 2010. Molecular diagnosis of hemoglobinopathies and thalassemia. Prenat Diagn 30: 608–610 [DOI] [PubMed] [Google Scholar]
- Li K, Zhu S, Russ HA, Xu S, Xu T, Zhang Y, Ma T, Hebrok M, Ding S. 2014. Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell Stem Cell 14: 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Liao B, Zhang H, Wang L, Shan Y, Xue Y, Huang K, Chen S, Zhou X, Chen Y, et al. . 2013. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J Biol Chem 288: 34671–34679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. . 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. . 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. 2011. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. 2013. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 691–693 [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. . 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson E, Bradley A, Kuehn M, Evans M. 1986. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature 323: 445–448 [DOI] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. . 2011. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8: 67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. 2012. A transcription activator-like effector toolbox for genome engineering. Nat Protoc 7: 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. 1985. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 317: 230–234 [DOI] [PubMed] [Google Scholar]
- Sun N, Zhao H. 2014. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng 111: 1048–1053 [DOI] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. 1986. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature 324: 34–38 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Wang C, Tang X, Sun X, Miao Z, Lv Y, Yang Y, Zhang H, Zhang P, Liu Y, Du L, et al. . 2012. TGFβ inhibition enhances the generation of hematopoietic progenitors from human ES cell-derived hemogenicendothelial cells using a stepwise strategy. Cell Res 22: 194–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. . 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM. 2006. Gene therapy: therapeutic gene causing lymphoma. Nature 440: 1123. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. 2013. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13: 659–662 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Muench MO, Fusaki N, Beyer AI, Wang J, Qi Z, Yu J, Kan YW. 2013. Blood cell-derived induced pluripotent stem cells free of reprogramming factors generated by Sendai viral vectors. Stem Cells Transl Med 2: 558–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Mali P, Huang X, Dowey SN, Cheng L. 2011. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood 118: 4599–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. 2003. Homologous recombination in human embryonic stem cells. Nat Biotechnol 21: 319–321 [DOI] [PubMed] [Google Scholar]