SUMMARY
Alternative splicing (AS) of pre-mRNA is utilized by higher eukaryotes to achieve increased transcriptome and proteomic complexity. The serine/arginine (SR) splicing factors regulate tissue- or cell-type-specific AS in a concentration- and phosphorylation-dependent manner. However, the mechanisms that modulate the cellular levels of active SR proteins remain to be elucidated. In the present study, we provide evidence for a role for the long nuclear-retained regulatory RNA (nrRNA), MALAT1 in AS regulation. MALAT1 interacts with SR proteins and influences the distribution of these and other splicing factors in nuclear speckle domains. Depletion of MALAT1 or overexpression of an SR protein changes the AS of a similar set of endogenous pre-mRNAs. Furthermore, MALAT1 regulates cellular levels of phosphorylated forms of SR proteins. Taken together, our results suggest that MALAT1 regulates AS by modulating the levels of active SR proteins. Our results further highlight the role for an nrRNA in the regulation of gene expression.
INTRODUCTION
Mammalian genomes harbor fewer protein-coding genes than initially anticipated, and gene numbers do not proportionally increase with increased genome complexity (for review, see Amaral et al., 2008; Mattick, 2009). Interestingly, numerous transcriptome analyses have identified RNAs that do not code for proteins, referred to as noncoding RNAs (ncRNAs) (Kapranov et al., 2002; van Bakel et al., 2010). Regulatory ncRNAs can be broadly classified into small (18–200 nt) and long ncRNAs (lncRNAs; 200 nt to >100 kb) (Prasanth and Spector, 2007). Although studies of small regulatory RNAs have dominated the field of RNA biology in recent years (Carthew and Sontheimer, 2009), a surprisingly wide array of cellular functions is also associated with lncRNAs (Mercer et al., 2009; Wilusz et al., 2009). Several lncRNAs are highly conserved, and their expression is developmentally and temporally regulated or restricted to particular tissues and organs, which further indicates their role in specific cellular processes (Guttman et al., 2009; Mercer et al., 2008; Ravasi et al., 2006).
Several studies have shown that specific RNAs are retained within the mammalian cell nucleus (nuclear-retained regulatory RNAs, nrRNAs) and are suggested to play structural roles or act as riboregulators (reviewed in Prasanth and Spector, 2007; Wilusz et al., 2009). Examples of some of the characterized long nrRNAs include Xist and Tsix in mammals and RoX RNAs in fruit flies, all of which are involved in dosage compensation (Prasanth and Spector, 2007); and MEN epsilon/β (NEAT1), implicated in paraspeckle structure maintenance (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). Recent genomic and RNA localization analyses have identified additional novel nrRNAs in mammalian cells, and the characterization of these is expected to reveal important gene-regulatory functions and roles associated with the complexity observed in multicellular organisms (Mercer et al., 2008; Sone et al., 2007; van Bakel et al., 2010).
The mammalian cell nucleus is compartmentalized into non-membranous subnuclear domains including nucleoli, nuclear speckles, paraspeckles, Cajal bodies, and promyelocytic leukemia (PML) bodies, which in turn are enriched in specific subsets of proteins and RNAs (Matera et al., 2009; Spector, 2006). Of particular interest in the present study are nuclear speckles (interchromatin granule clusters [IGCs] or SC35 domains), of which there are ~20–30 per nucleus. Nuclear speckles are highly dynamic subnuclear domains enriched with pre-mRNA splicing/processing factors (Hall et al., 2006; Lamond and Spector, 2003). Nuclear speckles do not represent major sites of transcription or splicing but rather are thought to be involved primarily in the assembly, modification, and/or storage of the pre-mRNA splicing machinery. It has been proposed that speckles are sites from where splicing factors are recruited to active sites of transcription (Lamond and Spector, 2003; Misteli, 2000). However, the mechanisms that regulate the cycling of splicing factors between nuclear speckles and sites of transcription have yet to be characterized.
Alternative splicing (AS) of pre-mRNAs is a key step in the regulation and diversification of gene function (Blencowe, 2006; Hallegger et al., 2010; Licatalosi and Darnell, 2010). It has recently been estimated that transcripts from more than 95% of human multi-exon-containing genes undergo AS, and the majority of these are variably expressed between different cell and tissue types (Pan et al., 2008; Wang et al., 2008). In general, AS is regulated by trans-acting protein factors, which include the small nuclear ribonucleoproteins (snRNPs), the serine/arginine-rich (SR) family of nuclear phosphoproteins (SR proteins), SR-related proteins, and the heterogeneous nuclear ribonucleoproteins (hnRNPs) (Blencowe, 2006; Long and Caceres, 2009). SR proteins have been extensively characterized as a class of RNA-binding proteins that generally function in constitutive splicing and AS in higher eukaryotic cells through their sequence-specific recognition of cis-acting exonic splicing enhancers and subsequent recruitment of other splicing factors to facilitate the assembly of the spliceosome (Lin and Fu, 2007; Long and Caceres, 2009). Cellular levels of SR family and SR-related proteins are tightly regulated, and changes in concentrations or phosphorylation of these factors can influence AS patterns of many pre-mRNAs (Bourgeois et al., 2004; Calarco et al., 2009; Long and Caceres, 2009; Stamm, 2008). However, the mechanisms cells utilize in order to establish active concentrations of these factors are not well understood.
In the present study we have examined the role of an abundant, long (>6.5 kb) mammalian nrRNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1; also referred to as Neat2) in the regulation of gene expression (Hutchinson et al., 2007; Wilusz et al., 2008). Originally identified based upon its overexpression in several cancers, the role of MALAT1 in normal cellular physiology remains unknown (Ji et al., 2003; Lin et al., 2006). Since MALAT1 is highly conserved among mammals and predominantly localizes to nuclear speckles, we hypothesize that it plays a critical role in one or more aspects of pre-mRNA metabolism. To test this hypothesis, we have analyzed the specific association of MALAT1 with pre-mRNA splicing factors, its role in the spatial distribution of splicing factors, and its involvement in the AS of endogenous pre-mRNAs. Our results demonstrate that MALAT1 interacts with SR splicing factors and modulates their distribution to nuclear speckles. Furthermore, we provide evidence suggesting that MALAT1 regulates AS of pre-mRNAs by controlling the functional levels of SR splicing factors.
RESULTS
MALAT1 nrRNA Localizes to Nuclear Speckles and Interacts with SR Splicing Factors
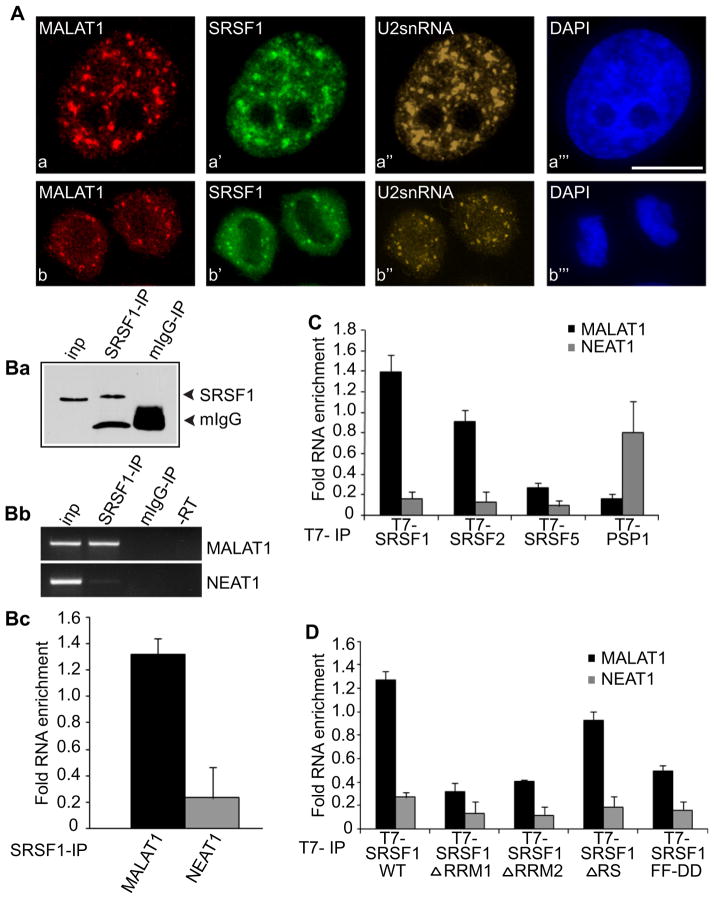
Dual RNA-FISH using probes specific for MALAT1 (Figures 1Aa and 1Ab) and U2-snRNA (Figures 1Aa″ and 1Ab″) and coimmunostaining using an antibody specific for the SR splicing factor, SRSF1 (earlier referred as SF2/ASF; Figures 1Aa′ and 1Ab′) (Manley and Krainer, 2010) in HeLa cells demonstrated enrichment of MALAT1 in nuclear speckles in interphase cells (Figures 1Aa–1Aa‴), as also reported recently (Clemson et al., 2009; Hutchinson et al., 2007). We also observe that MALAT1 concentrates in mitotic IGCs, a structural analog of nuclear speckles present in mitotic cells (Figures 1Ab–1Ab‴) (Prasanth et al., 2003).
Figure 1. MALAT1 Localizes to Nuclear Speckles and Interacts with SR Proteins.
(A) Co-RNA-FISH using probes against MALAT1 (Aa and Ab), U2 snRNA (Aa″ and Ab″) and immunostaining using an antibody against SRSF1 (Aa′ and Ab′) in interphase (Aa–Aa‴) and mitotic telophase HeLa cells (Ab–Ab‴). The DNA is counterstained with DAPI (blue; Aa‴ and Ab‴). The bar represents 10 μm.
(B) (Ba) Immunoprecipitation from HeLa cells using SRSF1 antibody or mouse serum followed by immunoblot with SRSF1 antibody. (Bb and Bc) RT-PCR or qPCR from the IP samples using MALAT1- or NEAT1-specific primers.
(C) qPCR from T7-IP (T7-SRSF1-WT-, T7-SRSF2-, T7-SRSF5-, and T7-PSP1-expressing cells) using MALAT1- or NEAT1-specific primers.
(D) qPCR from T7-IP (T7-SRSF1-WT, T7-SRSF1ΔRRM1, ΔRRM2, ΔRS, FF-DD-expressing cells) using MALAT1- or NEAT1-specific primers. Error bars in (Bc), (C), and (D) represent mean ± SD of three independent experiments. See also Figure S1.
To identify factors that interact with MALAT1 and potentially contribute to its localization in speckles, we addressed whether SR proteins, which are highly concentrated in speckle domains, interact with MALAT1. We first performed a binding site motif analysis to ask whether preferred binding sites for SRSF1 (Ray et al., 2009) are significantly enriched in one or more regions (using a sliding window of 1 kb) in human and mouse MALAT1 RNA, relative to regions of the same length within 100 randomly selected mRNAs from these species (see the Supplemental Experimental Procedures available online). This analysis identified significant enrichment for SRSF1-binding sites within the 5′ half of both human and mouse MALAT1, relative to the randomly selected mRNAs (p value = 1.0674e-217 [human] and p = 6.6882e-062 [mouse]) (Figures S1Aa and S1Ab). Similarly, ESEfinder (version 3, http://rulai.cshl.edu/tools/ESE3/) (Smith et al., 2006), a tool that searches for high-affinity SELEX-defined binding sites of various SR proteins, also revealed a concentration of SRSF1-binding sites in the 5′ half of mouse and human MALAT1 (28 and 34 sites in mouse and human MALAT1, respectively, threshold score ~3.9) (Figures S1Ac–S1Ad). Confirming that SRSF1 binds endogenous MALAT1, a recent study utilizing in vivo UV crosslinking of RNA followed by immunoprecipitation and high throughput sequencing (CLIP-Seq), demonstrated a direct interaction between SRSF1 and MALAT1 in HeLa cells (Sanford et al., 2009). Consistent with the computational predictions described above, the binding sites mapped in this study were primarily located within the 5′ half of MALAT1 (Figure S1Ag).
To investigate the specificity of the interaction between MALAT1 and SR proteins, we performed an RNA coimmunoprecipitation (RNA-IP) analysis using anti-SRSF1 antibody in formaldehyde crosslinked nuclear extracts from HeLa cells (Figure 1B). RT-PCR analyses of RNA-IP samples using primers specific to human MALAT1 revealed a specific interaction between MALAT1 and SRSF1 (Figures 1Bb and 1Bc). RNA-IP followed by RT-PCR in extracts from HeLa cells transiently expressing T7-tagged SR proteins (SRSF1, SRSF2 [SC35], SRSF3 [SRp20], SRSF5 [SRp40]) revealed interaction between MALAT1 and SRSF1, SRSF2, and SRSF3 (Figure 1C, Figures S1B and S1C). Surprisingly, SRSF5, which has a domain organization similar to that of SRSF1, consisting of two N-terminal RNA recognition motifs (RRMs) and a C-terminal RS domain, showed weak association with MALAT1 compared to other SR proteins tested (Figure 1C, Figure S1B). This difference was not due to a general lack of SRSF5-binding activity, since an in vivo reporter cell line assay revealed that the transiently transfected T7-SRSF5 was indeed functional in its ability to localize to a transcriptionally active gene locus and to interact with the reporter RNA (Figures S1D and S1E). Similarly, PSP1, an RNA-binding protein and a component of paraspeckles, also displayed a weak interaction with MALAT1 but at the same time showed a strong association with the paraspeckle-localized NEAT1 nrRNA (Figure 1C). The specificity of the association between SR proteins and MALAT1 was further supported by the observation that SR proteins showed weak interactions with NEAT1 (Figures 1B and 1C). These results demonstrate that a subset of SR proteins interact with MALAT1, and that multiple SRSF1 proteins bind specifically and directly to the 5′end of this nrRNA.
To address which domain(s) of SRSF1 mediates its interaction with MALAT1, we conducted RNA-IP followed by qPCR from extracts expressing several of T7-tagged SRSF1 mutants (SRSF1 ΔRRM1, ΔRRM2, and ΔRS) (Caceres et al., 1997). Deletion of either of the canonical RRM (RRM1) or the pseudo RRM (RRM2), but not the RS domain, resulted in a pronounced reduction in the interaction between SRSF1 and MALAT1 (Figure 1D). To further address the role of RRM1 in MALAT1 interaction, we also examined the association of an SRSF1 RRM1 mutant (SRSF1-FF-DD) reported to have reduced affinity for RNA and diminished activity in in vitro splicing assays (Caceres and Krainer, 1993). Our RNA-IP results also reveal a weak interaction between the SRSF1 FF-DD mutant and MALAT1 (Figure 1D). In SR proteins that contain a canonical RRM and a pseudo-RRM, both domains function together to contribute to RNA substrate specificity and binding affinity (Bourgeois et al., 2004). We observe that these RRMs contribute approximately equally to the interaction between SRSF1 and MALAT1.
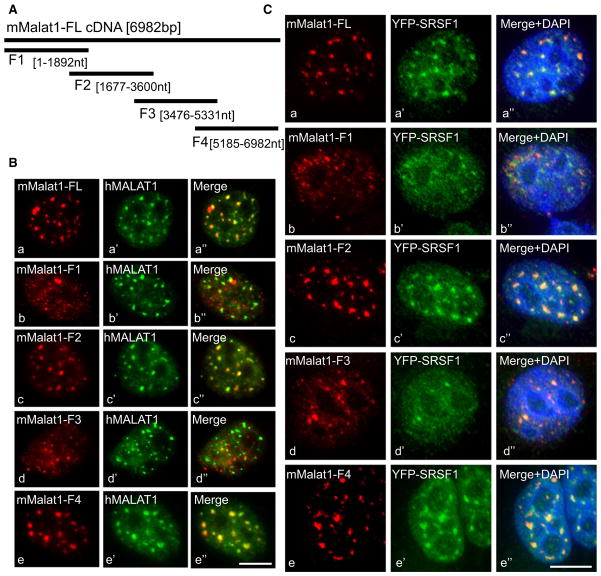
Independent Sequence Elements in MALAT1 Influence Its Distribution to Nuclear Speckles
Posttranscriptional modifications of RNA and/or differential association with specific nuclear-enriched proteins may be responsible for the nuclear retention of nrRNAs (Prasanth et al., 2005). In order to map the region(s) in MALAT1 involved in its localization at nuclear speckles and to identify the factor(s) that influences speckle localization of MALAT1, we generated eukaryotic expression plasmids encoding either mouse MALAT1 (mMALAT1) full-length cDNA or various mutant constructs containing partially overlapping regions of mMALAT1 cDNA (F1–F4; Figure 2A). RNA-IP using SRSF1 antibody in cells transiently expressing each of the MALAT1 mutants revealed that SRSF1 interacted with RNA transcribed from the F1, F2, and F3 regions (Figure S2A). These results are consistent with the detection of significant enrichment of SRSF1-binding sites, and SRSF1-CLIP data, where functional binding sites of SRSF1 are concentrated in the first 6 kb of the hMALAT1, corresponding to the F1–F3 region of mMALAT1 (Figure S1Ag) (Sanford et al., 2009).
Figure 2. MALAT1 RNA Contains Two Independent Nuclear Speckle-Localizing Motifs.
(A) Schematic representation of full-length mouse MALAT1 (6982 bp) and mutant constructs (F1–F4).
(B) Co-RNA FISH using mouse (Ba–Be)- and human (Ba′–Be′)-specific MALAT1 probes in HeLa cells that were transiently transfected with full-length (a) and mutant mMALAT1 constructs (F1 [Bb], F2 [Bc], F3 [Bd], and F4 [e]).
(C) RNA FISH using mMALAT1 probe (Ca–Ce) in HeLa cells expressing YFP-SRSF1 (Ca′–Ce′) that were transiently transfected with full-length mMALAT1 cDNA (Ca) and mutant constructs (F1 [b], F2 [c], F3 [d], and F4 [e]). The bar represents 10 μm. See also Figure S2.
Next, we examined the intracellular distribution of MALAT1 mutant RNAs. mMALAT1 constructs were individually transfected in HeLa cells, and RNA-FISH using probes that hybridize specifically to mMALAT1 was conducted to analyze the intracellular distribution of mMALAT1 full-length and mutant RNAs. The exogenously expressed mMALAT1 full-length RNA localized to nuclear speckles in HeLa cells and colocalized with endogenous human MALAT1 RNA (Figures 2Ba–2Ba″). MALAT1-F1 and F3 RNA showed punctate nuclear localization but did not localize to nuclear speckles (Figures 2Bb–2Bb″ and 2Bd–2Bd″). MALAT1-F2 and MALAT1-F4 RNAs independently displayed nuclear speckle distribution and colocalized with other nuclear speckle markers (Figures 2Bc–2Bc″ and 2Be–2Be″, and 2Cc–2Cc″ and 2Ce–2Ce″). The cellular distribution of exogenously expressed full-length and mutant mMALAT1 RNAs was similar even after depletion of endogenous hMALAT1 RNA by treatment with human MALAT1-specific antisense oligonucleotides, implying that the distribution of mMALAT1 in human cells is not measurably influenced by the levels of endogenous MALAT1 transcripts (Figure S2B). A BLAST comparison of sequences from the F2 and F4 regions of mMALAT1 did not reveal significant sequence similarity, indicating that either distributed common linear motifs, or possibly RNA secondary structure, could be responsible for their similar speckle distribution. Alternatively, association of MALAT1 F2 and F4 RNAs with independent nuclear speckle-localized factors could result in similar distributions.
We also analyzed the distribution of SR proteins in HeLa cells that overexpress mMALAT1. Cells expressing full-length, F2, and F4 RNAs continue to show speckle localization of SRSF1 (Figures 2Ca–2Ca″, 2Cc–2Cc″, and 2Ce–2Ce″). However, cells expressing F1 and F3 RNA fragments displayed more homogeneous distribution of SRSF1 (Figures 2Cb–2Cb″ and 2Cd–2Cd″). These results indicate that non-speckle-localized MALAT1 mutant RNAs F1 and F3 may act in a dominant-negative manner to affect the localization of SRSF1 in speckles.
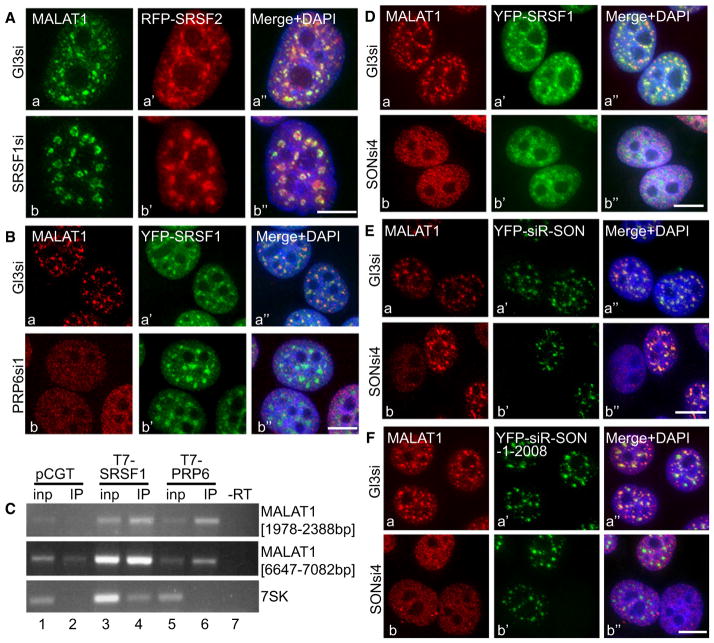
Multiple Nuclear Speckle-Associated Proteins Control the Speckle Distribution of MALAT1
In order to identify the protein component(s) that influences the speckle distribution of MALAT1, we tested the effects of knockdown of a representative set of nuclear speckle-associated splicing factors (SRSF1, SRSF2, B″-U2 snRNP, PRP6, and SON) on MALAT1 speckle distribution. In general, siRNA treatment achieved more than 80%–90% knockdown of the desired proteins (Figures S3A–S3B). MALAT1 RNA-FISH in SRSF1 (Figure 3A) or SRSF2-depleted (data not shown) HeLa cells continued to show prominent speckle localization of MALAT1. Similar results were also observed in mouse embryonic fibro-blasts that were derived from SRSF1 and SRSF2 knockout mice (data not shown). These findings indicate either that SR proteins are not required for the speckle localization of MALAT1 or that individual SR proteins regulate MALAT1 distribution in a redundant fashion. The nonspeckle association of SRSF1-interacting F1 and F3 mutant RNAs and the speckle localization of non-SRSF1 interacting F4 RNA further rules out the involvement of SRSF1 in the speckle distribution of MALAT1.
Figure 3. Nuclear Speckle-Associated Splicing Factors PRP6 and SON Influence the Distribution of MALAT1 in Nuclear Speckles.
(A) MALAT1 RNA-FISH (Aa and Ab) in control luciferase siRNA (GL3si; a–a″) and SRSF1 siRNA (b–b″)-treated HeLa cells expressing RFP-SRSF2 (a′–b′).
(B) MALAT1 RNA-FISH in control (Ba–Ba″) and PRP6 siRNA-1 (Bb–Bb″)-treated YFP-SRSF1 (Ba′–Bb′) stably expressing HeLa cells.
(C) RT-PCR from T7-antibody IP samples (lanes 2, 4, and 6) of HeLa cells expressing T7 (pCGT vector) alone (lanes 1 and 2), T7-SRSF1 (lanes 3 and 4) and T7-PRP6 (lanes 5 and 6), using MALAT1 primers. RT-PCR using 7SK primers were performed as a negative control.
(D) MALAT1 RNA-FISH (Da and Db) in control (Da–Da″) and SON siRNA4 (Db–Db″)-treated YFP-SRSF1 (Da′ and Db′)-expressing HeLa cells.
(E) MALAT1 RNA-FISH in control (Ea–Ea″) and SON siRNA4 (Eb–Eb″)-treated HeLa cells, which were transiently expressing siRNA4 refractory YFP-SON-FL (YFP-siR-SON; Ea′ and Eb′). Note that exogenously expressed SON restored the speckle distribution of MALAT1 (Eb–Eb″).
(F) MALAT1 RNA-FISH in control (Fa–Fa″) and SON siRNA4 (Fb–Fb″)-treated HeLa cells, which were transiently expressing siRNA4 refractory YFP-SON mutant (green; YFP-siR-SON-1-2008; Fa′ and Fb′). The bars in all the figures represent 10 μm. See also Figure S3.
Human PRP6 is a nuclear speckle-associated essential non-SR splicing factor that functions to bridge interactions between U5 and U4/U6 snRNPs during tri-snRNP assembly (Makarov et al., 2000). Depletion of PRP6 from cells using several independent siRNA oligos resulted in the reduced levels of MALAT1 in nuclear speckles and a more homogeneous nuclear distribution (Figure 3B, Figure S3C). However, PRP6 depletion did not disrupt nuclear speckles, as monitored by the speckle localization of YFP-SRSF1 (Figure 3B and Figure S3C). Furthermore, RNA-IP in nuclear extracts from cells that were transfected with T7-PRP6 cDNA followed by RT-PCR using MALAT1-specific primers confirmed an interaction between MALAT1 and PRP6 (Figure 3C and Figure S3D). Interestingly, RNA-IP followed by qPCR analyses in cells that were coexpressing T7-PRP6 along with any of the four MALAT1 mutants (F1 to F4) revealed that T7-PRP6 interacted with all of the four MALAT1 mutant RNAs, though RNA from F1 and F3 showed maximum interaction (Figure S3D). Both F1 and F3 fragments, when expressed in cells, did not localize to speckles, indicating that interaction with PRP6 alone is not sufficient for speckle association. Therefore we conclude that PRP6 is necessary but not sufficient for the speckle localization of MALAT1.
In addition to PRP6, we have also identified the involvement of the SR-related protein SON, another speckle protein, in the distribution of MALAT1 to nuclear speckles. SON has been proposed to act as a scaffold for the proper organization of nuclear speckle components (Sharma et al., 2010). Depletion of SON resulted in the redistribution of MALAT1 from nuclear speckles to a more homogeneous nuclear localization (Figure 3D and Figure S3E). To confirm the involvement of SON in the speckle localization of MALAT1, we performed an siRNA rescue assay. MALAT1 RNA-FISH was performed in SON siRNA-transfected HeLa cells expressing an RNAi-refractory version of YFP-SON (YFP-siR-SON) (Sharma et al., 2010) (Figure 3E). MALAT1 relocalized to nuclear speckles in cells depleted of endogenous SON but expressed YFP-siR-SON (Figures 3Eb–3Eb″). The SON protein contains an RS domain and a G patch and also has a C-terminal double-stranded RNA-binding domain (DSRBD) (Sharma et al., 2010). Rescue assays using the RNAi-refractory version of mutant-YFP-SON that lacks G patch and DSRBD (YFP-siR-SON-1-2008) demonstrated that even though the YFP-siR-SON-1-2008 by itself localized to nuclear speckles, it failed to recruit MALAT1 to nuclear speckles (Figure 3Fb–3Fb″). These results suggest that the speckle localization domain of SON is different from the region of SON that facilitates the recruitment of MALAT1 to speckles.
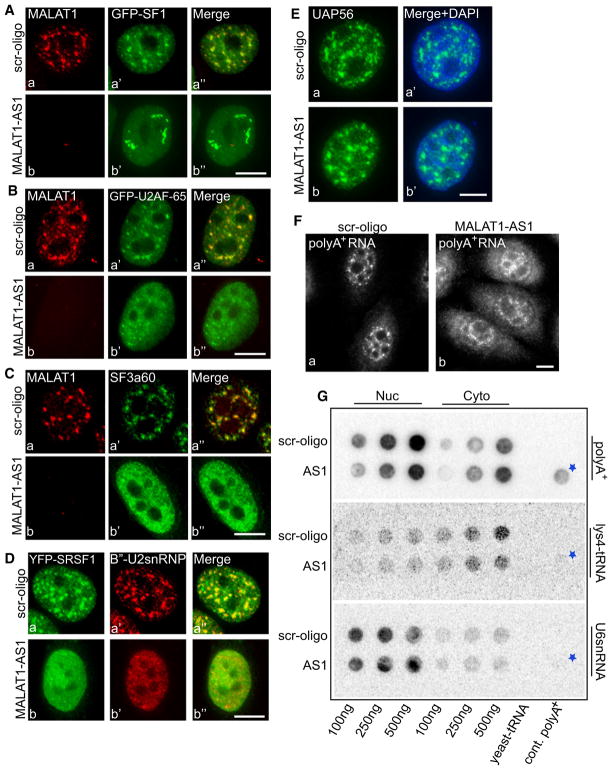
MALAT1 Modulates the Distribution of Splicing Factors at Nuclear Speckles
Long nrRNAs have been proposed to play critical roles in the regulation of gene expression and, likely intimately related to this role, in providing a structural framework for specific subnuclear domains (Prasanth and Spector, 2007; Wilusz et al., 2009). We examined the effect of MALAT1 depletion on the localization of various pre-mRNA processing factors in MALAT1-depleted cells after 48 hr of MALAT1-specific phosphorothioate modified antisense oligonucleotide treatment (Figure 4). RNA-FISH and qPCR assays revealed ~85%–90% loss of MALAT1 in cells after antisense oligonucleotide treatment (Figure 4 and Figure S4A). MALAT1 depletion resulted in decreased nuclear speckle association of several of the GFP-tagged or endogenous pre-mRNA splicing factors, including SF1 (Figure 4A), U2AF-65 (Figure 4B and Figure S4B), SF3a60 (Figure 4C and Figure S6B), and B″-U2snRNP (Figure 4D and Figure S4C). SF1 is a splicing factor that recognizes the branchpoint sequence and, together with U2AF, promotes binding of U2-snRNP to pre-mRNA, whereas SF3a60 and B″ proteins are integral components of U2-snRNP (Jurica and Moore, 2003). More than 60% of MALAT1-depleted cells also displayed decreased nuclear speckle distribution of various YFP-tagged SR proteins, including SRSF1 (Figures 4Da–4Db and Figure S4C; n = 500) and SRSF3 (data not shown). In most of the cases examined, the decrease in speckle association of splicing factors upon MALAT1 depletion was associated with an increase in the levels of a homogeneous nuclear pool (compare Figures 4Ba′ and 4Bb′, Figures 4Ca′ and 4Cb′, and Figure 4D). However, endogenous or GFP-UAP56 (Figure 4E and Figure S4D), a protein that is part of a splicing-dependent mRNA export complex, and SON (data not shown) continued to localize to nuclear speckles in MALAT1-depleted cells. Our results imply that MALAT1 is not a structural RNA, similar to what has been suggested earlier (Clemson et al., 2009), but that it modulates the speckle association of a subset of pre-mRNA splicing factors.
Figure 4. MALAT1 Is Not Required for Nuclear Speckle Structural Integrity but Influences the Distribution of Splicing Factors to Nuclear Speckles.
(A and B) MALAT1 RNA-FISH in control (scr-oligo, a–a″) and MALAT1 antisense oligo-treated (48 hr post antisense treatment; b–b″) HeLa cells expressing GFP-SF1 (A) or GFP-U2AF65 (B).
(C) MALAT1 RNA-FISH and SF3a60 immunolocalization in control (Ca–Ca″) and MALAT1-depleted (Cb–Cb″) HeLa cell.
(D) YFP-SRSF1 and B″-U2snRNP immunolocalization in scr-oligo (Da–Da″) and MALAT1-depleted (Db–Db″) HeLa cell.
(E) UAP56 immunolocalization in control (Ea–Ea′) and MALAT1-depleted (Eb–Eb′) HeLa cell.
(F) RNA-FISH using oligo-dT probe in control (Fa) and MALAT1 antisense oligonucleotides-treated (Fb) HeLa cells. The bars in all the figures represent 10 μm.
(G) RNA slot blot using oligo dT probe in control (scr-oligo) and MALAT1 antisense oligonucleotide-treated (AS1) HeLa cell nuclear (nuc) and cytoplasmic (cyto) extracts showed increased levels of cytoplasmic poly(A)+ RNA upon MALAT1 depletion (fold change, MALAT1 AS1/scr 100 ng = 0.63; 250 ng = 1.36; 500 ng = 1.6). See also Figure S4. The asterisk represents poly(A)+ RNA from control cells.
We also examined the distribution of nuclear poly(A)+ RNA in MALAT1-depleted cells. RNA-FISH using fluorescently labeled Oligo-dT probe in control (scr-oligo-treated) cells revealed speckle localization of poly(A)+RNA (Figure 4F). Oligo-dT probe also hybridized to the cytoplasmic pool of poly(A)+ RNA corresponding to exported mRNAs. Interestingly, MALAT1-depleted cells showed a moderate increase in the cytoplasmic pool of poly(A)+ RNA, as observed both by RNA-FISH as well as RNA slot blot analysis (Figures 4F and 4G). This suggests a possible role for MALAT1 in the nuclear retention of a subpopulation of poly(A)+ RNA.
Prolonged Depletion of MALAT1 Results in Aberrant Mitosis
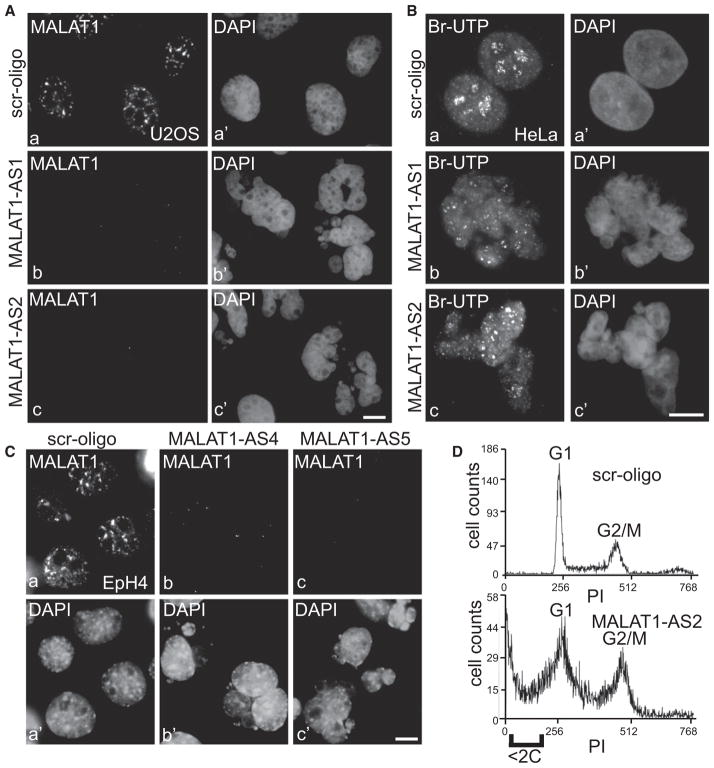
In order to understand the cellular function of MALAT1, cells treated with control and MALAT1 antisense oligonucleotides were monitored up to 72 hr for phenotypic analysis. Staining of DNA with DAPI revealed that a large percentage of MALAT1-depleted cells exhibited fragmented nuclei with each cell containing six to eight nuclei of different sizes (Figures 5A and 5B) (~45% in MALAT1 AS versus 5% in control oligo-treated cells; n = 1000). The nuclear fragmentation phenotype was significant after 72 hr of antisense treatment as compared to 48 hr (~15% in MALAT1 AS treated versus 6% in control cells; n = 1000), even though MALAT1 was depleted in more than 90% of cells within 24–48 hr of AS treatment. This result indicates that the breakdown of nuclei requires prolonged depletion of MALAT1. MALAT1 depletion in mouse mammary cells (EpH4) using mouse MALAT1-specific antisense oligo-nucleotides also revealed similar fragmented-nuclei phenotype (Figure 5C). In vivo Br-UTP transcription assay indicated comparable levels of transcription between control and MALAT1-depleted fragmented nuclei (Figure 5B), suggesting that the observed phenotype was mediated by changes in posttranscriptional events. Interestingly, in all the MALAT1-depleted cells examined, fragmented nuclei were formed due to chromosome segregation defects during mitosis and none of the interphase cells showed nuclear fragmentation phenotype without undergoing mitosis, as indicated by time-lapse live cell imaging in YFP-H2B-expressing cells (Movies S1 and S2; Figure S5). Flow cytometric analysis revealed that MALAT1 depletion resulted in increased cell death, with a large fraction of cells accumulating at G2/M boundary (Figure 5D).
Figure 5. MALAT1 Depletion Results in the Fragmentation of Cell Nuclei.
(A and C) MALAT1 RNA-FISH in U2OS (A) and EpH4 (C) cells that were transfected with control (scr-oligo; a–a′), human specific (A, AS1 and AS2; b–b′ and c–c′) or mouse-specific (C; AS4 and AS5; b–b′ and c–c′) MALAT1 antisense oligonucleotides (72 hr posttransfection). (B) In vivo Br-UTP transcription analyses in control
(Ba–Ba′) and MALAT1 antisense oligo-transfected (Bb–Bb′ and Bc–Bc′) HeLa cells. The bars in (A), (B), and (C) represent 10 μm. (D) Flow cytometric analyses revealed increased cell death (<2C DNA content) with increased G2/M population in MALAT1-depleted HeLa cells. See also Figure S5.
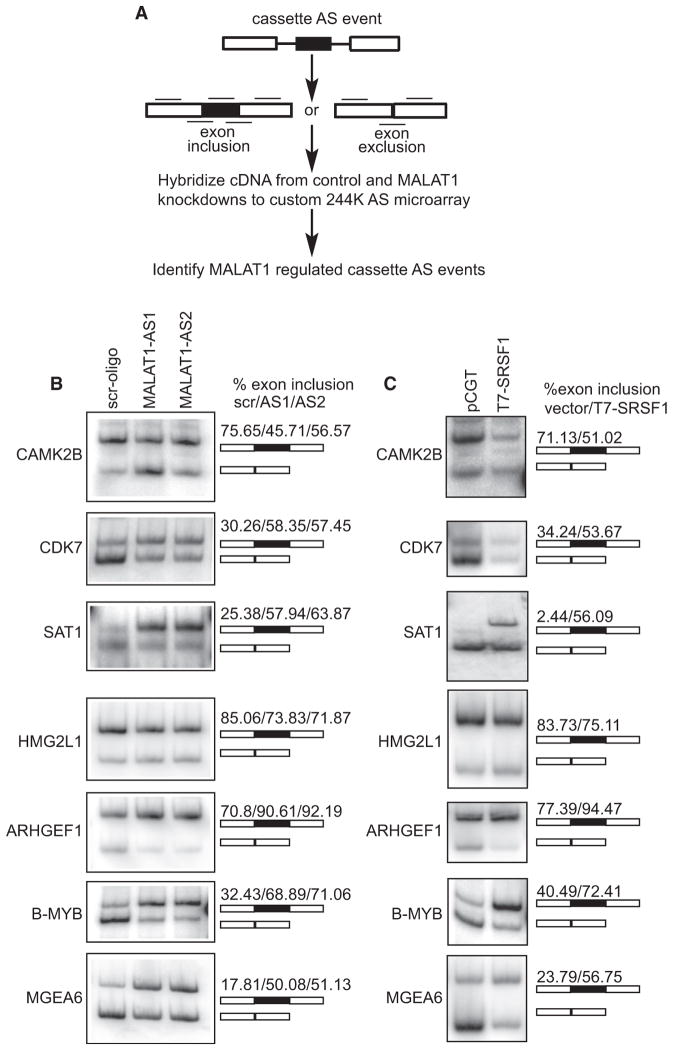
MALAT1 Modulates Alternative Splicing of Endogenous Pre-mRNAs and Regulates Cellular Levels of Phosphorylated SR Proteins
MALAT1 localizes to nuclear speckles and interacts with SR splicing factors, and its depletion results in the differential distribution of pre-mRNA splicing factors. Given the documented roles of SR proteins in splicing regulation, we hypothesized that MALAT1 plays an important role in AS by modulating the pools of active SR proteins and other splicing regulators. To identify AS events regulated by MALAT1, we hybridized labeled cDNA prepared from polyA+ RNA isolated (48 hr after transfection) from MALAT1-antisense oligo and GFP-antisense oligo-treated HeLa cells, to a custom AS microarray capable of profiling ~5782 human cassette AS events that are conserved between human and mouse (Figure 6A) (Calarco et al., 2007; Pan et al., 2004). Percent exon inclusion estimates were determined using the GenASAP algorithm (Shai et al., 2006). Events were initially filtered to include only those that showed constitutive exon (C1 and C2) probe values greater than the negative control probes and to include only the events ranked in the top 50th percentile by GenASAP. Out of 1286 events ranked by GenASAP in expressed genes (18.5%), 238 showed greater than 10% change in inclusion level upon MALAT1 depletion (Table S1). Semiquantitative RT-PCR analysis using primers specific for constitutive exons flanking several of these AS events (CAMK2B, CDK7, SAT1, HMG2L1, TLK2, PAX2, and ARHGEF1) was performed on RNA from control versus MALAT1-depleted cells (using two independent antisense oligonucleotides against MALAT1) and confirmed the microarray predictions (Figure 6B and data not shown). A similar number of AS events predicted not to change in the microarray data did not display significant changes by RT-PCR assays. Our results thus reveal MALAT1-dependent change in AS of several endogenous pre-mRNAs. Furthermore, MALAT1-depleted cells also displayed changes in AS of B-MYB and MGEA6 pre-mRNAs, events that were not represented by probes on our microarray (Figure 6B). These AS events were previously reported to be sensitive to cellular levels of SRSF1 (Karni et al., 2007). In most of the cases we tested, MALAT1 depletion resulted in increased levels of exon inclusion, an event that is often linked to the activity of SR proteins (Bourgeois et al., 2004; Long and Caceres, 2009). Interestingly, RT-PCR from cell extracts that transiently overexpress SRSF1 showed AS patterns similar to those observed upon MALAT1 depletion (Figure 6C). Finally, RNA-IP using SRSF1 antibody showed interaction between SRSF1 and mRNAs that showed changes in AS in MALAT1-depleted or SRSF1-overexpressed cells (data not shown). These results imply that MALAT1 modulates AS of endogenous pre-mRNAs by regulating the differential expression or activity of one or more SR splicing factors, including SRSF1.
Figure 6. MALAT1 Regulates AS of Pre-mRNAs.
(A) Schematic representation of the method used for the global analysis of MALAT1-regulated AS. Black lines designate exon body and exon junction probes used for analyzing AS on a custom 244K Agilent microarray.
(B and C) RT-PCR analysis using primers specific to exons in MALAT1 or to constitutive exons flanking SRSF1-regulated alternative exons in CAMK2B, CDK7, SAT1, HMG2L1, ARHGEF1, B-MYB, and MGEA6 mRNAs indicate changes in AS in MALAT1-depleted (6B) and SRSF1-overexpressed (6C) HeLa cells. Alternative exon-included (upper band) and -excluded bands (lower band) and the percentage change of alternative exon inclusion between scr. oligo/MALAT1-AS oligos (AS1 and AS2) observed is shown. See also Table S1.
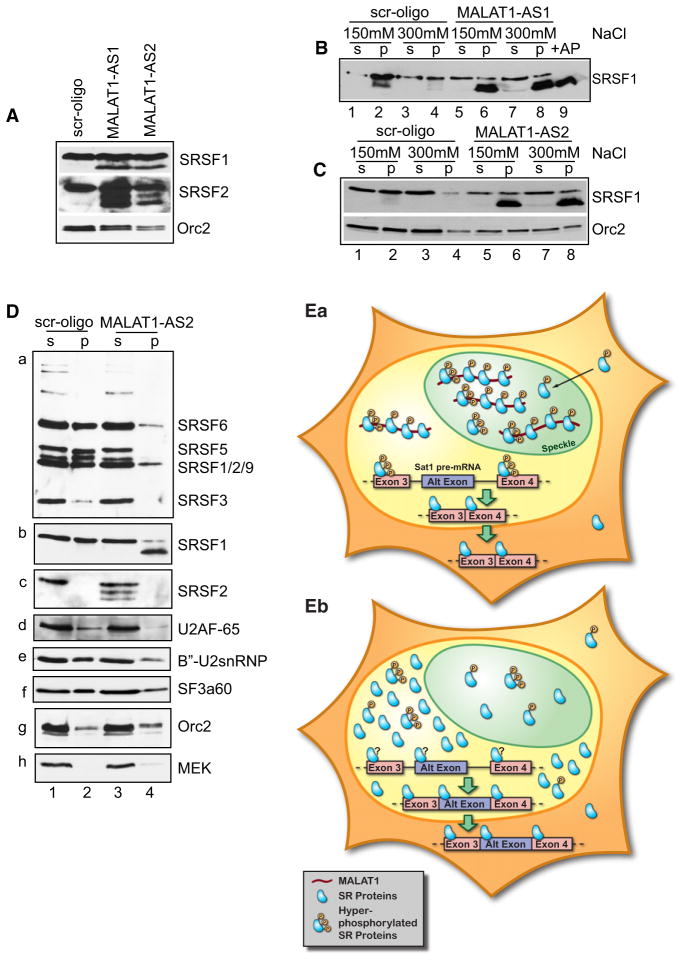
Alterations either in the relative concentrations or in RS-domain phosphorylation of SR proteins are known to modulate AS (Long and Caceres, 2009; Misteli et al., 1998; Stamm, 2008). To investigate the involvement of MALAT1 in SR protein regulation, we examined the levels of SR proteins in MALAT1-depleted cells. Immunoblotting using antibodies against SRSF1 and SRSF2 in MALAT1-depleted cell extracts showed significant increases in levels of SR proteins (Figure 7A). Furthermore, a fraction of SRSF1 and SRSF2 in MALAT1-depleted extracts displayed increased mobility in SDS polyacrylamide gels, which suggested that there is an increased fraction of the dephosphorylated forms of these SR proteins. We examined the relative distribution of different forms of SR proteins in MALAT1-depleted cells by cellular fractionation under different salt extraction conditions. Immunoblot analysis revealed that in MALAT1-depleted, salt-resistant cell extracts (150 mM and 300 mM NaCl; Figures 7B and 7C, lanes 6 and 8), a major fraction of SRSF1 consisted of the faster migrating form (Figures 7B and 7C, compare lanes 2 and 4 with lanes 6 and 8). The mobility of the faster-migrating fraction of SRSF1 was comparable to the pool of SRSF1 from total extracts that was dephosphorylated using phosphatase (+AP) (Figure 7B, compare lanes 6 and 8 with lane 9 and Figure S6A; compare lanes 6 and 8 with lane 9). These results demonstrate that a significant fraction of SRSF1 in MALAT1-depleted cells is present in dephosphorylated form. Immunoblot analyses using 3C5 (Figure 7Da) and mAB104 (data not shown) antibodies (both recognize phosphorylated form of SR proteins) revealed decreased levels of all of the phosphorylated SR proteins in the salt-resistant pellet fraction in MALAT1-depleted cells (Figure 7Da). Interestingly, unlike SRSF1, in MALAT1-depleted cells, all of the dephosphorylated SRSF2 was present in the salt-sensitive supernatant fraction, indicating differential extractability of various SR-proteins in MALAT1-depleted cells (Figure 7Dc). We also analyzed the relative distribution of several of the pre-mRNA splicing factors (U2AF-65, B″-U2snRNP and SF3a60) that showed decreased association with nuclear speckles upon MALAT1-depletion. The insoluble pellet fraction in MALAT1-depleted cells contained comparatively lesser amounts of all of the splicing factors tested (Figures 7Dd–7Df). To test whether the nuclear speckle disassociation of SRSF1 observed in a significant fraction of MALAT1-depleted cells is due to their dephosphorylation, we compared the nuclear speckle association of exogenously-expressed wild-type (WT) and a phosphomimetic mutant of SRSF1 (SRSF1-RD, where serines in the RS domain are substituted with aspartate residues) (Cazalla et al., 2002) in MALAT1-depleted cells. Immunolocalization studies revealed that both WT and SRSF1-RD mutant displayed homogenous distribution in MALAT1-depleted cells, indicating that the speckle disassociation of SRSF1 in MALAT1-depleted cells is not primarily due to its dephosphorylation but that both events could be regulated by MALAT1 (Figure S6B). Taken together, our results demonstrate that MALAT1 depletion increases the levels of cellular SR proteins and alters the distribution and the ratio of phosphorylated to dephosphorylated pools of SR proteins. Such changes most likely influence AS of pre-mRNAs and lead to corresponding changes in the expression of specific protein isoforms in cells.
Figure 7. MALAT1 Modulates the Cellular Levels and Phosphorylation of SR Proteins.
(A) Immunoblot assays in control (scr) and MA-LAT1-depleted total cell extracts (AS1 and AS2) revealed increased levels of SRSF1 and SRSF2 in MALAT1-depleted cells. Relative fold increase in the levels of SR proteins in MALAT1-depleted cells: SRSF1, AS1 2.07 and AS2 3.13; SRSF2, AS1 3.15 and AS2 1.52. Note that the MALAT1-depleted cell extracts contained extra bands of SR proteins that show fast mobility.
(B and C) Control (lanes 1–4) and MALAT1 anti-sense oligonucleotides (B, AS1; and C, AS2) transfected (lanes 5–8) HeLa cells were extracted using 150 and 300 mM NaCl salt into soluble supernatant (s; lanes 1, 3, 5, and 7) and insoluble chromatin pellet (p; lanes 2, 4, 6, and 8) fractions and immunoblotted using SRSF1 antibody. Lane 9 (B) represents total cell extract treated with Antarctic phosphatase (+AP).
(D) Immunoblot analyses of control and MALAT1-depleted extracts (300 mM NaCl extracted) with 3C5 (Da), SRSF1 (Db), SRSF2 (Dc), U2AF-65 (Dd), B″-U2snRNP (De), SF3a60 (Df) antibodies. Orc2 and MEK were used as loading controls. (E) Hypothetical model depicting the role of MA-LAT1 in AS regulation. (Ea) In normal cells, MA-LAT1, by associating with SR proteins in nuclear speckles and in the nucleoplasm, regulates their recruitment to the pre-mRNA, thereby regulating AS. Here we have shown SAT1 pre-mRNA as an example that undergoes alternate exon exclusion in normal cells. However, in MALAT1-depleted cells (Eb), cellular levels of SR proteins are increased and are also present predominantly in the dephosphorylated form, resulting in changes in AS of pre-mRNA. In case of SAT1 pre-mRNA, MALAT1 depletion results in the inclusion of an alternative exon containing weak splice sites. See also Figure S6.
DISCUSSION
Recent studies have revealed the existence of many lncRNAs in cells (Mercer et al., 2009; Wilusz et al., 2009). In the present study, we have examined the role of an abundant (RNA copy number in HeLa cells estimated to be ~2500 copies/cell [Figure S6C]) long noncoding nrRNA, MALAT1, in gene expression control. Phylogenetic analysis indicates that MALAT1 is highly conserved in mammals (Hutchinson et al., 2007; Lin et al., 2006). We demonstrate that MALAT1 localizes to nuclear speckles and interacts with several pre-mRNA splicing factors. Our studies clearly show that unlike other long nrRNAs, NEAT1 or Hsrω-n (Prasanth et al., 2000), MALAT1 does not play a structural role in the formation or maintenance of a nuclear domain but influences the distribution of pre-mRNA splicing factors to nuclear speckles. A recent study reported that mAb SRSF2 or SC35 immunoreactive nuclear speckles are unaltered in MALAT1-depleted cells (Clemson et al., 2009). The mAb SRSF2 antibody (Fu and Maniatis, 1990) preferentially recognizes the phosphorylated SR proteins (Prasanth et al., 2003). We demonstrate that MALAT1 depletion results in an increase in the dephosphorylated pool of SR proteins that display a more homogeneous nuclear distribution. Since the SRSF2 mAb primarily recognizes phosphorylated SR proteins, it likely detects a steady-state pool of residual phosphorylated SR proteins present in MALAT1-depleted speckles. We have shown mislocalization of speckle components and changes in AS of pre-mRNAs in MALAT1-depleted cells. It is possible that changes in AS of endogenous pre-mRNAs are a consequence of the mislocalization of pre-mRNA processing factors and that MALAT1 plays critical roles in the cycling of SR proteins between speckles and sites of transcription.
We have demonstrated that a significant fraction of SR proteins in MALAT1-depleted cells are present in dephosphorylated form. In general, a continuous phosphorylation/dephosphorylation cycle of SR proteins is required for pre-mRNA splicing and is also an important contributor to the regulation of AS patterns that determine the fate of pre-mRNAs. For instance, hyperphosphorylated SR domains influence the binding of SR proteins to pre-mRNA and regulate splice site selection by dictating protein-protein and protein-RNA interactions within the spliceosome, whereas partially dephosphorylated SR proteins support first step transesterification reactions (Cao et al., 1997; Xiao and Manley, 1997, 1998). The phosphorylation status of SR proteins also influences intranuclear trafficking of SR proteins between nuclear speckles and transcription sites (Caceres et al., 1997; Misteli et al., 1998). We hypothesize that MALAT1 regulates the cellular levels of phosphorylated SR proteins, thereby influencing the cellular ratio of phosphorylated versus dephosphorylated forms. By controlling the levels of phosphorylated SR proteins, MALAT1 not only modulates AS but also likely regulates other posttranscriptional gene-regulatory mechanisms linked to SR proteins, including RNA export, nonsense-mediated decay, and translation (Long and Caceres, 2009; Stamm, 2008). The increased cytoplasmic pool of poly(A)+ RNA observed in MALAT1-depleted cells could be due to the increased cellular levels of dephosphorylated SRSF1, since dephosphorylation of SRSF1 is critical for mRNP export and is also known to enhance binding of SRSF1 to cytoplasmic mRNA (Huang et al., 2004; Sanford et al., 2005). At present, it is not clear how MALAT1 depletion alters the ratios of phosphorylated to dephosphorylated SR proteins in the cell. It is possible that MALAT1 could modulate the activity of kinases (SRPKs or Clk/STY family), or of phosphatases (PP1 or PP2A), that modify SR proteins (Stamm, 2008). Recent studies revealed that both SRPK1 and PP1 influence AS by regulating SR protein phosphorylation (Shi and Manley, 2007; Zhong et al., 2009). Interestingly, we observed altered localization of SRPK1 in MALAT1-depleted cells, indicating the potential involvement of MALAT1 in SRPK1 activity (Figure S6D). Alternatively, MALAT1, by interacting with SR proteins could influence their stability. Increased cellular levels of SR proteins as well as changes in AS of pre-mRNAs (similar to that of SRSF1 overexpressing cells) in MALAT1-depleted cells further support this possibility. Future studies will address how MALAT1 modulates levels of phosphorylated SR proteins.
An important question arising from observations that SR proteins regulate AS is how this regulation is achieved in vivo in the context of specific cell or tissue type, or in response to extracellular signals. In the present study, we demonstrate that MALAT1, by interacting with a specific set of SR splicing factors, acts as a “molecular sponge” to regulate SR protein activity (Figure 7E). Such an interaction between MALAT1 and SR proteins could create a gradient of functionally competent SR proteins and further ensure that SR proteins are present at the right time, place, and concentration to regulate the AS of cellular pre-mRNAs.
EXPERIMENTAL PROCEDURES
Further detailed experimental procedures are described in the Supplemental Information.
Antisense Oligonucleotide and siRNA Treatment
Phosphorothioate internucleosidic linkage-modified antisense oligonucleotides (with five 2′-O-methoxyethyl nucleotides on the 5′ and 3′ ends and ten consecutive oligodeoxynucleotides to support RNase H activity) (AS1, GG GAGTTACTTGCCAACTTG; AS2, ATGGAGGTATGACATATAAT; AS4, AGGC AAACGAAACATTGGCA; AS5, CGGTGCAAGGCTTAGGAATT) were used to deplete human (AS1 and AS2) or mouse (AS4 and AS5) MALAT1 (Isis Pharmaceuticals, CA, USA). The oligonucleotides were transfected to cells two times (48 hr) or three times (72 hr) within a gap of 24 hr, at a final concentration of 100 nM, using Lipofectamine RNAi max reagent as per the manufacturer’s instructions (Invitrogen, USA). Depletion of SRSF1, SRSF2, SON, B″-U2snRNP, and PRP6 was performed by using double-stranded siRNAs (Dharmacon, USA; and IDT, USA) as previously described (Prasanth et al., 2004). The siRNA sequences will be provided upon request.
RNA Coimmunoprecipitation
RNA-IP was performed based on the protocol by Sun et al., with minor modifications (Sun et al., 2006). HeLa cells were transiently transfected with T7-tagged plasmid constructs (2–3 μg) (pCGT-vector, pCGT-SRSF1, SRSF1-ΔRRM1, SRSF1-ΔRRM2, SRSF1-ΔRS, SRSF2, SRSF3, SRSF5, PSP1, or PRP6) and RNA immunoprecipitation was performed utilizing reversible chemical crosslinking of RNA-protein interactions by formaldehyde followed by immunoprecipitation using T7 (Novagen, USA) or SRSF1 antibodies (mAb96; for endogenous IP). After IP, extracts were reverse cross-linked, total RNA was extracted using Trizol LS (Invitrogen, USA) and treated with RNase-free DNase I (Invitrogen, USA), and RT-PCR was conducted using either random-hexamer or oligo-dT primers as per the manufacturer’s instructions (Applied Biosystems, USA). PCR or qPCR was performed using specific set of primers (detailed in the Supplemental Information).
Alternative Splicing Microarray Analysis
AS events conserved between human and mouse represented on a customized human 244K microarray (Agilent) were identified as previously described (Calarco et al., 2007). Microarray design was also as previously described (Pan et al., 2004). Hybridization of Cy3- and Cy5-labeled cDNA synthesized from polyA+ RNA from MALAT1-antisense oligo treated or GFP-antisense oligo-treated HeLa cells was performed using a HS4800 Pro Hybridization Station (Tecan) at 42°C for 22 hr. Microarray slides were scanned with an Agilent microarray scanner at 5 μm resolution, and TIFF images were processed using Feature Extraction Software (Agilent). Processed data were then analyzed using the GenASAP algorithm (Shai et al., 2006).
The AS microarray data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession number GSE22963 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22963).
Supplementary Material
Acknowledgments
We would like to thank Drs. D. Bernard and A. Bessis (mouse MALAT1 plasmid); M. Carmo-Fonseca (GFP-SF1, U2AF65, U2AF35, UAP56); J. Caceres, J. Sanford, and A. Krainer (SRSF1 constructs); and R. Lührmann (PRP6 plasmid and pAb), J. Stevenin (SRSF2 mAb), and J. Nickerson (UAP56 pAb) for their kind gift of reagents. We thank Drs. M. Hastings, A. Krainer, J. Caceres, G. Carmichael, T. Misteli, and D. Spector for the valuable discussions; J. Sanford for sharing SRSF1 CLIP-seq data; and J. Kim, Y. Ge, and R. Zheng for technical help. We also would like to thank Drs. N. Aki-mitsu, M. Bellini, A. Lal, P. Newmark, and Prasanth lab members for critical reading of the manuscript. This work was supported by a grant UIUC-ICR-MCB (to K.V.P.) and grant NSF0843604 (to S.G.P.). A.T.W., S.M.F., and C.F.B. are employees of Isis Pharmaceutical Corp. They receive salary from the company and hold stock options.
Footnotes
Supplemental Information includes six figures, one table, two movies, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:10.1016/j.molcel.2010.08.011.
References
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Xing Y, Caceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear non-coding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 2010;277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large non-coding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2006;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Achsel T, Luhrmann R. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J Mol Biol. 2000;298:567–575. doi: 10.1006/jmbi.2000.3685. [DOI] [PubMed] [Google Scholar]
- Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macro-molecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC. Omega speckles—a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci. 2000;113:3485–3497. doi: 10.1242/jcs.113.19.3485. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Bio-technol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci USA. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENep-silon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai O, Morris QD, Blencowe BJ, Frey BJ. Inferring global levels of alternative splicing isoforms using a generative model of microarray data. Bioinformatics. 2006;22:606–613. doi: 10.1093/bioinformatics/btk028. [DOI] [PubMed] [Google Scholar]
- Sharma A, Takata H, Shibahara KI, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Spector DL. SnapShot: cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN varepsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “Dark Matter” transcripts are associated with known genes. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.