Abstract
The integration of longitudinal brain structure analysis with neurointensive care strategies continues to be a substantial difficulty facing the traumatic brain injury (TBI) research community. For patient-tailored case analysis, it remains challenging to establish how lesion profile modulates longitudinal changes in cortical structure and connectivity, as well as how these changes lead to behavioral, cognitive and neural dysfunction. Additionally, despite the clinical potential of morphometric and connectomic studies, few analytic tools are available for their study in TBI. Here we review the state of the art in structural and connectomic neuroimaging for the study of TBI and illustrate a set of recently-developed, patient-tailored approaches for the study of TBI-related brain atrophy and alterations in morphometry as well as inter-regional connectivity. The ability of such techniques to quantify how injury modulates longitudinal changes in cortical shape, structure and circuitry is highlighted. Quantitative approaches such as these can be used to assess and monitor the clinical condition and evolution of TBI victims, and can have substantial translational impact, especially when used in conjunction with measures of neuropsychological function.
Keywords: traumatic brain injury, neuroimaging, magnetic resonance imaging, diffusion tensor imaging, connectomics
1. Introduction
An important challenge to the neuroimaging of traumatic brain injury (TBI) involves elucidating how the lesion profile of a TBI subject modulates longitudinal changes in volumetrics, morphometrics and connectomics, and how these changes lead to behavioral, cognitive and neural dysfunction. Although considerable efforts have been devoted to task of clarifying the relationship between acute TBI lesion load and patient case evolution, it remains challenging to improve TBI clinical outcome by incorporating information about the specific deficits and injury profiles of patients into their treatment and rehabilitation routines. This is primarily because current neurological, neurosurgical and neuropsychological treatment protocols for TBI must too often rely upon exceedingly general knowledge of lesion type, location and degree of severity. Thus, the heterogeneity of brain injuries remains a barrier to the design and implementation of patient-tailored treatment and to the formulation of rehabilitation protocols which take advantage of individualized neuroimaging analyses. Although computed tomography (CT) and magnetic resonance imaging (MRI) have become standard in TBI care, few image processing techniques are tailored for TBI brains and the detailed lesion analysis TBI remains challenging.
The purpose of this review is three-fold. Firstly, it aims to discuss and highlight prior advances in structural and connectomic neuroimaging for the study of TBI. Its second purpose is to illustrate a set of newly-developed, patient-tailored approaches to the analysis of TBI neuroimaging volumes, including methods for segmenting gross TBI lesions semi-automatically and for quantifying the effects of TBI-related white matter (WM) atrophy1. Thirdly, it aims to illustrate how magnetic resonance imaging (MRI), diffusion tensor imaging (DTI) and advanced image processing methods can be integrated to study the longitudinal effects of severe TBI. Specifically, in three representative cases, the combined use of structural, morphometric and connectomic analysis to perform patient-tailored profiling of TBI-related changes in cortical curvature, area and thickness is demonstrated. In these three subjects with progressive lesion loads, the presence of brain atrophy is identified broadly over the cortex, including over cortical regions which appear to be healthy in both acute and chronic multimodal MRI/DTI scans. Widespread changes in cortical curvature and surface area over six months following brain trauma are described and quantified, pointing to the complex evolution of brain shape as a result of TBI. The techniques reviewed here can be used to test research hypotheses about brain atrophy and shape changes in TBI and to clarify how this phenomenon differentially affects brain regions depending on injury location. Because the use of quantitative approaches is typically preferable to that of qualitative ones when monitoring the clinical condition and evolution of TBI victims, such methods can have useful translational impact by aiding in the process of patient-tailored TBI profiling and rehabilitation.
2. Image acquisition protocols
The field of non-invasive human TBI neuroimaging has witnessed spectacular progress in the past 40 years, beginning with the use of early computed tomography (CT) systems in the early to mid-1970s2 and ending with today’s sophisticated multimodal neuroimaging methods3. By the mid-1980s, many neuroimaging experts and clinicians had acknowledged that CT suffered from substantial drawbacks compared to the emerging technique of magnetic resonance imaging (MRI), particularly because CT can underestimate cerebral injury severity, misevaluate cortical contusion extent and fail to identify diffuse axonal injury (DAI). By contrast, MRI is highly sensitive for detecting both hemorrhagic and non-hemorrhagic lesions, which allows one accurately to classify various types of traumatic lesions. Around the same time, the related though distinct technique of magnetic resonance spectroscopy (MRS) was becoming increasingly useful for the study of metabolic changes prompted by TBI, especially by highlighting the role of increased excitatory neurotransmitter signaling after brain injury4. Such a technique, when complemented by positron emission tomography (PET), allowed researchers for the first time to investigate physiological changes in TBI patients by assessing regional cerebral blood flow (rCBF), which can reflect neural system utilization as well as attempts by the brain to compensate for compromised function in lesion-affected areas5.
The subsequent advent and popularization of diffusion weighted imaging (DWI) and diffusion tensor imaging (DTI) in the early 1990s allowed researchers to characterize ischemic lesions as hyperintense areas with decreased apparent diffusion coefficient (ADC) values, thereby making the acute lesions adjacent to chronic infarcts readily apparent6. Additional information provided by MRI sequences developed around the same time, such as fluid-attenuated inversion-recovery (FLAIR), provided the welcome opportunity to visualize and quantify non-hemorrhaging edematous brain regions perfused by cerebrospinal fluid (CSF), particularly in the ischemic penumbra7. Because FLAIR is an inversion-recovery sequence designed to reduce CSF signal substantially, this technique enables very heavy T2 weighting without high signal and potential artifact from CSF and thereby makes cortical contusions and subdural hematomas much easier to detect than in conventional T1- and T2-weighted MRI8. With the introduction of T2*-weighted gradient recalled echo (GRE) MRI9, the ability to identify and map intracranial hemorrhage increased substantially, which also led to improved understanding of why hemorrhagic regions are likely to exhibit appreciable DAI10. Specifically, because the mechanical elasticity of vasculature is typically higher than that of axons, the presence of hemorrhages as obviated by GRE imaging is an excellent local indicator of DAI. Later on, the development of susceptibility weighted imaging (SWI) by Haake and colleagues improved the ability to detect micro-hemorrhages by exploiting the paramagnetic property of intravenous deoxyhemoglobin11.
One significant current challenge in the field of TBI neuroimaging relates to addressing the presence of motion artifacts by TBI patients in the MR scanner. This is particularly problematic during acute TBI scans, where it is difficult for awake patients to lie still or to follow the instructions of clinical staff. This challenge is aggravated by the typical need to acquire a variety of MRI scans involving different acquisition sequences in order to highlight distinct types of pathology. Though the advent of multiplexed turbo sequences can appreciably diminish the amount of time required for the acquisition of T1- and T2-weighted MRI, other scan types such as SWI and DTI can substantially increase the time length of MRI sessions, particularly when DTI acquisition involves a large number of gradient directions. For these reasons, the accuracy and validity of TBI neuroimaging analyses often depend upon the availability and effectiveness of robust motion correction algorithms for MR images. Given the current trend toward including novel and increasingly sophisticated MR sequences in TBI image acquisition protocols, it is to be expected that this need will only increase in the future.
Throughout the 1990s, as the number of MR sequences for TBI imaging increased, the need for standardization of image acquisition protocols became obvious. To address this necessity, the Defense Centers of Excellence within the US Defense and Veterans Brain Injury Center issued a clinical recommendation in 2013 pertaining to the US military’s preferred TBI MRI protocol for neuroimaging following mild TBI12. This protocol included, besides MPRAGE T1 and T2 imaging, structural sequences such as FLAIR and GRE T2, as well as diffusion imaging (DWI and DTI). In addition, a detailed list of sequence parameters was provided for both 1.5 T and 3.0 T MRI scanners. Despite such attempts at standardization, one significant challenge facing the TBI neuroimaging community today is the heterogeneity of imaging protocols across clinical sites, which continues to pose a substantial impediment to multi-center collaborations. One neuroinformatics effort aimed at responding to this problem is the Federal Interagency Traumatic Brain Injury Research (FITBIR) Informatics System (fitbir.nih.gov), which was created with the express purpose of sharing TBI neuroimaging data and of facilitating collaboration between TBI research laboratories. Because FITBIR is a recent effort, however, the need for protocol standardization and streamlined data sharing remains one of the most substantial current drawbacks to the implementation of large-cohort TBI studies and more effort should be dedicated by the TBI community to the task of joining such current efforts toward standardization and sharing.
To illustrate the current state of the art regarding TBI image acquisition protocols, we here illustrate the combined use of MRI, DTI and advanced image processing methods for the longitudinal study of severe TBI. For three representative TBI patients, MRI data were acquired using a protocol approved by the Institutional Review Board at the University of California, Los Angeles. Signed informed consent was obtained from each patient or from their legally authorized representative before the performance of any procedure. The first selected patient is a 17 year-old male pedestrian who suffered a TBI as a consequence of being hit by a moving vehicle. The patient was admitted to the Neurointensive Care Unit (NICU) of the Ronald Reagan UCLA Medical Center with a Glasgow Coma Score (GCS) of 3, and transferred from the unit with a Glasgow Outcome Scale-Extended (GOS-E) score of 2 (vegetative state). The GOS-E score at 6-month follow-up (GOS-6) was 4 (upper severe disability). For this patient, the acute and chronic scans were acquired 2 and 181 days after injury, respectively. The second patient is a 62 year-old female who was involved in a bicycle fall and who was admitted to the NICU with a GCS of 7. The GOS-E score upon discharge from the NICU was 2, and the GOS-6 score was 3 (lower severe disability). The acute and chronic scans were acquired 5 and 194 days after injury, respectively. The third patient selected is a 60 year-old male who acquired a TBI as a result of a fall. Whereas the GCS upon admission was 14, the GOS-E score at discharge from the NICU was 2 (vegetative state), and the GOS-6 score was 4. The acute and chronic scans were acquired 3 and 194 days after injury, respectively.
Our data acquisition protocol was previously described in detail13. Briefly, MR volumes were acquired at 3 T from each subject using a Siemens Trio TIM scanner (Siemens AG, Erlangen, Germany). MRI acquisition sessions were held both several days (acute baseline) as well as about 6 months (chronic follow-up) after the traumatic injury event and every subject was scanned using the same scanner for both acute and chronic time points. MR sequences included MP-RAGE T1-weighted imaging14, fluid-attenuated inversion recovery (FLAIR15), T2-weighted turbo spin echo (TSE, also known as fast spin echo, FSE16), gradient-recalled echo (GRE) T2-weighted images, susceptibility weighted imaging (SWI17) and DTI.
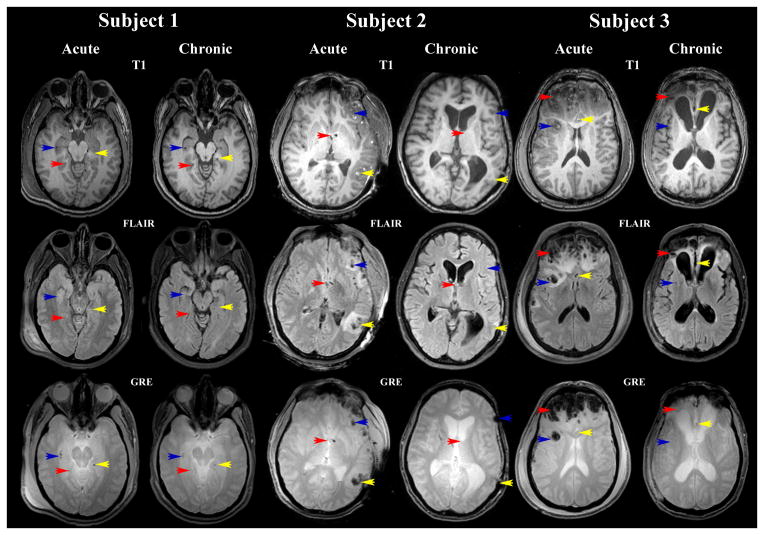
Sample MR images from each patient are displayed in Figure 1. For the first patient, inspection of these images reveals hemorrhages in the deep WM of the right hemisphere (RH), with peri-ventricular hemorrhages and edema in both hemispheres. Temporal lobe hemorrhage is observed in the RH, with contre-coup hemorrhagic lesions to the left hemisphere (LH) as well. Minor bleeding is observed in the medial aspect of both temporal lobes, although a larger number of micro-hemorrhages are seen in the RH. Patient 2 presents with extensive fronto-temporal edema and hemorrhage, especially in the LH, although trauma is also present contralaterally to a significant extent. MRI scans also reveal some hemorrhage throughout the posterior portion of the brain. Finally, whereas Patients 1 and 2 have comparatively light lesion loads, Patient 3 presents with the most severe amounts of hemorrhage and cerebral injury. For this patient, extensive bleeding exists in the frontal and temporal lobes of both hemispheres, both in the GM as well as in the WM. Modest peri-hemorrhagic edema is also noted.
Figure 1.
Sample MR images displayed in radiological convention for three TBI cases. The sequence types shown include T1, FLAIR and GRE. Red, yellow and blue arrows identify the locations of three different insults which are visible using all three sequences.
3. Tissue classification, volumetrics and morphometry
The task of performing automatic tissue classification in T1- and T2-weighted MRI volumes acquired from TBI patients is recognized as an exceptionally challenging problem in image processing. Whereas the accurate, automatic segmentation of healthy brains has become routine with the introduction and implementation of atlas-based probabilistic labeling and classification, such methods are highly prone to failure in the MRI volumes of patients with gross anatomic lesions31, 32. This is because gross pathology can introduce large differences in brain appearance between healthy MR volumes and lesion-impacted volumes, which make traditional atlas-based, probabilistic labeling algorithms prone to misclassification in conditions such as TBI.
Attempts to address the problem of lesion segmentation date as far back as the early 1990s, when techniques such as k-nearest neighbor18 and fuzzy C-means19 multispectral algorithms were first applied to MRI volumes exhibiting TBI or tumor lesions, subject to manual tracing for the removal of extracranial tissues from the segmentation. In fact, user supervision followed by manual corrections has been the norm for the TBI segmentation until only very recently3. Unsurprisingly, many of the earliest methods for user-assisted WM/GM tissue classification in the presence of TBI lesions were inspired by algorithms dedicated to the segmentation of images exhibiting lesions due to brain tumors20, 21, stroke22–24 or to multiple sclerosis25, 26. More recently, unified segmentation-normalization and fuzzy clustering algorithms have been combined with increased reliability in detecting and classifying injury-affected brain areas27. A multi-resolution binary level set method based on the Song-Chan algorithm has also been introduced for segmenting intracranial hematomas with results comparable to those obtained by human experts28. More advanced segmentation methods such as the SIENAX algorithm have been used to evaluate cortical thickness in TBI patient cohorts of small to moderate size, though many of these methods do not specifically account for non-diffeomorphic changes in TBI appearance across time29. Despite the fact that automated algorithms for the detection and quantification of TBI lesions have been proposed30, their use remains restricted to only a few neuroimaging centers and their robustness has not been validated extensively based on scans from large TBI populations.
Many of the earliest methods for TBI volumetric analysis relied almost exclusively on interactive, manual image editing and on time-consuming pixel counting in regions of interest18. Based on such methods, the assessment of simple quantitative metrics such as the total brain volume (TBV) and the ventricle-to-brain ratio (VBR) – which had been introduced in the 1970s – was marginally improved by using MRI instead of CT, for which such metrics had initially been developed. Simultaneously, the advent of novel MR sequences for TBI imaging such as FLAIR, SWI and T2-weighted GRE brought about the need for multimodal image processing algorithms which could provide richer quantitative descriptions of pathology and which could handle the ever-increasing number of volume types being routinely acquired in clinical settings. Following this line of research, Gerig and colleagues have proposed methods for automatic TBI volume segmentation based on subject-specific modification of atlas priors as well as based on outlier detection31, 32. One method developed by Niethammer et al.33 handles the complexities of longitudinal TBI lesion changes by introducing a geometric pathology shape model where lesion-related deformations are captured simultaneously with respect to deformations in the underlying image, thereby allowing one to visualize and to quantify hemorrhagic recession accurately.
For at least 10 years subsequent to the introduction of routine brain MRI scans, the ability to perform longitudinal analyses of TBI volumetrics and morphometrics was extremely rudimentary, being restricted mostly to the calculation of changes in brain volume, ventricle size, and lesion load 18. As previously stated, the application of automatic segmentation methods to TBI volumes has been and remains problematic, if only because time-consuming manual corrections must often be performed in the presence of gross pathology. Nevertheless, throughout the past 5 years, longitudinal studies of cortical atrophy due to TBI have become more common34, 35. Investigations of morphometric brain changes prompted by TBI have also been conducted using approaches such as tensor-based morphometry, which has highlighted localized volumetric losses in WM regions and in subcortical nuclei as well as ventricular volume increases across TBI cohorts of moderate size36. Template matching techniques have also been applied to visualize the spatial distribution of contusions in TBI patients, which can yield maps of brain locations which are most likely to suffer from atrophy in the event of a traumatic event37.
The typical absence of neuroimaging volumes acquired prior to the traumatic event constitutes a substantial and highly problematic challenge in the field of TBI imaging research. If available, such volumes could be highly beneficial for the purpose of longitudinal comparison by means of volumetric, morphometric and connectomic analyses. To date, however, very few case studies are available in which both pre- as well as post-TBI neuroimaging data were acquired. In one interesting study involving both MRI and PET, physiological changes were investigated in a TBI patient relative to those of his uninjured monozygotic (MZ) twin and against normal variability between co-twins in 10 comparably-aged, uninjured MZ twin pairs5. Such cases, however, remain exceptionally rare in the field of TBI research and the goal of reconstructing brain shape and connectivity patterns before TBI in the absence of imaging before injury remains extremely desirable though very problematic.
In the three sample subjects described in the previous section, non-hemorrhagic lesions adjacent to cerebrospinal fluid (CSF) were identified as hyper-intensities on FLAIR images, and segmentation quality was confirmed using GRE and TSE T2-weighted imaging. Non-hemorrhagic shearing lesions were defined as hyper-intense lesions that were visible primarily on T2-weighted or FLAIR images. SWI volumes were used to identify micro-hemorrhages which were poorly detectable or undetectable using other sequences. Segmentation and regional parcellation were performed using FreeSurfer38–40 following methodology described by Destrieux et al.41 As these methods were developed for the analysis of healthy-appearing brain volumes, additional computationally intensive steps were required, including user-supervised pathology identification and multiple iterative segmentation attempts. Because the cortical surface topology changes from the acute to the chronic time point, the cortical mesh for every subject at each time point was registered to an atlas generated over a large number of adult normal subjects as a function of the unit sphere39, 40, 42. Each mesh was non-rigidly morphed to the atlas by aligning sulcal-gyral patterns while minimizing shear and areal distortion. Parcellation of gyri and sulci was performed automatically using probabilistic labeling on the statistical atlas43. To visually represent morphometric changes in brain structure occurring between time points, three quantitative measures (cortical surface area, Gaussian curvature and cortical thickness) were extracted for each cortical location using freely-available methodologies38–40.
To investigate the longitudinal effects of TBI neuropathology within each patient, the structural profile of each subject was reconstructed in three dimensions and displayed using 3D Slicer software (www.slicer.org) to illustrate changes in each cortical structure. Once the value of each metric (cortical thickness, cortical area, Gaussian curvature) had been computed for each time point, the longitudinal difference between acute and chronic scans was plotted on the cortical surface. Because TBI pathology can alter WM/GM tissue contrast properties, the boundary between these two tissue types cannot always be extracted from T1-weighted imaging. For this reason, morphometrics associated with parcels which could not be segmented appropriately due to TBI-related loss of image contrast at the WM/GM boundary were discarded from the analysis.
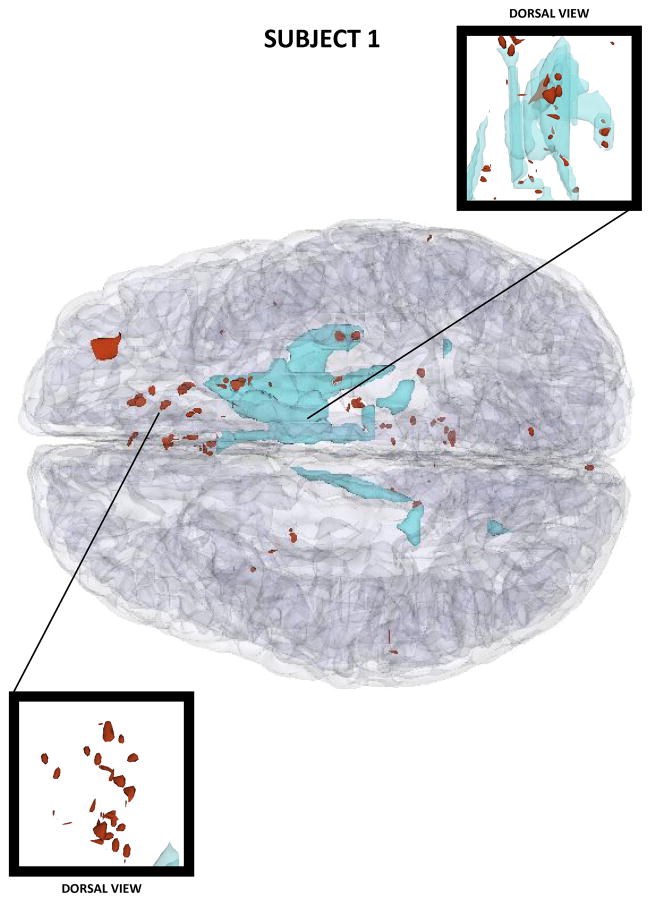
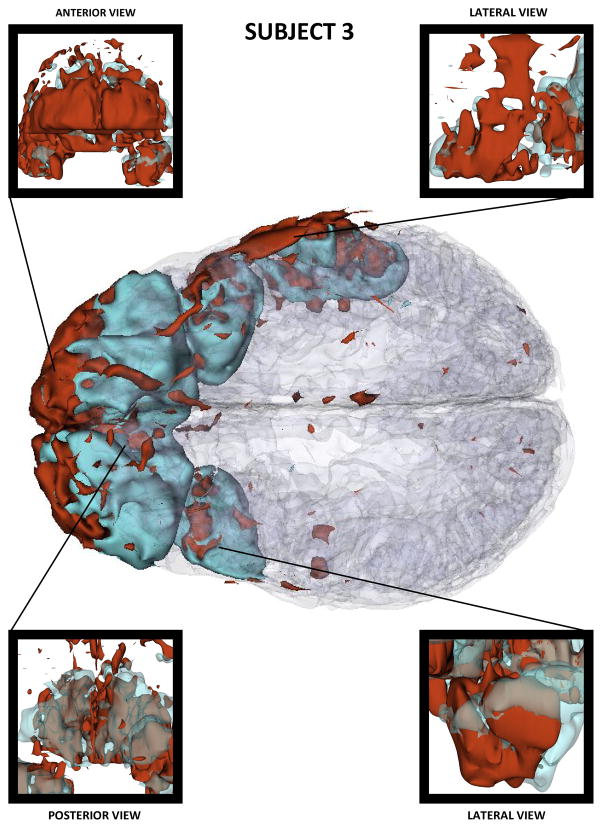
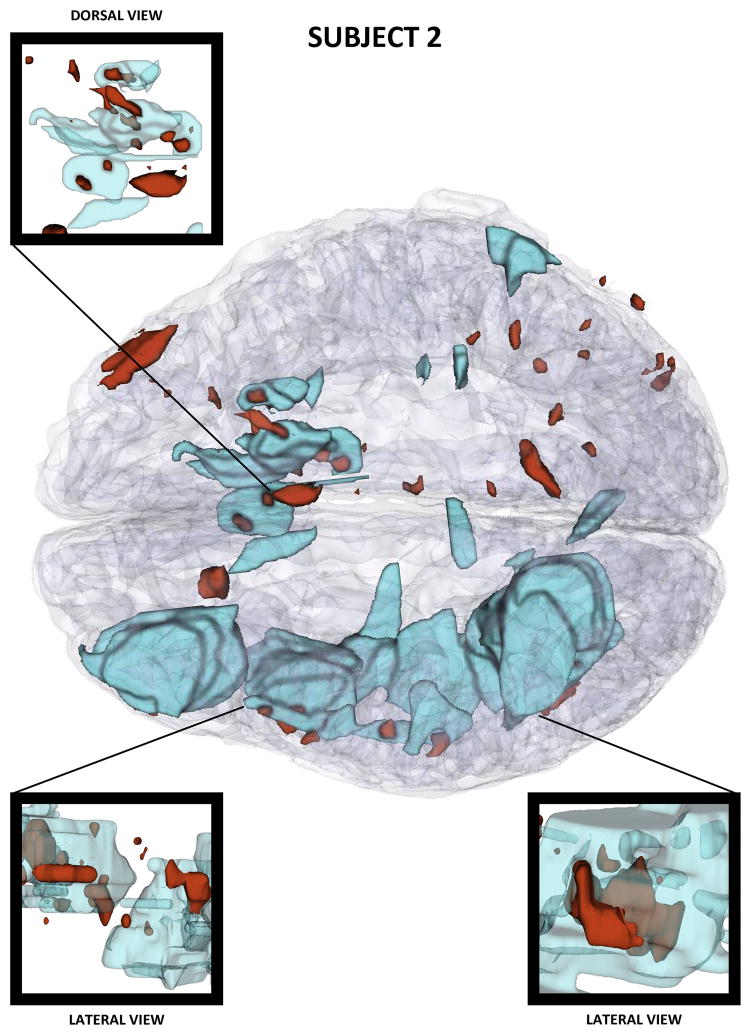
Figures 2–4 show three-dimensional (3D) models of both healthy-appearing GM as well as pathology (either hemorrhagic or non-hemorrhagic) in the three patients. These models reveal progressively larger lesion loads in each case, with Patient 1 presenting moderate amounts of bleeding in both hemispheres and Patient 3 suffering from extensive hemorrhage in both frontal lobes. Among the three cases presented, Patient 3 features the most profound amount of TBI pathology, with profuse bleeding and swelling in the frontal and temporal lobes of both hemispheres.
Figure 2.
Three-dimensional model of TBI anatomy for Patient 1 (acute baseline time point), as generated in 3D Slicer. Non-hemorrhagic and hemorrhagic lesions are displayed in cyan and red, respectively. The GM is shown using a transparent model.
Figure 4.
As in Figure 2, for Patient 3.
4. Brain connectivity and connectomics
Whereas the structural neuroimaging, tissue classification and morphometry of the TBI brain have been the focus of research for at least two decades, the effort dedicated to the study of TBI-related alterations in WM connectivity and connectomics has been more recent. Not long after the introduction of clinical MRI scanners, many researchers became aware that structural MR can only reveal DAI to a very limited extent and that more powerful imaging methods would be required to quantitate this important aspect of brain injury in humans44. In fact, it was not until the mid- to late-1990s that the use of DWI and DTI became common, and connectomics itself did not become a distinct area of scientific interest until around 200545. Although the spatial resolution of noninvasive neuroimaging continues to be far below that of hystopathological autopsy methods by many factors of magnitude, considerable advances have been made toward imaging and visualizating WM connections and their disruption after DAI.
Subsequent to the introduction of DWI and DTI, a number of early studies involving the application of these techniques to TBI analysis aimed to quantify changes in mean diffusivity (MD), apparent diffusion coefficient (ADC) and fractional anisotropy (FA) prompted by primary and secondary injuries46–50. Such studies highlighted the long-term effects of TBI upon brain connectivity, suggesting that all severities of TBI can result in axonal damage, whereas irreversible myelin damage is primarily apparent in moderate to severe TBI46. Importantly, DTI analysis has been found to provide objective means for explaining cognitive deficit patterns in TBI even in cases where the traumatic event had been sustained many years before the imaging evaluation51. In recent years, DWI and DTI have received increasing recognition as excellent noninvasive techniques for the purpose of evaluating injury in brain regions which are otherwise healthy appearing on conventional MRI scans52.
Although systematic analyses of brain connectivity alterations related to TBI have gained in prominence only throughout the past ~5 years, cross-sectional comparisons of DTI measures appear to converge upon the finding that TBI patients have decreased FA and increased MD compared to healthy control subjects over large portions of the brain53. Connectivity studies have also highlighted that WM degeneration plays a prominent role in the process of brain volume loss throughout the months following brain injury, and that connectomic studies are required to understand the relationship between degeneration patterns, on the one hand, and individual neural and cognitive deficit profiles, on the other hand3. In addition to WM atrophy and demyelination, DTI has also been used to quantify changes in neural tissue during recovery, with FA being observed to reach normal levels one year after trauma in patients with favorable outcome54.
The application of network theoretic approaches to the study of TBI connectivity is relatively recent. One fairly novel technique dubbed tract-based spatial statistics (TBSS) has been used to explore whether damage to specific white matter tracts is associated with particular patterns of cognitive impairment involving the commonly affected domains of memory, executive function and information processing speed55. This method revealed that the impact of WM changes upon cognitive function is likely to depend on damage to key pathways which link nodes in the distributed brain networks supporting high-level cognition. Another study by Achard et al.56 involving comatose TBI patients found evidence for radical reorganization of high degree hub nodes in the injured human brain, with global topological properties of complex brain networks being homeostatically conserved. These authors proposed that cortical regions which had been the hubs of healthy brain networks had lost their status as hubs as a consequence of injury, and that consciousness is likely dependent upon the anatomical location of hub nodes in human brain networks.
For the three sample TBI patients already described, processing workflows to compute inter-regional connectivity matrices were created using purpose-built software described elsewhere57. For each subject, the calculation of connectivity between regions involved identifying the location of each fiber tract extremity within the brain, followed by the delineation of the GM volume associated with each parcel. For each fiber connection between any two distinct parcels, the corresponding entry in the connectivity matrix of the brain was appropriately updated to reflect an increment in fiber count58, 59. By means of this, the connectivity matrix of the brain was generated for each subject, with entries which specified the total count of fibers between every pair of regions. Fibers whose lengths were shorter than 1.5 cm were excluded from the analysis to minimize the effect of fibers which had been artifactually generated by the tractography algorithm. Each connectivity matrix was normalized over the total number of fibers within that subject, and the percentage change in connectivity density between time points was then computed as the difference between connectivity matrix entries at each time point divided by the connectivity matrix entry at the acute (reference) time point.
Connectomic information was represented in a circular format60. Brain parcels denoting gyri or sulci were displayed within a circle of radially aligned elements called a connectogram, which contained information representing the two hemispheres symmetrically on the corresponding side of the vertical axis. Parcellated regions were assigned unique RGB colors listed exhaustively elsewhere61. Parcels were arranged within each connectogram lobe in the order of their locations along the antero-posterior axis of the cortical surface associated with the published FS normal population atlas41. An unambiguous abbreviation scheme was created to label each parcellation, as detailed elsewhere61. Within the outer circle which is associated with cortical parcels, five heat maps were created to encode one of five structural measures associated with each corresponding parcel. Proceeding towards the center of the connectogram, these measures are total GM volume, total area of the surface associated with the parcel, its mean cortical thickness, mean curvature and its brain connectivity per unit volume. The latter measure was computed as the density of fibers with endings within that parcel divided by the parcel’s total GM volume. The value of each structural measure was encoded using a color mapping which ranged from the minimum to the maximum of the data set. Specifically, the cortical thickness t with values in the interval [tmin, tmax] was normalized as t1 = (t − tmin)/(tmax − tmin). The value of t1 was associated with a single color; for example, hues at the extremities of the color map correspond to tmin and tmax, as needed. For the brain stem, cerebellum and subcortical structures, values for area, thickness and curvature were unavailable from FS and their appropriate heat map entries were drawn in white. Links were drawn between connectogram regions to illustrate connectivity density decreases whose values were more than two standard deviations below their mean (thus indicating appreciable decreases in connectivity between time points). Connections between regions whose connectivity could not be accurately computed due to the presence of TBI-related isotropy were discarded from the analysis, and heat map entries for structures which had been affected by pathology were drawn in gray. Further details on the methodology for generating connectograms and guidelines for their interpretation are available elsewhere61.
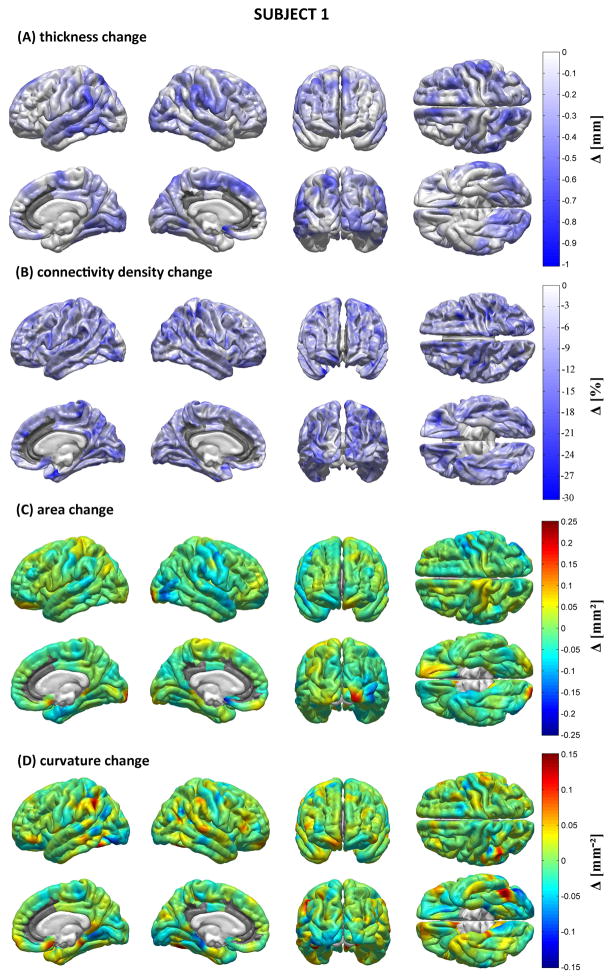
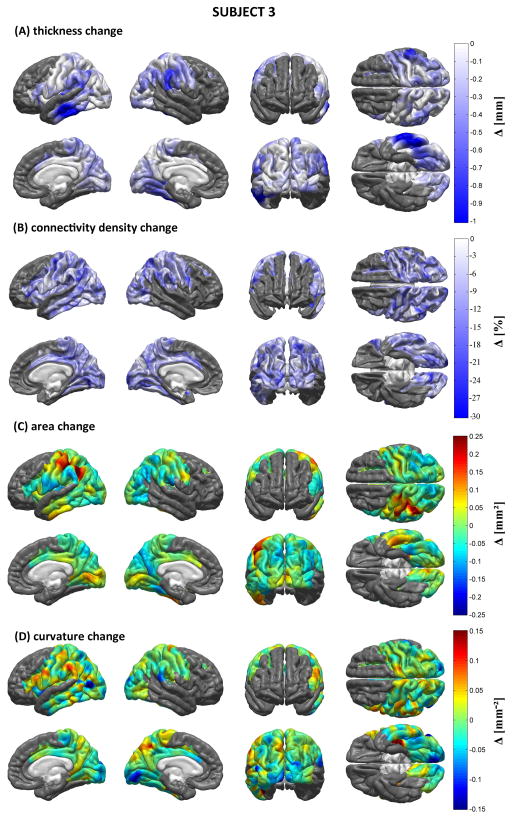
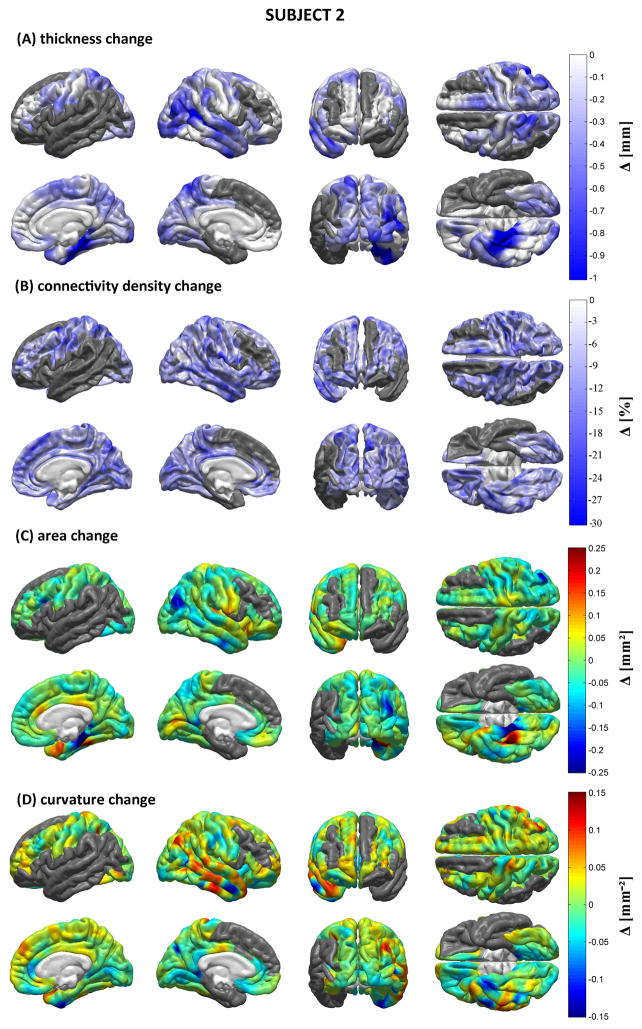
Figures 5–7 display the results of our structural, morphometric and connectomic analysis for each patient. These figures show changes in each measure (cortical thickness, connectivity density, local cortical area and Gaussian curvature) between the acute and chronic time points. Regions whose quantitative metrics could not be computed accurately due to TBI lesion-related loss of MR image contrast at the WM/GM boundary are grayed out. In Patient 1, widespread decreases in cortical thickness are observed in the right temporal lobe as well as bilaterally in the supramarginal gyri. The ventral aspects of both occipital lobes also appear to have been affected, as does the medial part of the left superior frontal gyrus. Areas affected by appreciable connectivity density changes include the orbitofrontal areas of both hemispheres, the postcentral sulcus of the LH, and the paracentral lobule of the RH, though the entire cortex appears to have been diffusely affected. Extreme changes in Gaussian curvature are observed for Jensen’s sulcus in the LH (adjacent to the left supramarginal gyrus), bilaterally in the parahippocampal gyri, as well as in the left inferior occipital gyrus. For Patient 2, decreases in cortical thickness are noteworthy in both hemispheres, the areas most affected being the lateral aspect of the right temporal lobe (particularly the superior temporal sulcus), where some extreme changes in Gaussian curvature are also observed. The supramarginal gyrus is also affected by atrophy in the RH, whereas the superior parietal lobule appears to have been affected bilaterally, though to a larger extent in the LH. Large connectivity density changes are observed in the lateral aspect of the right temporal lobe. The occipital lobes of both hemispheres have also experienced decreases in cortical thickness, though more appreciably in the RH where extreme changes in cortical surface area are also recorded. As in Patient 1, connectivity density losses are found to be diffuse—though not uniform—over the cortex. In Patient 3, the supramarginal gyri of both hemispheres have been notably affected by atrophy, as have the lateral and ventral aspects of the left temporal lobe, especially within the inferior temporal sulcus of the LH. Additionally, the supramarginal gyri exhibit large changes in Gaussian curvature and the left supramarginal gyrus is found to have suffered from a large decrease in surface area. By contrast, appreciable increases in surface area are seen in Jensen’s sulcus and in the postcentral sulcus (LH only). As in Patients 1 and 2, connectivity losses are diffusely present over the cortical surface.
Figure 5.
Morphometric analysis for Patient 1. Shown are longitudinal changes (Δ) in cortical thickness (A), connectivity density (B), cortical surface area (C) and Gaussian curvature (D) from the acute to the chronic time point. Regions whose metrics could not be computed accurately due to lesion-related loss of MR image contrast at the WM/GM boundary are grayed out.
Figure 7.
As in Figure 5, for Patient 3.
Table 1 summarizes the results of the volumetric analysis performed on the non-cerebral structures, in addition to the ventricular system (lateral, third and fourth ventricles). All subcortical structures are found to exhibit appreciable decreases in volume, whereas the ventricular system is found to exhibit a large volumetric increase in all three subjects, as typically encountered in TBI62, 63. Cerebellar atrophy is additionally identified in each subject, as documented by other studies as well64–67.
Table 1.
Longitudinal volumetric changes for the ventricular system and non-cerebral structures.
| patient | volume [cm3]
|
Δ [%] | ||
|---|---|---|---|---|
| acute | chronic | |||
| cerebellar WM | 1 | 25.55 | 25.17 | −1.5 |
| 2 | 28.17 | 23.36 | −17.1 | |
| 3 | 12.95 | 11.37 | −12.2 | |
|
| ||||
| cerebellar GM | 1 | 100.80 | 87.40 | −13.29 |
| 2 | 91.29 | 90.21 | −1.18 | |
| 3 | 53.03 | 52.72 | −0.58 | |
|
| ||||
| ventricular system | 1 | 13.68 | 29.40 | 114.9 |
| 2 | 20.37 | 69.61 | 241.7 | |
| 3 | 14.13 | 46.81 | 231.4 | |
|
| ||||
| thalamus | 1 | 15.59 | 12.33 | −20.9 |
| 2 | 13.31 | 11.43 | −14.1 | |
| 3 | 6.91 | 6.49 | −6.2 | |
|
| ||||
| caudate nucleus | 1 | 8.94 | 7.51 | −16.0 |
| 2 | 6.46 | 5.19 | −19.7 | |
| 3 | 3.66 | 3.51 | −4.1 | |
|
| ||||
| putamen | 1 | 13.40 | 10.94 | −18.3 |
| 2 | 10.19 | 8.07 | −20.7 | |
| 3 | 5.68 | 4.77 | −16.0 | |
|
| ||||
| pallidum | 1 | 4.18 | 3.66 | −12.5 |
| 2 | 3.01 | 2.19 | −27.3 | |
| 3 | 1.26 | 1.10 | −12.7 | |
|
| ||||
| hippocampus | 1 | 8.70 | 7.29 | −16.2 |
| 2 | 7.15 | 6.12 | −14.4 | |
| 3 | 3.98 | 3.71 | −6.7 | |
|
| ||||
| amygdala | 1 | 3.54 | 2.83 | −20.0 |
| 2 | 2.54 | 2.00 | −21.1 | |
| 3 | 1.53 | 1.31 | −14.2 | |
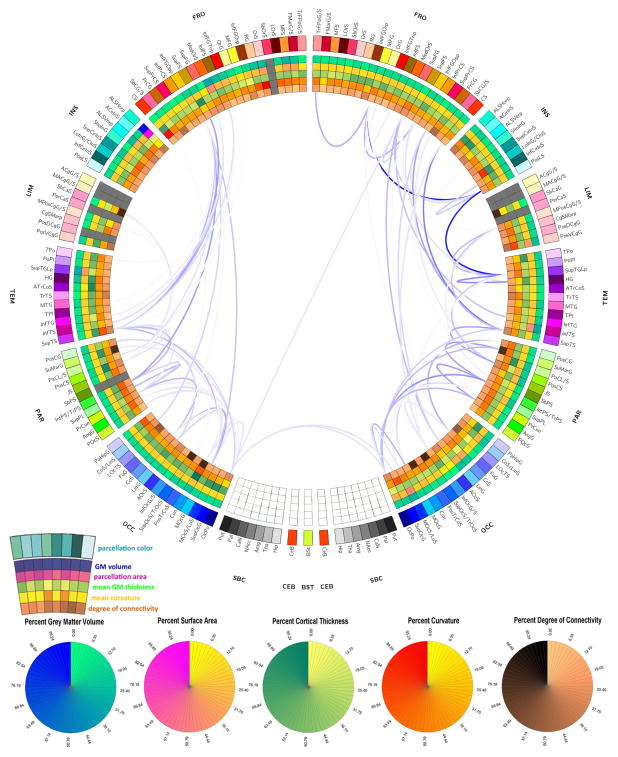
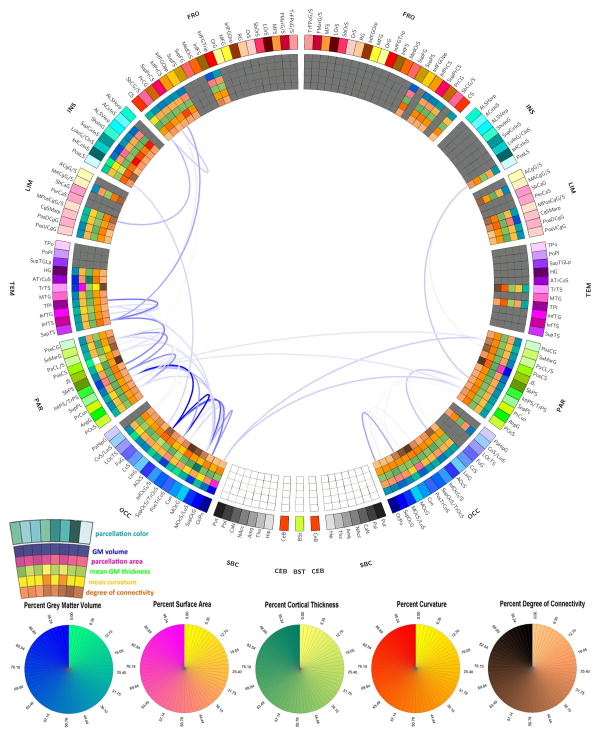
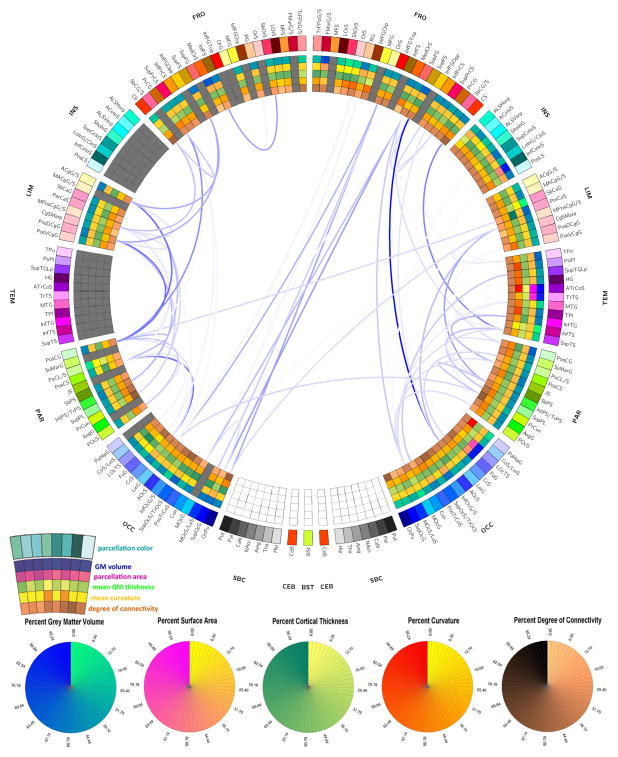
Figures 8–10 display the connectograms which illustrate the atrophy experienced by each patient 6 months after injury. In all cases, both intra- and inter-hemispheric connections appear to have been affected by substantial atrophy. In the case of Patient 1 (Figure 8), only 7 parcels were affected by gross pathology at the acute time point, and the extent of atrophy between regions was thus easier to calculate because the increase in isotropy associated with the presence of pathology was not as extensive in this patient compared to the other two. Generally speaking, Figure 8 reveals the presence of a WM atrophy pattern which is fairly symmetric with respect to the longitudinal fissure, with pronounced loss of connectivity between the frontal and temporal lobes, as well as between parietal and occipital cortices bilaterally. In Figure 9 corresponding to Patient 2, WM atrophy could not be computed for as many pairs of regions as in Patient 1 (Figure 8) because the left insula, left temporal lobe and many portions of the left and right frontal lobes were damaged by the primary TBI. Nevertheless, inspection of Figure 9 still reveals the presence of WM atrophy for connections between the limbic and parietal lobes (bilaterally) and between frontal and occipital regions. Finally, Figure 10 depicts the atrophy pattern experienced by Patient 3, where extensive portions of the frontal lobes, insulae, limbic and temporal lobes were affected by gross lesions at the acute time point. In this subject, appreciable atrophy is found primarily among WM fibers connecting the left parietal and occipital lobes.
Figure 8.
Connectogram representation of WM atrophy in Patient 1. The outermost ring shows the various brain regions arranged by lobe (FR – frontal; INS – insula; LIM – limbic; TEM – temporal; PAR –parietal; OCC – occipital; NC – non-cortical; BS – brain stem; CeB - cerebellum) and further ordered anterior-to-posterior. The set of five rings (from the outside going inward) use colors to encode the percentage change in the following five measures over 6 months (from the acute to the chronic time point): GM volume, parcel area, mean GM thickness, mean curvature, and degree of connectivity. A link is drawn between two regions if the connectivity density of the connection represented by that link has experienced a longitudinal decrease D which satisfies the inequality |D| ≥ μ (D) + 2σ (D), where μ (D) and σ (D) denote the mean and standard deviation of D over all reconstructed WM connections. Link transparency encodes |D| from its lowest value (faintest hue) to its highest one (darkest hue). Heat map entries for structures which had been affected by gross pathology are drawn in gray. For further details, see the Methods section.
Figure 10.
As in Figure 8, for Patient 3.
Figure 9.
As in Figure 8, for Patient 2.
5. Discussion
Brain injury confronts translational neuroimaging researchers with daunting methodological complexities, many of which remain unaddressed by the current state of the art. In many paradigms for the study of brain plasticity after injury, one is often interested in how specific damage to target locations in the nervous system affects brain structure and function68. Such changes in brain organization can share numerous features across subjects in controlled experiments where specific effects upon brain structure and function are sought in response to specific lesions whose nature and location in the peripheral nervous system is well defined (as, for example, in de-afferentation experiments). Hypothesis formulation in the context of such experiments is facilitated by the fact that brain areas dedicated to basic functions such as vision, olfaction and movement have been well studied in animal models as well as in man. By contrast, TBI poses significant investigative difficulties because (1) damage is often inflicted upon more than a single structural or functional area, (2) injuries can affect brain regions and structures whose detailed functions have not been studied and quantified rigorously, (3) inflicted injuries are heterogeneous across the TBI population, and (4) diffuse axonal injury (DAI) prompted by TBI affects the structure and function of various brain regions in ways which can be challenging to quantify reliably. For these reasons, TBI is considerably more challenging to study than other conditions of the diseased brain, both methodologically and conceptually. Partly for these reasons, there has been insufficient progress towards combining personalized case description and characterization with surgical and neuro-intensive care methods.
In many types of dementia, appreciable pathology-related commonalities in anatomic alterations are shared across diseased populations, and standardized methodologies can be formulated to situate patients within their cohorts for the purpose of statistical description and inference. In TBI, however, damaged gray matter (GM) areas differ across the TBI population, and WM connections are also affected differently depending upon the direction and force of impact as well as upon differences in cortical anatomy among subjects. The availability of methodologies for generating personalized TBI atrophy profiles can therefore be beneficial to the field of TBI clinical care.
The ability to generate longitudinal cortical atrophy profiles has become increasingly important in TBI research for the purpose of predicting long-term functional deficits and outcome. A study by MacKenzie et al.69 found that loss of consciousness is directly associated with the extent of brain atrophy 11 months after injury, and highlighted the importance of understanding the relationship between acute injury load, chronic atrophy profiles and functional deficits. Blatter et al.62, on the other hand, found appreciable correlation between brain atrophy and cognitive outcome, whereas Strangman et al.70 concluded that regional brain morphometry can predict memory rehabilitation outcome after TBIs of all severities. The latter study also found that the volumetrics of prefrontal and cingulate cortices could allow one to make accurate predictions of memory function outcome, and that the magnitude of the prediction range provided by regional brain volumes was in some cases almost equal to that provided by functional pretest scores on the outcome variable. In a sample of 20 children with TBI, Wilde et al.71 noted decreased cortical thickness in both frontal lobes at 3 months following injury, and additionally in the fusiform and lingual areas at 18 months post-injury, with some regional atrophy measures having the ability to predict behavioral regulation as quantified using neuropsychological testing.
The longitudinal effects of TBI upon brain structure are a topic which has received a considerable amount of attention throughout the past few decades62, 69, 72, 73. This interest has partly been motivated by the desire to understand how primary and secondary brain injuries modulate long-term loss and/or recovery of function, and how acute neurosurgical and neuropharmacological interventions can best be tailored to improve longitudinal trajectories in TBI patients. On the one hand, the ability to quantify the relative vulnerability of distinct brain regions may allow clinicians to classify case severity with a view towards long-term outcome prognosis and towards evaluating each patient’s potential for recovery. On the other hand, improved understanding of how WM integrity is affected by TBI could one day allow one to understand the details of TBI-related functional deficits and the mechanisms whereby information processing between different regions is affected by longitudinal atrophy.
To date, the clinical significance of injury-related changes in brain shape has not been explored comprehensively, and this review is the first to illustrate the calculation and analysis of longitudinal changes in local brain curvature and surface area. Cortical maps of these metrics may be of clinical interest because they can provide an appreciable amount of insight regarding local patterns of change. One possibility is that positive changes in Gaussian curvature suggest that the cortex locally exhibits sharper outward curving chronically compared to the acute stage. A plausible scenario for this involves the situation where GM at some location has been subjected to less atrophy compared to neighboring points, such that the point of interest ‘stands out’ of the surface compared to surrounding areas. Conversely, a negative change in curvature at some location can imply that the cortex has become flatter compared to surrounding areas. Such a situation could be due to increased atrophy at the location of interest compared to neighboring points, which would cause the surface to ‘sink in’. In the first case above, the increase in curvature may be due to greater neuronal loss surrounding the location of interest, whereas in the second case there is greater neuronal loss at the location of interest. Thus, cortical curvature analysis can reveal how TBI differentially affects brain shape across adjacent cortical locations. Because this type of insight cannot be obtained solely from knowledge of cortical thickness changes, curvature analysis can provide structural information which is complementary to cortical atrophy analysis. Changes in curvature may thus provide additional useful ways to conceptualize and quantify detailed changes in cortical shape, and could provide useful insight into TBI-related morphometry changes.
The availability of multiple longitudinal scans in a condition as temporally and spatially dynamic as TBI can lead to improvement in the accuracy of morphometric and volumetric calculations. In the three clinical examples presented, cortical morphometrics were computed from MRI data acquired at two time points (~3 days and ~6 months after injury). Additional intermediate longitudinal scans, however, can be incorporated into statistical models designed to estimate how clinical care in the acute phase can decrease the rate of atrophy in the long run. Such information would likely be beneficial for quantifying how atrophy modulates patient outcome, as well as for improving clinical decision-making.
6. Conclusion
In conclusion, we here reviewed the current state of the art in TBI neuroimaging and presented an approach for the quantification and visualization of cortical atrophy in the human brain by means of MRI/DTI image analysis. An important advantage of the methods presented is that they can provide timely information which may be useful for the formulation of patient-tailored rehabilitation protocols. When applied in conjunction with neuropsychological and neurophysiological measures, such an analysis could provide novel insights into how each TBI patient responds to injury and treatment, and may benefit the neuro-intensive care community by providing new ways to extract meaningful and clinically useful information from dynamic measurements of brain structure in TBI.
Figure 3.
As in Figure 2, for Patient 2.
Figure 6.
As in Figure 5, for Patient 2.
Acknowledgments
Funding: This work was supported by the National Alliance for Medical Image Computing (NA-MIC; www.na-mic.org), under NIH Roadmap Initiative grant 2U54EB005149, sub-award to J. D. V. H., by the NINDS, grant P01NS058489 to P. M. V., and R41NS081792, sub-award to J. D. V. H.
We acknowledge the assistance of Bo Wang, Marcel W. Prastawa, Guido Gerig, Stephen R. Aylward, Marc Niethammer, Ron Kikinis and of the staff in the Institute for Neuroimaging and Informatics at the University of Southern California.
Footnotes
Conflicts of interest: The authors report no actual or perceived conflict of interest.
References
- 1.Irimia A, Chambers MC, Torgerson CM, Filippou M, Hovda DA, Alger JR, et al. Patient-tailored connectomics visualization for the assessment of white matter atrophy in traumatic brain injury. Front Neurol. 2012;3:10. doi: 10.3389/fneur.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gentry LR, Godersky JC, Thompson B. MR imaging of head trauma: review of the distribution and radiopathologic features of traumatic lesions. AJR Am J Roentgenol. 1988;150(3):663–72. doi: 10.2214/ajr.150.3.663. [DOI] [PubMed] [Google Scholar]
- 3.Irimia A, Wang B, Aylward SR, Prastawa MW, Pace DF, Gerig G, et al. Neuroimaging of structural pathology and connectomics in traumatic brain injury: Toward personalized outcome prediction. Neuroimage Clin. 2012;1(1):1–17. doi: 10.1016/j.nicl.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faden AI, Demediuk P, Panter SS, Vink R. The role of excitatory amino acids and NMDA receptors in traumatic brain injury. Science. 1989;244(4906):798–800. doi: 10.1126/science.2567056. [DOI] [PubMed] [Google Scholar]
- 5.Kirkby BS, Van Horn JD, Ostrem JL, Weinberger DR, Berman KF. Cognitive activation during PET: a case study of monozygotic twins discordant for closed head injury. Neuropsychologia. 1996;34(7):689–97. doi: 10.1016/0028-3932(95)00147-6. [DOI] [PubMed] [Google Scholar]
- 6.Warach S, Gaa J, Siewert B, Wielopolski P, Edelman RR. Acute human stroke studied by whole brain echo planar diffusion-weighted magnetic resonance imaging. Ann Neurol. 1995;37(2):231–41. doi: 10.1002/ana.410370214. [DOI] [PubMed] [Google Scholar]
- 7.Ashikaga R, Araki Y, Ishida O. MRI of head injury using FLAIR. Neuroradiology. 1997;39(4):239–42. doi: 10.1007/s002340050401. [DOI] [PubMed] [Google Scholar]
- 8.Hajnal JV, Bryant DJ, Kasuboski L, Pattany PM, Decoene B, Lewis PD, et al. Use of Fluid Attenuated Inversion Recovery (FLAIR) Pulse Sequences in MRI of the Brain. J Comput Assist Tomo. 1992;16(6):841–4. doi: 10.1097/00004728-199211000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Atlas SW, Mark AS, Grossman RI, Gomori JM. Intracranial Hemorrhage - Gradient-Echo Mr Imaging at 1.5-T - Comparison with Spin-Echo Imaging and Clinical-Applications. Radiology. 1988;168(3):803–7. doi: 10.1148/radiology.168.3.3406410. [DOI] [PubMed] [Google Scholar]
- 10.Kuzma BB, Goodman JM. Improved identification of axonal shear injuries with gradient echo MR technique. Surg Neurol. 2000;53(4):400–2. [PubMed] [Google Scholar]
- 11.Reichenbach JR, Venkatesan R, Schillinger DJ, Kido DK, Haacke EM. Small vessels in the human brain: MR venography with deoxyhemoglobin as an intrinsic contrast agent. Radiology. 1997;204(1):272–7. doi: 10.1148/radiology.204.1.9205259. [DOI] [PubMed] [Google Scholar]
- 12.Defense_Centers_of_Excellence_[Internet] Neuroimaging following mild traumatic brain injury in the non-deployed setting. Silver Spring, MD: Defense and Veterans Brain Injury Center; 2013. Available from: http://www.DVBIC.org/ [Google Scholar]
- 13.Irimia A, Chambers MC, Alger JR, Filippou M, Prastawa MW, Wang B, et al. Comparison of acute and chronic traumatic brain injury using semi-automatic multimodal segmentation of MR volumes. J Neurotrauma. 2011;28(11):2287–306. doi: 10.1089/neu.2011.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mugler JP, 3rd, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magn Reson Med. 1990;15(1):152–7. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- 15.De Coene B, Hajnal J, Gatehouse P, Longmore D, White S, Oatridge A, et al. MR of the brain using fluid-attenuated inversion recovery (FLAIR) pulse sequences. AJNR Am J Neuroradiol. 1992;13(6):1555–64. [PMC free article] [PubMed] [Google Scholar]
- 16.Jones KM, Mulkern RV, Schwartz RB, Oshio K, Barnes PD, Jolesz FA. Fast spin-echo MR imaging of the brain and spine: current concepts. AJR Am J Roentgenol. 1992;158(6):1313–20. doi: 10.2214/ajr.158.6.1590133. [DOI] [PubMed] [Google Scholar]
- 17.Sehgal V, Delproposto Z, Haacke EM, Tong KA, Wycliffe N, Kido DK, et al. Clinical applications of neuroimaging with susceptibility-weighted imaging. J Magn Reson Imaging. 2005;22(4):439–50. doi: 10.1002/jmri.20404. [DOI] [PubMed] [Google Scholar]
- 18.Blatter DD, Bigler ED, Gale SD, Johnson SC, Anderson CV, Burnett BM, et al. MR-based brain and cerebrospinal fluid measurement after traumatic brain injury: Correlation with neuropsychological outcome. Am J Neuroradiol. 1997;18(1):1–10. [PMC free article] [PubMed] [Google Scholar]
- 19.Thatcher RW, Camacho M, Salazar A, Linden C, Biver C, Clarke L. Quantitative MRI of the gray-white matter distribution in traumatic brain injury. J Neurotraum. 1997;14(1):1–14. doi: 10.1089/neu.1997.14.1. [DOI] [PubMed] [Google Scholar]
- 20.Itti L, Chang L, Ernst T. Segmentation of progressive multifocal leukoencephalopathy lesions in fluid-attenuated inversion recovery magnetic. J Neuroimaging. 2001;11(4):412–7. doi: 10.1111/j.1552-6569.2001.tb00071.x. [DOI] [PubMed] [Google Scholar]
- 21.Xie K, Yang J, Zhang ZG, Zhu YM. Semi-automated brain tumor and edema segmentation using MRI. Eur J Radiol. 2005;56(1):12–9. doi: 10.1016/j.ejrad.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs MA, Mitsias P, Soltanian-Zadeh H, Santhakumar S, Ghanei A, Hammond R, et al. Multiparametric MRI tissue characterization in clinical stroke with correlation to clinical outcome - Part 2. Stroke. 2001;32(4):950–7. doi: 10.1161/01.str.32.4.950. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs MA, Zhang ZG, Knight RA, Soltanian-Zadeh H, Goussev AV, Peck DJ, et al. A model for multiparametric MRI tissue characterization in experimental cerebral ischemia with histological validation in rat - Part 1. Stroke. 2001;32(4):943–9. doi: 10.1161/01.str.32.4.943. [DOI] [PubMed] [Google Scholar]
- 24.Mitsias PD, Jacobs MA, Hammoud R, Pasnoor M, Santhakumar S, Papamitsakis NIH, et al. Multiparametric MRI ISODATA ischemic lesion analysis - Correlation with the clinical neurological deficit and single-parameter MRI techniques. Stroke. 2002;33(12):2839–44. doi: 10.1161/01.str.0000043072.76353.7c. [DOI] [PubMed] [Google Scholar]
- 25.Raff U, Rojas GM, Hutchinson M, Simon JH. Quantitation of T2 lesion load in patients with multiple sclerosis: A novel semiautomated segmentation technique. Acad Radiol. 2000;7(4):237–47. doi: 10.1016/s1076-6332(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 26.Johnston B, Atkins MS, Mackiewich B, Anderson M. Segmentation of multiple sclerosis lesions in intensity corrected multispectral MRI. Ieee T Med Imaging. 1996;15(2):154–69. doi: 10.1109/42.491417. [DOI] [PubMed] [Google Scholar]
- 27.Seghier ML, Ramlackhansingh A, Crinion J, Leff AP, Price CJ. Lesion identification using unified segmentation-normalisation models and fuzzy clustering. Neuroimage. 2008;41(4):1253–66. doi: 10.1016/j.neuroimage.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao CC, Xiao FR, Wong JM, Chiang IJ. A multiresolution binary level set method and its application to intracranial hematoma segmentation. Comput Med Imag Grap. 2009;33(6):423–30. doi: 10.1016/j.compmedimag.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Sidaros A, Skimminge A, Liptrot MG, Sidaros K, Engberg AW, Herning M, et al. Long-term global and regional brain volume changes following severe traumatic brain injury: A longitudinal study with clinical correlates. Neuroimage. 2009;44(1):1–8. doi: 10.1016/j.neuroimage.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 30.Hillary FG, Biswal BB. Automated Detection and Quantification of Brain Lesions in Acute Traumatic Brain Injury Using MRI. Brain Imaging Behav. 2009;3(2):111–22. [Google Scholar]
- 31.Wang B, Prastawa MW, Awate SP, Irimia A, Chambers MC, Vespa PM, Van Horn JD, Gerig G. Segmentation of serial MRI of TBI patients using personalized atlas construction and topological change estimation. IEEE International Symposium on Biomedical Engineering; San Diego, CA. 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B, Prastawa MW, Awate SP, Irimia A, Chambers MC, Vespa PM, Van Horn JD, Gerig G. In: Haynor DR, Ourselin S, editors. A patient-specific segmentation framework for longitudinal MR images of traumatic brain injury; Proc of SPIE; San Diego, CA, USA. 2012; pp. 831402–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niethammer M, Hart GL, Pace DF, Vespa PM, Irimia A, Van Horn JD, et al. Geometric metamorphosis. Med Image Comput Comput Assist Interv. 2011;14(Pt 2):639–46. doi: 10.1007/978-3-642-23629-7_78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding K, de la Plata CM, Wang JY, Mumphrey M, Moore C, Harper C, et al. Cerebral Atrophy after Traumatic White Matter Injury: Correlation with Acute Neuroimaging and Outcome. J Neurotraum. 2008;25(12):1433–40. doi: 10.1089/neu.2008.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irimia A, Chambers MC, Alger JR, Filippou M, Prastawa MW, Wang B, et al. Comparison of Acute and Chronic Traumatic Brain Injury Using Semi-Automatic Multimodal Segmentation of MR Volumes. J Neurotraum. 2011;28(11):2287–306. doi: 10.1089/neu.2011.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Avants B, Patel S, Whyte J, Coslett BH, Pluta J, et al. Structural consequences of diffuse traumatic brain injury: A large deformation tensor-based morphometry study. Neuroimage. 2008;39(3):1014–26. doi: 10.1016/j.neuroimage.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine B, Kovacevic N, Nica EI, Cheung G, Gao F, Schwartz ML, et al. The Toronto traumatic brain injury study - Injury severity and quantified MRI. Neurology. 2008;70(10):771–8. doi: 10.1212/01.wnl.0000304108.32283.aa. [DOI] [PubMed] [Google Scholar]
- 38.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–55. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 39.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis - II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 40.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis - I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 41.Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53(1):1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischl B, Sereno MI, Tootell RBH, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human Brain Mapping. 1999;8(4):272–84. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, et al. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 44.Gentry LR. Imaging of Closed-Head Injury. Radiology. 1994;191(1):1–17. doi: 10.1148/radiology.191.1.8134551. [DOI] [PubMed] [Google Scholar]
- 45.Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. Plos Comput Biol. 2005;1(4):245–51. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraus MF, Susmaras T, Caughlin BP, Walker CJ, Sweeney JA, Little DM. White matter integrity and cognition in chronic traumatic brain injury: a diffusion tensor imaging study. Brain. 2007;130:2508–19. doi: 10.1093/brain/awm216. [DOI] [PubMed] [Google Scholar]
- 47.Schlaug G, Renga V, Lindenberg R, Nair D. Diffusion Tensor Imaging as a Predictor of Corticospinal Tract Integrity and Recovery Potential in Stroke Patients. Stroke. 2009;40(4):E123-E. [Google Scholar]
- 48.Zakaria N, Kallakuri S, Cavanaugh J, Shen YM, Yu YJ, Kou ZF, et al. Assessing the potential of diffusion tensor imaging as a non-invasive tool for detecting diffusion axonal injuries in a rodent model of traumatic brain injury. J Neurotraum. 2007;24(7):1233. [Google Scholar]
- 49.Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage. 2002;17(3):1429–36. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- 50.Song SK, Sun SW, Ju WK, Lin SJ, Cross AH, Neufeld AH. Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. Neuroimage. 2003;20(3):1714–22. doi: 10.1016/j.neuroimage.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Nakayama N, Okumura A, Shinoda J, Yasokawa YT, Miwa K, Yoshimura SI, et al. Evidence for white matter disruption in traumatic brain injury without macroscopic lesions. J Neurol Neurosur Ps. 2006;77(7) doi: 10.1136/jnnp.2005.077875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galloway NR, Tong KA, Ashwal S, Oyoyo U, Obenaus A. Diffusion-Weighted Imaging Improves Outcome Prediction in Pediatric Traumatic Brain Injury. J Neurotraum. 2008;25(10):1153–62. doi: 10.1089/neu.2007.0494. [DOI] [PubMed] [Google Scholar]
- 53.Bendlin BB, Ries ML, Lazar M, Alexander AL, Dempsey RJ, Rowley HA, et al. Longitudinal changes in patients with traumatic brain injury assessed with diffusion-tensor and volumetric imaging. Neuroimage. 2008;42(2):503–14. doi: 10.1016/j.neuroimage.2008.04.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sidaros A, Engberg A, Sidaros K, Liptrot MG, Herning M, Petersen P, et al. Diffusion tensor imaging during recovery from severe traumatic brain injury and relation to clinical outcome: a longitudinal study. Brain. 2008;131:559–72. doi: 10.1093/brain/awm294. [DOI] [PubMed] [Google Scholar]
- 55.Kinnunen KM, Greenwood R, Powell JH, Leech R, Hawkins PC, Bonnelle V, et al. White matter damage and cognitive impairment after traumatic brain injury. Brain. 2011;134(Pt 2):449–63. doi: 10.1093/brain/awq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Achard S, Delon-Martin C, Vertes PE, Renard F, Schenck M, Schneider F, et al. Hubs of brain functional networks are radically reorganized in comatose patients. Proc Natl Acad Sci U S A. 2012;109(50):20608–13. doi: 10.1073/pnas.1208933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Horn JD, Irimia A, Torgerson CM, Chambers MC, Kikinis R, Toga AW. Mapping Connectivity Damage in the Case of Phineas Gage. Plos One. 2012;7(5) doi: 10.1371/journal.pone.0037454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagmann P, Cammoun L, Gigandet X, Gerhard S, Grant PE, Wedeen V, et al. MR connectomics: Principles and challenges. J Neurosci Meth. 2010;194(1):34–45. doi: 10.1016/j.jneumeth.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 59.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen V, et al. Mapping the structural core of human cerebral cortex. Plos Biol. 2008;6(7):1479–93. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irimia A, Chambers MC, Torgerson CM, Van Horn JD. Circular representation of human cortical networks for subject and population-level connectomic visualization. Neuroimage. 2012;60(2):1340–51. doi: 10.1016/j.neuroimage.2012.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blatter DD, Bigler ED, Gale SD, Johnson SC, Anderson CV, Burnett BM, et al. MR-based brain and cerebrospinal fluid measurement after traumatic brain injury: correlation with neuropsychological outcome. AJNR Am J Neuroradiol. 1997;18(1):1–10. [PMC free article] [PubMed] [Google Scholar]
- 63.Levin HS, Meyers CA, Grossman RG, Sarwar M. Ventricular enlargement after closed head injury. Arch Neurol. 1981;38(10):623–9. doi: 10.1001/archneur.1981.00510100051007. [DOI] [PubMed] [Google Scholar]
- 64.Spanos GK, Wilde EA, Bigler ED, Cleavinger HB, Fearing MA, Levin HS, et al. cerebellar atrophy after moderate-to-severe pediatric traumatic brain injury. AJNR Am J Neuroradiol. 2007;28(3):537–42. [PMC free article] [PubMed] [Google Scholar]
- 65.Gale SD, Johnson SC, Bigler ED, Blatter DD. Trauma-induced degenerative changes in brain injury: a morphometric analysis of three patients with preinjury and postinjury MR scans. J Neurotrauma. 1995;12(2):151–8. doi: 10.1089/neu.1995.12.151. [DOI] [PubMed] [Google Scholar]
- 66.Park E, McKnight S, Ai J, Baker AJ. Purkinje cell vulnerability to mild and severe forebrain head trauma. J Neuropathol Exp Neurol. 2006;65(3):226–34. doi: 10.1097/01.jnen.0000202888.29705.93. [DOI] [PubMed] [Google Scholar]
- 67.Caeyenberghs K, Leemans A, Geurts M, Linden CV, Smits-Engelsman BC, Sunaert S, et al. Correlations between white matter integrity and motor function in traumatic brain injury patients. Neurorehabil Neural Repair. 2011;25(6):492–502. doi: 10.1177/1545968310394870. [DOI] [PubMed] [Google Scholar]
- 68.Kaas JH, Qi HX, Burish MJ, Gharbawie OA, Onifer SM, Massey JM. Cortical and subcortical plasticity in the brains of humans, primates, and rats after damage to sensory afferents in the dorsal columns of the spinal cord. Exp Neurol. 2008;209(2):407–16. doi: 10.1016/j.expneurol.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.MacKenzie JD, Siddiqi F, Babb JS, Bagley LJ, Mannon LJ, Sinson GP, et al. Brain atrophy in mild or moderate traumatic brain injury: a longitudinal quantitative analysis. AJNR Am J Neuroradiol. 2002;23(9):1509–15. [PMC free article] [PubMed] [Google Scholar]
- 70.Strangman GE, O’Neil-Pirozzi TM, Supelana C, Goldstein R, Katz DI, Glenn MB. Regional brain morphometry predicts memory rehabilitation outcome after traumatic brain injury. Front Hum Neurosci. 2010;4:182. doi: 10.3389/fnhum.2010.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilde EA, Merkley TL, Bigler ED, Max JE, Schmidt AT, Ayoub KW, et al. Longitudinal changes in cortical thickness in children after traumatic brain injury and their relation to behavioral regulation and emotional control. Int J Dev Neurosci. 2012;30(3):267–76. doi: 10.1016/j.ijdevneu.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gale SD, Johnson SC, Bigler ED, Blatter DD. Nonspecific white matter degeneration following traumatic brain injury. J Int Neuropsychol Soc. 1995;1(1):17–28. doi: 10.1017/s1355617700000060. [DOI] [PubMed] [Google Scholar]
- 73.Cullum CM, Bigler ED. Ventricle size, cortical atrophy and the relationship with neuropsychological status in closed head injury: a quantitative analysis. J Clin Exp Neuropsychol. 1986;8(4):437–52. doi: 10.1080/01688638608401333. [DOI] [PubMed] [Google Scholar]