Abstract
Macrophages, when activated by IFN-γ and TLR signaling, elicit innate immune responses. IFN regulatory factor 8 (IRF8) is a transcription factor that facilitates macrophage activation and innate immunity. We show that, in resting macrophages, some IRF8 is conjugated to small ubiquitin-like modifiers (SUMO) 2/3 through the lysine residue 310. SUMO3-conjugated IRF8 failed to induce IL12p40 and other IRF8 target genes, consistent with SUMO-mediated transcriptional repression reported for other transcription factors. SUMO3-conjugated IRF8 showed reduced mobility in live nuclei and bound poorly to the IL12p40 gene. However, macrophage activation caused a sharp reduction in the amount of SUMOylated IRF8. This reduction coincided with the induction of a deSUMOylating enzyme, sentrin-specific peptidase 1 (SENP1), in activated macrophages. In transfection analysis, SENP1 removed SUMO3 from IRF8 and enhanced expression of IL12p40 and other target genes. Conversely, SENP1 knockdown repressed IRF8 target gene expression. In parallel with IRF8 deSUMOylation, macrophage activation led to the induction of proteins active in the SUMO pathway and caused a global shift in nuclear protein SUMOylation patterns. Together, the IRF8 SUMO conjugation/deconjugation switch is part of a larger transition in SUMO modifications that takes place upon macrophage activation, serving as a mechanism to trigger innate immune responses.
Small ubiquitin-like modifiers (SUMO) are composed of ∼100 aa (∼12 kDa) and covalently conjugated to the lysine (K) residues of substrate proteins through three-step enzymatic reactions (1). Among three SUMO molecules present in vertebrates, SUMO2 and SUMO3 share ∼95% amino acid identity, whereas SUMO1 is only ∼50% identical with SUMO2/3. SUMO molecules are conjugated to substrate proteins carrying a consensus motif ψKXE and related sequences (2). SUMO conjugation is reversible, in that the SUMO moiety can be removed by SUMO-deconjugating enzymes of the sentrin-specific peptidase (SENP) family (3, 4). SUMO conjugation and deconjugation are a process conserved throughout eukaryotes and are involved in diverse regulatory activities. Many transcription factors are SUMOylated, which in general results in the loss of transactivation function. As an interesting feature of SUMO modification, it has been noted that a relatively small fraction of total proteins is SUMOylated at any time point, yet causing profound transcriptional repression (5). Partially explaining this enigma, SUMO-mediated transcriptional repression is often attributed to the recruitment of histone deacetylases, repressive histone methyltransferases, and corepressors, presumably leading to the formation of repressive chromatin (6, 7).
SUMO pathways are activated by a variety of stress, including ionizing radiation, heat shock, and oxidative stress, which causes global alterations in SUMOylated proteins (8–11). SUMOylation is influenced by other signaling events such as ubiquitination and phosphorylation (2). Innate immune responses against infectious pathogens involve SUMOylation. We previously reported that IFN regulatory factor (IRF) 3 and IRF7, members of the IRF family, are SUMOylated following viral infection (12, 13). SUMOylated IRF3 and IRF7 repress type I IFN transcription, which contributes to the attenuation of excess IFN responses. Other IRF members, including IRF1, are also SUMOylated by E3 ligases of the protein inhibitor of activated STAT (PIAS) family, resulting in inhibition of IRF1 transactivation (14, 15). Additionally, TLR pathway activation triggers SUMOylation of TRAF family member-associated NF-κB activator to regulate type I IFN induction (16). Furthermore, PIAS family proteins regulate NF-κB activation and alter the function of natural regulatory T cells (17, 18). Supporting a link between SUMO modification and host defense, we previously showed that the Ebola virus anti-IFN protein VP35 inhibits type I IFN transcription by prematurely SUMOylating IRF7 (19).
IRF8 is another member of the IRF family expressed in macrophages and dendritic cells (DCs) (20). IRF8 directs differentiation of myeloid progenitor cells to functional macrophages (21). Similarly, IRF8 stimulates development of DC subsets and controls production of IL12p40 and type I IFNs (20, 22). IRF8 activates other factors, including chemokines, transcription factors, and those involved in many antipathogenic activities (23–26). IRF8 also enhances CIITA transcription, thereby increasing MHCII expression and Ag presentation (27, 28). Supporting the critical importance of IRF8 in host defense, point mutations in IRF8 are associated with severe immunodeficiency and increased susceptibility to infectious pathogens in humans and mice (22, 29, 30). Various posttranslational modifications influence the activity of IRF8. For example, phosphorylation and dephosphorylation alter IRF8's functions, affecting innate immune responses and leukemia pathogenesis (26, 31). IRF8 is conjugated to ubiquitin through an E3 ligase TRIM21 that alters IRF8's ability to regulate IL12p40 transcription positively and negatively (32, 33).
This study began with our observation that IRF8 was SUMOylated in resting macrophages. We show in this study that the lysine (K) at 310 is a major SUMOylation site in IRF8, and SUMO conjugation abrogates IRF8's ability to stimulate transcription of its target genes, including IL12p40 and IFN-β. A defining observation was that the levels of SUMOylated IRF8 decreased upon macrophage activation triggered by IFN-γ/TLR stimulation. The change in IRF8 SUMOylation coincided with a global shift in the nuclear protein SUMOylation and the induction of enzymes that regulate SUMO pathways. Among multiple SUMO-deconjugating enzymes, SENP1 was induced most robustly in macrophages after stimulation and removed SUMO from IRF8, leading to the reversal of SUMO-mediated repression of target gene transcription. Our results are consistent with the paradigm in which deSUMOylation causes derepression of key genes to allow progression of cellular maturation, examples of which have been reported for monocytes, erythroid cells, and keratinocytes (34–36).
Materials and Methods
Plasmids and vectors
pcDNA3.1 vectors (Invitrogen) containing mouse IRF8 cDNA or IRF8 tagged to GFP (GFP-IRF8), CFP-IRF8, or Flag-IRF8 and those for T7- or V5-tagged SUMO1, SUMO2, SUMO3, and SUMO3GA were described (19, 32, 33). pcDNA3.1 vector for mutant Flag-IRF8 was constructed by site-directed mutagenesis. SUMO3 fusions of IRF8 are described in Supplemental Fig. 2. pcDNA 3.1 vector for a murine full-length Senp1 with or without hemagglutinin (HA) tag was constructed using pCMV-SPORT6-mSENP1 (Invitrogen). The catalytic mutant C599S was constructed by site-directed mutagenesis from the above clone (37). The plasmids for GFP-SUMO3, YFP-SUMO3, and YFP-SUMO3GA were constructed from pEGFP-C1 or pEYFP-C1 vector by inserting PCR-amplified SUMO fragments into the XhoI and HindIII sites of the vector. Senp1 short hairpin RNA (shRNA) vector was constructed by inserting the following oligomers, 5′-gatccccTTCCACTCCAGCGTCAGGC ttcaagagaGCCTGACGCTGGA-GTGGAAttttta-3′, 5′-agcttaaaaaTTCCACTCCAGCGTCAGGCtctcttgaa-3′, and 5′-GCCTGACGCTGGAGTGGAAggg-3′ (si299 sequence) into pSuper-Puro vector (Oligoengine). Retroviral pMSCV-puro vectors for wild-type (wt) IRF8 and GFP-IRF8 were described, and those for GFP-SUMO3, SENP1, and the C599S mutant were constructed by standard cloning procedures (22, 38) (Clontech).
Cells, coimmunoprecipitation, and SUMOylation assays
RAW 264.7 (RAW), NIH3T3, and 293T cells (American Type Culture Collection) were maintained in DMEM with 10% FBS or 10% donor serum (for NIH3T3). IRF8−/− macrophage cell line, CL2 cells were cultured in RPMI 1640 and 10% FBS plus rM-CSF (5 ng/ml) (32). Bone marrow (BM)-derived macrophages were obtained by culturing BM mononuclear cells in the presence of 20 ng/ml M-CSF (Invitrogen) for 5–6 d. BM-derived IRF8−/− DCs were cultured in the presence of Flt3L for ∼4 wk. These cells exhibited a DC progenitor-like property and differentiated into DCs after IRF8 transduction. Detailed procedures and the properties of these DCs will be presented elsewhere (P. Tailor, unpublished observations). Procedures for quantitative RT-PCR (qRT-PCR), in vivo SUMOylation, and coimmunoprecipitation assays were performed, as described (19).
Fluorescence resonance energy transfer and fluorescence recovery after photobleaching
Fluorescence resonance energy transfer (FRET) analyses were performed with NIH3T3 cells transfected with CFP-IRF8 and/or YFP-SUMO3 or YFP-GA for 30 h on a Zeiss microscope Axiovert 200 with the Axiovision FRET software (39). Fluorescence recovery after photobleaching (FRAP) assays were performed with NIH3T3 cells transduced with pMSCV vector for GFP-IRF8 or GFP-IRF8 fused to SUMO1 or SUMO3, as described (38). Detailed procedures are in Supplemental Fig. 3.
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) analysis was performed essentially as described (19). Briefly, CL2 cells expressing GFP-SUMO3, GFP-IRF8, or GFP-IRF8-SUMO3 from the pMSCV vector were cross-linked with 1% formaldehyde for 10 min and sonicated in Misonix Sonicator 3000. Chromatin preparations corresponding to 0.2 × 106 cells were incubated with rabbit anti-GFP Ab (Abcam) or control rabbit IgG bound to Dyna-beads protein G (Invitrogen) overnight at 4°C. Immunoprecipitated chromatin was decross-linked, and purified DNA was analyzed by quantitative PCR using appropriate primers. The percentage input was calculated as 2−[cycle threshold (CT) IP sample − CT input].
Results
IRF8 is conjugated to SUMO2/3 in resting macrophages
To determine whether IRF8 is SUMOylated in macrophages, immunoprecipitation analysis was performed with nuclear extracts from unstimulated RAW264.7 (hereafter RAW) cells or RAW cells stimulated with IFN-γ or IFN-γ plus CpG (IFN-γ/CpG). Materials precipitated with anti-IRF8 Ab were blotted with anti-SUMO2/3 Ab. As shown in Fig. 1A (left panels), in unstimulated cells, a fraction of IRF8 consistently exhibited SUMO2/3 reactivity, which migrated more slowly than the rest of IRF8 (∼62 kDa versus ∼51 kDa), indicating conjugation of SUMO2/3 to IRF8. However, the levels of SUMO 2/3-linked IRF8 fell noticeably when cells were stimulated with IFN-γ or IFN-γ/CpG. The total IRF8 levels, however, increased after stimulation, as expected (32, 33). The detailed time course analysis in Fig. 1A (right panels) showed very similar results, in that IRF8 was SUMOylated before stimulation, which was attenuated upon macrophage activation. Those data indicate that IRF8 is modified by SUMO2/3 in resting macrophages, but macrophage activation prompts a reduction in SUMOylated IRF8. We did not detect conjugation of SUMO1 to IRF8, possibly suggesting that IRF8 may be predominantly modified by SUMO2 or SUMO3 in RAW cells under these conditions.
Figure 1.
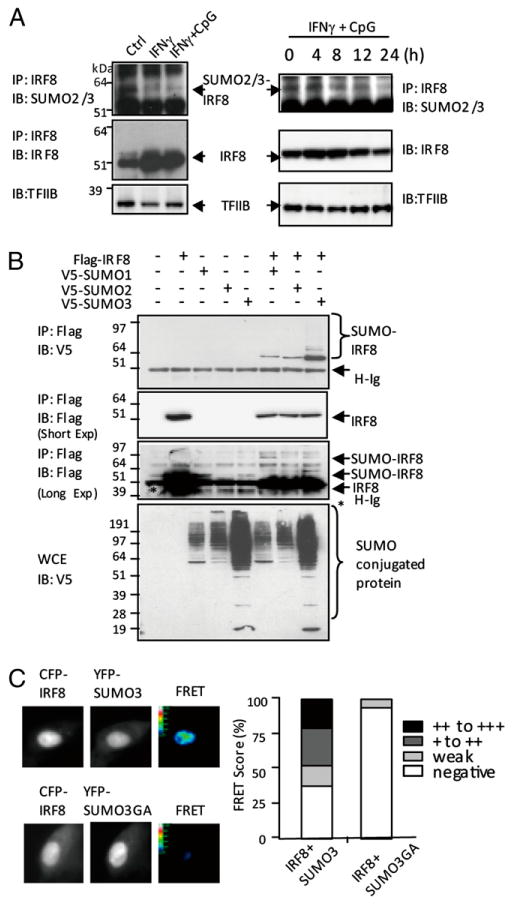
IRF8 SUMOylation status in resting and activated macrophages. (A) Left panels, Nuclear extracts (500 μg) from unstimulated RAW cells or stimulated with IFN-γ (150 U/ml) for 8 h or IFN-γ overnight, followed by CpG (150 ng/ml) for 6 h, were immunoprecipitated with Ab for IRF8 and blotted against SUMO2/3 or IRF8. Extracts were blotted for TFIIB for loading control. Right panels, Unstimulated RAW cells (0) or stimulated with IFN-γ overnight, followed by CpG for indicated times and nuclear extracts, were tested, as above. TFIIB was tested as a loading control. (B) 293T cells (1 × 106) were transfected with Flag-IRF8 (1 μg), V5-SUMO1, SUMO2, or SUMO3 for 30 h, and extracts were precipitated with anti-Flag Ab and blotted with anti-V5 or anti-Flag Ab. The lower middle panel depicts results of a longer exposure revealing multiple SUMOylated IRF8 bands. The bottom panel indicates immunoblot of WCE with anti-V5 Ab. (C) NIH3T3 cells (1 × 106) were transfected with 1 μg CFP-IRF8 and YFP-SUMO3 or SUMO3GA for 30 h and tested for a real-time interaction of IRF8 with SUMO3 by FRET analysis. Only SUMO3, not SUMO3GA, gave FRET signals (upper panel). In the lower panel, FRET signals were quantified by scoring fluorescent intensity of ∼200 cells, according to the modified Youvan method.
To confirm that IRF8 is a SUMO substrate, immunoprecipitation experiments were carried out in 293T cells transfected with Flag-tagged IRF8 (Flag-IRF8) and V5-tagged SUMO1, SUMO2, or SUMO3. Data in Fig. 1B showed that all three SUMO proteins are conjugated to IRF8 (upper panel). A single, SUMO-conjugated IRF8 band was predominant after a short exposure (upper middle panel). However, additional, multiple SUMO-conjugated bands were visible upon a longer exposure (lower middle panel), suggesting that polymeric SUMO chains were formed on IRF8, although a covalent linkage with other proteins may also have occurred (40). In these assays, SUMO3 consistently gave more robust IRF8 modification than SUMO1 and SUMO2, the basis of which has not been studied further. As expected, immunoblot of whole-cell extracts (WCE) revealed that proteins other than IRF8 were also conjugated to V5-SUMOs (lower panel). Based on these data and the structural similarity between SUMO2 and SUMO3, we tested mostly the activity of SUMO3 in the current study.
FRET analysis detects transfer of fluorescent energy from a donor to an acceptor that takes place in two proteins of close physical proximity. This assay has been used to study SUMO conjugation events in vitro and in vivo (41). To further solidify IRF8 SUMO conjugation in vivo, NIH3T3 cells were transfected with CFP-IRF8 and YFP-SUMO3, or YFP-mutant SUMO3 (SUMO3GA), in which the C-terminal amino acids GG were mutated to AA, eliminating the SUMOylation activity (42). In microscopic inspection in Fig. 1C (left panel), we detected FRET signals when CFP-IRF8 was coexpressed with YFP-SUMO3, but not YFP-SUMO3GA. We quantified the microscopic observations by ranking FRET signals into four levels, as recommended (39). More than 200 CFP- and YFP-positive cells were ranked (Fig. 1C, right panel). About 50% of cells showed FRET signals with varying scores. In contrast, nearly 90% cells expressing YFP-SUMO3GA were FRET negative. These results further support a close physical link between IRF8 and SUMO3.
Identification of K310 as a major IRF8 SUMO conjugation site
IRF8 has several amino acid motifs that conform to or resemble the consensus SUMO site, ΨKXE. To identify a functional SUMO conjugation site in IRF8, the K residues in the motifs were mutated to arginine (R), and the resultant mutants were tested for SUMO1 or SUMO3 conjugation. Results in Fig. 2 showed that mutation of K310 abolished conjugation of both SUMO1 and SUMO3, whereas mutation of K72 did not. Immunostaining analysis showed that both wt IRF8 and the IRF8 K310R mutant localized to the nucleus along with SUMO3, confirming that the absence of SUMO conjugation was not due to dysregulated localization of the mutant (Supplemental Fig. 1A). We also tested additional four K residues that may serve as SUMO conjugation sites as assessed by a National Center for Biotechnology Information database. Mutation of these sites had no effect on IRF8 SUMO conjugation (Supplemental Fig. 1B, summary in Supplemental Fig. 1C). These data show that K310 is a single, main SUMO conjugation site in IRF8.
Figure 2.
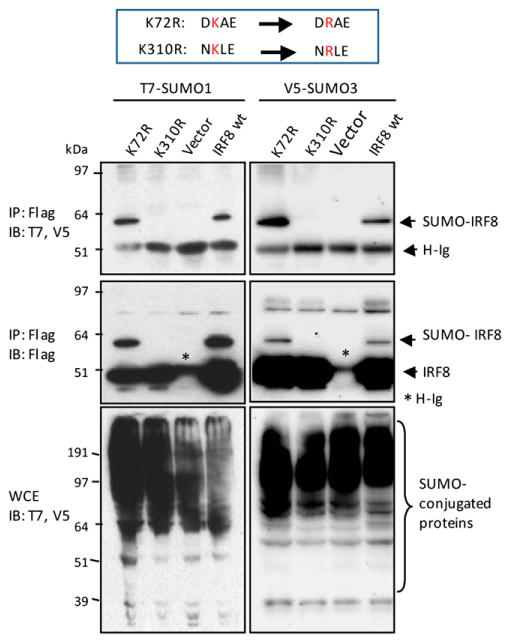
Identification of a SUMOylation site in IRF8. The 293T cells transfected with Flag-IRF8 mutants (K72R or K310R) and T7-SUMO1 or V5-SUMO3 were immunoprecipitated with anti-Flag Ab and blotted with anti-T7 and anti-V5 Ab (upper middle panel) or anti-Flag Ab (lower middle panel). Vector alone and wt Flag-IRF8 (wt) were tested as controls. The bottom panel indicates immunoblot analysis of WCE.
SUMO3 conjugation inhibits IRF8 target gene transactivation
To assess the functional consequence of IRF8 SUMOylation, we tested wt IRF8, the K310R mutant, and two additional IRF8-SUMO conjugate constructs for the ability to stimulate Il12b (IL12p40) and Ifnb (IFN-β) promoters. For the IRF8-SUMO conjugates, SUMO3 was covalently conjugated to either the N terminus or C terminus of IRF8 (SUMO3-IRF8 or IRF8-SUMO3, respectively; see diagram in Supplemental Fig. 1D). When transfected in 293T cells, IRF8-SUMO3 retained the SUMO moiety and migrated more slowly than IRF8, but SUMO3-IRF8 was reverted to the size of unconjugated IRF8 within 30 h, presumably due to the cleavage of SUMO from IRF8, indicating that the former was more stable than the latter (see immunoblot data in Supplemental Fig. 1E). Molecularly constructed SUMO conjugates such as these have been widely used for studying the functional significance of SUMOylation, mostly because of improved stability (43). In luciferase assays performed in RAW cells, wt IRF8 enhanced activities of both reporters by 2- to 5-fold, with a further increase after IFN-γ/CpG stimulation (Fig. 3A). K310R gave modest, but consistently higher luciferase activities (25–50% increase) in both reporters. The SUMO3-IRF8 conjugate, from which SUMO3 was removed during culture, gave reporter activity comparable to wt IRF8. In contrast, the stable SUMO conjugate, IRF8-SUMO3, did not stimulate either reporter. Next, these IRF8 constructs were stably expressed in IRF8−/− macrophages (named CL2) and tested for mRNA expression of Il12b and Ccl9, IRF8 targets, that are important for innate immune responses (Fig. 3B) (19, 32, 33). Wt IRF8 enhanced mRNA expression of both genes before and after IFN-γ/CpG stimulation. The K310R mutant further increased the expression, although moderately, suggesting that SUMOylation represses IRF8 transactivation. Supporting this view, K310R increased expression of additional IRF8 targets, Pml and Ciita, over wt IRF8 by 40–60% (Supplemental Fig. 1F). IRF8-SUMO3, in contrast, failed to enhance expression of these genes. IRF8-SUMO3 repression of gene expression was not due to a nonspecific effect, because this construct did not repress induction of TNF-α by IFN-γ/TLR (Supplemental Fig. 1G). Similarly, when stably expressed in IRF8−/− DCs, wt IRF8 enhanced expression of Il12b and Ifnb mRNA after CpG stimulation, but IRF8-SUMO3 did not (Fig. 3C) (22). Furthermore, wt IRF8, but not IRF8-SUMO3, stimulated surface expression of MHC II on DCs upon CpG stimulation (Fig. 3D). When tested in RAW cells that express endogenous IRF8, IRF8-SUMO3 repressed IFN-γ/CpG induction of Il12b and Ifnb, indicating that IRF8-SUMO3 repressed endogenous IRF8 as well (Supplemental Fig. 1H). To study whether IRF8-SUMO3 negatively affects activity of endogenous IRF8, these results indicate that SUMO3 conjugation abrogates IRF8's capacity to stimulate target gene expression.
Figure 3.
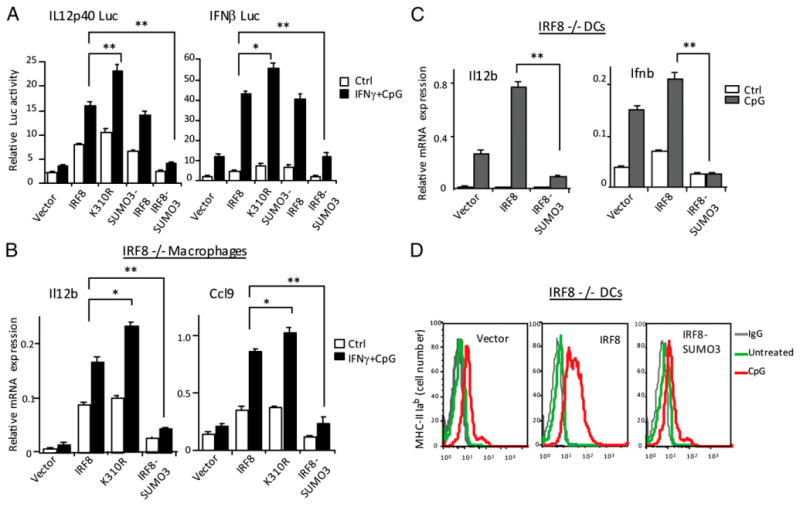
SUMO3 conjugation represses transactivation of IRF8 target genes. (A) RAW cells (1 × 106) were transfected with 600 ng indicated luciferase reporters, 800 ng pcDNA-IRF8 vectors, and 60 ng pRL-TK vector for 30 h and stimulated with IFN-γ overnight and CpG for 6 h. Reporter activity was normalized by Renilla luciferase activity. Values represent the average of three assays ± SD. Statistical significance was tested by Student t test; *p < 0.01, **p < 0.005. (B) IRF8−/− macrophages (CL2) were transduced with pMSCV vectors for wt IRF8, K310R mutant or SUMO3-IRF8, or IRF8-SUMO3 for 5 d and stimulated with (filled bars) or without (open bars) IFN-γ and CpG. Levels of Il12b and Ccl9 transcripts were measured by qRT-PCR. Values represent the average of three determinations ± SD. Statistical significance was tested by Student t test; *p < 0.01, **p < 0.005. (C) IRF8−/− DCs were transduced with the above vectors for 5 d and stimulated with (filled bars) or without (open bars) CpG for 7 h and Il12b and Ifnb transcript levels were measured, as above. Statistical significance was tested by Student t test; **p < 0.005. (D) IRF8−/− DCs transduced with indicated vectors were stimulated with or without CpG for 36 h, and expression of MHCII (I-Ab) was detected by flow cytometry. The gray lines indicate signals by control IgG.
SUMO conjugation impairs genome-wide mobility of IRF8
SUMO conjugation alters intranuclear localization of some transcription factors (44, 45). We checked localization of GFP-IRF8 and GFP-IRF8-SUMO3 in the nucleus of living cells, both unconjugated and SUMO3-conjugated IRF8 distributed evenly in the nucleus, indicating that SUMO3 conjugation did not alter the uniform distribution pattern reported for IRF8 (32) (Fig. 4A). GFP-SUMO3, not conjugated to IRF8, tested as a control also localized mostly in the nucleus, whereas free GFP distributed both in the cytoplasm and the nucleus. We then asked whether SUMOylation affects an interaction of IRF8 with chromatin by FRAP analysis. Most transcription factors, including IRF8, are highly mobile within the nucleus and bind to chromatin with a rapid, on-and-off mode, which reflects a global, genome-wide chromatin-scanning activity of the transcription factor, which is critical for their function (38, 46). NIH3T3 cells expressing GFP-IRF8 or GFP-IRF8-SUMO3 were briefly photobleached within a small area in the nucleus, and fluorescence recovery was measured for the subsequent 28 s. Cells expressing free GFP or GFP-SUMO3 were also tested as controls. Fig. 4B depicts the recovery profile for each construct obtained from 15 separate cells. Free GFP recovered almost instantly following photobleach (blue). Although more slowly than free GFP, GFP-IRF8 also recovered rapidly, showing a t1/2 of ∼2 s (red) (38). GFP-IRF8 regained original fluorescence intensity fully by 16 s. In contrast, GFP-IRF8-SUMO3 showed a dramatically slower recovery, reclaiming only ∼75% of the original fluorescence even at the end of measurement (black). GFP-SUMO3, in contrast, showed the slowest mobility (green) particularly evident in an early stage, suggesting its tendency to bind to chromatin. In line with our results, SUMO1 was reported to move very slowly in a similar FRAP assay (44). These results indicate that SUMO3 conjugation markedly reduces IRF8's genome-wide mobility. Consistent with these data, we found that conjugation of SUMO1 also dramatically reduced IRF8's real-time mobility (Supplemental Fig. 2B).
Figure 4.
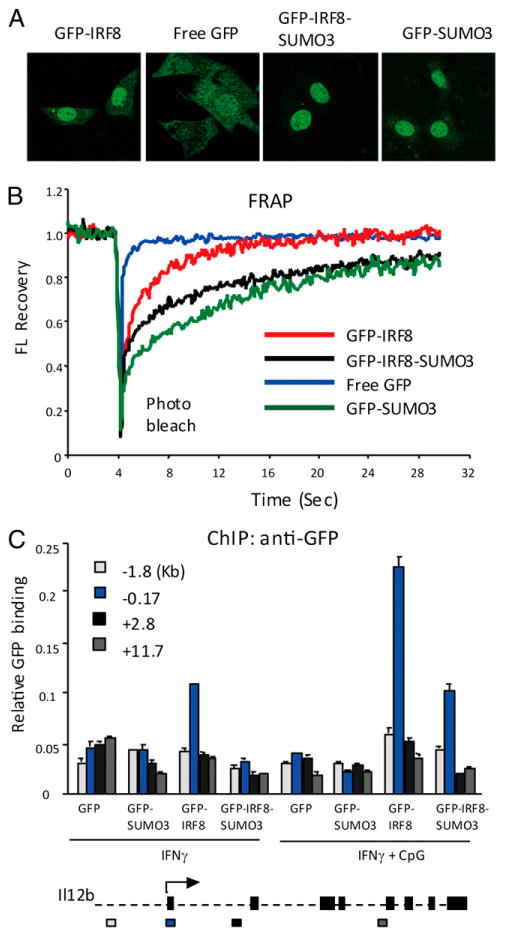
SUMO3 conjugation reduces genome-wide mobility of IRF8 and target-binding activity. (A) NIH3T3 cells were transduced with pMSCV vectors for GFP alone, GFP-SUMO3, wt GFP-IRF8, or GFP-IRF8-SUMO3, and localization of each construct was viewed by live cell imaging. (B) In FRAP analysis, a small region within the nucleus was bleached at indicated time, and the recovery of fluorescence (FL) signals was plotted against time (sec). The recovery curves represent the average of 15 separate cells. See reduced recovery of GFP-IRF8-SUMO1 in Supplemental Fig. 3. (C) CL2 cells were transduced with indicated pMSCV vectors and stimulated with IFN-γ or IFN-γ/CpG. Binding of these constructs to the indicated regions of the Il12b gene was tested by ChIP using anti-GFP Ab. Values represent the average of three determinations ± SD. The bottom diagram indicates the exon-intron organization of Il12b. The arrow marks the TSS.
SUMOylated IRF8 binds poorly to the Il12b promoter
Having observed inhibition of global IRF8 mobility by SUMO conjugation, it was of interest to assess whether SUMO conjugation affects binding of IRF8 to specific target sites. ChIP analysis was performed for the Il12b gene in CL2 macrophages stably expressing GFP-IRF8 or GFP-IRF8-SUMO3. Anti-GFP Ab was used to detect binding of IRF8 to four regions within the Il12b gene, that is, 1.8 kb upstream promoter, the transcription start site (TSS), and two coding regions (see Fig. 4C for the Il12b gene map and the location of the primers used for ChIP in the bottom). After IFN-γ stimulation, GFP-IRF8 bound at/near the TSS, and this binding was further increased upon stimulation with CpG. Binding of GFP-IRF8-SUMO3 to the TSS was lower than that of GFP-IRF8, although consistently above background. Neither GFP-IRF8 nor GFP-IRF8-SUMO3 bound to other regions of Il12b. The TSS-restricted binding was expected, because a target site of IRF8 was identified in the proximal promoter in the gene (22). As expected, free GFP and GFP-SUMO3 did not bind to any of Il12b regions tested, validating specificity of IRF8 binding. These results indicate that SUMO3 conjugation reduces, but does not abolish IRF8's ability to bind to a target site. Consistent with this view, IRF8-SUMO3 bound less efficiently to another target, Cst3, than wt IRF8 (data not shown).
IFN-γ and TLR stimulation triggers a global shift in SUMOylated nuclear proteins in macrophages
We next asked whether macrophage activation changes SUMOylation of proteins other than IRF8. This question was asked, because we previously observed that IFN-γ/TLR stimulation of macrophages causes a large increase in ubiquitinated nuclear proteins (47). To detect SUMO2/3-conjugated nuclear proteins, extracts from unstimulated or stimulated RAW cells were blotted with anti-SUMO2/3 Ab (Fig. 5A). SUMO2/3-conjugated proteins were markedly increased after stimulation by IFN-γ/CpG, IFN-γ/LPS, and Newcastle disease virus. The increase was particularly noticeable at 14–24 h. The levels of transcription factor II B (TFIIB), tested as a loading control, remained unchanged. Stimulation by CpG or LPS alone led to only a meager increase in SUMOylated proteins, whereas IFN-γ stimulation alone resulted in a more substantive increase, in line with the observations on ubiquitinated proteins (Supplemental Fig. 3A) (47). A similar, but less dramatic increase was detected for SUMO1-conjugated nuclear proteins after IFN-γ/TLR stimulation (Supplemental Fig. 3B). Together, a large number of nuclear proteins are conjugated to SUMO after RAW macrophage cell activation. The global shift in SUMOylated proteins observed in this study is reminiscent of stress-induced changes in SUMOylated proteins reported in other cell types, in which some proteins are newly SUMOylated, whereas others are deSUMOylated after stress (8–11).
Figure 5.
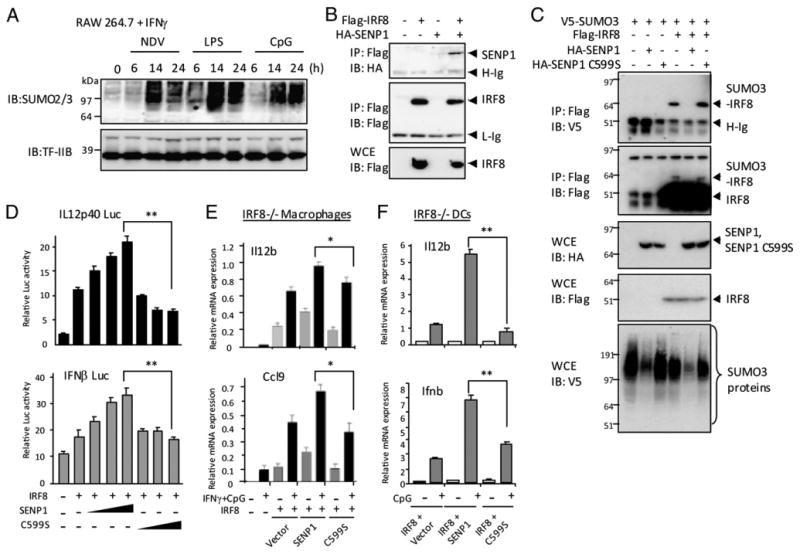
Macrophage activation triggers a global change in SUMOylated nuclear proteins, and the SENP1 enhances IRF8 transactivation. (A) RAW cells were stimulated with IFN-γ overnight, followed by stimulation by Newcastle disease virus, LPS, or CpG for indicated time. A total of 30 μg nuclear extracts was resolved in 4–12% Nu PAGE and blotted with anti-SUMO2/3 Ab. 0, indicates no treatment. (B) 293T cells were cotransfected with 2 μg pcDNA vector for Flag-IRF8 and 2 μg HA-Senp1 and 500 μg lysates were immunoprecipitated with anti-Flag Ab and blotted with anti-HA Ab (upper panel) or Flag (middle panel). In the lower panel, WCE (20 μg) were blotted with Ab for Flag. (C) 293T cells were transfected with 0.5 μg pcDNA vector for V5-SUMO3, 1 μg Flag-IRF8, and 1 μg HA-SENP1 or HA-C599S. A total of 500 μg lysates was immunoprecipitated with anti-Flag Ab and blotted with anti-V5 or anti-Flag Ab. In the two lower panels, WCE (20 μg) were immunoblotted with indicated Ab. (D) RAW cells were transfected with luciferase reporters for the Il12b or IFNb promoters, increasing amounts of pcDNA vectors for SENP1 or C599S (200, 400, 800 ng), IRF8 (800 ng), and pRL-TK and stimulated with IFN-γ/CpG, and luciferase activity was measured, as above. The values represent the average of three determinations ± SD. (E) CL2 macrophages were transduced with pMSCV vectors for IRF8 and SENP1 or the C599S mutant for 5 d and stimulated with or without IFN-γ/CpG. Expression of indicated transcripts was tested by qRT-PCR, as above. (F) IRF8−/− BM DCs (106 cells) were transduced with indicated vectors for 5 d and stimulated with CpG for 4 h, and expression of indicated transcripts was tested, as above. The values represent the average of three determinations ± SD. Statistical significance was tested by Student t test; *p < 0.01, **p < 0.005.
SENP1 is induced in macrophages after IFN-γ/CpG stimulation and deconjugates SUMO3 from IRF8
In light of the results that IRF8 SUMOylation decreased after IFN-γ/CpG stimulation, despite the global increase in SUMOylated nuclear proteins (Fig. 1A), we considered the possibility that IRF8 is one of those proteins that are deSUMOylated upon macrophage activation, examples found in other types of stress (8–11). Before testing whether SUMO was enzymatically removed from IRF8, we asked whether expression of Senp genes that encode SUMO-deconjugating enzymes alters after macrophage activation (3, 4, 48). We tested all seven Senp transcripts expressed in RAW cells. As shown in Supplemental Fig. 3C, that among all seven Senp transcripts, Senp1 mRNA was most robustly induced in RAW cells after IFN-γ/CpG stimulation: other Senps (Senp2–Senp6) were also induced, although weakly, although Senp7 and Senp8 were not. Immunoblot results shown in Supplemental Fig. 3D indicated that SENP1 protein was not detectable before stimulation, but induced after stimulation. In BM-derived macrophages, Senp1 mRNA was also induced along with other Senps after stimulation (Supplemental Fig. 3E). Given the global increase in SUMOylated proteins after macrophage activation (Fig. 5A), we also tested expression of genes involved in SUMO conjugation. As shown in Supplemental Fig. 3E, IFN-γ/TLR stimulation strongly induced all four SUMO E3 ligases of the PIAS family, Pias1, Pias2, Pias3, and Pias4. Furthermore, ubiquitin-conjugating enzyme E2I and all three Sumo genes were also induced after IFN-γ/TLR stimulation.
Thus, IFN-γ/TLR stimulation enhanced expression of genes encoding factors that catalyze both SUMOylation and deSUMOylation.
SENP1 has been shown to interact with a substrate protein, as detected by coimmunoprecipitation assays (49). We performed coimmunoprecipitation experiments with cells expressing Flag-IRF8 and HA-SENP1. As shown in Fig. 5B, IRF8 and SENP1 were coprecipitated when expressed together, supporting the possibility that IRF8 may serve as a SENP1 substrate. We next tested whether SENP1 catalyzes deSUMOylation of IRF8. Flag-IRF8 was coexpressed with wt HA-SENP1 or C599S, a SENP1 mutant lacking the catalytic activity, along with V5-SUMO3, and coimmunoprecipitation experiments were performed with anti-Flag Ab (37). As shown in Fig. 5C, in the absence of SENP1, Flag-IRF8 was readily conjugated to SUMO3. However, SUMO3 conjugation was virtually absent when SENP1 was coexpressed. Consistent with an enzymatic basis of this effect, the C599S mutant did not affect Flag-IRF8 SUMO3 conjugation (upper two panels). Immunoblot data in lower three panels showed that SENP1 and IRF8 were properly expressed and that SENP1, but not the C599S mutant, globally reduced SUMOylated proteins in the cells. Thus, SENP1 can remove SUMO3 from its substrate, IRF8. Reinforcing deSUMOylating activity of SENP1, polymeric SUMO chains linked to the IRF8-SUMO3 conjugate were also removed from IRF8 by SENP1, but not by the mutant (Supplemental Fig. 4A).
Ectopic SENP1 expression enhances IRF8 transactivation
To study whether SENP1 alters repressive activity of SUMOylated IRF8, we performed reporter assays with the Il12b and Ifnb promoters (Fig. 5D). The activity of both promoters was enhanced by transfection of IRF8, as expected. Cotransfection of wt SENP1 further enhanced activity of both reporters in a dose-dependent manner. However, the C599S mutant did not enhance activity of either reporter, indicating that SENP1 enhanced reporter activity by reducing SUMO conjugation. Supporting this idea, SENP1 had no effect on reporter activity by the K310R mutant (Supplemental Fig. 4E). To further investigate the effect of SENP1 expression on IRF8 function, SENP1 was stably introduced in IRF8−/− CL2 macrophages along with IRF8 by retroviral transduction, and expression of endogenous IRF8 target genes was tested. Results in Fig. 5E showed that IRF8 alone enhanced Il12b and Ccl9 transcripts, and cotransduction of SENP1, but not C599S, further increased their expression. The effect of SENP1 coexpression was highly reproducible, and consistently seen with samples prepared separately. Cotransduction of IRF8 and SENP1, but not C599S in IRF8−/− DCs also led to increased expression of Il12b and Ifnb mRNAs (Fig. 5F). Thus, SENP1 increases IRF8's ability to transactivate target genes, presumably by deSUMOylating IRF8.
SENP1 knockdown weakens IRF8 transactivation
To ascertain whether SENP1 promotes IRF8 transactivation, we knocked down Senp1 expression by shRNA and tested its effect on IRF8 target gene expression. Immunoblot analysis in Supplemental Fig. 4B showed that SENP1 protein levels were markedly reduced when Senp1 shRNA vector, but not scrambled shRNA (ctrl), was coexpressed in NIH3T3 cells. Senp and control shRNA were then stably expressed in CL2 macrophages along with IRF8, and tested for their effects on Il12b and Ccl9 induction. Results in Fig. 6A showed that Senp1 shRNA reduced induction of both genes by ∼30–45% relative to control shRNA or vector alone. Similarly, Senp1 shRNA reduced levels of Il12b and Ifnb induction approximately by half in DCs (Fig. 6B). Induction of additional IRF8 targets, Ciita, Ccl9, and Irf7, was also reduced in the presence of Senp1 shRNA (Supplemental Fig. 4C, 4D). These results further support the idea that SENP1 relieves IRF8 of SUMO-mediated transcriptional repression.
Figure 6.
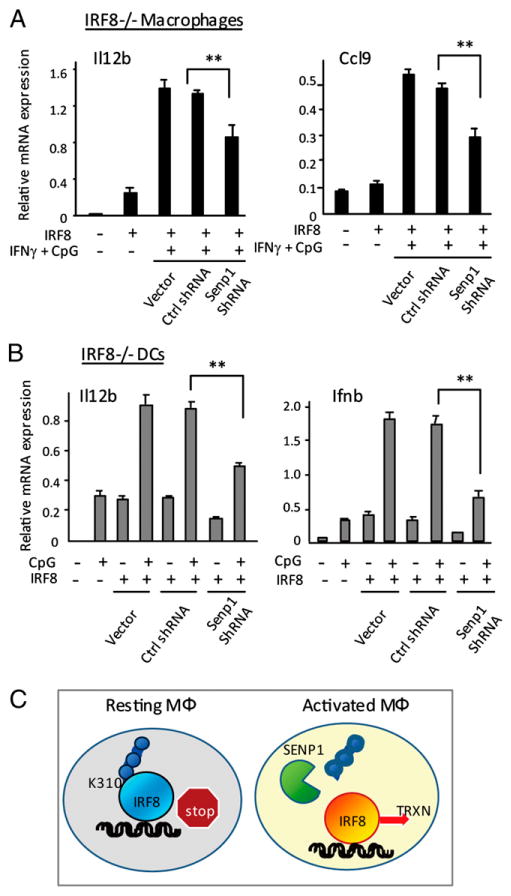
SENP1 knockdown inhibits IRF8 transactivation. (A) CL2 cells were transduced with pMSCV vector for wt IRF8, along with empty vector, control shRNA, or Senp1 shRNA pSuper vector, and cultured for 5 d, followed by stimulation with IFN-γ/CpG. Expression of indicated genes was measured by qRT-PCR, as above. (B) IRF8−/− BM DCs (106 cells) were transduced with viral vectors, as above, and stimulated with CpG for 4 h. The values represent the average of three determinations ± SD. Statistical significance was tested by Student t test; **p < 0.005. (C) SUMO conjugation-deconjugation switches IRF8 function: a model. SUMO-conjugated IRF8 present in resting macrophages acts as a repressor. Activation of macrophages with IFN-γ or IFN-γ/TLR stimulates SENP1 expression. SENP1 then removes SUMO from IRF8, converting it to a transcriptional activator. This SUMO switch is critical for the induction of IRF8-regulated genes.
Discussion
IRF8 plays a pivotal role in eliciting innate immune responses in activated macrophages and DCs (22, 29, 30, 32). We show that, in unstimulated RAW macrophages, IRF8 was SUMOylated, in a state unable to transactivate target genes. IRF8 became deSUMOylated after IFN-γ/TLR stimulation, coinciding with transactivation. The change in IRF8 SUMOylation took place in the wake of larger changes in SUMO-conjugated nuclear proteins, which was set off upon macrophage activation: RAW cell activation caused a global increase in SUMOylated proteins, accompanying induction of Sumo 1, 2, 3, the E2 enzyme ubiquitin-conjugating enzyme E2I, and all four Pias E3 ligases. Paralleling with these changes, however, there was a dramatic induction of multiple Senp deSUMOylating enzymes. The simultaneous induction of enzymes that catalyze opposite reactions indicates that SUMO modifications occur in a bidirectional fashion during macrophage activation, where some proteins are newly SUMOylated, whereas others are deSUMOylated. Clearly, IRF8 belongs to the latter. Both events are likely to be relevant to the execution of innate immune responses. Somewhat analogous to our observations, Deyrieux and Wilson (34) reported that cellular SUMOylation dynamic plays a critical role in keratinocyte differentiation. During Ca2+-induced keratinocyte differentiation enzymes of SUMOylation pathways, E1, E2, and E3 as well as SENP1 are induced to globally alter SUMOylation of many proteins that is required for proper keratinocyte differentiation. In contrast, a variety of stress, including heat shock, oxidative stress, and genotoxic stress, prompts changes in SUMOylation of many proteins, which most likely contribute to cells' stress responses (8, 10, 11, 50). Macrophage activation may share aspects of stress response, a view supported by the production of reactive oxygen species and NO in activation macrophages (23–25).
SUMOylation of IRF8
In RAW cells, a portion of IRF8 was conjugated to SUMO, mostly SUMO2/3 through the K310 residue. Coimmunoprecipitation analysis indicated that IRF8 can conjugate both monomeric and polymeric SUMO peptides, similar to other proteins (40, 51). SUMOylated IRF8 was associated with transcription repression, as judged by enhanced transactivation by the K310R mutant and diminished activation by the IRF8-SUMO3 conjugate, a conclusion consistent with the large body of literature (1, 6, 7, 48). FRAP analysis found that SUMO conjugation impairs genome-wide mobility of IRF8, an in vivo parameter that signifies functional integrity of a transcription factor (38, 46). The reduced IRF8 mobility may be partly accounted for by the property of SUMO3 itself, because GFP-SUMO3 exhibited a very slow mobility as well. The slow mobility of SUMO3 noted in this work is in agreement with a report on SUMO1 (44). In ChIP analysis, binding of the IRF8-SUMO3 conjugate to the Il12b target site was weaker than that of wt IRF8. The weakened target binding may be accounted for by the assembly of multiple repressive factors and poor genome-wide mobility (6, 7). In addition, epitope sequestration by assembled factors cannot be ruled out for reduced target binding.
IRF8 SUMO deconjugation by SENP1
SUMOylated IRF8 levels fell following macrophage activation, coinciding with marked induction of SENP1. Supporting the role for this enzyme in processing SUMOylated IRF8, SENP1 removed SUMO3 from IRF8 in a manner dependent on its catalytic activity. SENP1 could reduce monomeric and polymeric SUMO3 from IRF8. Moreover, SENP1 altered the functional property of IRF8, because SENP1 ectopic expression enhanced promoter activity and mRNA expression of IRF8 target genes. Supporting a direct link between deSUMOylation and IRF8 transactivation, SENP1 enhancement of IRF8 activity was also dependent on the intact catalytic activity. Moreover, the enhancing effect of SENP1 was reversed by shRNA-based Senp1 knockdown. These results led us to suggest a model in which SENP1, by virtue of its deSUMOylating activity, switches IRF8 from a repressor to an activator, a change critical for functional macrophage activation by IRF8 (Fig. 6C). At present, the mechanism by which SENP1 selectively deSUMOylates IRF8 among other proteins remains elusive. It is possible that a ready physical interaction between the two proteins as observed by our coimmunoprecipitation assays may be a basis of substrate selectivity. Given that other SENPs besides SENP1 were also induced after IFN-γ/TLR stimulation, albeit less strongly, it is conceivable that they also take part in deSUMOylation of IRF8 or more likely other substrates. SENP members have different activities, ranging from SUMO maturation to chain editing to SUMO deconjugation (3, 4). SENPs are also differentially distributed in the nucleus. Some of these SENPs are shown to relieve SUMO-mediated transcriptional repression. For example, SENP1 deconjugates SUMO from HDAC1 and p300 to relieve repression (48, 49). Sirtuin 1 is constitutively SUMOylated, the state that confers full deacetylase activity. Sirtuin 1 is deSUMOylated by SENP1 after UV radiation or by oxidative stress, leading to in-activation of its histone deacetylase activity and increased apo- ptosis (49). SENPs are involved in other regulatory processes. Senp1−/− mice are embryonic lethal due to the dysregulation of HIF-1α stability, which is controlled by SUMOylation (48, 52). Moreover, Huang et al. (7, 53) recently demonstrated that SENP3 plays a role in regulating inflammatory gene expression in a model in which SUMOylated liver X receptor transrepresses NF-κB–activated genes by recruiting the NCOR complex. This trans-repression was relieved by SENP3 recruited to the target genes by interacting with liver X receptor. Another recent study showed that SENP3 is also involved in p300 deSUMOylation and HIF-1α transactivation (54).
Our proposition that IRF8 deSUMOylation is causally linked to its functional activation in macrophages gains further credence in view of an emerging paradigm in which SENPs play an integral role in promoting maturation of hematopoietic cells. Tillmanns et al. (35) showed that the maintenance of myeloid progenitor cells depends on MafB repression through its SUMOylation. In that report, a MafB mutant lacking SUMOylation site prompts premature macrophage differentiation. Furthermore, GATA1, a transcription factor that regulates the globin gene switch, is deSUMOylated by SENP1 during RBC development (36). GATA1 deSUMOylation is shown to be a critical step for erythrocyte formation, as evidenced by the absence of erythropoiesis in Senp1−/− fetal liver cells (36).
In summary, by its deSUMOylating activity, SENP1 switches repressive IRF8 to a potent transactivator, an event fundamental to the onset of innate immune responses in macrophages.
Acknowledgments
We thank M. Kuehn (National Cancer Institute, National Institutes of Health) and T. Kubota (National Institute of Infectious Diseases, Tokyo, Japan) for critical reading of the manuscript and advice on experiments, and J. McNally and T. Karpova in the Fluorescence Imaging Facility (National Cancer Institute, National Institutes of Health) for assistance in FRAP experiments.
This work was supported by the intramural programs of the National Institute of Child Health and Human Development and the National Institutes of Health intramural AIDS Targeted Antiviral program.
Abbreviations used in this article
- BM
bone marrow
- ChIP
chromatin immunoprecipitation
- DC
dendritic cell
- FRAP
fluorescence recovery after photobleaching
- FRET
fluorescence resonance energy transfer
- HA
hemagglutinin
- IRF
IFN regulatory factor
- PIAS
protein inhibitor of activated STAT
- qRT-PCR
quantitative RT-PCR
- SENP
sentrin-specific peptidase
- shRNA
short hairpin RNA
- SUMO
small ubiquitin-like modifier
- TFIIB
transcription factor II B
- TSS
transcription start site
- WCE
whole-cell extract
- wt
wild-type
Footnotes
The online version of this article contains supplemental material.
Disclosures: The authors have no financial conflicts of interest.
References
- 1.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 2.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartek J, Hodny Z. SUMO boosts the DNA damage response barrier against cancer. Cancer Cell. 2010;17:9–11. doi: 10.1016/j.ccr.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 9.Flick K, Kaiser P. Proteomic revelation: SUMO changes partners when the heat is on. Sci Signal. 2009;2:pe45. doi: 10.1126/scisignal.281pe45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Tempé D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 12.Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota T, Matsuoka M, Xu S, Otsuki N, Takeda M, Kato A, Ozato K. PIASy inhibits virus-induced and interferon-stimulated transcription through distinct mechanisms. J Biol Chem. 2011;286:8165–8175. doi: 10.1074/jbc.M110.195255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa K, Yokosawa H. PIAS3 induces SUMO-1 modification and transcriptional repression of IRF-1. FEBS Lett. 2002;530:204–208. doi: 10.1016/s0014-5793(02)03486-5. [DOI] [PubMed] [Google Scholar]
- 15.Han KJ, Jiang L, Shu HB. Regulation of IRF2 transcriptional activity by its sumoylation. Biochem Biophys Res Commun. 2008;372:772–778. doi: 10.1016/j.bbrc.2008.05.103. [DOI] [PubMed] [Google Scholar]
- 16.Renner F, Saul VV, Pagenstecher A, Wittwer T, Schmitz ML. Inducible SUMO modification of TANK alleviates its repression of TLR7 signalling. EMBO Rep. 2011;12:129–135. doi: 10.1038/embor.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Tahk S, Yee KM, Fan G, Shuai K. The ligase PIAS1 restricts natural regulatory T cell differentiation by epigenetic repression. Science. 2010;330:521–525. doi: 10.1126/science.1193787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Yang Y, Chernishof V, Loo RR, Jang H, Tahk S, Yang R, Mink S, Shultz D, Bellone CJ, et al. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, Ozato K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, Kato T, Hotta C, Nishiyama A, Kurotaki D, Yoshinari M, Takami M, Ichino M, Nakazawa M, Matsuyama T, et al. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PLoS One. 2011;6:e25812. doi: 10.1371/journal.pone.0025812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tailor P, Tamura T, Morse HC, III, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 24.Xiong H, Zhu C, Li H, Chen F, Mayer L, Ozato K, Unkeless JC, Plevy SE. Complex formation of the interferon (IFN) consensus sequence-binding protein with IRF-1 is essential for murine macrophage IFN-gamma-induced iNOS gene expression. J Biol Chem. 2003;278:2271–2277. doi: 10.1074/jbc.M209583200. [DOI] [PubMed] [Google Scholar]
- 25.Alter-Koltunoff M, Ehrlich S, Dror N, Azriel A, Eilers M, Hauser H, Bowen H, Barton CH, Tamura T, Ozato K, Levi BZ. Nramp1-mediated innate resistance to intraphagosomal pathogens is regulated by IRF-8, PU.1, and Miz-1. J Biol Chem. 2003;278:44025–44032. doi: 10.1074/jbc.M307954200. [DOI] [PubMed] [Google Scholar]
- 26.Kautz B, Kakar R, David E, Eklund EA. SHP1 protein-tyrosine phosphatase inhibits gp91PHOX and p67PHOX expression by inhibiting interaction of PU.1, IRF1, interferon consensus sequence-binding protein, and CREB-binding protein with homologous Cis elements in the CYBB and NCF2 genes. J Biol Chem. 2001;276:37868–37878. doi: 10.1074/jbc.M103381200. [DOI] [PubMed] [Google Scholar]
- 27.Tsujimura H, Tamura T, Ozato K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J Immunol. 2003;170:1131–1135. doi: 10.4049/jimmunol.170.3.1131. [DOI] [PubMed] [Google Scholar]
- 28.Smith MA, Wright G, Wu J, Tailor P, Ozato K, Chen X, Wei S, Piskurich JF, Ting JP, Wright KL. Positive regulatory domain I (PRDM1) and IRF8/PU.1 counter-regulate MHC class II transactivator (CIITA) expression during dendritic cell maturation. J Biol Chem. 2011;286:7893–7904. doi: 10.1074/jbc.M110.165431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turcotte K, Gauthier S, Tuite A, Mullick A, Malo D, Gros P. A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia-like syndrome in BXH-2 mice. J Exp Med. 2005;201:881–890. doi: 10.1084/jem.20042170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fragale A, Stellacci E, Ilari R, Remoli AL, Lanciotti A, Perroti E, Orsatti R, Hahnel V, Rehli M, Ozato K, Battistini A. Critical role of IRF-8 in negative regulation of TLR3 expression by SHP-2 activity in human myeloid dendritic cells. J Immunol. 2011;186:1951–1962. doi: 10.4049/jimmunol.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, III, Ozato K. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- 33.Kim JY, Ozato K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. J Immunol. 2009;182:2131–2140. doi: 10.4049/jimmunol.0802755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deyrieux AF, Wilson VG. In vitro culture conditions to study keratinocyte differentiation using the HaCaT cell line. Cytotechnology. 2007;54:77–83. doi: 10.1007/s10616-007-9076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tillmanns S, Otto C, Jaffray E, Du Roure C, Bakri Y, Vanhille L, Sarrazin S, Hay RT, Sieweke MH. SUMO modification regulates MafB-driven macrophage differentiation by enabling Myb-dependent transcriptional repression. Mol Cell Biol. 2007;27:5554–5564. doi: 10.1128/MCB.01811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu L, Ji W, Zhang H, Renda MJ, He Y, Lin S, Cheng EC, Chen H, Krause DS, Min W. SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J Exp Med. 2010;207:1183–1195. doi: 10.1084/jem.20092215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Chau SF, Lam KH, Chan HY, Ng TB, Au SW. Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease. Biochem J. 2006;398:345–352. doi: 10.1042/BJ20060526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laricchia-Robbio L, Tamura T, Karpova T, Sprague BL, McNally JG, Ozato K. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. Proc Natl Acad Sci USA. 2005;102:14368–14373. doi: 10.1073/pnas.0504014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 41.Stankovic-Valentin N, Kozaczkiewicz L, Curth K, Melchior F. An in vitro FRET-based assay for the analysis of SUMO conjugation and iso-peptidase cleavage. Methods Mol Biol. 2009;497:241–251. doi: 10.1007/978-1-59745-566-4_16. [DOI] [PubMed] [Google Scholar]
- 42.Chen WY, Lee WC, Hsu NC, Huang F, Chung BC. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1) J Biol Chem. 2004;279:38730–38735. doi: 10.1074/jbc.M405006200. [DOI] [PubMed] [Google Scholar]
- 43.Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ET. SUMO-specific protease 2 is essential for suppression of pol-ycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chalkiadaki A, Talianidis I. SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol Cell Biol. 2005;25:5095–5105. doi: 10.1128/MCB.25.12.5095-5105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nayak A, Glöckner-Pagel J, Vaeth M, Schumann JE, Buttmann M, Bopp T, Schmitt E, Serfling E, Berberich-Siebelt F. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J Biol Chem. 2009;284:10935–10946. doi: 10.1074/jbc.M900465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dundr M, Misteli T. Measuring dynamics of nuclear proteins by photobleaching. Curr Protoc Cell Biol. 2003;Chapter 13(Unit 13.5) doi: 10.1002/0471143030.cb1305s18. [DOI] [PubMed] [Google Scholar]
- 47.Kim JY, Anderson ED, Huynh W, Dey A, Ozato K. Proteomic survey of ubiquitin-linked nuclear proteins in interferon-stimulated macrophages. J Interferon Cytokine Res. 2011;31:619–628. doi: 10.1089/jir.2011.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh ET. SUMOylation and de-SUMOylation: wrestling with life's processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 51.Jaffray EG, Hay RT. Detection of modification by ubiquitin-like proteins. Methods. 2006;38:35–38. doi: 10.1016/j.ymeth.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 52.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang C, Han Y, Wang Y, Sun X, Yan S, Yeh ET, Chen Y, Cang H, Li H, Shi G, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009;28:2748–2762. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]


