Abstract
Genomic approaches continue to provide unprecedented insight into the microbiome, yet host immune interactions with diverse microbiota can be difficult to study. We therefore generated a microbial microarray containing defined antigens isolated from a broad range of microbial flora to examine adaptive and innate immunity. Serological studies with this microarray show that immunoglobulins from multiple mammalian species exhibit unique patterns of reactivity, while exposure of animals to distinct microbes induces specific serological recognition. While adaptive immunity exhibited plasticity toward microbial antigens, immunological tolerance limits reactivity toward self. We discovered that several innate immune galectins exhibit specific recognition of microbes that express self-like antigens, leading to direct killing of a broad range of gram negative and positive microbes. Thus, host protection against microbes appears to represent a balance between adaptive and innate immunity to defend against evolving antigenic determinants while protecting against molecular mimicry.
INTRODUCTION
While infectious disease typically represents a breach in host immunity by an individual organism, hosts battle continuously with resident microbiota 1–4. Indeed, the microbiome represents a previously underappreciated influence on numerous biological processes, including subsequent immunity to newly acquired microbes. Although genomic approaches underscore the diversity of the microbiome, many questions remain regarding host interactions with the microbiome itself 5–7. While previous studies examine host factor interactions with a variety of microbial determinants 8,9, a wide variety of innate and adaptive immune factors appear to specifically target microbes through recognition of cell surface glycans 10. Indeed, as the unique localization and density of microbial glycans on the surface of individual microbes directly facilitates host interactions, glycan determinants have long been recognized as key structures recognized by host immune factors 11,12. However, despite the importance of host-microbial glycan interactions, the specificity of host immune factors with carbohydrate-binding activity remains only partially defined.
As microbial glycans often represent the unique antigenic determinants used to define specific strains within microbial species 12, examination of host factors against a diverse range of microbial glycans may not only identify unique host interactions with individual microbes, but may also provide key insight into the structural motifs required for these interactions. Recent studies demonstrate that synthetic microbial glycans or glycans directly harvested from microbes coupled in an array format can be used to characterize serological specificity for distinct microbes 13–23, demonstrating that glycans can be readily harvested from a variety of microbes and examined in parallel to evaluate host-microbial interactions. As host immune factors work in concert to interact with a variety of microbial determinants, yet the individual structural motifs required for the binding of many host immune factors remain unknown, we isolated a diverse set of previously defined microbial glycans from a broad range of microbial genera to provide a platform for defining host factor interactions with specific microbial glycans. Microbial glycans harvested in this manner were coupled in an array format, generating a microbial glycan microarray (MGM) composed of distinct and defined microbial glycans, to interrogate the binding preferences and specificity of host immune factors with carbohydrate binding activity.
Using this format, we found that sera from distinct species displayed unique reactivity to a range of microbial antigens, while challenge with specific microbes resulted in enhanced reactivity toward the inoculated microbe, which demonstrated the accessibility of glycans coupled in this array format. Unexpectedly, several host innate immune galectins displayed exquisite specificity for microbial antigens resembling self-like antigens. Importantly, galectins not only bound intact microbes expressing various self-like antigens, but these innate immune lectins also decreased the viability of target microbes. Overall, these studies suggest that adaptive and innate immune factors work in concert to provide immunity against a broad range of microbial genera.
RESULTS
Generation of a microbial glycan microarray
In an effort to generate a platform designed to determine the binding specificity of host immune factors toward distinct carbohydrate antigens isolated from diverse microbial flora, we utilized highly purified and previously characterized bacterial polysaccharides (BPS) isolated from a broad range of microbes as outlined previously 24,25. To examine the printing efficiency of isolated BPS, we first examined the BPS of Pseudomonas aeruginosa O2 (PA O2) printed on amine reactive N-hydroxysuccinimide (NHS) activated glass slides. Interrogation of bound PA O2 BPS with anti-PA O2 anti-sera in this array format demonstrated a concentration dependency of recognition, with saturation occurring at BPS concentrations of ~125 µg/ml (Supplementary Results, Supplementary Fig. 1a). Thus, the remaining library of previously purified and highly characterized BPS isolated from a diverse range of microbial species was immobilized by printing BPS over a range of concentrations to facilitate optimal detection of host-pathogen interactions as a microbial glycan microarray (MGM) (Fig. 1a).
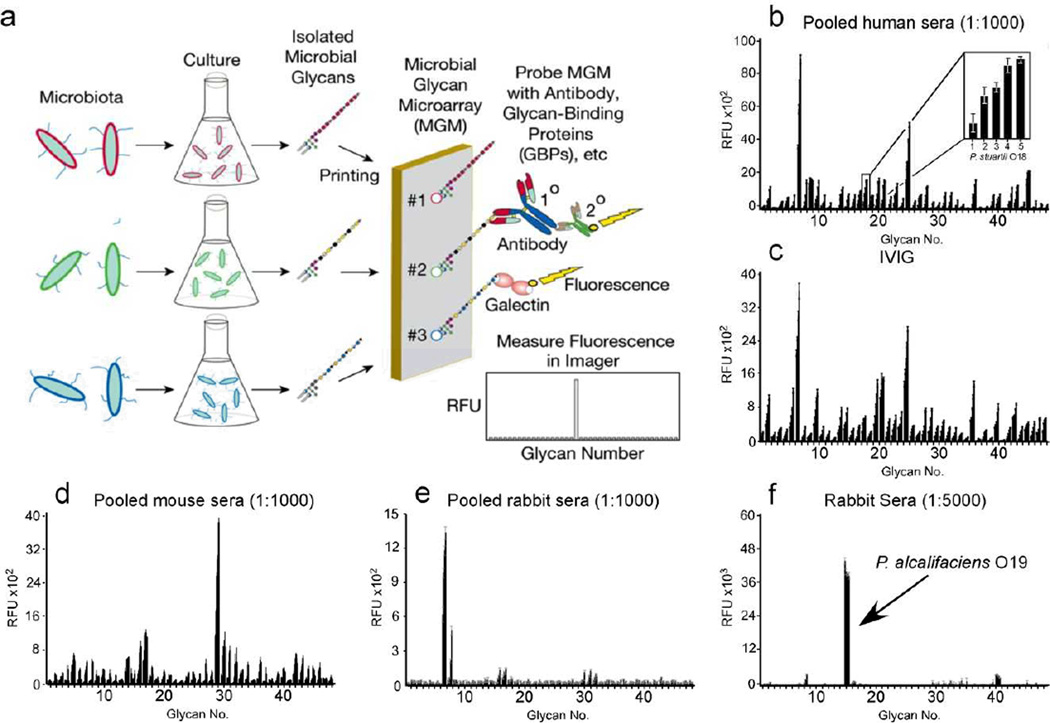
Figure 1. Production and validation of MGM and recognition of microbial glycan structures by sera.
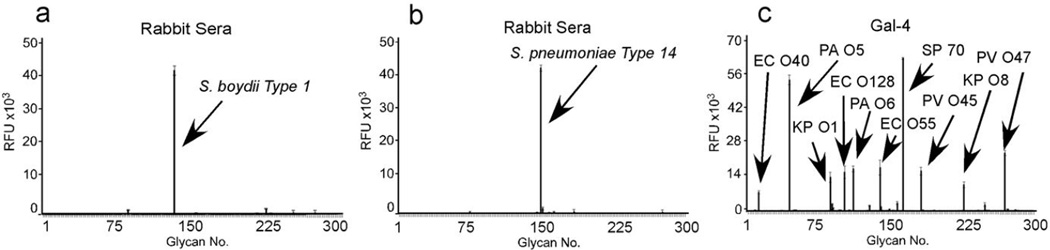
(a) Schematic of MGM design and utility. (b) MGM data obtained after incubation with pooled normal human sera at 1:1000 followed by labeled anti-IgG and anti-IgM). b inset: magnification of antibody interactions with increasing printed concentration of Providencia stuartii O18 BPS (31.25, 62.5, 125, 250, 500 µg/mL). (c) MGM probed with 200 µg/ml IVIG pooled from ~10,000 human donors followed by detection with anti-IgG. (d,e) Detection of pooled normal mouse sera at 1:1000 (d) or pooled normal rabbit sera at 1:1000 (e) with anti-mouse or anti-rabbit IgG, respectively. (f) MGM data obtained after incubation with 1:5000 dilution of sera from rabbits challenged with the indicated bacterial species followed by detection with anti-rabbit IgG. See Supplementary Dataset 1 for complete microarray data.
Using this platform, we examined host interactions with diverse microbial glycan antigens. We first examined pooled sera isolated from five healthy human volunteers. Examination of human sera for IgG demonstrated a distinct pattern of reactivity on the MGM (Fig. 1b and Supplementary Fig. 1). By contrast, while no IgM reactivity could be detected at a similar dilution (Supplementary Fig. 1), a broad range of IgM reactivity distinguishable from IgG could be detected at lower dilutions (Supplementary Fig. 1). Importantly, commercially available intravenous immunoglobulin (IVIG), a pooled source prepared from over 10,000 healthy human volunteers, provided a similar IgG pattern (Fig. 1c and Supplementary Fig. 1) as observed following analysis of pooled sera from five healthy human volunteers. These results suggest that microbial antigens remain accessible following isolation and printing in a microarray format and that IgM and IgG in human sera display characteristic reactivity toward particular microbial antigens.
As different mammalian species may be exposed to distinct microbes, we next tested whether sera isolated from multiple mammalian species display differential reactivity toward the various antigenic determinants on the MGM. This is especially important when considering that most current genomic approaches lack the capacity to identify individual strains within the microbiome and previous studies demonstrate that exposure to specific microbes can result in discrete changes in seroreactivity toward microbial glycans 26,27,28. Examination of serum IgG from mice and rabbits exhibited strikingly distinct patterns of reactivity from each other and from human IgG, with unique reactivity toward different species and different strains within the same microbial species (Fig. 1d,e, and Supplementary Fig. 1), further illustrating the importance of examining multiple microbial strains within a species. To determine whether exposure to an individual microbe affects reactivity to the MGM, we examined sera isolated from rabbits following inoculation with specific microbes. While testing of high dilutions of sera isolated from pre-immune recipients displayed low MGM reactivity (Supplementary Fig. 1), high titer seroreactivity toward the individual glycan antigen isolated from the inoculated microbes could be detected on the MGM following microbial exposure, whereas little reactivity to unrelated antigens was observed (Fig. 1f and Supplementary Fig. 1). These results demonstrate that adaptive immunity displays a high degree of plasticity and specificity and that the MGM easily and sensitively permits examination of host-microbiota interactions against very specific antigenic determinants represented on the array.
The MGM accurately predicts host-microbial interactions
While adaptive immunity provides critical host protection, innate immunity represents the first line of defense against potential pathogens 29. As a result, we evaluated the binding specificity of innate immune factors toward the microbial glycans on the MGM. To accomplish this, we examined several members of the galectin family, as recent studies suggest that galectin family members recognize a diverse set of microbial glycans 30–33. Examination of galectin-3 (Gal-3), galectin-4 (Gal-4) and galectin-8 (Gal-8) on the MGM demonstrated a unique specificity for one particular antigen on the array (Fig. 2a–c), the antigenic determinant of Providencia alcalifaciens O5 (PA O5). These results strongly suggest that unlike most innate immune factors, galectins appear to possess the unique ability to specifically recognize a specific subset of microbes. To determine whether recognition of the O antigen of PA O5 on the MGM actually predicts real interactions with the intact microbe, we explored binding of Gal-3, Gal-4, or Gal-8 to PA O5. Consistent with their binding prolife on the MGM, Gal-3, Gal-4, and Gal-8 bound PA O5 (Fig. 2d–f), yet failed to recognize PA O19, a related strain also printed on the MGM, but not recognized by Gal-3, Gal-4 and Gal-8 (Supplementary Fig. 2). These results demonstrate that binding interactions detected on the MGM can accurately predict previously unrecognized interactions between innate immune factors and microbes.
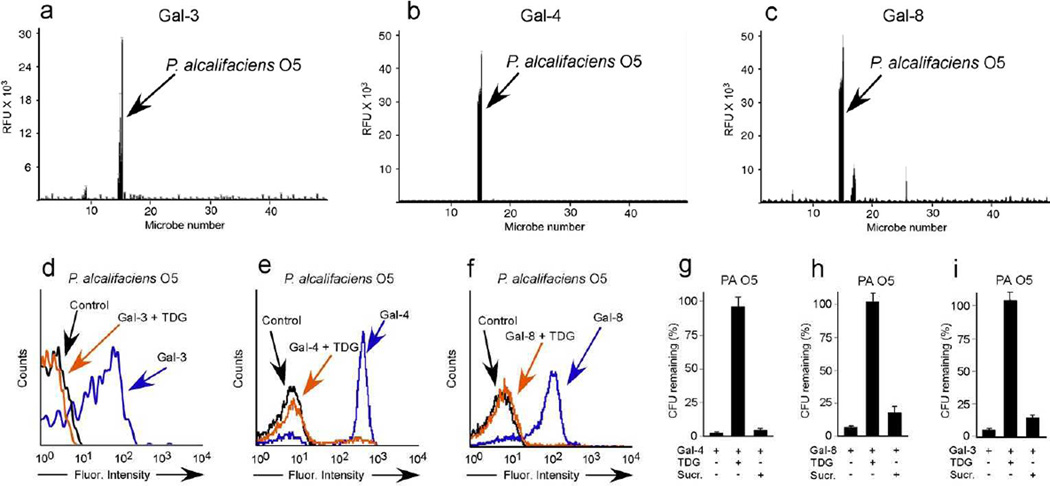
Figure 2. MGM identifies new bacterial targets for galectin binding and killing.
MGM data obtained after incubation with fluorescently tagged Gal-3 (a), Gal-4 (b), and Gal-8 (c) at ~1.5µM. See Supplemetnary Dataset 1 for complete microarray data. Flow cytometric analysis of P. alcalifaciens O5 (PA O5) after incubation with Gal-3 (d), Gal-4 (e), or Gal-8 (f) at ~0.1µM with or without inclusion of 20mM TDG. Quantification of PA O5 after addition of 5µM Gal-4 (g), Gal-8 (h) or Gal-3 (i) with or without addition of 50mM TDG or sucrose (Sucr). n=2–3 in 1 representative experiment of 3; error bars represent means ± 1 s.d.
In an effort to understand the remarkable proclivity of Gal-3, Gal-4, and Gal-8 for PA O5 among all the microbes represented on the MGM, we examined the type of O antigen expressed by PA O5 and compared this to the composition of other antigens represented on the array. As this array specifically employed previously characterized carbohydrate antigens, we cataloged these structures and assessed whether PA O5 might possess a determinant unique from other microbial glycans on the array. Of all the O antigens on the array, PA O5 represented the only O antigen with a common mammalian glycan signature, the α-Gal epitope (Supplementary Table 1). Consistent with this, previous studies suggested that several galectins, in particular Gal-4 and Gal-8, specifically recognize and kill E. coli O86:B7 (EC O86), which expresses a human blood group B-like antigen, providing a potential mechanism whereby blood group B positive individuals protect themselves against molecular mimicry 34. The ability of Gal-3, Gal-4, and Gal-8 to also recognize PA O5 raises the possibility that galectins may target additional microbes expressing alternative self-like antigens as a broader form of protection against molecular mimicry.
Galectins inhibit the viability of target microbes
To determine whether galectin recognition provides immunity against PA O5, we examined whether galectins can directly alter PA O5 viability. Gal-4 and Gal-8 not only recognized PA O5, but binding resulted in loss of bacterial viability, providing a mechanism whereby Gal-4 and Gal-8 may confer specific immunity against this microbe (Fig. 2g,h, and Supplementary Fig. 2). Surprisingly, Gal-3 recognition of PA O5 also resulted in loss of bacterial viability, similar to that observed for Gal-4 and Gal-8, whereas similar to previous results 34, Gal-3 failed to alter EC O86 viability when evaluated in parallel (Fig. 2i and Supplementary Fig. 2). These results suggest that different galectins may display differential effects on the viability of microbes with self-like antigens. Inclusion of TDG, an inhibitor of galectin-carbohydrate interactions, completely prevented Gal-3, Gal-4, and Gal-8 binding and loss of viability of PA O5 (Fig. 2d–i), confirming that the effects require specific antigen recognition. Importantly, Gal-3, Gal-4, and Gal-8 failed to alter the viability of PA O19 (Supplementary Fig. 2), which demonstrated that galectin-induced effects require distinct recognition of self-like antigens. Taken together, these results demonstrate that the MGM powerfully predicts previously unrecognized immune factor-microbial interactions with specific insight into the structural motifs likely required for glycan binding. In doing so, these data demonstrate that Gal-3, Gal-4, and Gal-8 display antibody-like specificity for microbial antigens that display molecular mimicry.
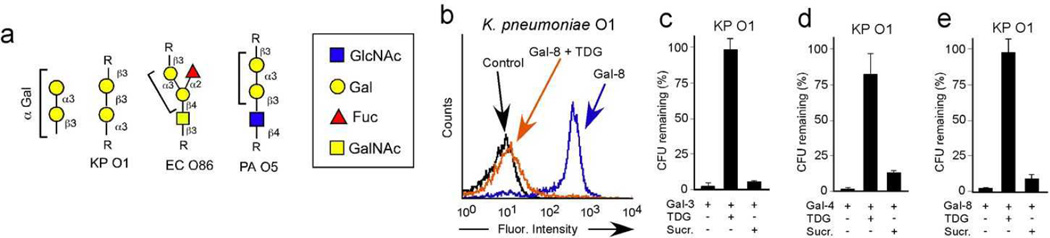
To build upon the insight into galectin-microbial glycan binding specificity obtained from the MGM, we next determined whether galectins recognize a broader range of microbes bearing the same self-like antigen predicted by the MGM. To examine this, we turned to a relatively new searchable database of antigenic structures from a diverse range of pathogenic species (http://csdb.glycoscience.ru/bacterial/) and interrogated the database for similar structures as occur on PA O5. Using this approach, we predicted that Klebsiella pneumoniae O1 (KP O1), which expresses a nearly identical antigenic structure as expressed by PA O5 35, could be a potential target of galectins (Fig. 3a). Consistent with this prediction, galectins recognized KP O1 and binding was blocked in the presence of TDG, strongly suggesting that binding reflected recognition of the unique KP O1 antigen (Fig. 3b). Importantly, similar to the inability of galectins to recognize PA O19, galectins also failed to recognize a related strain of KP O1, KP O4, which does not express a self-like antigen 35 (Supplementary Fig. 2). Similar to the effect of these innate immune lectins on PA O5, galectins caused a decrease in viability of KP O1, while they did not alter the viability of KP O4 (Fig. 3c–e and Supplementary Fig. 2). Inclusion of TDG prevented galectin-induced loss of viability (Fig. 3c–e). These results demonstrate that the binding specificity of galectins obtained following interrogation on the MGM accurately predicts galectin interactions with key microbial glycan features even when presented on completely different microbial species.
Figure 3. Bacterial structural database provides new bacterial targets for galectin binding and killing.
(a) Schematic representation of glycan structures on the glycan array paired with similar structures found on indicated bacterial strains. (b) Flow cytometric analysis of K. pneumoniae O1 (KP O1) after incubation with ~0.1µM Gal-8 with or without inclusion of 20mM TDG. Quantification of KP O1 after addition of 5µM Gal-3 (c), Gal-4 (d) or Gal-8 (e) with or without addition of 50mM TDG or sucrose (Sucr). n=2–3 in 1 representative experiment of 3; error bars represent means ± 1 s.d.
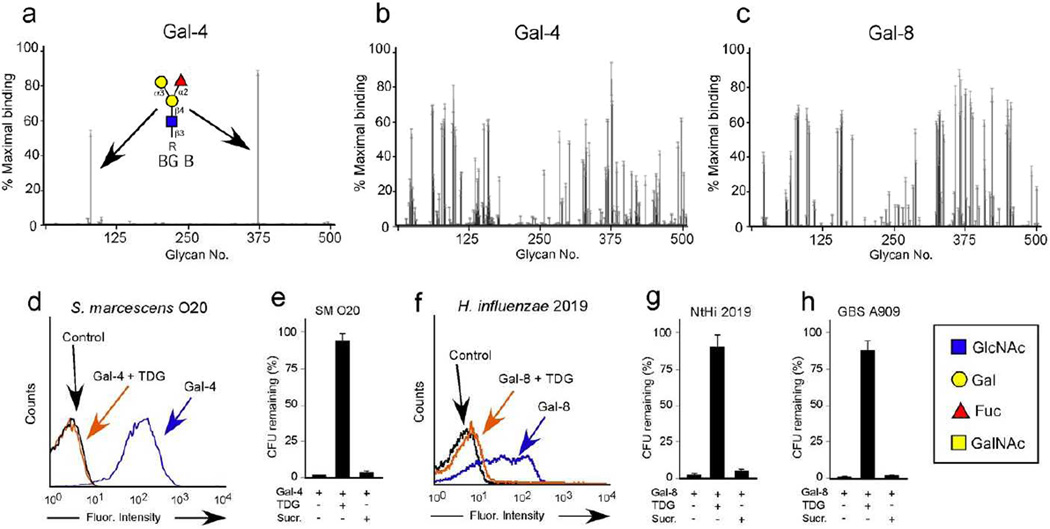
The ability of galectins to recognize additional microbes with non-blood group self-like antigens stands in contrast with previous results suggesting that galectins specifically recognize blood group antigens 34. Indeed, while galectins display high specificity for blood group antigens when examined at low concentrations (Fig. 4a), galectins recognized glycans containing the exact same glycan motif identified in PA O5 and KP O1, the α-Gal antigen, in addition to other commonly occurring self-like antigens at higher concentrations (Fig. 4b,c) 36, suggesting that galectins may target additional microbes expressing alternative self-like antigenic structures. To extend the collective insight obtained from the MGM and the mammalian glycan microarray, we searched for additional unique microbial genera that express either α-Gal or β-galactose (β-Gal) epitopes, very common motifs recognized by galectins on the mammalian glycan array (Fig. 4a–c). Using this approach, we identified Serratia marcescens and nontypeable Haemophilus influenzae (NtHi) 2019 as microbes that express the α-Gal and β-Gal antigens, respectively. Importantly, Gal-4 displayed significant recognition of S. marcescens, which resulted in loss of viability (Fig. 4d,e). Similarly, Gal-8 recognized NtHi 2019 and caused loss of viability, while inclusion of TDG inhibited galectin binding and effects on microbial viability (Fig. 4f,g). Gal-4 failed to recognize a strain of S. marcescens that does not express either α- or β-Gal antigens (Supplementary Fig. 2). Given the ability of galectins to cause loss of viability of multiple gram-negative species, we next determined whether galectins might also target gram-positive organisms that bear alternative self-like antigens. Similar interrogation of known microbial structural motifs among gram-positive microbes revealed that Streptococcus agalactiae (GBS A909), a commonly occurring gram-positive organism, expresses the self-like sialylated lactosamine antigen. Similar to the ability of galectins to decrease viability of gram-negative bacteria expressing self-like antigens, Gal-8 effectively caused loss of viability of GBS A909 (Fig. 4h). Taken together, these results demonstrate that galectins possess the unique ability to bind and affect the viability of a variety of microbes that express self-like antigens across a range of gram-negative or gram-positive microbes, providing broad protection against microbial mimicry.
Figure 4. Galectins bind and kill a broad range of new bacterial targets.
(a–c) CFGv4.2 glycan microarray assayed with 0.2µM Gal-4 (a), 5µM Gal-4 (b), and 5µM Gal-8 (c). Error bars represent means ± 1 s.d. See Supplementary Dataset 2 for complete microarray data. (d) Flow cytometric analysis of S. marcescens O20 (SM O20) after incubation with ~0.1µM Gal-4 with or without inclusion of 20mM TDG. (e) Quantification of SM O20 after addition of 5µM Gal-4 with or without addition of 50mM TDG or sucrose (Sucr). (n=2–3 in 1 representative experiment of 3). (e) Flow cytometric analysis of H. influenzae 2019 (NtHi 2019) after incubation with ~0.1µM Gal-8 with or without inclusion of 20mM TDG. (f) Quantification of NtHi 2019 after addition of 5µM Gal-8 with or without addition of 50mM TDG or Sucr. (n=2 in 1 representative experiment of 3). (g) Quantification of Streptococcus agalactiae A909 (GBS A909) after addition of 5µM Gal-8 with or without addition of 50mM TDG or Sucr.
Galectins bind multiple microbes with self-like antigens
The ability of galectins to target multiple microbial species that express unique mammalian antigens, while failing to bind or kill similar strains with non-mammalian glycan antigens, suggests that galectin-mediated innate immunity may be specific to microbes with these antigenic characteristics. However, to more formally examine galectin microbial specificity, a larger test library is needed. To accomplish this, we expanded the MGM to include antigens isolated from additional microbial genera, including gram-positive organisms. Analogous to the LPS O-antigens, capsular polysaccharides (CPS) provide discrete markers of distinct strains within gram-positive microbial species 37. Similar to native BPS, intact LPS, or CPS isolated from a broad range of microbial species was efficiently coupled to the array and could be specifically recognized by sera when printed separately (Supplementary Fig. 3) or together in a single array format containing nearly 300 unique antigenic determinants (Fig. 5a,b). Having developed an expanded MGM populated with microbes with known mammalian antigens, we next examined the specificity of galectins for these microbial antigens. Gal-4 displayed distinct recognition of a diverse array of microbes on the expanded MGM, including specific strains of gram-negative Klebsiella pneumoniae (KP), Escherichia coli (EC), Providencia alcalifaciens (PA), Proteus vulgaris (PV) and gram-positive Streptococcus pneumoniae (SP) (Fig. 5c). While each of these antigenic determinants varied in composition, each represented a unique mammalian carbohydrate determinant (Supplementary Fig. 4–6). Thus, similar to antibodies, galectins appear to display selective reactivity toward discrete microbial antigens. However, unlike antibodies, galectins have an unprecedented ability to specifically recognize a variety of antigens that are only related by their individual similarity to a distinct type of core mammalian antigen (Supplementary Fig. 4–6), which represent similar glycan motifs recognized by galectins on the mammalian glycan microarray and in other studies 38. These results illustrate the novel ability of galectins to specifically recognize self-like antigens on a broad range of microbes.
Figure 5. Expansion of MGM to include gram positive and additional gram negative microbial antigens.
(a–d) Expanded MGM data obtained after incubation with a 1:5000 dilution of sera from rabbits challenged with Providencia alcalifaciens O19 (a) or Streptococcus pneumoniae Type 14 (b) or 10µM Gal-4 (c). EC O128 = Escherichia coli O128. EC O40 = Escherichia coli O40. PV O45 = Proteus vulgaris O45. PA O5 = Providencia alcalifaciens O5. PA O6 = Providencia alcalifaciens O6. KP O1 = Klebsiella pneumonia O1. EC O55 = Escherichia coli O55. SP 70 = Streptococcus pneumoniae type 70. PV O47 = Proteus vulgaris O47. See Supplementary Dataset 3 for complete microarray data.
Galectins fail to alter mammalian cell viability
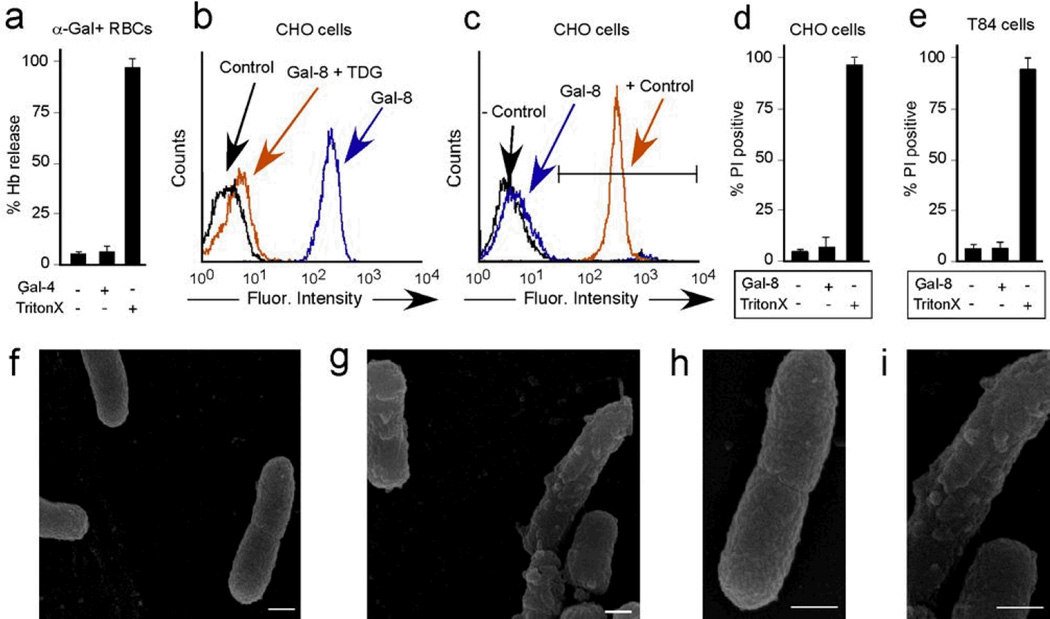
The ability of galectins to apparently exclusively recognize self-like antigens stands in stark contrast to other innate and adaptive immune factors 29. Indeed, discrimination of self from non-self at the level of ligand recognition of determinants unique to pathogens represents a fundamental paradigm within immunology concerning immune factor specificity for target pathogens 29. However, in contrast to most immune factors, galectins recognized the same pattern of glycans on the mammalian array as the self-like antigen structures bound on the MGM, strongly suggesting that galectins recognize very similar structures on microbes and mammalian cells. As a result, galectins may actually induce similar changes, such as loss of membrane integrity, in mammalian cells with similar carbohydrate structures (Supplementary Fig. 7). However, while galectins induced significant loss in the viability of diverse microbial genera, incubation of galectins with rabbit erythrocytes or CHO cells, which express α-Gal or lactosamine terminal glycans respectively, did not induce detectable changes in membrane integrity (Fig. 6a–d and Supplementary Fig. 7). Galectins also failed to alter the membrane integrity of epithelial cells, further suggesting that galectins can be secreted from and bind host cells without inducing deleterious effects, while retaining the ability to specifically target microbes with similar glycan structures (Fig. 6e). Consistent with this, significant changes in membrane architecture accompanied galectin-induced loss of bacterial viability (Fig. 6f–i), further demonstrating that galectins possess a unique ability to discriminate pathogens from self while recognizing very similar antigenic structures.
Figure 6. Galectin binding to eukaryotic cells does not alter cell viability.
(a) Quantification of hemoglobin release from rabbit erythrocytes after incubation with 5µM Gal-4 or 1% Triton X (n=2–3 in 1 representative experiment of 3). (b) Flow cytometric analysis of WT CHO cells after incubation with ~0.1µM Gal-8 with or without inclusion of 20mM TDG. (c) Flow cytometric analysis of PI positive WT CHO cells after incubation with ~0.1µM Gal-8 or 1% Triton X (+ control). (d) Quantification of percent PI+ WT CHO cells after incubation with 5µM Gal-8 or 0.1% Triton X (n=2 in 1 representative experiment of 3). (e) Quantification of percent PI+ T84 epithelial cells after incubation with 5 µM Gal-8 or 0.1% Triton X (n=3 in 1 representative experiment of 3). (f–i) Scanning electron microscopy images (15,000×) of KP O1 after incubation with PBS (f) or 5µM Gal-8 (g). Increased magnification of panels f (h) and g (i). Scale bars = 500 nm.
DISCUSSION
Analysis of immunological factors on the MGM provided important insight concerning the seroreactivity of different species toward a broad range of microbiota and demonstrated that galectins provide exquisitely specific immunity against a broad range of microbes expressing self-like antigens. These results suggest that antibodies and innate immune factors work in a complementary fashion to provide overall host protection against microbiota. As pathogens evolved elaborate structures to avoid innate immunity, vertebrates evolved an equally impressive mechanism of combating antigenic diversity among pathogens. Indeed, adaptive immunity has the capacity to respond to an infinite number of antigenic structures, as evidenced by the broad seroreactivity on the MGM. However, in order for adaptive immunity to retain plasticity while avoiding self-reactivity, the removal of self-reactive cells and selection of peripheral tolerance mechanisms leads to critical tolerance that allows immunological distinction of self from non-self 39. While elimination of self-reactive immune cells reduces the probability of autoimmunity, a fitness cost would be anticipated due to the reduced ability of an individual to respond to self-like antigens on pathogens. However, compensatory mechanisms at the level of innate immunity may exist whereby individuals protect themselves against molecular mimicry.
The targeted innate immune activity of galectins stands in contrast to all other previously described innate or adaptive immune factors. While most immune factors discriminate self from non-self by recognizing molecular motifs unique to microbes 40,41, the ability of galectins to not only recognize, but apparently exclusively recognize microbes that express self-like antigens, represents an remarkable form of host-microbial interaction. Indeed, most immune factors utilize recognition of distinct microbial determinants to specifically target their antimicrobial activity and thus reduce potentially deleterious off target effects toward host cells 40,41. Thus, the ability of galectins to recognize the same antigenic determinant on microbes and mammalian cells, but only kill microbes, represents a unique form of immunity. Furthermore, the ability of galectins to recognize and kill a microbe through engagement of a distinct antigenic determinant represents the first example of an antimicrobial with the capacity to intrinsically and specifically target an individual microbe, which represents a new paradigm in antimicrobial activity.
The ability of these lectins to specifically and intrinsically target self-like antigens not only illustrates the utility of the MGM in examining host-microbial interactions, but also likely reflects structural simplicity of early pathogen antigens prior to the evolution of adaptive immunity. As the most evolutionary ancient lectins currently known, galectin predecessors likely provided a key form of soluble immunity from the beginning of all metazoans. Indeed, galectins appear to possess unique roles in host pathogen interactions in a diverse range of species 42. However, increased diversity of microbial antigens likely outstripped the ability of galectins to likewise accommodate all microbial antigens, which likely set the stage for selection of adaptive immunity. However, while adaptive immunity evolved a singular ability to recognize diverse antigenic species, the ability of galectins to accommodate self-like antigens likely provided an important evolutionary substrate to complement the reduced ability of adaptive immunity to target pathogens bearing self-like antigens. Thus, the ability of galectins to specifically target pathogens bearing self-like antigens complements adaptive immunity and provides unprecedented antibody-like immunity against microbial mimicry.
Material and Methods
Preparation of bacterial polysaccharides (BPS)
LPS was isolated from dried bacterial cells by hot phenol–water extraction and purified by treatment with DNAse and RNAse and ultracentrifugation. In the event that BPS were released from LPS, each isolated LPS was subjected to mild acid hydrolysis using 2% acetic acid to remove the Lipid A portion leaving the Inner Core, Outer Core and O-antigen (BPS). Isolated supernatants were fractionated on a Sephadex G-50 column in 0.05 M pyridinium acetate buffer pH 4.5. This protocol resulted in highly purified bacterial polysaccharides that were employed to elucidate the structure of each glycan by NMR analysis as outlined previously 24,25. Aliquots of the samples used to interrogate the glycan structure were then lyophilized for storage. Prior to printing microbial glycans, lyophilized samples were dissolved in printing buffer (300 mM phosphate buffer + 0.005% Tween-20, pH 8.4) to a stock concentration of 1 mg/mL. Stock samples were further diluted into 96-well microplates prior to printing via two-fold serial dilutions using a BioMek 1000 printing robot (Beckman/Coulter).
Printing of the Microbial Glycan Microarray (MGM)
10 uL of diluted glycan antigens were deposited into 384-well microplates via a pipetting robot (BioMek 2000, Beckman Coulter) and then spotted using a MicroGrid II (Digilab) microarray printing robot equipped with solid steel Stealth microarray pins (SMP4B, TeleChem) onto SlideH (Schott/Nexterion) or CodeLink (GE Healthcare) NHS-ester activated microscope slides. Each pin delivers ~1 nL of dissolved sample per spot deposition and 6 replicate spots of each sample were imprinted on each array. Arrays were printed at ~55% Relative Humidity (RH) and allowed 30min of immobilization at 100% RH before being desiccated at RT. Following desiccation, printed arrays were treated with a blocking step to inactivate remaining NHS-ester groups on the array surface by immersion in 50 mM ethanolamine in 50 mM borate buffer for 1hr on a rotary shaker. Following blocking, slides were rinsed in ddH20 and dried by centrifugation prior to incubation/analysis. MGMv1 contained 5 concentrations of each sample at 31.25, 62.5, 125, 250, 500 µg/mL as either BPS or BPS plus spacer. MGMv2 contained only one printed concentration of glycan at 500 µg/mL as isolated BPS, whole LPS, or whole CPS.
Analysis of binding to the MGM
Printed MGMs were assessed for functionality using sera of humans, mice, rabbits and immunized rabbits as indicated, or a series of innate immune proteins, as indicated (rabbit sera was a kind gift from Dr. Rosalski). The IVIG preparation used in this study was Sandoglobulin (CSL Behring, Bern, Switzerland) and was extensively dialyzed to remove stabilizing components, as previously described 43. Normal human sera were isolated from healthy human volunteers by venipuncture and collection of whole blood into sterile serum tubes, following an Emory University IRB-approved protocol. Isolation of mouse and rabbit sera was accomplished using similar blood-drawing protocols. Isolated sera were then frozen at −80°C until further use. Sera analyzed on the array represents sera pooled from five individual donors prior to analysis on the array. Preparation of galectins is outlined below. To analyze binding on MGM a similar protocol as outlined previously was employed 44–47. Briefly, primary antibody or antisera was applied directly to the array surface, incubated for 1h in a humidified chamber and washed in TSM Binding Buffer (Tris buffer/Salts/Metal ions (TSM) - 20 mM Tris-HCl, 150 mM sodium chloride, 2 mM calcium chloride, 2 mM magnesium chloride, pH 7.4). The slide was then incubated with either Alexa488-streptavidin for detection of biotinylated proteins, with species specific secondary antibodies (Alexa Fluor488-anti-IgM or Alexa Fluor647-anti-IgG, Invitrogen) to the primary IgG or IgM for 1h at room temperature followed by washing and detection of fluorescence with a microarray scanner (Scan Array Express, PerkinElmer Lifer Sciences). The integrated spot intensities were determined using Imagene software (BioDiscovery).
Preparation of recombinant human galectins
Gal-3, Gal-4 and Gal-8 were prepared as outlined previously 46. Briefly, purified recombinant galectins by affinity chromatography on lactosyl-sepharose and eluted with 100mM lactose in PBS plus 2-mercaptoethanol (2-ME). Prior to derivatization, 2-ME was removed from galectin samples using a PD-10 gel filtration column (GE Healthcare), followed by addition of lactose (100 mM final concentration) to help maintain galectin stability and reduce the likelihood of adduct formation at or near the carbohydrate recognition domain (CRD). All galectins were biotinylated by incubating 3–5 mg/mL of galectin with 2 mM EZ-linkTM Sulfo-NHS-LC-Biotin (Sulfosuccinimidyl-6-(biotinamido) hexanoate) (Pierce) for 2h at 4°C. Unconjugated EZ-linkTM Sulfo-NHS-LC-Biotin and free lactose was separated from derivatized protein using a PD-10 gel filtration column. Derivatized protein was re-purified on lactosyl-sepharose and eluted with 100mM lactose in PBS plus 2-mercaptoethanol (2-ME) in order to eliminate any inactive protein resulting from derivitization, followed by removal of 2-ME using a PD-10 gel filtration column (GE Healthcare) prior to glycan microarray or direct bacterial binding analysis.
Mammalian glycan array preparation and analysis
The Consortium for Functional Glycomics (http://www.functionalglycomics.org/) provided glycan microarrays (v4.2) prepared as described previously 45,47. For galectin recognition of glycans on the printed glycan microarray, slides were incubated with 0.2 µM or 5 µM Gal-4, or 5 µM Gal-8 in TSM binding buffer + 14 mM 2-ME for 1h at room temperature in a dark humid chamber. The slide was washed by successive immersion in TSM containing 0.05% Tween 20 (4 times) and TSM (4 times). The slide was incubated with Alexa Fluor-488-streptavidin. After 1h at room temperature in a dark humid chamber, we washed the slide by successive immersion in TSM containing 0.05% Tween 20 (4 times), TSM (4 times), and water (4 times). The slide was dried by microcentrifugation and an image of bound fluorescence was obtained using a microarray scanner (Scan Array Express, PerkinElmer Lifer Sciences). Integrated spot intensities were determined using Imagene software (BioDiscovery).
Bacterial strains
E. coli O86:B7 was kindly provided by Dr. P. George Wang (Georgia State University). P. alcalifaciens O5 and P. alcalifaciens O19 were provided by Dr. Yuriy Knirel (ND Zelinsky Institute of Organic Chemistry, Moscow, Russia). K. pneumoniae O1, K. pneumoniae O2a, K. pneumoniae O2a,c, and K. pneumoniae O4 as well as S. marcescens O20, were provided by Dr. Chris Whitfield (University of Guelph). Each was grown and maintained in aerobic conditions at 37°C using Luria Burtani (LB) culture media (Fisher). H. influenzae 2019 was provided by Dr. Michael Apicella (The University of Iowa). This strain as well as β-lactamase positive, control H. influenzae, obtained from the Emory University Clinical Microbiology lab, was grown and maintained in 5% CO2 conditions at 37°C using Brain Heart Infusion Media (Bacto, Becton Dickenson, Franklin Lakes, NJ) supplemented with 10 mg/mL Hemin (Sigma Aldrich, St. Louis, MO) and 10 mg/mL NADH (Sigma Aldrich). The S. marcescens control strain (ATCC#14756) as well as S. agalactiae A909 (ATCC#BAA-1138) were purchased from ATCC. S. marcescens control strain ATCC #14756 was grown in identical conditions to S. marcescens O20. S. agalactiae A909 was grown and maintained in standard aerobic conditions at 37°C using Todd Hewitt Broth (THB) (Bacto, Becton Dickenson, Franklin Lakes, NJ).
Flow cytometric and eukaryotic viability analysis
For galectin binding, bacteria or eukaryotic cells were washed twice in PBS at 4°C and incubated with biotinylated Gal-3, Gal-4, or Gal-8 at concentrations between 1–5 µg/ml at 4°C for 1h. As controls, cells were incubated with 20 mM thiodigalactoside (TDG) along with the galectins. Following incubation, cells were washed three times and incubated with Alexa Fluor 488 streptavidin or Alexa Fluor 633 streptavidin (Molecular Probes, Life Technologies, Grand Island, NY) at 4°C for 1h. Cells were washed twice, followed by resuspension in 400 µL PBS for analysis by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ). Flow analysis was accomplished using CellQuest software (BD Biosciences, Franklin Lakes, NJ). Within each independent experiment, duplicate assays were performed with 1 representative plot shown, and each independent experiment was performed at least twice. Analysis of membrane integrity following galectin engagement was accomplished by incubating CHO cells or rabbit red blood cell (RBCs) (Lampire Biological Laboratories, Pipersville, PA) with the indicated galectin or 0.1% Triton X at 37°C for 1h. CHO cells were then stained with the membrane impermeable DNA binding dye, propidium iodide, followed by flow cytometric examination. T84 epithelial cells were examined in an analogous manner by examining the potential uptake of propidium iodide following incubation 0.1% Triton X or galectin for 1h. In contrast, RBC supernatants were examined for hemolysis following galectin or Triton X incubation by measuring absorbance at 540nm.
Assaying microbicidal activity
When assaying potential anti-microbial effects of galectins, each strain was grown to an OD600 of ~ 0.1, followed by incubation with the indicated concentrations of each galectin for 2h at 37°C with shaking. Following incubation with each respective galectin, the number of viable bacteria was determined by dilution plating and CFU enumeration. Either TDG (50 mM) or sucrose (50 mM) was incubated with the galectin when indicated for 10 minutes prior to incubation with the bacteria.
Preparation of samples for Scanning Electron Microscopy
K. pneumoniae O1 was grown to OD600 of ~ 0.1, was incubated for 30min with PBS control or 5 µM Gal-8 at 37°C, after which 20 mM lactose was added to halt treatment and reduce agglutination. Bacteria were washed 2× with PBS to remove debris. Droplets containing either untreated or Gal-8-treated bacteria were placed onto poly-L-Lysine treated silicon chips and allowed to settle, then fixed overnight in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4. Samples were washed with distilled water, followed by post-fixation in 1% osmium tetroxide. The samples were washed with distilled water followed by dehydration using an ascending ethanol gradient to three changes of 100% ethanol. The ethanol was exchanged with hexamethyldisilizane (HMDS) via three changes of HMDS then allowed the HMDS level to drop and the samples allowed to dry completely overnight. Samples were mounted onto SEM stubs and sputter coated with either gold or chromium and viewed them in the DS130 SEM (ISI-TOPCON) using in-lens imaging. The displayed images were viewed at 20,000× magnification.
MGMv1 and CFG mammalian glycan array data included in this study will be uploaded to http://www.functionalglycomics.org/.
Supplementary Material
Acknowledgements
This work was supported by National Blood Foundation and NIH LRP Grants to S.R.S., R01AI050143 and P01HL107151NIH to J.C.P., and HL085607, P41GM103694, and R24GM098791 to R.D.C. and U54GM62116 to the Consortium for Functional Glycomics. S.V.G. was supported by grants from the Swiss National Science Foundation (No. 310030_135734) and from CSL Behring AG, Bern, Switzerland. L.C.R was supported by (n° BEX 9320/13-0 - CAPES Foundation, Ministry of Education Brazil). We thank Tongzhong Ju, Xuezheng Song, Yingchun Wang and Sandra Cummings for helpful discussion. We also thank Liz Robison and Janani Rangarajan for help printing the various versions of the MGM. Consortium for Functional Glycomics array data can be accessed at http://www.functionalglycomics.org.
Abbreviations
- MGM
microbial glycan microarray
- BPS
bacterial polysaccharides
- LPS
lipopolysaccharide
- CPS
capsular polysaccharides
- IVIG
intravenous immunoglobulin
- TDG
thiodigalactoside
- PI
propidium iodide
- CHO
Chinese Hamster Ovary
Footnotes
Author contributions: S.R.S., C.M.A. and R.M. planned the project along with R.D.C. and J.C.P. S.R.S., C.M.A, R.M., J.H.M. carried out array experiments and anaylsis. S.R.S., C.M.A., J.P.G. and A.J.N. carried out bacterial binding and killing experiments. S.R.S., C.M.A., and L.C.R. carried assays with eukaryotic cells. R.M., O.B., N.R., J.H.M., S.V.G, D.F.S and Y.K. provided critical bacteria and array reagents and support. S.R.S., C.M.A., J.C.P. and R.D.C. wrote the manuscript, which was additionally edited and commented on by the other authors.
Competing interest statement: The authors declare no competing financial interest.
References
- 1.Strugnell RA, Wijburg OL. The role of secretory antibodies in infection immunity. Nat Rev Microbiol. 2010;8:656–667. doi: 10.1038/nrmicro2384. [DOI] [PubMed] [Google Scholar]
- 2.Shanahan F. The colonic microflora and probiotic therapy in health and disease. Curr Opin Gastroenterol. 2011;27:61–65. doi: 10.1097/MOG.0b013e328340076f. [DOI] [PubMed] [Google Scholar]
- 3.Gevers D, et al. The human microbiome project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10:e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mezzasoma L, et al. Antigen microarrays for serodiagnosis of infectious diseases. Clin Chem. 2002;48:121–130. [PubMed] [Google Scholar]
- 9.Davies DH, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 11.Costerton JW, Irvin RT, Cheng KJ. The bacterial glycocalyx in nature and disease. Annu Rev Microbiol. 1981;35:299–324. doi: 10.1146/annurev.mi.35.100181.001503. [DOI] [PubMed] [Google Scholar]
- 12.Peterson PK, Quie PG. Bacterial surface components and the pathogenesis of infectious diseases. Annu Rev Med. 1981;32:29–43. doi: 10.1146/annurev.me.32.020181.000333. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 14.Parthasarathy N, DeShazer D, England M, Waag DM. Polysaccharide microarray technology for the detection of Burkholderia pseudomallei and Burkholderia mallei antibodies. Diagn Microbiol Infect Dis. 2006;56:329–332. doi: 10.1016/j.diagmicrobio.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parthasarathy N, et al. Application of carbohydrate microarray technology for the detection of Burkholderia pseudomallei, Bacillus anthracis and Francisella tularensis antibodies. Carbohydr Res. 2008;343:2783–2788. doi: 10.1016/j.carres.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Thirumalapura NR, Morton RJ, Ramachandran A, Malayer JR. Lipopolysaccharide microarrays for the detection of antibodies. J Immunol Methods. 2005;298:73–81. doi: 10.1016/j.jim.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Baader J, et al. Polysaccharide microarrays with a CMOS based signal detection unit. Biosens Bioelectron. 26:1839–1846. doi: 10.1016/j.bios.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Blixt O, Hoffmann J, Svenson S, Norberg T. Pathogen specific carbohydrate antigen microarrays: a chip for detection of Salmonella O-antigen specific antibodies. Glycoconj J. 2008;25:27–36. doi: 10.1007/s10719-007-9045-0. [DOI] [PubMed] [Google Scholar]
- 19.Kamena F, et al. Synthetic GPI array to study antitoxic malaria response. Nat Chem Biol. 2008;4:238–240. doi: 10.1038/nchembio.75. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, et al. Photogenerated glycan arrays identify immunogenic sugar moieties of Bacillus anthracis exosporium. Proteomics. 2007;7:180–184. doi: 10.1002/pmic.200600478. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg H, et al. Structural basis for langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J Mol Biol. 405:1027–1039. doi: 10.1016/j.jmb.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arthur CM, Cummings RD, Stowell SR. Using glycan microarrays to understand immunity. Curr Opin Chem Biol. 2014;18C:55–61. doi: 10.1016/j.cbpa.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knirel YA, et al. Human tandem-repeat-type galectins bind bacterial non-betaGal polysaccharides. Glycoconjugate journal. 2014;31:7–12. doi: 10.1007/s10719-013-9497-3. [DOI] [PubMed] [Google Scholar]
- 24.Kocharova NA, et al. Structure of the O-polysaccharide of Providencia stuartii O4 containing 4-(N-acetyl-L-aspart-4-yl)amino-4,6-dideoxy-D-glucose. Carbohydr Res. 2004;339:195–200. doi: 10.1016/j.carres.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Kocharova NA, et al. Structure of the O-polysaccharide of Providencia alcalifaciens O19. Carbohydr Res. 2004;339:415–419. doi: 10.1016/j.carres.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Springer GF, Horton RE, Forbes M. Origin of antihuman blood group B agglutinins in germfree chicks. Ann N Y Acad Sci. 1959;78:272–275. doi: 10.1111/j.1749-6632.1959.tb53110.x. [DOI] [PubMed] [Google Scholar]
- 27.Springer GF, Horton RE. Blood group isoantibody stimulation in man by feeding blood group-active bacteria. J Clin Invest. 1969;48:1280–1291. doi: 10.1172/JCI106094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuczynski J, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 30.Kohatsu L, Hsu DK, Jegalian AG, Liu FT, Baum LG. Galectin-3 induces death of Candida species expressing specific beta-1,2-linked mannans. J Immunol. 2006;177:4718–4726. doi: 10.4049/jimmunol.177.7.4718. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier I, Sato S. Specific recognition and cleavage of galectin-3 by Leishmania major through species-specific polygalactose epitope. J Biol Chem. 2002;277:17663–17670. doi: 10.1074/jbc.M201562200. [DOI] [PubMed] [Google Scholar]
- 32.John CM, et al. Galectin-3 binds lactosaminylated lipooligosaccharides from Neisseria gonorrhoeae and is selectively expressed by mucosal epithelial cells that are infected. Cell Microbiol. 2002;4:649–662. doi: 10.1046/j.1462-5822.2002.00219.x. [DOI] [PubMed] [Google Scholar]
- 33.Quattroni P, et al. Galectin-3 binds Neisseria meningitidis and increases interaction with phagocytic cells. Cell Microbiol. 2012;14:1657–1675. doi: 10.1111/j.1462-5822.2012.01838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stowell SR, et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinogradov E, et al. Structures of lipopolysaccharides from Klebsiella pneumoniae. Eluicidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the O chains. J Biol Chem. 2002;277:25070–25081. doi: 10.1074/jbc.M202683200. [DOI] [PubMed] [Google Scholar]
- 36.Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nat Rev Immunol. 2008;8:874–887. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yother J. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol. 2011;65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 38.Hirabayashi J, et al. Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochim Biophys Acta. 2002;1572:232–254. doi: 10.1016/s0304-4165(02)00311-2. [DOI] [PubMed] [Google Scholar]
- 39.Klinman NR. The"clonal selection hypothesis" and current concepts of B cell tolerance. Immunity. 1996;5:189–195. doi: 10.1016/s1074-7613(00)80314-3. [DOI] [PubMed] [Google Scholar]
- 40.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 41.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 42.Kamhawi S, et al. A role for insect galectins in parasite survival. Cell. 2004;119:329–341. doi: 10.1016/j.cell.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 43.von Gunten S, et al. Intravenous immunoglobulin contains a broad repertoire of anticarbohydrate antibodies that is not restricted to the IgG2 subclass. J Allergy Clin Immunol. 2009;123:1268–1276. doi: 10.1016/j.jaci.2009.03.013. e1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westphal OKJ. Bacterial lipopolysaccharide. Extraction with phenol-water and further applications of the procedure. Methods Carbohydrate Chemistry. 1965;5:83–91. [Google Scholar]
- 45.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stowell SR, et al. Ligand reduces galectin-1 sensitivity to oxidative inactivation by enhancing dimer formation. J Biol Chem. 2009;284:4989–4999. doi: 10.1074/jbc.M808925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stowell SR, et al. Galectin-1-2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem. 2008;283:10109–10123. doi: 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.