Abstract
The BET family proteins recognize acetylated chromatin through their two bromodomains, acting as transcriptional activators or tethering viral genomes to the mitotic chromosomes of their host. The structural mechanism for how the N-terminal bromodomain of human BRD2 (BRD2-BD1) deciphers the mono-acetylated status of histone H4 tail was recently reported. Here we show the crystal structure of the second bromodomain of BRD2 (BRD2-BD2) in complex with the di-acetylated histone H4 tail (H4K5ac/K12ac). To our surprise, a single K5ac/K12ac peptide interacts with two BRD2-BD2 molecules simultaneously: the K5ac residue binds to one BRD2-BD2 molecule while the K12ac residue binds to another. These results provide a structural basis for the recognition of two different patterns of the histone acetylation status by a single bromodomain.
Structured summary
MINT-7989882, MINT-7989824, MINT-7989846, MINT-7989865: H4 (uniprotkb:P62805) binds (MI:0407) to BRD2 (uniprotkb:P25440) by surface plasmon resonance (MI:0107)
MINT-7989539: H4 (uniprotkb:P62805) and BRD2 (uniprotkb:P25440) bind (MI:0407) by X-ray crystallography (MI:0114)
Keywords: BET family, Bromodomain, Cell cycle, Chromatin, Crystal structure, Papilloma virus, Transcription
1. Introduction
The histone code, defined by combinatorial modifications of the flanking histone tails, represents eukaryotic regulatory mechanisms of DNA-mediated events through structural alteration of the chromatin and/or association of trans-acting factors onto it [1]. Among known modifications of the core histones, acetylation of the histone tails and its recognition is a typical hallmark for transcriptional activation from chromatin DNA [2]. The acetylated N-terminal tails of the histones are specifically recognized by a bromodomain which is a conserved structural domain composed of approximately 110 amino acids [3]. The bromodomain is present in many chromatin-associated factors, including nuclear histone acetyltransferases (HATs), chromatin-remodeling factors, and the bromodomains and extra-terminal domain (BET) family of nuclear proteins [3,4].
The BET family of nuclear proteins has the unique architecture with two tandem bromodomains and a conserved extra-terminal domain [3–6]. Through its bromodomains, the BET protein BRD2 selectively recognizes the acetylated lysine 12 (K12) of histone H4 for transcriptional activation [7], while another BET protein BRDT most prefers to recognize the K5/K8-di-acetylated histone H4 [8]. In human cells, BRD2- and BRD3-associated chromatin is significantly enriched in K5- and K12-acetylated histone H4 [9]. Importantly, the BET proteins BRD2 and BRD4 associate with acetylated chromatin throughout the cell cycle [7,10], while other non-BET bromodomain family members dissociate from the chromosomes during mitosis [11]. This mitotic retention on chromosomes is the unique feature of the BET proteins, which can be utilized by papilloma virus, and also presumably by Kaposi's sarcoma-associated herpesvirus, for tethering their genomes to the mitotic chromosome of their host, and for propagating them during the cell division [12,13]. Recently, structural basis of the recognition of K5/K8-di-acetylated histone H4 tail by the N-terminal bromodomain of BRDT was reported, in which both the acetyllysine residues bind to one binding pocket of BRDT bromodomain [14]. We reported the structural basis for the N-terminal bromodomain of BRD2, BRD2-BD1, to decipher the histone code by means of recognizing K12-acetylated histone H4 tail [15,16]. However, structural basis how the second C-terminal bromodomain of BET family proteins recognize an acetylation status of histone H4 has been unclear, except a few NMR analyses [17,18].
To further gain insight into the structural basis for the recognition of the BET bromodomains and acetylated histone H4, we determined the crystal structure of the second C-terminal bromodomain of BRD2, BRD2-BD2, in complex with a histone H4 tail peptide which is di-acetylated at K5 and K12 (H4K5ac/K12ac). Surprisingly, the K5/K12-di-acetylated H4 tail peptide interacted with two BRD2-BD2 molecules simultaneously. Based on the structural and biochemical analyses, we discuss a possible role of the second bromodomain of BRD2 for recognition of active chromatin containing K5- and K12-acetylated histone H4.
2. Materials and methods
2.1. Peptides and proteins
The K5/K12-di-acetylated histone H4 tail peptide (residues 1–15; SGRGK(Ac)GGKGLGK(Ac)GGA) used for the crystal structure analysis, and relevant histone tail peptides, H4 (1–20) and H3 (10–27), biotinylated at C-termini used for the binding analysis, were purchased from Toray Research Center (Kamakura, Japan). Expression and purification of the BRD2-BD2 protein were performed as described elsewhere [19].
2.2. Crystallization and data collection
The BRD2-BD2 protein in complex with the di-acetylated histone H4 tail peptide was crystallized by co-crystallization method, by mixing the peptide and protein solutions (10:1 molar ratio) and incubating them at 4 °C for about 4 h before setting up the crystallization experiment. The crystals were obtained from 30% PEG MME 2000 and 50 mM Tris–HCl, pH 8.0, as thin plates of 20–30 μm thick. These crystals belong to P21 space group and containing four BD2 molecules and two peptides in an asymmetric unit. A complete dataset up to 2.30 Å resolution was collected at the beamline AR-NW12A Photon Factory, Japan, and processed and scaled using the HKL2000 program suite [20].
2.3. Structure determination and refinement
The crystal structure of the BRD2-BD2 protein in complex with the di-acetylated histone H4 tail peptide was solved by the molecular replacement method, using the BRD2-BD1 structure [15] as a model, with the MOLREP program incorporated in CCP4. The protein chains were fitted manually using the graphics program O [21]. The model was refined with CNS [22], using a maximum likelihood target that included amplitude and phase probability distributions. NCS restraints (200 kcal mol−1 Å−2) were maintained between the NCS molecules throughout the refinement. During the course of fitting, σA-weighted electron density maps clearly revealed the electron density for the acetylated histone H4 tail peptides. The program REFMAC5 from the CCP4 suite [23] was used in the final steps of refinement. With the NCS restraints which were imposed throughout the refinement, the final complete structure at 2.30 Å resolution consists of 847 protein residues, 28 peptide residues, 412 water molecules, gave a final R-factor of 20.4% and an Rfree of 26.8%. The stereochemistry of the protein chains is good, as assessed with PROCHECK [24]. Refinement statistics are summarized in Table 1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank. The PDB ID is 2E3K.
Table 1.
Summary of data collection and refinement statistics.
| BRD2-BD2 + H4K5ac/K12ac | |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.0 |
| Space group | P21 |
| Unit cell (Å, °) | a = 45.83, b = 128.59, c = 45.85, β = 104.7 |
| Resolution (Å) | 50–2.30 (2.38–2.30) |
| Redundancya | 3.7 (3.7) |
| Unique reflections | 22 434 |
| Completeness (%) | 97.7 (97.9) |
| Rmerge (%)b | 4.8 (16.6) |
| Refinement statistics | |
| Resolution (Å) | 20–2.30 |
| σ cutoff | 0 |
| Reflections | 21 151 |
| No. protein residues | 847 |
| No. peptide residues | 28 |
| No. water molecules | 412 |
| Rcryst (%)c | 20.4 |
| Rfree (%)d | 26.8 |
| Average B factors | 43.69 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.023 |
| Bond angles (°) | 1.87 |
Numbers in parentheses are values in the highest resolution shell.
Rmerge = Σ |Iobs − <I>|/Σ <I> summed over all observations and reflections.
Rcryst = Σ ‖Fobs| − |Fcalc‖/Σ |Fobs|.
Rfree was calculated with 5% of data omitted from refinement.
2.4. Surface plasmon resonance binding assays
The binding affinities of the BRD2-BD2 proteins against the H4 or H3 tail peptides were examined on a Biacore 3000 (GE Healthcare) with PBS (pH 7.4) as the running buffer. Biotinylated H4 tail peptides, corresponding to the N-terminal region from 1 to 20 of histone H4, or biotinylated H3 tail peptides, corresponding to the region from 10 to 27 of histone H3, were immobilized on flow cells #2–#4 of a sensor chip SA (GE Healthcare). Amounts of the immobilized peptides were within the range of 650–930 resonance units, and the binding of the BRD2-BD2 protein to each peptide-immobilized sensor chip was normalized to each theoretical maximal resonance unit (Rmax). Control surfaces with d-biotin attached were used to correct for refractive index differences between the samples and the running buffer. Dissociation constants (KD) for the interaction were calculated with the steady-state affinity model of the BIAevaluation 4.1 software (GE Healthcare).
3. Results and discussion
3.1. Overall structure of BRD2-BD2 in complex with the K5/K12-di-acetylated H4 tail
We solved the crystal structure of the second bromodomain of human BRD2 (BRD2-BD2: residues 348–455; Fig. 1A and B) in complex with a histone H4 tail peptide (residues 1–15) which is di-acetylated at K5 and at K12. The crystals are of P21 space group, with four bromodomain molecules, and two peptides in an asymmetric unit (Fig. 1C). The BRD2-BD2 structure contains a left-handed α-helical bundle formed by four α-helices (i.e. αZ, αA, αB, and αC) (Fig. 1B), which is similar to the solution structures of BRD2-BD2 [17] and BRD4-BD2 [18], crystal structures of BRD2-BD1 [15,16], BRDT-BD1/BD2 [14], and other bromodomain structures [25–30]. An extended long loop, ZA, connecting the helices αZ and αA, and another loop, BC, connecting the helices αB and αC make a deep cleft with a cavity in the middle of the pocket, which defines the binding site for acetylated histone tail (Fig. 1B and C). The hydrophobic core of the domain is stabilized mainly by the conserved hydrophobic residues, and additionally, by some conserved hydrophilic residues.
Fig. 1.
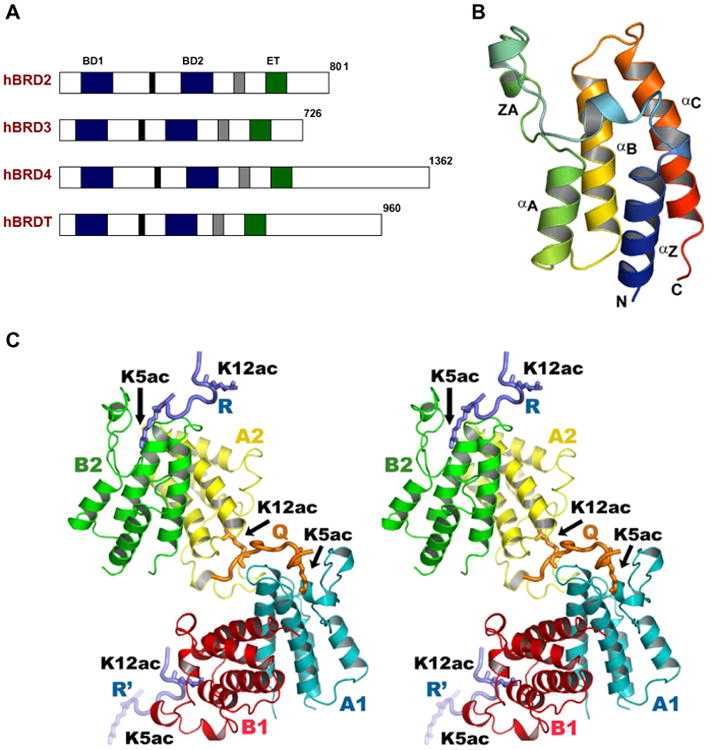
BRD2-BD2 interaction with acetylated histone H4 tails. (A) Schematic representations of the BRD proteins. The bromodomains (BD1 and BD2) and the extra-terminal domain (ET) are indicated by blue and green boxes, respectively. Two other conserved regions are shown as black and gray boxes. (B) A ribbon diagram of the BRD2-BD2 structure, colored from blue (N-terminus) to red (C-terminus). (C) Stereo-view of the BRD2-BD2 complex. The BRD2-BD2 and H4K5ac/K12ac tail peptides are shown by ribbon models, and the side-chains of H4K5ac and H4K12ac are indicated by stick models. The color codes for the BRD2-BD2 dimers and the H4 tails are consistent in all figures unless otherwise stated. All structure figures were generated with Pymol (http://www.pymol.org/).
The refined structure showed that BRD2-BD2 formed two homodimers in the asymmetric unit. In the crystal, the weak dimeric interface arises from the helices αB and αC (Fig. S1). In this weak dimeric interface, no direct intermolecular interactions have been observed but through buried water molecules. Our in vivo co-immunoprecipitation studies did not show any self-association between the BD2 molecules (data not shown), and the NMR study reported by Shi and her colleagues also suggests that BRD2-BD2 is a monomer in solution [17]. In addition, although the crystal structure of BRDT-BD2 possesses similar NCS arrangement in the asymmetric unit, BRDT-BD2 is a monomer in solution [14]. Taken together, we consider that BRD2-BD2 may also be a monomer in solution.
3.2. Recognition of the K5/K12-di-acetylated histone H4 tail by BRD2-BD2
The crystal structure analysis clearly showed that the di-acetylated H4 tail peptide bound to BRD2-BD2. The difference Fourier map unambiguously detected the electron density for the acetylated H4 tail peptide in the cleft region of the BD2 protein (Fig. S2A and B). We observed several noteworthy features from this structural analysis: First, the BRD2-BD2 protein recognized acetyl-lysine 5 (K5ac) as well as acetyl-lysine 12 (K12ac) of the H4 tail peptide. Second, besides its recognition for the acetyl-lysine at different positions, it recognized them simultaneously through different BRD2-BD2 molecules. Third, extensive interactions were observed between the BRD2-BD2 protein and many residues of the H4 tail peptide (described below).
3.3. Interaction of the di-acetylated H4 peptide chain Q with BRD2-BD2
In the structure of the complex, the H4 peptide Q binds into the long cleft region, which is produced by the monomers A1 and A2 of A1B1 and A2B2, respectively, with a non-specific extended conformation (Fig. 1C). The side-chain of K5ac sits in the deep hydrophobic pocket of the molecule A1, formed by the residues P371, F372, V376, L381, L383, C425, and V435 (Fig. 2A). The acetyl group of K5ac is positioned in the cavity in such a way that the carbonyl group is hydrogen bonded to the Nδ2 of N429, as well as indirectly to the side-chain of Y386 through a water molecule (Fig. 2A), which is essentially conserved in other bromodomain structures [3,5]. A hydrogen bond network between the water molecules exists on one side of the K5ac side-chain. The side-chain atom Nζ of K5ac forms a water-mediated hydrogen bond with the carbonyl group of P371. Similar modes of interactions are also observed for the side-chain K12ac, which binds to the monomer A2 (Fig. 2B). The charge distribution on the cavity surface is complementary to the aliphatic moiety (Fig. S3).
Fig. 2.
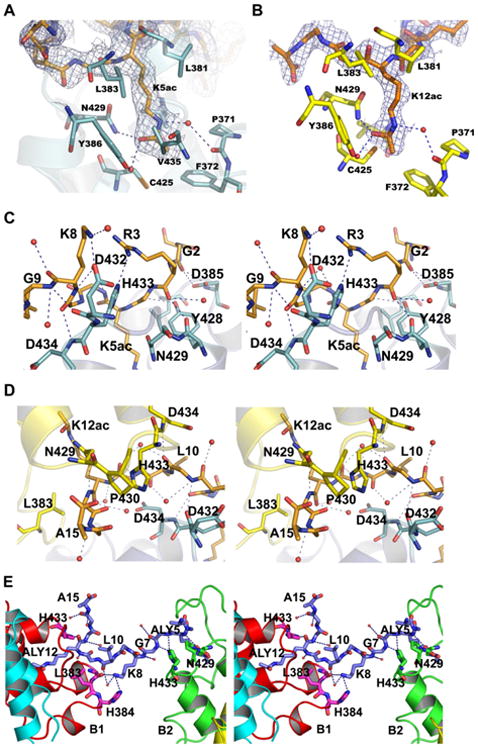
The BRD2-BD2 dimers interact with di-acetylated H4 tails, Q and R. (A) A representative cartoon diagram of K5ac binding region. (B) A representative cartoon diagram of K12ac binding region. The acetyl-lysine binding interaction is very similar both in K5ac and K12ac. (C) A close-up view (in stereo) of the H4 tail Q near the K5ac binding region. (D) A close-up view (in stereo) of the H4 tail Q near the K12ac binding region. (E) A close-up view (in stereo) of the H4 tail, R interaction with BD2. The interacting residues and water molecules in (A–E), are shown by sticks and balls, respectively.
The di-acetylated H4 tail peptide extensively interacts with its protein partners, A1 and A2 molecules, as well as with many water molecules (Fig. 2C, D and S2A). The first 8 residues of the peptide associate with the chain A1, and the residues from 9 to 15 interact with A2 of the A2B2 chains (Fig. S2A). The main-chain carbonyl group of G2 possesses a bifurcated hydrogen bond interaction with the atoms Oδ1 and Oδ2 of D385 (Fig. 2C). The backbone carbonyl group of R3 contributes a main-chain-main-chain interaction with Y428, and also forms a hydrogen bond with a water molecule. Nevertheless, the guanidyl group of R3 (the NH1 atom) contributes a substantial interaction with Nε2 of H433 (Fig. 2C). A hydrogen bond is formed between the carbonyl group of the residue, G4, and the hydroxyl group of Y428. In addition, carbonyl–carbonyl dipolar interaction is also observed between G4 and Y428 (Fig. 2C). Although the residue G6 does not have any electrostatic interaction with the protein atoms, it forms hydrogen bond interactions with water molecules. For G7, the backbone amide group exhibits a weak interaction with the Nδ1 atom of H433, however, the main-chain amino group of D434 contributes a hydrogen bond to its carbonyl group. A salt link is also observed between K8 and the residue D432.
The backbone amide and carbonyl groups of G9 are hydrogen bonded to the carbonyl group of D432 of the molecule A1, and the main-chain amide group of D434 of the molecule A2, respectively (Fig. 2C). Moreover, it also electrostatically interacts with two water molecules. The flexible residues G11 and G13 make electrostatic interactions with Nδ1 of H433 and Oδ1 of N429 in the molecule A2 (Fig. 2D). The mode of side-chain interaction of K12ac is well conserved with that of K5ac. The last amino acid, A15, of the H4 tail peptide is hydrogen bonded to the main-chain nitrogen atom of P430.
3.4. Interaction of the second di-acetylated H4 peptide chain R with BRD2-BD2
The electron density for the H4 peptide chain R was also clearly visible, except for the first two residues, S1 and G2, and for the side-chain of R3 (Fig. S2B). This H4 peptide interacts with B2 of A2B2 and B3 of the symmetry-related A3B3 (A1′B1′) molecules (Fig. 2E). G4 hydrophobically interacts with Y428 and P430 of the B2 molecule. The main-chain nitrogen atom of K5ac makes a hydrogen bond interaction with the side-chain atom, Oδ1 of N429. The interaction for K5ac side-chain atoms with the BD2 protein are well maintained as found in the peptide chain Q. The next residue, G6, forms a hydrogen bond to Nε2 of H433. In addition to contributing the electrostatic interaction with water molecule, K8 also makes hydrogen bonds with the residues G382 and H384 in the molecule A3. The carbonyl group of K12ac makes contact with H433. Furthermore, G13 and G14 sit in the hydrophobic pocket created by the residues, W370 and Y373.
Interestingly, the peptide chain Q has more interactions with BRD2-BD2 residues than the interactions observed in the peptide chain R. In the complex with the peptide Q, the axis of the helical bundle of the bromodomain A2 is approximately 120° with respect to that of the bromodomain A1, and the arrangement of orientation makes in such a way that the peptide-binding sites of the bromodomain A1 and A2 are close together. Whereas in the peptide R complex case, the binding sites of the bromodomain B2 and B3 (symmetry molecule, B1′) are relatively wide apart as well as the helical bundle axes are nearly perpendicular to each other. When measured the distances between Cα of K5ac and K12ac, we observed the corresponding distances of 16.27 and 19.41 Å in the chains Q and R, respectively. In both the Q and R chains, the backbone torsion angles (Φ, ψ) at K5ac and K12ac are approximately (−110°, 5°) and (−90°, 120°), respectively, which correspond to the β-strand region of the Ramachandran plot. The two peptides possess non-specific flexible secondary structures which appeared mainly due to the presence of many glycine residues in the peptide, particularly, the peptide R maintain nearly fully extended conformation compared to the chain Q (Fig. S4). The extensive interactions with the K12/K5-di-acetylated histone H4 tail, as observed in the present study, may help BRD2 associate with mitotic chromosomes [7,10].
3.5. Binding analysis of BRD2-BD2 and acetylated histone tail peptides
The N429A (N382A in the reference 17) mutation reportedly reduces the binding affinity with the K12-acetylated H4 peptide, and L381A (L334A) and L383A (L336A) mutants also moderately affect the binding efficiency with the acetylated H4 peptide in NMR titration studies [17]. From our present study, it is clear that N429 interacts with K12- and K5-acetylated H4 tail peptide. Besides, the region from L381-H384 clasps the acetylated H4 tail peptide on one side of the peptide-binding cleft region (Fig. 2A and B). The corresponding bromodomain in BRD4, BRD4-BD2, also recognizes the H4 tail di-acetylated at K5 and K12 [10,18]. Remarkably, the highly conserved residues D385, Y429, N429, D432 and H433 of BRD2-BD2, which are directly interacting with the H4 peptide chain, markedly affect the binding efficiency for the H4K5ac/K12ac peptide in solution [17].
We examined interaction between BRD2-BD2 and histone H4 tail peptide in solution by surface plasmon resonance (SPR) analysis. BRD2-BD2 bound to K5-, K8-, or K12-acetylated H4 tail peptide at almost similar affinities (Fig. 3A). The KD values of the SPR interactions, which were estimated from the range of the protein concentration from 0 to 320 μM, are approximately 180, 280, and 300 μM to the K5-, K8-, and K12-acetylated H4 tail peptides, respectively. On the contrary, the BRD2-BD2 did not bind to the K16-acetylated H4 tail peptide (Fig. 3A). Huang et al. reported that BRD2-BD2 binds H4K12ac with a KD of 2.9 mM in their NMR titration analysis [17], which is much weaker than our observed data. This discrepancy is presumably due to the difference of the methods analyzed in both studies. When we examined the interaction between BRD2-BD2 and the K5/K12-di-acetylated H4 tail peptide, we observed almost as twice-efficient binding as compared to the mono-acetylated H4 tails (Fig. 3A). The KD value of this interaction to the di-acetylated H4 peptide was estimated to be approximately 140 μM, suggesting that two molecules of BRD2-BD2 may simultaneously bind to one molecule of the immobilized K5/K12-di-acetylated H4 tail peptide, with least cooperativity. These results reveal that BRD2-BD2 binds to the histone H4 tail through the acetylated lysine at K5, K8 or K12, but not through K16. As the SPR analysis suggests BRD2-BD2 also recognizes H4K8ac in unknown manner, further structural studies will be required to understand how BRD2-BD2 recognizes H4K8ac. We also examined interaction between BRD2-BD2 and histone H3 tail peptides. Intriguingly, BRD2-BD2 substantially bound to the K18-acetylated histone H3 tail peptide (residues 10–27), while it did not bind to the H3K23ac peptide (Fig. 3B). The estimated KD value of 180 μM observed for the BRD2-BD2 – H3K18ac interaction is similar to the value of 251 μM observed for the binding of BRDT-BD2 to H3K18ac by isothermal titration calorimetry [14]. Taken together, these results indicate the preferential binding of BRD2-BD2 to the K18-acetylated H3 tail as well as to the K5/8/12-acetylated H4 tails.
Fig. 3.
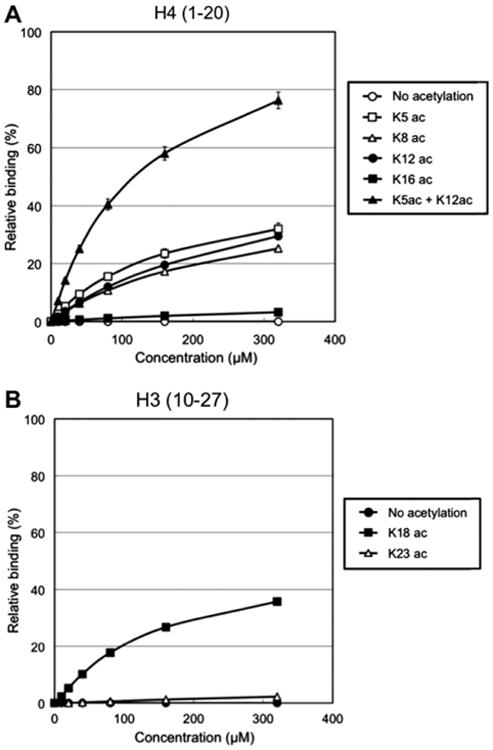
Surface plasmon resonance binding analysis of BRD2-BD2 and the acetylated histone tails. (A) Binding analysis between BRD2-BD2 and histone H4 tails. Dosedependent binding results ranging from 0 to 320 μM of the protein concentration were shown. The binding activity to the BRD2-BD2 protein was compared between the hypoacetylated (open circle), K5-acetylated (open square), K8-acetylated (open triangle), K12-acetylated (filled circle), K16-acetylated (filled square), and K5/K12-di-acetylated (filled triangle) histone H4 tail peptides (residues 1–20). Binding score was evaluated regarding the theoretical maximal resonance unit (Rmax) of the each peptide as 100%. (B) Binding analysis between BRD2-BD2 and histone H3 tails. The binding activity to the BRD2-BD2 protein was compared between the hypoacetylated (filled circle), K18-acetylated (filled square), and K23-acetylated (open triangle) histone H3 tail peptides (residues 10–27).
3.6. Structural comparison with BRDT-BD2
Recently, the crystal structures of BD1 and BD2 of mouse BRDT (mBRDT) were reported [14]. The overall structure of BRDT-BD2 is almost identical to that of BRD2-BD2. Superposition of the molecule A of BRDT-BD2 onto the molecule A of BRD2-BD2 gave r.m.s. deviation of 0.5 Å for all Cα atoms (Fig. 4A). Intriguingly, the oligomer formation in BRDT-BD2 is identical to that found in the BRD2-BD2 structure (Fig. 4A). BRDT-BD2 recognizes histone H3 tail acetylated at K18 (H3K18ac) [14]. We observed that the amino acids and structures in the binding sites of both BD2 bromodomains are almost conserved and identical (Fig. 4B and S5), implying that BRDT-BD2 may also recognize K5ac and/or K12ac of the histone H4 tail. Conversely, BRD2-BD2 recognized H3K18ac as demonstrated in Fig. 3B. When we compared H3K18ac and H4K5ac/K12ac peptides in the structures of respective complexes, R17 of histone H3 and R3 of histone H4 are located at equivalent positions pointing towards the conserved residues, D354 and H355 (in mBRDT-BD2), and D432 and H433 (in BRD2-BD2), respectively. Hence the two residues, H3-R17 and H4-R3, may play critical roles for specific recognition of the acetylated histone tails by BRDT-BD2 and BRD2-BD2.
Fig. 4.
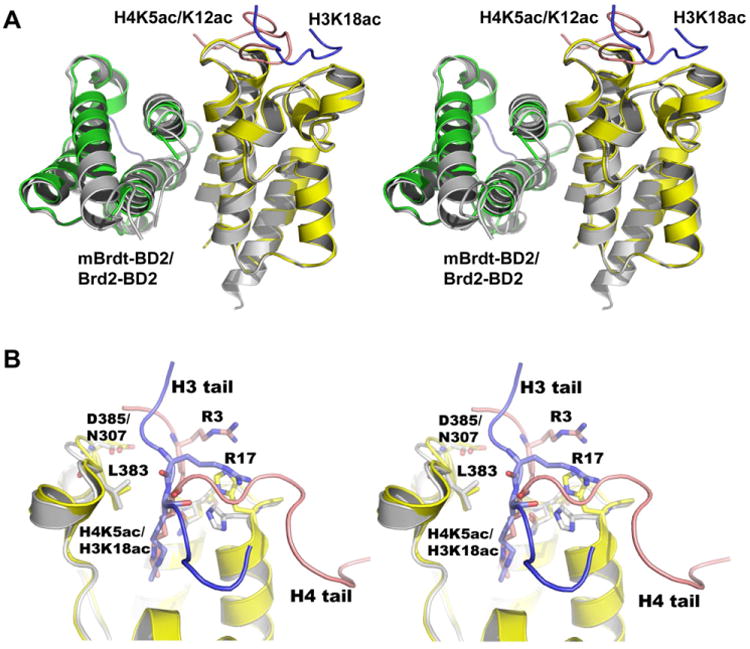
Comparison between BRD2-BD2 and mBRDT-BD2 complexes. (A) Stereo-view of the superposition of the BRD2-BD2 chains (colored in green and yellow) on to the mBRDT-BD2 chains (colored in gray) over Cα-atoms corresponding to the equivalent structured regions. The H3 and H4 tail peptides are colored in blue and orange, respectively. (B) A close-up stereo-view near the acetyl-lysine binding regions. The residues, Arg3 of histone H4 and Arg17 of histone H3 (shown by sticks) are located at equivalent position in the respective complexes. The conserved residues of the respective BD2 molecules, responsible for the peptide interactions are shown by sticks.
3.7. Structural comparison with other bromodomains
The left-handed four-helical domain of the BRD2-BD2 protein is conserved in the bromodomain family [25-30]. The overall structures of these bromodomains are similar; however, the acetylated histone H4 tail-binding site possesses major structural variations, particularly in the ZA and BC loop regions (data not shown). Recently we reported the structural basis of BRD2-BD1 for the recognition of K12-acetylated H4 tail [16]. When we superposed the BRD2-BD2 complex with the BRD2-BD1 complex for main-chain atoms of the bromodomains, the overall structures of the BD1 and BD2 are nearly similar to each other (average r.m.s. deviation value of ∼1.0 Å). However, an intriguing feature is observed when analyzed the peptide-binding regions of BD1 and BD2 by superimposing the histone H4 tail peptides with respect to their acetyl-lysines. In the case of BD2 recognition for K5ac, the peptide runs from N- to C-terminal direction with respect to the ZA loop of the bromodomain in one monomer (Fig. S6A) whereas it runs in the opposite direction for K12ac interaction with respect to ZA loop in another monomer (Fig. S6B). It suggests that BRD2-BD2 recognizes the peptide direction depending upon the interaction of K5ac and K12ac. It is interesting to note that the peptide direction in the BD1 complex is opposite to that found in the BD2 complex, with respect to K12ac position (Fig. S6B), suggesting that, for the K12ac binding, the BD1 and BD2 bromodomains recognize the H4 peptide in different manner.
Sequence comparison between BRD2-BD1 and BRD2-BD2 shows approximately 44% of sequence identity between them (Fig. S5). The BD2 bromodomain interacts with the H4 peptide through the residues from the ZA and BC loops, but in the BD1 complex, the interaction occurred through the residues mainly from the BC loop. The residues in the vicinity of the acetyl-lysine binding region are highly conserved; however, some non-conserved residues are observed in the peptide-binding regions, for example, H433/D160, P430/K157, and H384/P111 (Fig. S5). Thus, significant numbers of non-conserved residues which are responsible for the peptide interaction are the key components for discriminating acetylated H4 tail recognition between BD1 and BD2 in different manner.
Sequence comparisons from BRD2 and BRD4 indicate that the corresponding bromodomains (i.e. BRD2-BD1 vs BRD4-BD1; BRD2-BD2 vs BRD4-BD2) are highly homologous (75% sequence identity) as compared to the non-equivalent bromodomains, BD1 vs BD2 (Fig. S5). The H4 tail-interacting residues are highly conserved between the equivalent bromodomains, suggesting that the structures of BRD4-BD1 and BRD4-BD2 are similar to that of BRD2-BD1 and BRD2-BD2, respectively.
Our present study and previous reports [15,16] indicated that the BRD2-BD1 and -BD2 bromodomains both recognize K12ac and K5ac of histone H4. However, further structural and biophysical studies on how the tandem BET bromodomains, BD1 and BD2, simultaneously recognize multi-acetylated histone tails will aid understanding of the mechanism for deciphering and protecting the acetylation histone code on the chromatin.
Supplementary Material
Acknowledgments
We thank Drs. M. K. Jang, K. Nakano and C. Shang for initial experiments of this study, S. Wakatsuki and N. Igarashi (AR-NW12A, Photon Factory, KEK) for help in data collection, and T. Nakayama and A. Ishii for clerical assistance. This work was supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO) of Japan to T.U., by Grants-in-Aid for Young Scientists from Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to T.U. (Nos. 19790083 and 22790101), and by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses of MEXT to S.Y.
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2010.08.013.
Contributor Information
Shigeyuki Yokoyama, Email: yokoyama@biochem.s.u-tokyo.ac.jp.
Balasundaram Padmanabhan, Email: bpadmanabhan@hotmail.com.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 4.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 5.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 6.Lin YJ, Umehara T, Inoue M, Saito K, Kigawa T, Jang MK, Ozato K, Yokoyama S, Padmanabhan B, Güntert P. Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 2008;17:2174–2179. doi: 10.1110/ps.037580.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci USA. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 12.Platt GM, Simpson GR, Mittnacht S, Schulz TF. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 14.Morinière J, Rousseaux S, Steuerwald U, Soler-López M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Müller CW, Petosa C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, Uda-Tochio H, Hamana H, Terada T, Adachi N, Matsumoto T, Tanaka A, Horikoshi M, Ozato K, Padmanabhan B, Yokoyama S. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J Biol Chem. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- 16.Umehara T, Nakamura Y, Jang MK, Nakano K, Tanaka A, Ozato K, Padmanabhan B, Yokoyama S. Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J Biol Chem. 2010;285:7610–7618. doi: 10.1074/jbc.M109.062422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H, Zhang J, Shen W, Wang X, Wu J, Wu J, Shi Y. Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails. BMC Struct Biol. 2007;7:57. doi: 10.1186/1472-6807-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Wang X, Zhang J, Huang H, Ding B, Wu J, Shi Y. Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails. Biochemistry. 2008;47:6403–6417. doi: 10.1021/bi8001659. [DOI] [PubMed] [Google Scholar]
- 19.Umehara T, Wakamori M, Tanaka A, Padmanabhan B, Yokoyama S. Purification, crystallization and preliminary X-ray diffraction of the C-terminal bromodomain from human BRD2. Acta Crystallogr. 2007;F63:613–615. doi: 10.1107/S1744309107028473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Jones TA, Zou JY, Cowan SW, Kijeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;D47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski R, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallgr. 1993;26:283–291. [Google Scholar]
- 25.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 26.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- 27.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 29.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 30.Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


