Abstract
Rare variant tests have been of great interest in testing genetic associations with diseases and disease-related quantitative traits in recent years. Among these tests, the sequence kernel association test (SKAT) is an omnibus test for effects of rare genetic variants, in a linear or logistic regression framework. It is often described as a variance component test treating the genotypic effects as random. When the linear kernel is used, its test statistic can be expressed as a weighted sum of single-marker score test statistics. In this paper, we extend the test to survival phenotypes in a Cox regression framework. Because of the anticonservative small-sample performance of the score test in a Cox model, we substitute signed square-root likelihood ratio statistics for the score statistics, and confirm that the small-sample control of type I error is greatly improved. This test can also be applied in meta-analysis. We show in our simulation studies that this test has superior statistical power except in a few specific scenarios, as compared to burden tests in a Cox model. We also present results in an application to time-to-obesity using genotypes from Framingham Heart Study SNP Health Association Resource.
Keywords: Cox proportional hazard model, likelihood ratio test, rare variant analysis, variance component test
Introduction
Rare genetic variants may account for some of the unexplained heritability in previous genome-wide association studies (GWAS) [Eichler et al., 2010]. Single-marker tests, which are commonly used in GWAS, have very little power to detect association with rare genetic variants with small-to-moderate effect sizes. In recent years, rare variant tests that aggregate information from multiple genetic markers within prespecified gene regions have been proposed. One class of rare variants tests is the burden test (BT), which collapses genotypes from multiple rare variants into a summary genetic burden score and tests the association between the trait of interest and the burden score [Li and Leal, 2008; Madsen and Browning, 2009; Morgenthaler and Thilly, 2007; Morris and Zeggini, 2010]. In practice, this test can be performed as a Wald test, a score test, or a likelihood ratio test (LRT), with or without weights. Morris and Zeggini suggested use of the LRT in BTs [Morris and Zeggini, 2010]. BTs are most powerful when the proportion of causal variants is high and all causal variants have the same direction of effects in the prespecified gene region tested. They have very little power when causal variants with both protective and detrimental effects are present in the test region [Wu et al., 2011].
Alternatively, other tests are performed without collapsing genotypes from multiple rare variants [Neale et al., 2011; Pan, 2009; Wu et al., 2011]. The sequence kernel association test (SKAT) is one of these tests [Wu et al., 2011]. It was developed as a score test on the variance component parameter for the genetic random effects in linear and logistic mixed effects models. The test statistic can be written as a weighted sum of single-marker score test statistics when using the linear kernel, which can be applied in meta-analyses [Lee et al., 2013; Lumley et al., 2012]. SKAT has several advantages over competing rare variant tests: it is powerful when both protective rare variants and detrimental rare variants are present; the score test requires fitting the null regression model only once; P-values are computed analytically.
However, most rare variant tests to date were developed for binary or quantitative outcomes, and little attention has been paid to rare variant tests for time-to-event outcomes. Cai et al. [2011] developed a kernel machine approach to test the pathway effect on survival outcomes, and Lin et al. [2011] extended this approach to test single nucleotide polymorphism (SNP) sets of common genetic variants. These approaches may also be applied as a rare variant test. However, the necessity of resampling to calculate P-values increases the computational burden and prevents the popular use of these approaches in genome-wide analyses.
In this article, we derive an SKAT score statistic for survival outcomes in a Cox proportional hazard model framework. Assuming linear kernels, we rewrite this SKAT score statistic as a weighted sum of single-marker score test statistics, which can be easily obtained in standard statistical packages. However, it is well known that the score test in the Cox model may be anticonservative when the effective sample size is small [Fleming et al., 1987]. Thus, we propose replacing single-marker score test statistics by corresponding LRT statistics. We illustrate in details how this approach can be applied in meta-analysis, without having access to individual level data.
In our simulation studies, we compare the SKAT approaches with BTs from the Cox model. We demonstrate that our SKAT approach has higher power than BTs from the Cox model when causal variants with both protective and detrimental effects are present in the test region, or when the association signal is sparse. Finally, we illustrate our approach by analyzing a time-to-obesity phenotype measured in the Original and Offspring Cohorts from the Framingham Heart Study (FHS), using genotypes from SNP Health Association Resource (SHARe).
Methods
SKAT in the Cox Proportional Hazard Model
We first define notations used throughout this section. Let yi = (ti, δi), i = 1, 2,…, n be independent time-to-event observations with time ti and event/censoring indicator δi, Xi, and Gi be row vectors of p covariates and q genotypes for individual i, then the Cox proportional hazard model is
where β is p fixed effects for the covariates, and γ is q random effects for the genotypes, with mean 0 and variance σ2Iq. W is a diagonal weight matrix. We are interested in testing H0: σ2 = 0 vs. H1: σ2 > 0.
Let X be an n × p matrix with rows Xi, G be an n × q matrix with rows Gi, r be a vector of martingale residuals estimated from the null model
then the SKAT statistic is
Let Σ be the covariance matrix of the vector WGTr under the null hypothesis, then
where λj are the eigenvalues of Σ, and are independent chi-square distributions with 1 degree of freedom. Please see Appendix A for the derivation.
We also show in Appendix A that if zj are single-marker score test statistics (which follow a standard normal distribution under the null hypothesis), the SKAT statistic can be written as
where Σjj are the diagonal elements of the matrix Σ. In this formulation, we can replace score test statistics zj by the square root of corresponding single-marker LRT statistics. These two approaches are asymptotically equivalent, but may have different performance in small samples.
Meta-Analysis
One advantage of writing the SKAT statistic as the weighted sum of single-marker test statistics is the straightforward extension to meta-analysis, when individual level data are not available. Assuming there are K cohorts in the meta-analysis, we only need covariance matrices Σ(k) from each cohort, and single-marker test statistics zj(k) for each genetic variant from each cohort. Then the summary SKAT statistic is
We note that single-marker test statistics zj(k) should be signed square root of LRT statistics to reflect the directions of effects in different cohorts. Assuming the cohorts are independent, then under the null hypothesis, Q follows a weighted sum of independent chi-square distributions with 1 degree of freedom, with weights equal to the eigenvalues of
BTs in the Cox Proportional Hazard Model
In the Cox proportional hazard model
if γ is a vector with all elements γ0, then testing the genotypic effects is H0: γ0 = 0 vs. H1: γ0 = ≠ 0. This is a BT with the collapsed genetic burden score (weighted sum of genetic variants) , where Gij is the j th element of vector Gi for individual i, and Wjj is the j th diagonal element of the weight matrix W. The test can be performed as a Wald test, a LRT, or a score test.
Simulation Studies
We performed simulation studies to evaluate the empirical type I error rates and empirical power for four statistical tests: 1. SKAT in the Cox proportional hazard model, using LRT statistics from single-marker tests (Cox SKAT LRT); 2. SKAT in the Cox proportional hazard model, using score test statistics from single-marker tests (Cox SKAT Score); 3. BT in the Cox proportional hazard model, using LRT (Cox BT LRT); 4. BT in the Cox proportional hazard model, using score test (Cox BT Score). In all simulation studies we used Wu weights [Wu et al., 2011], which are the beta distribution density function with parameters 1 and 25, evaluated at the minor allele frequency (MAF).
Type I Error
For each parameter setting, we simulated 4,000 genotype datasets with a sample size of 2000 and 20 biallelic genetic markers with MAF randomly sampled from Unif (0.005, 0.05). The linkage disequilibrium (LD) correlation between adjacent markers was fixed at r = 0.5, and decays as an autoregressive model with order 1 for farther markers. For each genotype dataset, 10,000 phenotype datasets including covariates: age ~ N (50, 52), and sex ~ Bernoulli (0.5) were simulated.
The baseline (age = 50, sex = 0) survival time was simulated from a Weibull (2, 2) [Bender et al., 2005]. Assuming proportional hazards, the survival time for an individual with covariates age and sex was simulated from
where U was randomly sampled from Unif (0, 1).
We simulated four censoring schemes for the censoring time C: 1. C = ∞, no censoring; 2. C ~ Unif (0, 10); 3. C ~ Unif (0, 5); 4. C ~ Unif (0, 2). Then we calculated the event time ti = min(Ti, Ci) and event/censoring indicator δi = I(Ti ≤ Ci).
Power
For each parameter setting, we simulated 100 genotype datasets, and 10,000 phenotype datasets for each genotype dataset. We followed the same procedure as in type I error simulations to simulate genotype datasets and covariates. The baseline (age = 50, sex = 0) survival time was also simulated from a Weibull (2, 2). Assuming proportional hazards, the survival time for an individual with covariates age and sex, and genotypes gj(j = 1, 2, …, q) was simulated from
We varied the proportion of causal markers from 20% to 50% and 80%, and we simulated both same and opposite directions of effects. Causal markers were randomly selected for each phenotype replicate, and γj = 0 for neutral markers. For causal markers, the effect size is
where MAFj is the MAF of marker j, D is the genotype correlation matrix for the 20 markers, and v is a vector indicating the directions of causal markers in each replicate. The constant c was fixed at 0.01 in all scenarios. The censoring scheme was the same as in type I error simulations. Empirical power was calculated at the significance level of 0.001.
We also performed two additional simulation studies to compare the methods in scenarios when the association signal is sparse. We simulated 20 biallelic genetic markers as in previous simulation studies, with only two of them causal in the same direction. We also simulated four biallelic genetic markers with correlation between adjacent markers fixed at r = 0.5, and decays as an autoregressive model with order 1 for farther markers, with two causal markers with the same direction of effect.
Results
Type I Error Simulations
Empirical type I error rates of four methods at significance levels 0.05, 10−3, 10−4, and 2.5 × 10−6 from simulation studies are presented in Table 1. The Cox SKAT Score is conservative at low alpha levels when there is no censoring or when the proportion of censoring is low or modest, but anticonservative when the proportion of censoring is high. This is also evident in supplementary Figure S1, where points corresponding to Cox SKAT Score P-values are below the reference line of uniform distribution, and more apparently in Figure 1, where points corresponding to Cox SKAT Score P-values are above the reference line. The Cox BT Score is generally anticonservative at low alpha levels in all censoring scenarios, as also seen in supplementary Figures S1–S3 and Figure 1.
Table 1.
Relative empirical type I error rates from simulation studies. Each entry represents the proportion of P-values less than corresponding alpha level from 40 million simulation replicates, divided by alpha
| Alpha | Censoring | Median censor % | Cox SKAT LRT | Cox SKAT Score | Cox BT LRT | Cox BT Score |
|---|---|---|---|---|---|---|
| 0.05 | ∞ | 0 | 1.02 | 1.00 | 1.01 | 1.01 |
| Unif (0, 10) | 17.5% | 1.02 | 1.00 | 1.01 | 1.02 | |
| Unif (0, 5) | 35.0% | 1.02 | 1.00 | 1.01 | 1.02 | |
| Unif (0, 2) | 74.2% | 1.05 | 1.01 | 1.01 | 1.01 | |
| 10−3 | ∞ | 0 | 1.02 | 0.85 | 1.02 | 1.11 |
| Unif (0, 10) | 17.5% | 1.03 | 0.86 | 1.02 | 1.13 | |
| Unif (0, 5) | 35.0% | 1.00 | 0.87 | 1.02 | 1.15 | |
| Unif (0, 2) | 74.2% | 0.98 | 1.17 | 1.04 | 1.22 | |
| 10−4 | ∞ | 0 | 1.03 | 0.76 | 1.02 | 1.27 |
| Unif (0, 10) | 17.5% | 1.03 | 0.79 | 1.05 | 1.32 | |
| Unif (0, 5) | 35.0% | 0.98 | 0.81 | 1.03 | 1.34 | |
| Unif (0, 2) | 74.2% | 0.91 | 1.57 | 1.07 | 1.67 | |
| 2.5 × 10−6 | ∞ | 0 | 1.11 | 0.65 | 1.01 | 1.72 |
| Unif (0, 10) | 17.5% | 1.10 | 0.92 | 1.14 | 2.01 | |
| Unif (0, 5) | 35.0% | 0.86 | 0.73 | 0.79 | 2.20 | |
| Unif (0, 2) | 74.2% | 0.73 | 3.03 | 1.05 | 3.25 |
Figure 1.
Quantile–Quantile plot for type I error simulation results from high proportion of censoring scenario. P-values from 40 million simulation replicates using four methods are plotted against expected P-values (uniform distribution on (0, 1)). The censoring time was randomly sampled from a uniform distribution on (0, 2), corresponding to 74.2% median censoring proportion in 40 million replicates. We simulated 20 genetic variants and the total sample size was 2000.
In all censoring scenarios, Cox SKAT LRT and Cox BT LRT have empirical type I error rates close to corresponding significance levels, at all four alpha levels. Score tests have inflated type I error rates in certain scenarios. In subsequent power simulation studies, we only compared Cox SKAT LRT and Cox BT LRT.
Power Simulations
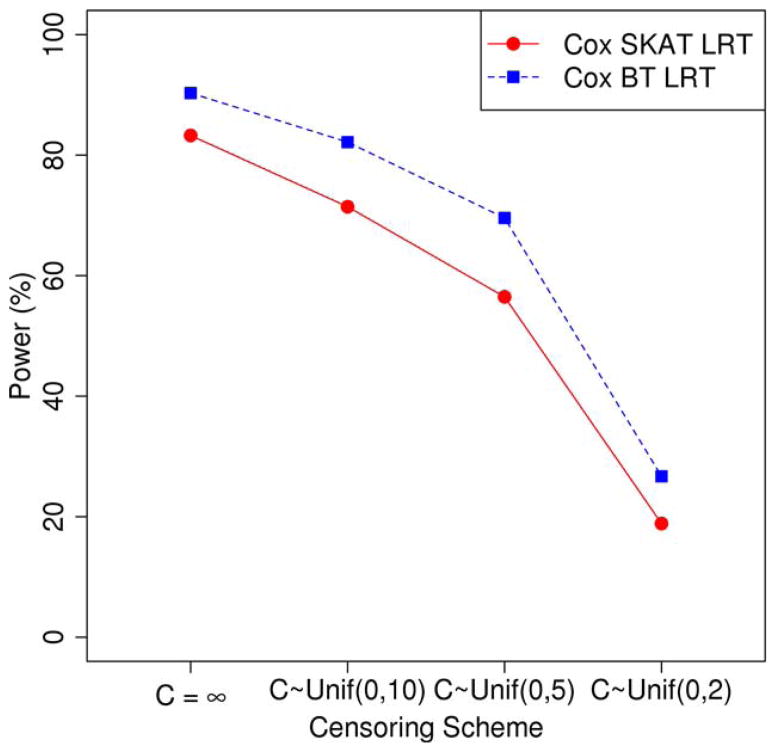
We present empirical power results from 1 million simulation replicates in the scenario where 50% of total genetic variants are causal in Figures 2 and 3. Figure 2 indicates that Cox BT LRT is more powerful than Cox SKAT LRT when all causal variants have the same direction of effects, and Figure 3 suggests that Cox BT LRT has almost no power when 50% of causal variants have positive effects and the other 50% have negative effects.
Figure 2.
Power simulation results from 10 positively associated and 10 neutral genetic variants. Empirical power evaluated at the significance level of 0.001. The total sample size was 2000.
Figure 3.
Power simulation results from five positively associated, five negatively associated, and 10 neutral genetic variants. Empirical power evaluated at the significance level of 0.001. The total sample size was 2000.
We also present in supplementary Figures S4 and S5 empirical power results from 1 million simulation replicates in the scenario where 80% of total genetic variants are causal, and in supplementary Figures S6 and S7 empirical power results from the scenario where 20% of total genetic variants are causal. The conclusions are the same as in Figures 2 and 3: Cox SKAT LRT is most powerful when causal variants have different directions of effects, but slightly less powerful than Cox BT LRT when all causal variants have positive effects.
When only 2 of 20 (10%) genetic variants included in the test are causal, Cox SKAT LRT outperforms Cox BT LRT, even when causal variants have the same direction of effects (supplementary Figure S8), although the difference is small. Cox SKAT LRT has similar power with Cox BT LRT when 2 of 4 genetic variants included in the test are causal with the same direction of effects (supplementary Figure S9).
Application to Framingham Heart Study Data
The Original Cohort from the FHS was initiated in 1948 and included 5,209 participants from the town of Framingham, MA, with roughly equal numbers of men and women. These Original Cohort participants have undergone physical evaluations for cardiovascular disease and related risk factors roughly every 2 years. In 1971, the Offspring Cohort, consisting of 5,124 offspring from Original cohort members and their spouses, were recruited in the study. The Offspring Cohort participants have attended physical exams approximately every four years. We performed a genome-wide sliding window analysis in unrelated individuals from the FHS Original and Offspring Cohorts to illustrate our method and investigate its performance on real data. We have collected age and body mass index (BMI) data from 27 physical examinations for the Original Cohort and from eight physical examinations for the Offspring Cohort. We excluded individuals with BMI >30 at baseline and calculated time-to-obesity as the phenotype, where obesity was defined as having BMI >30. We selected 1,629 unrelated individuals with genotypes available in SHARe (955 cases and 674 controls), and performed the analysis using Cox SKAT LRT and Cox BT LRT, adjusting for sex, baseline age, cohort and first 10 principal components [Price et al., 2006]. We used a sliding window method to define the region to be analyzed. For each window of width 100 kb, with 50 kb each overlapping with the previous and subsequent windows, we included all SNPs regardless of MAF or annotation information. We used Wu weights [Wu et al., 2011] in the analysis.
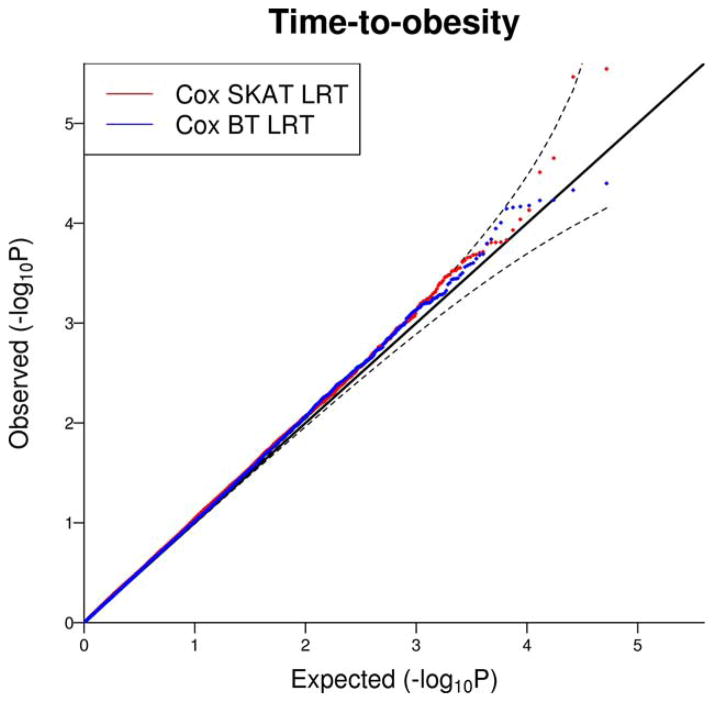
We obtained 55,616 windows in total. After removing 3,353 windows with 0 or 1 genetic variant, we had results from 52,263 windows, with the number of genetic variants ranging from 2 to 93 with median 17. We did not find any genome-wide significant associations at the significance level of 1.0 × 10−6. In Figure 4, we present the P-values from this analysis, and we can see that points are very close to the reference line of uniform distribution.
Figure 4.
Quantile–Quantile plot for the sliding window analysis on time-to-obesity in Framingham Heart Study. P-values from Cox SKAT LRT and Cox BT LRT are plotted against expected P-values (uniform distribution on (0, 1)). Unrelated individuals were selected from the original and offspring cohorts in Framingham Heart Study. Genotypes from SNP Health Association Resource were used in the genome-wide sliding window analysis with width 100 kb.
In addition to the genome-wide sliding window analysis, we performed a candidate gene study to test the association between obesity risk and 8 genes previously reported [Speliotes et al., 2010] to be associated with BMI and biologically meaningful: MC4R, BDNF, SH2B1, POMC, GIPR, HMGCR, TUB, and HMGA1. We used the same individuals as in the genome-wide sliding window analysis, and performed Cox SKAT LRT and Cox BT LRT adjusting for the same covariates. We restricted our analysis to SNPs within 50 kb of each gene and used Wu weights [Wu et al., 2011]. We present the candidate gene study results for the eight genes in Table 2. After correcting for multiple testing using a Bonferroni procedure, we failed to detect any association at the family-wise significance level of 0.05 (experiment-wise significance level of 0.05/8 = 0.00625). The lowest P-value from Cox SKAT LRT was 0.0084 (compared to 0.049 from Cox BT LRT) from HMGA1, which had 11 SNPs in our sample.
Table 2.
Candidate gene study results on time-to-obesity in Framingham Heart Study. Start and stop positions on NCBI Build 36. SNPs within 50 kb of the gene were included
| Gene | Chromosome | Start | Stop | N SNPs | Cox SKAT LRT | Cox BT LRT |
|---|---|---|---|---|---|---|
| MC4R | 18 | 56189544 | 56190981 | 12 | 0.42 | 0.18 |
| BDNF | 11 | 27633018 | 27677756 | 17 | 0.85 | 0.94 |
| SH2B1 | 16 | 28782725 | 28793027 | 3 | 0.22 | 0.22 |
| POMC | 2 | 25237226 | 25245063 | 14 | 0.40 | 0.36 |
| GIPR | 19 | 50863342 | 50877557 | 4 | 0.81 | 0.81 |
| HMGCR | 5 | 74668855 | 74693681 | 27 | 0.30 | 0.57 |
| TUB | 11 | 8016756 | 8084228 | 33 | 0.82 | 0.20 |
| HMGA1 | 6 | 34312628 | 34321986 | 11 | 0.0084 | 0.049 |
Discussion
In this paper, we propose an extension of SKAT to rare genetic variant analysis using a Cox proportional hazard model to analyze time-to-event outcomes. Such outcomes are common in genetic association studies, and the extension of SKAT to Cox regression is important because it provides an omnibus, flexible, and computationally easy way to test the association between a survival outcome and a set of genetic markers in a gene or genomic region. We show in our simulation studies that SKAT using score statistics has inflated type I error in the Cox proportional hazard model, when analyzing rare genetic variants. We propose an alternative, SKAT using likelihood ratio statistics from single-marker tests, to substitute for the score statistics. Asymptotically, the LRT is equivalent to the score test, but with a limited sample size and low minor allele frequencies, the LRT performs better than the score test in attaining the correct type I error rates when using Cox SKAT. In practice, the score test is expected to have the advantage of taking less time than the LRT, as the model only needs to be fit once. In the sliding window analysis on chromosome 1 of our real data example, Cox SKAT Score takes 504 sec CPU time on a single computing node on our cluster, while Cox SKAT LRT takes 1,575 sec CPU time on the same node.
Our formulation of the SKAT statistic also facilitates rare variant analysis on time-to-event outcomes in the context of large-scale multicohort meta-analysis. This is desired in meta-analysis consortia where researchers can share analysis results but generally have no access to individual level data from another cohort. Meta-analysis can be easily performed using analysis results from multiple cohorts. The test statistic and null distribution are equivalent to the single cohort test when there is only one cohort.
Although the kernel machine score test on survival outcomes proposed by Cai et al. [2011] is a general and flexible approach which can take different kernels, statistical significance is evaluated by resampling. Moreover, the performance of this test in rare genetic variant analysis has not been investigated before. This general kernel machine score test takes martingale residuals from the null model, and it is easy to show that the first term in their test statistic is equivalent to Cox SKAT score test when a linear kernel is used. However, as we show in this paper, when analyzing rare genetic variants, Cox SKAT score test suffers from inflated type I error if P-values are computed analytically, although the resampling procedure should still be valid. Alternatively, when using the LRT version of Cox SKAT, we can still compute P-values analytically, and we have shown in this paper that it maintains correct type I error rates in various scenarios. Recently, Lee et al. [2012] proposed a small sample adjustment procedure for SKAT, based on a higher moments matching method. It works well for unbalanced case-control designs. This method could potentially be adapted to Cox SKAT when analytical P-values are conservative.
For survival outcomes, SKAT is less powerful than BTs in our simulation studies when all causal variants have the same direction of effects and the proportion of causal variants is not small, but more powerful otherwise. This is in line with findings on continuous and dichotomous outcomes by Wu et al. in the original SKAT paper [Wu et al., 2011], and also by Chen et al. in SKAT for quantitative outcomes in related individuals [Chen et al., 2013].
We did not find any genome-wide significant associations with time-to-obesity in our analysis of FHS Original and Offspring Cohorts. One reason might be that the genotypes we used from SHARe were from SNP arrays that were originally designed for GWAS, and genotyped genetic variants were very sparse. However, we were able to confirm from our genome-wide sliding window analysis that Cox SKAT LRT does not have elevated type I error rates in this real data example, as the P-values are very close to a uniform distribution. We did not find significant associations in our candidate gene study of eight genes either. We hope to revisit this example when whole-genome sequencing data become available in FHS.
With recent technology advances in next-generation sequencing, rare genetic variants have become of great interest in genetic association studies, and SKAT with the linear kernel has proven to be a powerful and computationally efficient rare variant analysis approach in analyzing quantitative and dichotomous outcomes. Our approach proposed in this paper is a direct extension of SKAT with linear kernel. Compared with the general kernel machine approach proposed by Cai et al. [2011] and Lin et al. [2011], it is easier in computation as it calculates P-values analytically, although it loses the flexibility of using other kernels.
Supplementary Material
Acknowledgments
This research was partially supported by NIH awards R01 DK078616, U01 DK85526, and K24 DK080140. A portion of this research was conducted using the Linux Clusters for Genetic Analysis (LinGA) computing resources at Boston University Medicine Campus. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195). This work was partially supported by a contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278).
Appendix A
Derivation of the SKAT Statistic in the Cox Proportional Hazard Model
The log partial likelihood with respect to β and γ (using Efron’s method for ties) is
where
Let τ1 ≤ τ2 ≤ ··· ≤ τl be ordered failure times, for any failure time τj, let
be the index within ties at that failure time (0 ≤ mj ≤ nτj− 1). Let P be an n × l matrix with elements
Then is the cumulative hazard for individual i at time ti, and δi − ei(β, γ) is the martingale residual for known β, γ. Let V = diag(e1, e2,…, en) − PPT, X be an n × p matrix with rows Xi, G be an n × q matrix with rows Gi, δ and e be column vectors with elements δi and ei. Some calculation shows
The log likelihood with respect to β and σ2 can be written as
Let
then
Similar with the case in continuous and dichotomous outcomes, we take twice the first term as the SKAT statistic
By the central limit theorem we have
let
then under the null hypothesis
where λj are the eigenvalues of Σ, and are independent chi-square distributions with 1 degree of freedom.
Alternatively, for each single genetic marker gj (which are columns of the matrix G), the score test statistic (scalar) is
The SKAT statistic can be written as a weighted sum of single-marker test statistics. This is another way of expression.
Footnotes
Supporting Information is available in the online issue at wileyonlinelibrary.com.
References
- Bender R, Augustin T, Blettner M. Generating survival times to simulate Cox proportional hazards models. Stat Med. 2005;24:1713–1723. doi: 10.1002/sim.2059. [DOI] [PubMed] [Google Scholar]
- Cai T, Tonini G, Lin X. Kernel machine approach to testing the significance of multiple genetic markers for risk prediction. Biometrics. 2011;67:975–986. doi: 10.1111/j.1541-0420.2010.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet Epidemiol. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TR, Harrington DP, O’Sullivan M. Supremum Versions of the Log-Rank and Generalized Wilcoxon Statistics. J Am Stat Assoc. 1987;82:312–320. [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X NHLBI GO Exome Sequencing Project – ESP Lung Project Team. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Teslovich TM, Boehnke M, Lin X. General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet. 2013;93:42–53. doi: 10.1016/j.ajhg.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Cai T, Wu MC, Zhou Q, Liu G, Christiani DC, Lin X. Kernel machine SNP-set analysis for censored survival outcomes in genome-wide association studies. Genet Epidemiol. 2011;35:620–631. doi: 10.1002/gepi.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley T, Brody J, Dupuis J, Cupples LA. Meta-analysis of a rare-variant association test. 2012 http://stattech.wordpress.fos.auckland.ac.nz/files/2012/11/skat-meta-paper.pdf.
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genet Epidemiol. 2009;33:497–507. doi: 10.1002/gepi.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Mägi R, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.