Abstract
The etiology of many brain diseases remains allusive to date after intensive investigation of genomic background and symptomatology from the day of birth. Emerging evidences indicate that a third factor, epigenetics prior to the birth, can exert profound influence on the development and functioning of the brain and over many neurodevelopmental syndromes. This chapter reviews how aversive environmental exposure to parents might predispose or increase vulnerability of offspring to neurodevelopmental deficit through alteration of epigenetics. These epigenetic altering environmental factors will be discussed in the category of addictive agents, nutrition or diet, prescriptive medicine, environmental pollutant, and stress. Epigenetic alterations induced by these aversive environmental factors cover all aspects of epigenetics including DNA methylation, histone modification, non-coding RNA, and chromatin modification. Next, the mechanisms how these environmental inputs influence epigenetics will be discussed. Finally, how environmentally altered epigenetic marks affect neurodevelopment is exemplified by the alcohol-induced fetal alcohol syndrome. It is hoped that a thorough understanding of the nature of prenatal epigenetic inputs will enable researchers with a clear vision to better unravel neurodevelopmental deficit, late onset neuropsychiatric diseases, or idiosyncratic mental disorders.
Keywords: DNA methylation, Histone modification, Chromatin, Environmental hazards, Epigenetic medicine, Gene and environmental interaction, Fetal alcohol syndrome
1. INTRODUCTION
Recent progress on epigenetics begins to provide unprecedented insight how the formation of our nervous system is affected by the environmental inputs in life history dated long before birth, or even generations back (Figure 1). These environmental memories are recorded chemically in the form of epigenetic codes, deposited on the top of DNA or histones (for review see (Jaenisch & Bird, 2003; Portela & Esteller, 2010)). Evolutionarily, to avoid endless accumulation of epigenetic marks with each new cycle of life, during formation of zygotes and germ line, most of these memories are erased, but not entirely (Saitou, Kagiwada, & Kurimoto, 2012). Epigenetics codes in the form of DNA methylation, histone tail modification, or chromatin conformation can critically affect gene transcription by e.g. altering the 3-D DNA conformation to dictate transcription factor binding. Thus, depending on how much altered epigenetic codes are retained within a generation or several generations, this epigenetic memory may affect brain development or functioning through mis-regulation of gene transcription.
Figure 1.
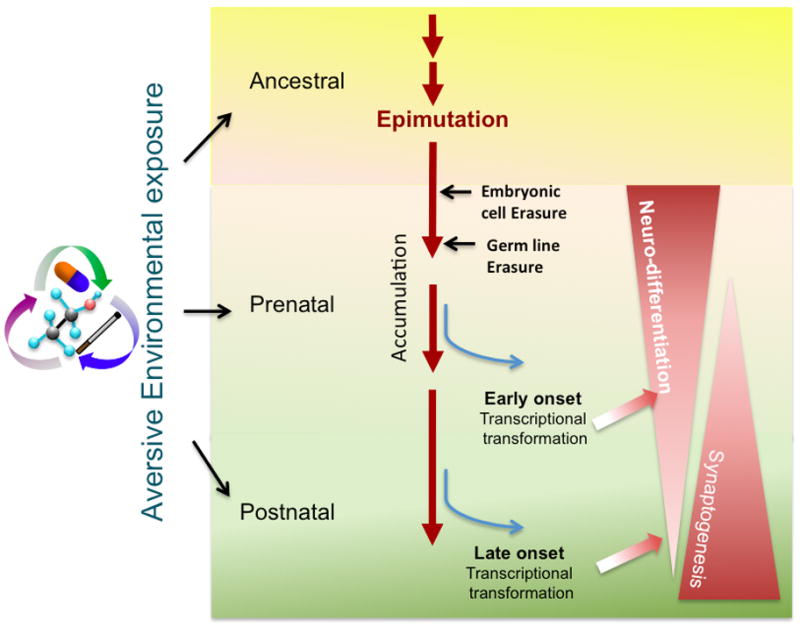
The aversive environment exposure (e.g. alcohol, nicotine, air pollution) can be recorded in the form of epigenetics throughout ancestral (including parental), prenatal, and postnatal stages. These altered epigenetics (epimutation) on the top of DNA or histone can be accumulated over time but can also be erased twice in a lifetime during zygotic stage and germ line maturation. The persisting epimutation when reached a critical threshold may alter gene transcription to affect the neural development. Depending on stage of life, the earlier onset may affect neural differentiation and brain formation. The continuous increment of epimutation or lasting influence of epigenetic changes may further affect synaptogenesis and together affect the brain function and neuropsychiatric disease at given time of the life.
There are three categorical stages of life history when epigenetics are registered--- ancestral (including parents), prenatal, and postnatal stages. Among these stages, prenatal epigenetic registration is the most eminent and profound influence on the formation or fine-tuning of the nervous system during development. Thus, environmental inputs prior to the birth have been reported to impose epigenetic entries, many of which are carried throughout the life of somatic cells, including those in the brain. This review will focus on epigenetic influences by various types of environmental factors specifically at prenatal stage. From a clinical point of view, this review advocates that many contributing factors and mechanisms of neurodevelopmental deficit, including late onset mental or psychiatric diseases (e.g. autism or schizophrenia), may have been seeded before birth beyond the default of the parental genetics.
Section 2 is devoted to eminent environmental factors including substances of abuse, prescription medications, pollutants, diets, and stress which are found to alter the epigenetics of offspring of exposed parents; the nature and types of epigenetics altered in the cells and organs will be reviewed. Section 3 elucidates how environmental factors might chemically alter epigenetics; thus leading to a better understanding of how other environmental factors might exert yet to be found influence. Section 4 elaborates that epigenetics evolved during neurodevelopment as an intrinsic program. Environmental factors, by altering epigenetics at individual genes or via the epigenetic program of differentiating neuroprogenitor cells, may alter the course of neurodevelopment. This will be demonstrated using alcohol exposure during pregnancy (fetal alcohol syndrome) as a model. The fetal alcohol syndrome is taken as an example, because it has a systemic influence of epigenetics in every forms and which has been found potentially transgenerational. Further, alcohol is one of the most abused environmental factors which affect men and women at all ages and has been dated back to pre-historical era.
We hope, in the following sections, via the illustration and discussion how the environmental factors long before our birth can be chemically recorded on DNA, histones, and other forms to alter the intrinsic epigenetic program and thereby altering gene expression in canonical neurodevelopment pathways, will increase our awareness and understanding how epigenetic memory pre-recorded prior to the birth might be a important causal predisposition to mental and neurological diseases in the life later.
2. Manifestation of Environmental Factors
This section starts with an illustration of most documented environmental factors that are found to affect epigenetic marks. They are categorized by their nature and ways of accessibility to our system and life, including addictive substances, diet, prescriptive drug, environmental pollutants, and the last but not least -- emotional stress. Their effects on altering epigenetics to affect early neural development start long ago when our parents or ancestors were subject to averse environmental exposure. The manifestations of averse environmental factors are summarized in Table 1 and Figure 2 and briefly discussed below. The current listing is likely the tip of the iceberg as more candidate factors are expected to emerge in the future. Through understanding their nature, we hope to perceive what other environmental factors might be candidate epigenetic modifiers and how they may access epigenetics via common or new mechanisms.
Table 1.
Epigenetic Change and Phenotype after Prenatal Exposure to Environmental Agents
| Change in Epigenetics | ||||
|---|---|---|---|---|
| Agent | DNA methylation | Histone modification | miRNA | Offspring phenotype correlated with epigenetic change |
| Alcohol | 5mC (Garro et al., 1991; Otero et al., 2012; Ouko et al., 2009; Wolff et al., 1998) 5hmC (Chen et al., 2013) |
H3Kac (Pal-Bhadra et al., 2007, Shukla et al., 2007) H3K4me, H3K9me, (Pal-Bhadra et al., 2007); H3K27me (Subbanna et al., 2013) |
miR148, miR152 (Kutay et al., 2012), miR21, miR153, miR335 (Sathyan et al., 2007) | Delayed maturation and reduced size of hippocampus (Chen et al., 2013) Reduced neuron cell number, cortical plate, thickness, delayed formation in the neural tube, forebrain, hindbrain (F. C. Zhou et al., 2011) |
| Nicotine | 5mC (Breton et al., 2009; J. Z. Maccani et al., 2013; Suter et al., 2010; Suter et al., 2011) | H3ac (Levine et al., 2011) | miR16, miR21, miR146a (M. A. Maccani et al., 2010) | Birth weight reduction (Suter et al., 2011) Increased aggression, locomotion in adult male (Yochum et al., 2014) |
| Cannabis | N/A | H3K4me, H3K9me (DiNieri et al., 2011) | N/A | Increased opiate reward sensitivity in adult (DiNieri et al., 2011) |
| Cocaine | 5mC (Bae & Zhang, 2005; K. D. Meyer et al., 2009; Novikova et al., 2008; H. Zhang et al., 2007) | N/A | N/A | Increased apoptosis in the term fetal heart; cardiac remodeling (K. Meyer, H. Zhang, & L. Zhang, 2009) |
| Opioid | 5mC (Chorbov et al., 2011) | N/A | N/A | N/A |
| Methamphetamine | 5mC (Itzhak et al., 2014) | N/A | N/A | Enhanced cocaine reward and hyper-locomotion; reduced conditional fear (Itzhak et al., 2014) |
| Folic acid (Deficiency) | 5mC (Gueant et al., 2013) | H3ac (Akchiche et al., 2012) | miR124(Kerek et al., 2013) miR302a (Liang et al., 2012) |
Growth retardation, reduced brain size(Kerek et al., 2013) |
| Caffeine | 5mC (Buscariollo et al., 2014; D. Xu et al., 2012) | N/A | N/A | Growth retardation (D. Xu et al., 2012) Altered cardiac function and morphology in adult mice (Buscariollo et al., 2014) |
| Valproic acid (VPA) | N/A | H3ac (Balmer et al., 2012; Monti, Polazzi, & Contestabile, 2009) | N/A | Reduced birth rate; reduced sociability and social preference (K. C. Kim et al., 2011) |
| γ-hydroxybutyrate (GHB) | N/A | H3ac (C. Klein et al., 2009) | N/A | N/A |
| Arsenic | 5mC (Intarasunanont et al., 2012; Kile et al., 2012; Pilsner et al., 2012; Xie et al., 2007) | H3ac (Cronican et al., 2013) | let 7a, miR16, miR17, miR20a, miR20b, miR26b, miR96, miR98, miR107, miR126, miR195, and miR-454 (Rager et al., 2013) | Impaired spatial and episodic memory, as well as fear conditioning performance (Cronican et al., 2013) |
| Lead | 5mC (Bihaqi et al., 2011; Pilsner et al., 2009; Schneider et al., 2013) | H3ac, H3K4me (Bihaqi et al., 2011) | N/A | Enhanced neurodegeneration in primate (Bihaqi et al., 2011) |
| Cadmium | 5mC (Castillo, Ibanez, Guajardo, Llanos, & Ronco, 2012; Kippler et al., 2013; A. P. Sanders et al., 2013) | N/A | N/A | Reduced birth weight and height (Castillo et al., 2012) |
| Methyl mercury | 5mC (Bose et al., 2012; Onishchenko et al., 2008) | H3ac, H3K27me (Onishchenko et al., 2008) | N/A | Depression like behavior (Onishchenko et al., 2008); |
| Bisphenol A | 5mC (Yaoi et al., 2008) | H3ac (Yaoi et al., 2008) | N/A | Delay the perinatal chloride shift in cortical neurons (Yeo et al., 2013) |
| Stress | 5mC (Champagne & Curley, 2009; Darnaudery & Maccari, 2008; Heim & Binder, 2012; Szyf, 2013) | H3ac, H3K9me H3K27me (Dalton et al., 2014; Reus et al., 2013) |
miR16 (Bai et al., 2012), miR9, miR29a, miR124, miR132, miR212(Uchida et al., 2011) | Induces depressive-like behaviors; altered response to aversive environments (Champagne & Curley, 2009; Franklin et al., 2010) |
Figure 2.
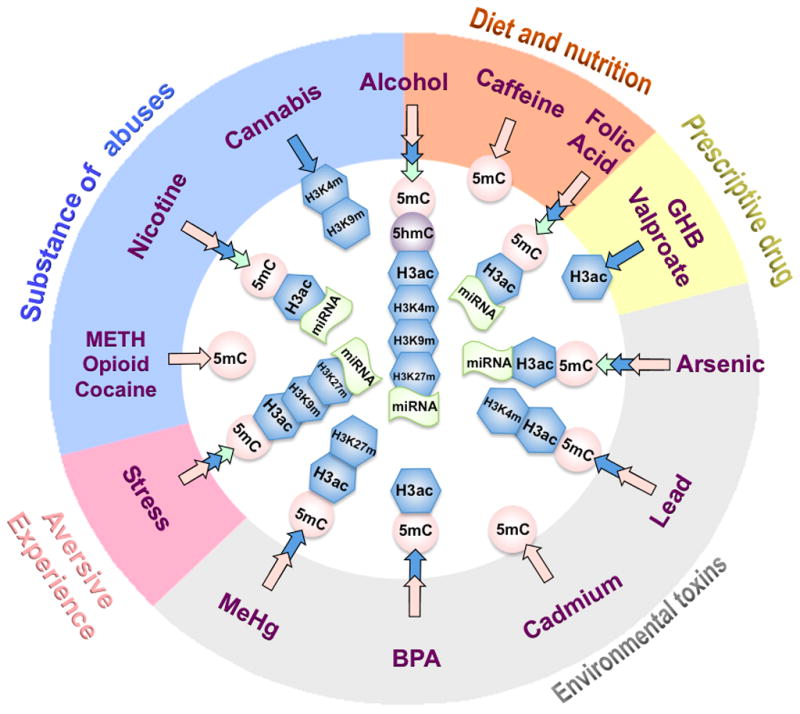
Epigenetic entry of aversive environmental factors. This figure provides information for the epigenetic modifications that have shown to be altered by different types of environmental factors prior to birth. These factors are divided into five categories depending on their nature and ways of access, including substance of abuse, diet and nutrition, prescriptive drugs, environmental toxins and stress. Some factors may cross the categories, e.g. Alcohol belongs to both substance of abuses and diet. The effected epigenetic marks including DNA methylation (5mC, 5hmC) and histone modification (H3ac, H3K4me, etc…) are illustrated. For subtypes altered miRNAs, see Table 1.
2.1 Substance of abuses
2.1.1 Drinking
Alcohol, like sugar, has been associated with every form of life since the beginning of evolution. Life forms depend on sugar to survive. Since alcohol is not a commodity for survival, evolution has maintained tenacious alcohol-seeking behavior through other means, such as pleasure. If the hedonic propensity does not eternally maintain alcohol-seeking behavior, evolution added another layers of reinforcement -- addiction. Alcohol has been found to instill multiple marks of epigenetic change. The nature and mechanisms of how this ubiquitous molecule can alter epigenetics will be discussed in Section 3, and how alcohol epigenetic changes might affect neurodevelopment manifesting as fetal alcohol spectrum disorder (FASD) will be discussed in Section 4. The following is a list of evident alcohol epigenetic changes. Excessive alcohol drinking not only affects individuals, but has also been known to affect their offspring. Since alcohol is inseparable from life throughout evolution, and alcohol affects life throughout different stages of their development, it is likely through mutations, epimutation and natural selection, the “alcohol” trait may have been genetically incorporated as a genomic signature or recorded on the epigenome as epigenetic memory.
Alcohol exposure during pregnancy can lead to Fetal Alcohol Spectrum Disorder (FASD). Studies have demonstrated that alcohol induces a wide range of epigenetic alterations. Recent reviews illustrate that alcohol is in a unique position to alter multiple levels of epigenetics (Resendiz, Chen, Ozturk, & Zhou, 2013) (Ungerer, Knezovich, & Ramsay, 2013). In essence, excessive alcohol exposure induces DNA methylation changes in sperm, embryos, or developing brain (Garro, McBeth, Lima, & Lieber, 1991; Otero, Thomas, Saski, Xia, & Kelly, 2012; Ouko et al., 2009; Wolff, Kodell, Moore, & Cooney, 1998). At epigenomic level, alcohol induces bi-directional changes of DNA methylation at cytosine (5-methylcytosine;5mC) by both hyper- and hypo-methylation (Liu, Balaraman, Wang, Nephew, & Zhou, 2009; F. C. Zhou et al., 2011a). Alcohol further affects cellular 5mC and the recently found second form of methylation 5-hydroxymethylcytosine (5hmC) (Guo, Su, Zhong, Ming, & Song, 2011; Ito et al., 2011; Kriaucionis & Heintz, 2009; Tahiliani et al., 2009) during embryonic and brain development (Chen, Ozturk, & Zhou, 2013) And, prenatal alcohol interferes the intrinsic DNA methylation program (DMP) (see Section 4) (F. C. Zhou, 2012; F. C. Zhou, Chen, & Love, 2011). Alcohol also increases histone 3 acetylation globally (J. S. Kim & Shukla, 2006; Pal-Bhadra et al., 2007; Shukla, Lee, Park, & Aroor, 2007), and alters miRNA expression in neural stem cells in a cell-type and stage specific manner (Miranda, 2012; Wang et al., 2009). Transgenerational epigenetic changes were also found with parental alcohol exposure leading to hypomethylation; specifically at imprinting gene H19 CpGs in F1 generation sperm DNA and F2 offspring brain (Knezovich & Ramsay, 2012) and non-imprinting gene proopiomelanocortin (Pomc) over F2 and F3 generations (Govorko, Bekdash, Zhang, & Sarkar, 2012). More studies are being conducted to validate the findings.
2.1.2 Smoking
According to recent Centers for Disease Control and Prevention (CDC)’s surveillance study, 14% of women smoke during pregnancy (Tong, 2013). Maternal smoking during pregnancy significantly affects fetal growth, preterm delivery and infants’ long term health (Hackshaw, Rodeck, & Boniface, 2011). Several studies use human placenta to examine epigenetic changes in women smoking during pregnancy and identified genome-wide (Suter et al., 2011) and gene specific (J. Z. Maccani, Koestler, Houseman, Marsit, & Kelsey, 2013) alteration of DNA methylation, as well as reduced miRNA expression (M. A. Maccani et al., 2010). A separate study analyzed buccal cell samples from prenatal tobacco-exposed children and found lower methylation level in the transposable element, AluYb8. Alu insertions cause genome instability and has been implicated in disease pathogenesis (Ade, Roy-Engel, & Deininger, 2013; Rowold & Herrera, 2000). Therefore, it would be interesting to see if these methylation changes by nicotine alters the transcription or even transposability of AluYb8.
2.1.3. Cocaine
Cocaine abuse in pregnant women is prevalent in the United State and is associated with a wide variety of fetal deficits including premature birth, cardiac defects, attention deficit disorder and so on (Ackerman, Riggins, & Black, 2010; Bae & Zhang, 2005; Frassica, Orav, Walsh, & Lipshultz, 1994; Friedman, 1995; Rotta & Cunha, 2000; H. Zhang, Darwanto, Linkhart, Sowers, & Zhang, 2007). Differential methylation were detected at the CpG islands of selected genes and were correspondingly associated with altered gene expression (Novikova et al., 2008).
For the effect on brain development, several studies looked at epigenetic regulation in cocaine reinforcement and identified brain-derived neurotropic factor (BDNF) as a target of epigenetic modifiers. For example, a self-administration cocaine rat model displayed increased H3 acetylation and decreased methyl CpG binding protein 2 (MeCP2) association with BDNF promoter IV in the medial prefrontal cortex (mPFC) of cocaine treated rats. This is associated with increased BDNF protein expression and Bdnf exon IV-containing transcripts (Sadri-Vakili et al., 2010). Whether epigenetic modification is involved in trans-generational behavior alterations is still unknown. Studies are needed to elucidate the phenotypic outcome of cocaine use on brain development. One study showed that while DNA methylation and gene expression profile changed, no phenotypic difference was observed in the hippocampus of early postnatal offspring of the control and cocaine-exposed mothers (Novikova et al., 2008). Another study demonstrated that paternal cocaine administration leads to impaired memory in the female offspring and causes hyperactivity in male offspring (He, Lidow, & Lidow, 2006).
In contrast, the impact of cocaine on fetal cardiac development is better studied. Studies show that pregnant rats exposed to cocaine results in myocardiocyte apoptosis in the term fetal heart (Bae & Zhang, 2005), and disrupted ischemic preconditioning –induced cardioprotection in adult offspring because of reduced protein kinase Cε (PKCε) expression (K. D. Meyer, H. Zhang, & L. Zhang, 2009). The same research group further determined the decreased PKCε expression is associated with increased DNA methylation at the SP1 binding sites in the PKCε promoter that reduced the recruitment of SP1 to PKCε promoter.
2.1.4 Cannabis
Cannabis, the major ingredient in marijuana, is the most common abused drug in pregnant women in the US (Ebrahim, 2003). Following its recent legalization in multiple states of USA in 2013, greater population is expected to expose to this drug. In utero exposure to cannabis has been shown to restrict fetal growth, reduce head circumference (El Marroun et al., 2009), and alter fetal behavior (Day & Richardson, 1991; Huizink, 2013). DiNieri et al. demonstrated the epigenetic relevance of cannabis (DiNieri et al., 2011). They showed dopamine receptor D2 (DRD2) mRNA expression was reduced in human ventral striatum of fetus maternally exposed to cannabis. Using a rat model to further investigate this finding, they reported that decreased Drd2 is associated with repressive chromatin marks (reduced H3K4, increased H3K9) and decreased Polymerase II binding, suggesting the long-term transcription machinery for Drd2 was disrupted. Since DRD2 involves in regulation of addiction (Thanos, Michaelides, Umegaki, & Volkow, 2008), this study provides a link between prenatal drug exposure and adulthood addiction through the retention of epigenetic memory.
2.1.5 Opioid
Opium, an old analgesic medication that is highly addictive, has proliferated into many chemical forms and is widely used today. A wide range of drugs fall into the category opioid, including hydrocodone, oxycodone, morphine, tramadol and so on (Abuse, 2011). According to a US multicenter study, approximate 10.7% of 8,527 infants screened were exposed to cocaine or opioid (Lester et al., 2001). It has been shown that prenatal opioid exposure causes more severe phenotype than cocaine exposure on the offspring nervous system (Das, Poole, & Bada, 2004). So far, there is no published report on the epigenetic effect of prenatal exposure to opioid. However, it has been reported that sperm DNA from opioid addicts have increased methylation at the Opioid receptor, mu1 (OPRM1) promoter region (Chorbov, Todorov, Lynskey, & Cicero, 2011). Oprm1 is important for controlling drug dependence through regulating the dopamine pathway (Chefer, Denoroy, Zapata, & Shippenberg, 2009). Taken together, opioid influence could be potentially transmitted through sperm to the next generation.
2.1.6 Methamphetamine (METH)
Methamphetamine (METH) is a stimulant and one of the major abused drugs worldwide. The United Nations Office on Drugs and Crime (http://www.unodc.org/wdr/en/ats.html) reported that, globally around 33.8 million people aged 15–64 had used Amphetamine-type stimulants (ATS) in 2011, in which, methamphetamine accounted for 71% of ATS. According to study, 5.2% of women in high METH prevalent region of the US used METH during pregnancy (Arria et al., 2006). Prenatal exposure to METH has been determined to lead to oxidative stress in embryonic brain and postnatal neurodevelopmental, cognitive and behavioral defects (Jeng, Wong, Ting, & Wells, 2005; Kwiatkowski, Roos, Stein, Thomas, & Donald, 2013). Recently, it is demonstrated that differential methylated region (DMR) in hippocampal DNA of adolescent offspring resulted from both prenatal METH exposure and differential maternal care (Itzhak, Ergui, & Young, 2014). The hypermethylated DMRs are enriched for “cerebral cortex GABAergic interneuron differentiation” gene ontology, and the hypomethylated DMRs are highly associated with “embryonic development”. Furthermore, the exposed F1 male displayed enhanced response to cocaine-conditioned reward and increased locomotion activity, while both F1 male and female show less response to conditioned fear. Together, these data suggest prenatal METH exposure could alter the DNA methylation profile of offspring and lead to abnormal behavior.
2.2. Diet and nutrition
Dietary and nutrition disparity during fetal stage has long been considered as a critical environmental factor, which may exert long lasting effect on development. Emerging evidence indicated that ancestral dietary and nutrition disparity can influence progeny phenotype through epigenetics. Gaining evidence shows specific dietary facors can alter DNA methylation globally and at specific gene levels (demonstrated below). A major entry point to epigenetics is the involvement of these dietary factors in pathways that interact with methyl donor. These factors include alcohol, as previously discussed. Additional elements include folic acid, caffeine, polyphenols, phytoestrogen genistein, and flavonoids. Here, we will discuss folic acid and caffeine as examples and discuss the outcome of maternal folate deficiency and/or caffeine exposure on fetal epigenetics.
2.2.1 Folic acid
Dietary folic acid is recommended for women during pregnancy because of its profound effect on reduced the risk of fetal neural tube defects (Berry et al., 1999; Centers for Disease, 1991). It has been reported that an estimated 50% of women before and 66% during pregnancy have folic acid supplementation (Hoyo et al., 2011). The direct influence of folic acid on epigenetics is through the methionine pathway that generates methyl donor for DNA and histone methylation. Nevertheless, folic acid deficiency has been also related to increased histone deacetylase (HDAC) (Akchiche et al., 2012) and decreased expression of miRNAs that are involved in apoptosis and cell proliferation (Tryndyak, Ross, Beland, & Pogribny, 2009). Gueant et al. provided a comprehensive review on folate and fetal epigenetic programming (Gueant, Namour, Gueant-Rodriguez, & Daval, 2013).
2.2.2 Caffeine
Caffeine is the most popular psychoactive substance that can be found in coffee, tea, soft drink, and energy drinks (Frary, Johnson, & Wang, 2005). It is well known that heavy maternal consumption of caffeine is linked to spontaneous abortion, intrauterine growth restriction (IUGR) (Kuczkowski, 2009), as well as fetal neurodevelopmental and behavioral deficits (Browne, 2006; Nehlig & Debry, 1994). Recent studies have started to look at its impact on epigenetics Xu et al. (2012) showed that DNA methylation was increased in the 11 β-hydroxysteroid dehydrogenase type 2 (11β-HSD-2) promoter in fetal hippocampal neuron from a rat model in utero exposure to caffeine. An increase of 11β-HSD-2 methylation is correlated with reduced 11β-HSD-2 expressions. 11β-HSD-2 is known to catalyze the conversion of cortisol to cortisone. Reduced levels of placental 11β-HSD-2 may lead to an increased exposure of glucocorticoid in fetus. Therefore, these data suggested that cocaine retards fetal development through epigenetic programing of the hypothalamus-pituitary-adrenal (HPA) via the 11β-HSD-2 gene.
2.3. Prescriptive medicine
A number of prescriptive medications have been found to have the side effect of directly changing epigenetics, and through which to affect development, particularly neurodevelopment. Many of them are no longer used in clinic practice, but medications with similar chemical and physical properties are being introduced by pharmaceutical industry. It is important to raise awareness in this category.
2.3.1 Valproic acid (VPA)
Valproic acid is a widely used antiepileptic drug for the treatment of seizures and bipolar disorder (Loscher, 2002). Several studies demonstrated higher risks with VPA (~10% infant growth defect) than use with other drugs (2.8%) during the fist trimester of pregnancy (Wyszynski et al., 2005), indicating the potency of VPA on fetal development. It is also an inhibitor of histone deacetylase (HDAC) (Gottlicher et al., 2001). Due to this effect, VPA treatment in human embryonic stem (ES) cells caused global genomic increase of H3 acetylation, while genes disrupting acetylation were down-regulated. Furthermore, VPA inhibited ES cell differentiation into neuronal precursor cell (Balmer et al., 2012), suggesting early neural fate decision is disturbed.
2.3.2 γ-hydroxybutyrate (GHB)
GHB is an endogenous inhibitory transmitter in human (Nelson, Kaufman, Kline, & Sokoloff, 1981) and is used in anesthesia, insomnia, and depression treatment (Mamelak, Scharf, & Woods, 1986). Despite its medical application, GHB has been abused for more than 20 years and commonly known as a “date-rape drug” or a “party drug” (O’Connell, Kaye, & Plosay, 2000). To date, teratogenic effect has not been reported for GHB. A women addicted to GHB gave birth to a healthy full term baby (Buonocore, Bracci, & Weindling, 2010). However, several studies have revealed its function in regulating epigenetic modification (C. Klein, Kemmel, Taleb, Aunis, & Maitre, 2009; Q. Lu et al., 2004). GHB has been shown to inhibit histone deacetylase, leading to accumulation of acetylated histones in adult rat brain after administration of pharmacological dose of GHB (C. Klein et al., 2009). Studies are needed to determine the effects of GHB prenatal exposure on fetal epigenetic, postnatal neurodevelopment and behavior change.
2.4. Environmental toxins and pollutants
This category of environmental factors represents the largest group, and ever-increasing candidates are being found.
2.4.1 Arsenic
Arsenic is one the major pollutants in drinking water worldwide, especially in Bangladesh, India, China and Taiwan (Meharg, 2005). It has been linked with multiple types of cancer by epidemiologic evidence (Celik et al., 2008; Straif et al., 2009). Several studies identified Arsenic effect on epigenetics in somatic cells. A detailed review of these studies can be found in Reichard and Puga (Reichard & Puga, 2010). It is now known that the epigenetic influence by arsenic can be transmitted to the next generation. Fetal brain samples of prenatal arsenic exposed mice showed global reduction of acetylation at H3K9, while adult exposure displayed impaired memory and fear conditioning performance. (Cronican et al., 2013). Cord blood samples from arsenic exposed human maintained higher amounts of arsenic and revealed aberrant DNA methylation in a gene specific manner (Kile et al., 2012; Koestler, Avissar-Whiting, Houseman, Karagas, & Marsit, 2013). Furthermore, miRNAs with increased expression associated with higher urine arsenic have been identified (Rager et al., 2013). However, how these epigenetic changes correlate with previously identified disrupted fetal neuronal development (Chattopadhyay, Bhaumik, Nag Chaudhury, & Das Gupta, 2002) still needs to be explored.
2.4.2 Lead
Lead toxicity causes significant health problems in the US and worldwide. More than 250,000 children age before 5 years have higher blood lead levels than the CDC reference level based on a 2012 report from the CDC (Kuehn, 2012). Prenatal exposure to lead interrupts neuronal development plasticity, especially to the rapid growing neonatal brain (Bellinger, 2005; T. Sanders, Liu, Buchner, & Tchounwou, 2009; White et al., 2007). Therefore, it is important to know the mechanisms for lead neurotoxicity. The effects of lead on epigenetics is mostly associated with aberrant DNA methylation and altered expression of methylation-associated enzymes such as DNMTs and MECP2 (Bihaqi, Huang, Wu, & Zawia, 2011; Pilsner et al., 2009; Schneider, Kidd, & Anderson, 2013) although the reason still needs to be elucidated. Senut et al. provided a detailed review on lead exposure and epigenetics of fetal brain development (Senut et al., 2012).
2.4.3 Cadmium
Cadmium exposure is mostly though consumption of vegetables, drinking water, seafood, and though tobacco smoking that has severe impacts on global human health (Olsson et al., 2002). Prenatal exposure to cadmium has been shown to lead to altered thymocyte development (Hanson, Brundage, Schafer, Tou, & Barnett, 2010), aberrant immune response (Holaskova, Elliott, Hanson, Schafer, & Barnett, 2012), and decreased birth weight in a sex-specific manner (Kippler et al., 2013). The sex-specific toxicity is also correlated with differential methylation between girls and boys based on cord blood sample from exposed newborns and blood from 4–5 year children of Bangladesh (Kippler et al., 2013). A similar study conducted in the US also identified differential methylated genes in leukocytes from mother-baby pairs (A. P. Sanders et al., 2013). Interestingly, those genes set were largely distinct between maternal and fetal DNA, indicating different pathways are misregulated between mother and children.
2.4.4 Methyl mercury
Human exposure to methyl mercury (MeHg) is mostly through the consumption of ocean fish (Mergler et al., 2007). Therefore, high levels of MeHg exposure can be found in fish-eating population throughout the world, including people in the United States, Japan, Germany, (Pirrone & Mahaffey, 2005). For example, about 5–10% of women of childbearing age in the US have exceeding levels of MeHg (McDowell et al., 2004), and 73.7% of women in Japan of this age are exposed to high levels of MeHg (Yasutake A, 2004). It is known that MeHg can be transferred from the mother to fetus via neutral amino acid transportation across the placenta (Kajiwara, Yasutake, Adachi, & Hirayama, 1996). Neonatal and infants expose to MeHg by intake of milk from the mother. Prenatal exposure to MeHg lead to severe fetal neurological disorders including cerebral palsy, mental retardation, primitive reflexes, dysarthria, and hyperkinesia (Harada, 1995; Marsh et al., 1980; Myers & Davidson, 1998). Several studies have been performed to tease out the effects MeHg exposure on fetal epigenetics. An animal study exposed mice to MeHg from embryonic day 7 (E7) to postnatal day 7 (P7) found the offspring at 12-week old exhibited repressed chromatin marks (DNA hypermethylation, decreased histone acetylation, increased H3K27 trimethylation) at the Bdnf promoter IV (Onishchenko, Karpova, Sabri, Castren, & Ceccatelli, 2008). This altered epigenetic states was associated with decreased Bdnf expression and depression like behavior. Furthermore, they showed that fluoxetine (an antidepressant) treatment altered depression behavior, restored Bdnf expression, and increased H3 acetylation (Onishchenko et al., 2008). A different study revealed MeHg inhibited neural stem cell proliferation and induced senescence-associated markers. This decreased cell proliferation phenotype was confirmed in the subgranular zone in the hippocampi of adult mice prenatally exposed to low doses of MeHg (Bose, Onishchenko, Edoff, Janson Lang, & Ceccatelli, 2012). In addition, global DNA methylation and Dnmt3b expression were decreased in the parental NSC exposed to MeHg and in the daughter cell under MeHg free condition.
2.4.5 Bisphenol A (BPA)
Bisphenol A is widely used for plastic bottle, protective lining of metal-based food and beverage containers. Laboratory studies showed that adulthood BPA exposure affects normal neurodevelopment, sexually dimorphic behaviors, and hyperactivity disorder (Palanza, Gioiosa, vom Saal, & Parmigiani, 2008; vom Saal et al., 2007). Prenatal exposures to low doses of BPA lead to acceleration of neurogenesis and aberrant neuronal migration (Nakamura et al., 2006). From the epigenetic perspective, prenatal BPA exposure altered DNA methylation both gene-specifically (Yeo et al., 2013) and globally, but in a dose dependent manner (J. H. Kim et al., 2014). Yeo et al. showed that BPA induced MECP2 association and reduced H3K9 acetylation at the potassium chloride cotransporter 2 (Kcc2) promoter, which is associated with decreased Kcc2 expression and leading to delayed perinatal chloride shift in the cortical neurons. Their result provides a possible explanation for the epigenetic mechanism of neurotoxicity due to BPA exposure.
Many new environmental hazards have suspected to affect biological response through altering epigenetics, including nickel (Zoroddu et al., 2010), chromium (X. Zhou, Li, Arita, Sun, & Costa, 2009), plastic derived endocrine disruptor [Bis(2-ethylhexyl) phthalate, dibutyl phthalate] (Manikkam, Tracey, Guerrero-Bosagna, & Skinner, 2013), but their prenatal effects particularly in neurodevelopment still need to be elucidated.
E. Stress and Aversive Life Experience
Parental emotional stress has been known to impact the emotional abnormality and behavioral development of offspring. For example, studies show that maternal post-traumatic stress disorder (PTSD) can be transmitted across generations (Yehuda & Bierer, 2008; Yehuda, Schmeidler, Wainberg, Binder-Brynes, & Duvdevani, 1998) and is associated with dysregulation of neurobiological systems, such as the hypothalamus-pituitary- adrenal (HPA) axis (Boscarino, 1996; Mason, Giller, Kosten, Ostroff, & Podd, 1986; McGowan, 2013). Stress is implicated in the development of mental disorders and other adverse outcomes such as low birth weight (Reed, 2012), poor maternal-infant bonding (Muzik, Marcus, & Flynn, 2009), and infant sleep and behavioral problems at 18 months (Hairston et al., 2011).
Maternal PTSD (McGowan, 2013), maternal separation (Lewis, Staudinger, Scheck, & Olive, 2013), depression (Dalton, Kolshus, & McLoughlin, 2014; Heim & Binder, 2012), maternal care (Champagne & Curley, 2009), childhood adversity or early life stress (Franklin et al., 2010; Murgatroyd et al., 2009; Roth, Lubin, Funk, & Sweatt, 2009), and paternal stress (Rodgers, Morgan, Bronson, Revello, & Bale, 2013), are environmental inputs that have been shown to affect the epigenetic profile of the offspring. The most prominent mechanism for trans-generational epigenetic regulation in response to prenatal stress is via the hypothalamic-pituitary-adrenal (HPA) axis. It is known that changes in glucocorticoid levels influence the HPA axis as well as alter fetal brain development. In addition, altered cortisol levels are associated with changes in glucocorticoid receptor activity (Matthews, 2002). Indeed, glucocorticoid receptor (GR) gene (Nr3c1) has altered DNA methylation and gene expression in several human studies and animal models after prenatal exposure to stress (Hompes et al., 2013; McGowan et al., 2009; Oberlander et al., 2008; Tyrka, Price, Marsit, Walters, & Carpenter, 2012). Epigenetic changes in genes other than Nr3c1 have also been reported. For example, increased miR-16 expression (Bai et al., 2012) and increased promoter DNA methylation of BDNF (Roth et al., 2009) were found in separate studies, but both changes result in a reduction in brain derived neurotropic factor (BDNF) mRNA expression, suggesting BDNF may play a role in regulating offspring behavior in response to prenatal stress exposure.
3. Mechanisms of Environmentally Induced Epigenetic Changes
In this section, the epigenetic mechanisms mediated by prenatal exposure to environmental deleterious factors is discussed based on epigenetic categories (Figure 3), as well as giving examples of altered genes associated with epigenetic changes (Table 2).
Figure 3.
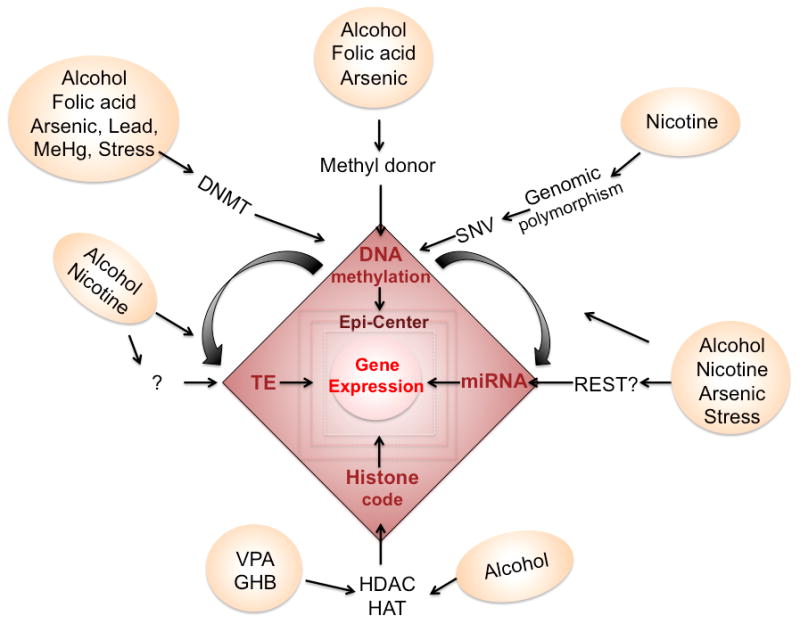
Mechanism of converting epigenetic marks. Each aversive environmental factor take specific entry to alter the epigenetics, e.g. through altering DNA methyltransferase (DNMT) and changing methyl donor availability to affect DNA methylation, changing histone deacetylase (HDAC) or histone acetyltrasnferase (HAT) to alter histone code, or through altering DNA methylation on regulatory regions of transposon element (TE) or on miRNA to affect TE and miRNA. To this date, there are missing information how environmental factors might directly affect the TE or miRNA. The RE1 silencing transcription factor (REST), which regulates the expression of miRNA, may be a potential pathway. Finally, the genomic polymorphism or C-T mutation resulted single nucleotide variant (SNV) can also affect the CpG methylation.
Table 2.
Genes altered by environmental agents
| Gene | Gene name | Epigenetic change | Agent | Reference |
|---|---|---|---|---|
| BDNF | Brain derived neurotropic factor | DNA methylation ↑ at whole gene | Alcohol | (Maier, Cramer, West, & Sohrabji, 1999) |
| DNA methylation ↑ at promoter VI | Nicotine | (Toledo-Rodriguez et al., 2010) | ||
| MECP2 expression ↓ No change in methylation level |
Lead | (Stansfield, Pilsner, Lu, Wright, & Guilarte, 2012) | ||
| DNA methylation ↑; H3K27me3 ↑; H3ac↓ at promoter IV | Methyl mercury | (Onishchenko et al., 2008) | ||
| DNA methylation ↑ at promoter IV & IX | Stress | (Roth et al., 2009) | ||
| RUNX3 | Runt-related transcription factor 3 | Site specific change in DNA methylation | Nicotine | (J. Z. Maccani et al., 2013) |
| CYP1A1 | cytochrome P450, family 1, subfamily A, polypeptide 1 | DNA methylation at XRE (transcription factor) binding site ↓ | Nicotine | (Suter et al., 2010) |
| DRD2 | Dopamine receptor D2 | H3K9me2 ↑ H3K4me3 ↓ RNApol II binding ↓ |
Cannabis | (DiNieri et al., 2011) |
| DRD4 | Dopamine receptor D4 | DNA methylation ↑ | Alcohol | (H. Zhang et al., 2013) |
| 11b-HSD-2 | 11β-hydroxysteroid dehydrogenase type 2 | DNA methylation at promoter ↑ | Caffeine | (D. Xu et al., 2012) |
| In the placenta: promoter methylation↑, Dnmt3a ↑; In fetal hypothalamus: promoter methylation↓, methylation at exon1 ↑; no change in fetal cortex | Stress | (Jensen Pena, Monk, & Champagne, 2012) | ||
| OPRM1 | Opioid receptor, mu 1 | DNA methylation ↑ | Alcohol | (H. Zhang et al., 2013) |
| DNA methylation at promoter ↑ | Opioid | (Chorbov et al., 2011) | ||
| PAX6 | Paired box 6 | H3K4me3/H3K27me3 ratio <1 (silenced chromatin) | Valproic acid | (Balmer et al., 2012) |
| KCC2 | Potassium chloride cotransporter 2 | MECP2 binding ↑ H3K9ac ↓ |
Bisphenol A | (Yeo et al., 2013) |
| MAOA | Monoamine oxidase A | DNA methylation ↑ | Alcohol | (Philibert, Plume, Gibbons, Brody, & Beach, 2012) |
| GABRB3 | GABA A receptor B3 | DNA methylation ↑ | Alcohol | (H. Zhang et al., 2013) |
| NR2B | NMDA receptor subunit 2b | CpG Site specific hypomethylation H3K9Ac ↑, H3K9Me ↓ |
Alcohol | (Qiang, Denny, Lieu, Carreon, & Li, 2011; H. Zhang et al., 2013) |
| H19/GF2 | DNA methylation ↓ | Alcohol | (Haycock & Ramsay, 2009) | |
| NEUROD6 | Neuronal differentiation 6 | Promoter methylation ↑ | Alcohol | (Liu et al., 2009) |
| POMC | Pro-opiomelanocortin | Promoter methylation ↑ | Alcohol | (Govorko et al., 2012) |
3.1. On DNA methylation
Alterations in DNA methylation are the most common type of epigenetic change that is found upon prenatal exposure to hazardous environmental agents. One of the key causal reasons is availability of methyl donor within the cellular environment. Alcohol, dietary folate, and arsenic directly regulate methylation through the alteration of the methionine pathway. Methionine is the precursor of an active methyl donor, S-adenosylmethionine (SAM). Alcohol has been shown to have multiple effects on methionine synthesis through inhibiting metabolic enzymes (Halsted et al., 1996), and methionine adenosyltransferease (MAT) (S. C. Lu et al., 2000). Dietary folate is a cofactor of methionine synthase, which converts homocysteine to methionine. Moreover, arsenic, which is a widespread source of contamination in drinking water, consumes SAM and glutathione to produce dimethylarsenic acid for detoxification and facilitating excretion (Drobna et al., 2005). The second causal alteration of DNA methylation is through regulating the expression of DNA methyltransferases (DNMTs). This has been found in a number of studies that exposed fetus to lead (Bihaqi et al., 2011) (Schneider et al., 2013), MeHg (Bose et al., 2012), and alcohol (Govorko et al., 2012; Perkinsa, Lehmanna, Lawrencea, & Kelly, 2013).
Alterations of DNA methylation are dynamic and stage dependent, and they occur both globally and gene specifically. For example, Novikova et al. used a mouse model to demonstrate the effects of maternal cocaine exposure during the second and third trimester and showed that global DNA methylation was decreased in hippocampal pyramidal neurons at postnatal day 3 (P3) but increased at P30 of the male offspring exposed to cocaine (Novikova et al., 2008). When examining gene specific methylation change, they found more CGI (CG island) and repetitive elements were hypomethylated at P3 and less CGI-associated promoters showed altered methylation state at P30. These results suggest that maternal cocaine exposure affects the offspring not only at the newborn stage but persists into prepubertal period, although the effect may be reduced during development.
The DNA methylation changes recorded in the epigenome can be carried in germ cells and if escaped from erasure can be transmitted to next generation. A well-documented case reported that the DNA methylation and disease states caused by the insecticide Vinclozolin were transmitted across 3–4 generations through sperm epigenetic mechanisms (Anway, Leathers, & Skinner, 2006; Skinner, Anway, Savenkova, Gore, & Crews, 2008). Another study looked at early life stress and its transmissible impacts and provided evidence that stress-induced epigenetic memory is retainable across generations (Franklin et al., 2010). This study showed that maternal separation in F0 animals leads to altered promoter methylation of Mecp2, CB1, and Crfr2 in the F1 and F2 sperm, and in F2 female brain as well as persistent behavioral change. The altered methylation was also associated with changes in gene expression (Franklin et al., 2010).
3.2. On Histone modification
The second most common altered epigenetic mark is histone modification. This includes histone acetylation, methylation, and other types of modification on different residues of histone subunits. Histone acetylation is generally considered as an open chromatin mark, while histone methylation could be either an open or condensed mark depending on the residue of methylation. For example, H3K4 methylation is considered as a gene activation signal, whereas H3K9 and H3K27 methylation is correlated with transcription repression (Y. Zhang & Reinberg, 2001).
Many of the environmental factors affect chromatin structure through direct interaction with enzymes that modified histone codes. The most prominent one is inhibition of histone deacetylase (HDAC) or histone acetyl transferase (HAT). For example, Valproic acid (VPA) and Trichostatin A (TSA; antifungal antibiotic) are well-known histone deacetylase (HDAC) inhibitors (Gottlicher et al., 2001; Vigushin et al., 2001). Previously, γ-hydroxybutyrate (GHB), which is structurally related to VPA and TSA, has also been classified as a HDAC inhibitor by increasing histone acetylation and reducing HDAC activity (C. Klein et al., 2009). Not only do these HDAC inhibitors lead to a global increase of H3 acetylation, but also indirectly increase the ratio of H3K4me3/H3K27me3, and to down-regulate genes that suppress acetylation (Balmer et al., 2012; Gurvich et al., 2005; C. Klein et al., 2009). The third mechanism is the alteration of synthesis of the enzymes that modify histone code. For example, alcohol has been shown to reduce the level of histone acetyltransferease and lead to decreased H3 and H4 acetylation in a rat FASD model (W. Guo et al., 2011).
3.3. On miRNA and other non-coding RNA
Other than DNA methylation and histone code, differential expression or methylation of miRNA and long non-coding RNA (ncRNA) has also been associated with prenatal exposure to alcohol, nicotine, arsenic and emotional stress (Dalton et al., 2014; M. A. Maccani et al., 2010; Rager et al., 2013). However, the direct causal mechanism on changes to these non-coding RNA is largely unclear. The known causes are alterations of DNA methylation on non-coding regions (Chen, Shin, Thamotharan, & Devaskar, 2014; Laufer et al., 2013) or alterations of RNA methylation on these non-coding RNAs (Xhemalce, Robson, & Kouzarides, 2012). For example, Laufer et al. identified ncRNAs that harbor differential DNA methylation and have altered ncRNA expression resulting from fetal alcohol exposure. Interestingly, some of these ncRNAs colocalize with imprinting regions that are related to neurological disorders (Laufer et al., 2013). It is proposed that increased promoter methylation for these ncRNAs link to decreased expression of ncRNAs (Vrba, Munoz-Rodriguez, Stampfer, & Futscher, 2013), similar to the general central dogma of gene regulation by DNA methylation. Additionally, a class of miRNAs contains upstream RE1 silencing transcription factor (REST) elements to regulate the miRNA expression (Fineberg, Kosik, & Davidson, 2009). Furthermore, some of the REST containing miRNAs also reciprocally target the REST complexes such as MeCP2 (Conaco, Otto, Han, & Mandel, 2006). Therefore, miRNAs and the REST complexs form a feedback loop to regulate each other (M. E. Klein et al., 2007; Yamakuchi & Lowenstein, 2009). For instance, MeCP2 induces miR-132 transcription by activating the neurotropic factor (BDNF), which promotes miR-132 expression. However, miR-132 also targets the long 3′UTR of MeCP2 transcripts, which is specifically present in differentiated neural tissue but not other tissues (M. E. Klein et al., 2007). Therefore, miR-132 regulates the translation of MeCP2 and their interaction control the balanced production at both sides in a tissue-specific manner.
3.4. On Transposable elements
Transposable elements (TEs) are repetitive and movable DNA sequences that are also called “jumping genes” for their ability to move and insert into the genome (Ade et al., 2013; Chenais, Caruso, Hiard, & Casse, 2012; Rowold & Herrera, 2000). Two of the most abundant transposable elements are the non-Long terminal repeat element Long Interspersed Elements-1 (LINE-1) and the nonautonomous Short Interspersed Elements Alu elements, which account for 42% of all TEs in the human genome (Lander et al., 2001). The richness of LINE-1 and Alu elements commonly lead to non-allelic homologous recombination that further induce genomic instability such as deletion/insertion, translocation, inversion or duplication (Konkel & Batzer, 2010).
Similar to non-coding RNA, one known mechanism for regulating transposable element is though DNA methylation (Chenais et al., 2012; Reiss, Zhang, Rouhi, Reuter, & Mager, 2010). Indeed, alterations of LINE-1 and Alu at methylation levels have been reported in several studies of prenatal exposure to deleterious factors such as alcohol (Wilhelm-Benartzi et al., 2012), smoking (Breton et al., 2009) and arsenic (Intarasunanont et al., 2012). For example, human placenta of maternal alcohol and nicotine-exposed fetus harbor differential methylation on LINE-1 and AluYb8 elements (Wilhelm-Benartzi et al., 2012). The reason why differential methylation of TEs is measured with fetal alcohol versus smoking exposure is still unknown. However, it has been shown in plants that methylation of TE inhibits transposition (Hollister & Gaut, 2009). Therefore, is it possible that TE methylation in response to prenatal environmental exposure alters genomic stability introduced by TE transposition. This altered genomic stability further determines the intrinsic genetic regulation response to external exposure.
4. Effect of Epigenetic Alterations on Neurodevelopment
4.1. Epigenetic Functional Concerns
How environmental factors change epigenetics to alter the neurodevelopment and function of the nervous system are core interests of biologists and clinicians. It should be noted first that many of the epigenetic changes may remain silent. However, epigenetic alteration can be accumulated over life, thus inherited and acquired epigenetic changes can reach the critical mass that tips the balance of DNA methylation or histone modification, leading to compaction or relaxation of the 3-D DNA conformation and changing the accessibility of the transcription factors. This may be relevant clinically to late onset disease, or idiosyncratic psychiatric disorders. Second, it is still yet to be elucidated what level and what loci of the epigenetic changes entail a transcriptional transformation, e.g. how may CpG methylation changes and their distance to the transcription action center affect gene expression. Current attention focuses in promoter regions where particularly functional transcription factors or other collaborative epigenetic complexes reside. However emerging understanding of the distribution and role of the epigenetic marks expand the horizons of the epicenters, such as enhancer, silencer, and alternative splicing determination sites. Further, new epigenetic functional roles are derived from the recent understanding of dynamics and diversity of the two DNA methylation forms. Although 5mC’s association with suppression of transcription is well accepted, the role of recently found 5hmC was ambivalent at the time of discovery (Bhutani, Burns, & Blau, 2011) and was considered an intermediate state of demethylation. We have recently reported that 5hmC is not an intermediate product but remains throughout life in the brain (Chen, Damayanti, Irudayaraj, Dunn, & Zhou, 2014). Recent research has begun to shed some light on the potential role of 5hmC. It has been found that 5mC and 5hmC, are diversely distributed in the genome where 5mC occurs preferentially in the promoter/enhancer regions, while 5hmC is increased in the intragenic gene body regions of genes undergoing active transcription in mouse embryonic stem (ES) cells (Stroud, Feng, Morey Kinney, Pradhan, & Jacobsen, 2011; Wu et al., 2011; Yildirim et al., 2011) and mouse cerebellum (J. U. Guo et al., 2011; Pastor et al., 2011; Wu et al., 2011). 5mC and 5hmC are also distributed in proximity to different categories of histone codes respectively, in ES cells (Pastor et al., 2011; Stroud et al., 2011; Williams et al., 2011; Y. Xu et al., 2011). We further demonstrated that 5mC and 5hmC are differentially associated with euchromatin versus heterochromatin. During neural development in early brain formation, 5mC preferentially binds to MBD1 and MeCP2 colocalizes with histone H3K9 trimethylation (H3K9me3) and H3K27me3, and is ultimately packed into heterochromatin during and after neural differentiation. 5mC is likely to organize a more selective transcription during differentiation (Chen, Damayanti, et al., 2014). In contrast, 5hmC preferentially binds to MBD3, colocalized with the euchromatin histone H3K4me2, and gradually translocates to the euchromatin, which demonstrates a transition to transcription needed for differentiation. Thus, 5mC and 5hmC are each functionally unique in the epigenetic landscape. This additional layer of complexity in the epigenome is more efficient when driving the complex journey of development, as well as more responsive to environmental inputs (Chen et al., 2013; Resendiz et al., 2013; F. C. Zhou, 2012). Perhaps the most conclusive evidence that we have observed is the co-localization of 5hmC with PolII in the euchromatin regions throughout neuronal differentiation, and the transient and complete dissociation of 5mC and PolII throughout chromatin in nucleus (Chen, Damayanti, et al., 2014).
4.2. Intrinsic Epigenetic Program
DNA methylation, throughout development, is not static or random, but rather “orderly” with a clear pattern. This orderly pattern is preserved over evolutionary time. Such an evolving epigenetic pattern that constitutes a “memory” (Bird, 2002) may be a way for maintenance of stable cellular identities (De Carvalho, You, & Jones, 2010; Deaton & Bird, 2011). The DNA methylation dynamics are observed as early as zygote formation by starting with global demethylation to the entire genome except the silenced X chromosome (Saitou et al., 2012), and from here a new wave of DNA methylation re-establishes methylation selectively among house keeping and tissue specific gene cohorts. This orderly methylation of gene cohorts has been hypothesized as a driving force for directing cellular restriction spatiotemporally in tissue and organ development. This orderly DNA methylation transformation during tissue or organ development is referred to here as DNA methylation program (DMP) [reviewed see (F. C. Zhou, 2012)].
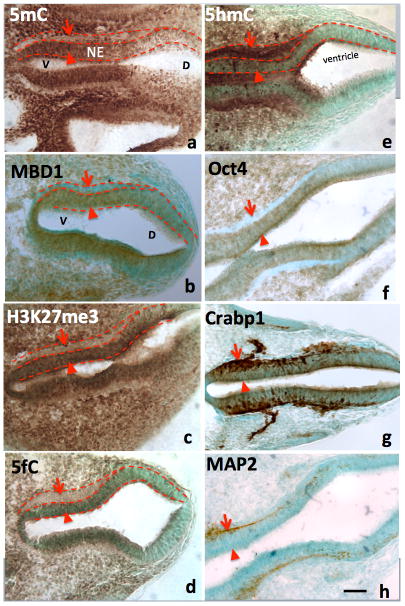
During early neural tube development, the DNA methylation and demethylation are dynamically progressing along with neural differentiation (Figure 4). Many of the neuroprogenitor cells near the ventricle bear low level of DNA methylation marks. The neural tubes expand their cell number through proliferation but with absence of DNMTs. Thus, new born cells do not bear much DNA methylation in actively proliferating ventricular zone and dorsal neural tube (F. C. Zhou et al., 2011). They acquire DNA methylation of both 5mC and 5hmC when the proliferation ceases, to begin their restriction (cell fate progression), and head for migration. The 5mC and 5hmC within the nuclei are initially distributed diffusely, but gradually polarize as the precursor cells differentiate and migrate. The 5mC-rich chromatin and 5hmC-rich chromatin become mosaic and correspond to the heterochromatic (DAPI-dense) and the euchromatic (DAPI-sparse) compartments respectively, within the nuclei (Chen, Damayanti, et al., 2014). The DNA methyltransferase 1 and 3 (DNMT1 and 3) escalate in parallel with these DNA methylations. The appearance of 5hmC and demethylation are accompanied with the enzyme ten-eleven transferase (Tet1/2). The DNA methylation binding protein e.g. MBD1 appears 1–2 days (in mouse) after the arrival of 5mC and 5hmC (F. C. Zhou et al., 2011). The demethylation into 5-carboxylcytosine (5caC) and 5-formylcytosine (5fC) were also found days (in mouse) after the 5hmC. Thus, at neurulation-stage, 5mC, 5hmC, MBD1, and DNMT1 exhibited a distinct temporal DMP pattern that coincided with neural differentiation in the neural tube (F. C. Zhou et al., 2011). In spatial pattern, as the neural tube develops in the AP-axis, the brainstem differentiation progresses first and progresses rostrally and caudally -- the DMP follow the progression of the neural axis. In the dorsoventral division, the ventralis differentiates earlier than the dorsalis, so as the DMP. DNMT is similarly distributed to 5mC spatiotemporally in neural axial and dorso-ventral patterns (F. C. Zhou et al., 2011). MBD1 shows a similar dorso-ventral pattern to that of 5mC but appears a day late in gestation. The progression of the DMP is demonstrated genome-wide. During neural stem cell differentiation, a signature epigenomic program is diversification of DNA methylation, in which many moderately methylated genes became hypo- and hyper-methylated (F. C. Zhou et al., 2011a). This is consistent with the understanding that many multipotent genes were turned off, e.g. Oct 4; and many cell specific genes were turned on, e.g. MAP2 (F. C. Zhou et al., 2011a).
Figure 4.
From Chen et al., Frontiers in Genetics (Chen, Damayanti, et al., 2014). The differential role of 5mC versus 5hmC in the DNA methylation program is inferred by their association with phenotypic marks during the making of neural tube. At early neurulation (E10), a sequential escalation of 5mC and 5hmC occurred, where 5mC appeared first at neuroepithelial (A, NE) layer (arrowheads) to prepare the differentiation, and faded as the NE cells differentiated and migrate away from ventricle (A, arrows). At this time, the distribution of the DNA methylation binding protein, MBD1, has exactly the same pattern as that of 5mC (B). Its function is depicted by the suppressive histone mark, H3K27me3, which distributed (C) similar to that of 5mC and MBD1. The demethylated metabolite 5fC also matched with 5mC. In contrast, the 5hmC appeared at differentiating neurons (E, arrowheads) and continued to increase in differentiated neurons (E, arrows). This program also occurred at ventro- (left) dorsal (right) gradation (A–H) where 5mC (A) preceded 5hmC (E). The role of 5mC and 5hmC on neural tube cell differentiation is further depicted by the differentiation phenotypic markers in a spatially precise manner. The role of the 5mC is demonstrated by the appearance of the Oct4 in the NE layer (F, arrowhead) and the loss of Oct4 in differentiated neurons (F, arrow). The role of 5hmC is noted by the stage-wise neuronal marker Crabp1 (G) and MAP2 (H) distributed in a spatially correlated manner. V: ventral; D: dorsal.
As the neural tube vesicles turn into brain regions, the cellular 5mC and 5hmC are highly correlated with the neural progenitor cells and their progression during differentiation throughout brain regions. Similar to the neural tube, throughout developing brain where neuroprogenitor cells are differentiating, 5mC appears first followed by 5hmC. As the DMP progresses further, many of the differentiating neurons containing a high level of DNA methylation will lose their 5mC and 5hmC after arriving in target regions of the brain when transitioning to active wiring and synaptogenesis. It is also apparent that the DNA demethylation marks, 5caC and 5fC, have existed along with the brain formation. This methylation and demethylation cycle can repeat over the progression of differentiation. The progression of DMP is cell stage-dependent. Thus, within a brain region, the late differentiating neuroprogenitor cells initiate the early DMP while the more differentiated neurons advance further in DMP. This is demonstrated in the hippocampus during early development where newly arrived neurons are distributed in orderly layers (Chen et al., 2013). This indicates that DMP includes DNA methylation and demethylation, which are prevailing throughout brain development. This likely explains the active repression, e.g. cell cycle genes, or multipotent genes Nanog and Oct4, and activation of neural determining or differentiation gene cohorts, e.g. Ascl1, Ngn1, and Sox1.
5hmC also demonstrates an epigenomic change in the developing hippocampus and cerebellum. When comparing early postnatal and one-year-old mice, it was found that the acquisition of 5hmC was developmentally programmed (Szulwach et al., 2011). As elucidated earlier, the correlation of 5hmC to activation of transcription is gaining more support. Furthermore, the 5hmC levels were inversely correlated with methyl-CpG–binding protein 2 (MeCP2) in the cerebellum and hippocampus during early postnatal development (Szulwach et al., 2011). Thus, the DMP includes 5mC and 5hmC dynamics, and methylation-demethylation transition spatiotemporally during development.
The cellular patterning of histone codes, as indicated in Section 4.2 is closely synchronized with DNA methylation during neural development (Chen, Damayanti, et al., 2014). 5mC is progressively associated with H3K9me3 and H3K27me3 in heterochromatin, and 5hmC is progressively associated with H3K4me2 in euchromatin as the neuroprogenitor cells claim their neural fate and migrate toward target sites. Together methylcytosines determine the 3-D conformation of the DNA to accommodate the binding of TF onto specific NT binding sites. The histone complex is also organized by the epigenetic regulator. Polycomb group (PcG) proteins associated with gene repression recruit H3K27me3 and H3K9me3 to maintain multipotency of the neuroprogenitor cells and inhibit differentiation, while trithorax group (TrxG) proteins associated with gene repression recruit H3K4me3 mark. Their interplays switch the bivalent states of proneuronal genes and multipotency genes such as neurogenins (Ngns), Pax6, Sox, Nkx2.2 and Ascl1 (Mikkelsen et al., 2007; Prozorovski et al., 2008) [review see (Nakashima, 2004)] and subsequently determine the fate and differentiation of the neuroprogenitor cells in the neuroepithelial layer. Epigenetic regulation of neurodevelopment is also regulated by small non-coding RNAs such as microRNAs (miRNAs) that are known to regulate neural stem cell fate by targeting mRNA at post-transcription level and chromatin-remodeling pathways during embryonic and adult neurogenesis [review see (Lopez-Ramirez & Nicoli, 2013)]. The timing and regulation of the miRNA formation in the neuroepithelial cells relevant to neuronal differentiation remain allusive. Advance in this area will greatly increase our understanding of how miRNA mediate the orderly evolving neuron.
4.3. Alcohol Drinking, Epigenetics Phenotypes, and FASDs
The altered epigenetics reaching critical mass has been demonstrated to affect wide-spectrum of tissue and organ development including the brain, their functions, and long lasting consequence. Since parental alcohol drinking, among many aversive environmental agents, induces epigenetic alterations and these epigenetic consequences have been studied in depth, the exemplifications below serve as a model for analyses of other environmental agents. As indicated above, parental or prenatal alcohol exposure results in a wide range of epigenetic changes including DNA methylation, histone modification and miRNA in the growing offspring. These epigenetic alterations affect multiple level and stage of the development.
4.3.1 Effect on germ line and placenta
First, prior to the conception, alcohol exposure is reported to increase DNA demethylation at normally hypermethylated imprinting control sites of the H19 locus in sperm (Ouko et al., 2009), and during pregnancy alcohol affects placenta DNA methylation (Haycock, 2009), which are likely to contribute to altering the critical gene expression dosages required for normal prenatal development and placenta supportive functions. Furthermore, the early alteration of DNA methylation may interfere with the default histone coding and chromatin conformation required during normal development (Knezovich & Ramsay, 2012).
4.3.2 Effect on embryonic and early brain development
The effect of prenatal alcohol-induced epigenetics alterations on development at embryonic stage is richly demonstrated in the literature. A high-throughput DNA-methylation immunoprecipitation analysis indicated that a binge-like alcohol exposure alters the methylation profile of over a thousand genes at early neural development (Liu et al., 2009; F. C. Zhou et al., 2011a). These alterations were further shown to be associated with a change in the expression of several genes, including those required for neural specification and neural growth (F. C. Zhou et al., 2011). Genomic analysis of neural stem cells (NSC) has revealed that alcohol retards moderately methylated genes from going toward hypermethylation and hypomethylation necessary for differentiation, indicating alcohol’s disruption of the neuroprogenitor DNA methylation program (F. C. Zhou et al., 2011a; F. C. Zhou et al., 2011). The evidence of altered phenotypes associated with alcohol’s DNA methylation was demonstrated by the use of agouti viable-yellow (Avy) mouse mutant, which contains a methylation-sensitive element within the Avy locus to dictate coat color dynamics (Duhl, Vrieling, Miller, Wolff, & Barsh, 1994; Wolff et al., 1998). It was found that parental alcohol exposure resulted in modification of DNA methylation and increase coat color diversity from yellow (unmethylated) to pseudoagouti (highly methylated) in the offspring (Kaminen-Ahola et al., 2010).
A key effect of maternal alcohol exposure on embryonic development is the perturbation of intrinsic DMP. Alcohol exposure during early fetal development (at mid-gestation), which is known to cause fetal alcohol spectrum disorders, altered the density and distribution of the DNA methylation marks. The alcohol exposure delayed the temporal and spatial cellular DNA methylation program in the neural tube that parallels with the retarded neural tube growth and decreased neuronal formation. The embryonic growth retardation is similar to direct inhibition of DNA methylation with 5-aza-cytidine (5-AZA). (F. C. Zhou et al., 2011). The prenatal alcohol exposure also continues to retard the DNA methylation program of 5mC and 5hmC dynamics in developing brain. As normal development proceed in mouse E15 to first week of life, the characteristic DMP, including 5mC, 5hmC and their binding proteins MBD1 and MBD3, lead the hippocampal neuronal differentiation and maturation spatiotemporally as indicated by their phenotypic marks in the Cornu Ammonis area 1 and dentate gyrus at perinatal stage. Alcohol hindered the acquisition and progression of methylation marks, and altered the chromatin translocation of these marks in the nucleus, which was correlated with developmental retardation (Chen et al., 2013).
Alcohol abuse profoundly affects histone modification in adult stage (D’Addario et al., 2013; Shukla & Aroor, 2006; Shukla et al., 2008). Parental alcohol exposure also impacts the emotional stress regulation system via histone modifications, as evidenced by altered histone 3 methylation and acetylation of POMC cells (Govorko et al., 2012). Excessive ethanol at a level attained by alcoholics was found to suppress the expression of four miRNAs, miR-9, 21, -153, and -335; whereas a lower ethanol concentration, attainable during social drinking, induced miR-335 expression (Balaraman, Tingling, Tsai, & Miranda, 2013; Sathyan, Golden, & Miranda, 2007). These miRNA can mediate apoptosis (e.g. miR-21), regulate neural progenitor cell, or cell cycle genes (e.g. miR-21, -153, and -335).
4.3.3 Potential late onset effect
Derived from fetal alcohol syndrome, we recently advocated the conception of “Epigenetic Medicine” which prescribes that many of the prenatal alcohol-induced abnormalities are not manifested or recognized until later life (Resendiz et al., 2013), such as mood disorders, schizoaffective disorders (Burd, Klug, Martsolf, & Kerbeshian, 2003; Famy, Streissguth, & Unis, 1998; O’Connor et al., 2002), epilepsy (Bonthius et al., 2001; O’Malley & Barr, 1998), and cancer (Murugan, Zhang, Mojtahedzadeh, & Sarkar, 2013; Polanco, Crismale-Gann, Reuhl, Sarkar, & Cohick, 2010). These late onset phenotypes may be activated as further accumulated epigenetic alterations beset by the initial environmental insult, reach their threshold. A major challenge in delineating the epigenetic background of these phenotypes is that they are not easily defined in a genetic context. Nonetheless, preliminary advances have been made in the epigenetic examination of stress regulating genes and their ties to anxiety disorders in adulthood. Similarly, though not yet validated in fetal alcohol models, promising epigenetic gene targets have emerged for schizophrenia (Kirkbride et al., 2012), depression (Sabunciyan et al., 2012), and various cancers (Murugan et al., 2013; Polanco et al., 2010).
4.3.4 Transgenerational effect
The transgenerational heritability of epigenetic traits that contribute to the phenotypes of disease states in the offspring is documented by the insecticide Vinclozolin which was transmitted across generations through sperm epigenetic mechanisms (Anway et al., 2006; Skinner et al., 2008). FASD features in the absence of maternal alcohol consumption where the fathers were alcoholics are being reported (Abel, 2004). Rats with chronic drinking were found to have lower than normal DNMT mRNA levels in sperm (Bielawski, Zaher, Svinarich, & Abel, 2002). As indicated earlier, parental alcohol can change DNA methylation in sperms (Haycock, 2009; Ouko et al., 2009). Another study reported hypomethylation specifically at H19 CpGs in F1 generation sperm DNA and F2 offspring brain (Knezovich & Ramsay, 2012). Prenatal alcohol has also been linked to inherited epigenetic aberrations on non-imprinted genes. Neurons containing POMC-derived peptides have been functionally compromised in patients with a family history of alcoholism, raising the possibility that the alcohol effects on the POMC system may be transmissible across generations (Zimmermann et al., 2004). In animal models, parental alcohol exposure increased the POMC DNA methylation of basal and stress paradigm of both males and females of the F1 generation, and further persisted in the F2 and F3 progeny of the established male germline (Govorko et al., 2012). These studies point to the potential heritability of alcohol-induced epigenetic abnormalities and associated gene functionality across generations. More studies are needed to confirm the genetic site, nature, and stability of reprogrammed epigenetics in the distant offspring. The outcomes could prove useful in furthering our understanding of many late-onset diseases or diseases of unknown etiology and their treatments.
Acknowledgments
The work is supported by the M. W. Keck Foundation, National Institute of Health AA016698, P60 AA07611, and Indiana University Collaborative Research Grant to FCZ. We like to thank Dr. Stephen Mason for editing the manuscript.
References
- Abel E. Paternal contribution to fetal alcohol syndrome. Addict Biol. 2004;9(2):127–133. doi: 10.1080/13556210410001716980. discussion 135–126. [DOI] [PubMed] [Google Scholar]
- Ackerman JP, Riggins T, Black MM. A review of the effects of prenatal cocaine exposure among school-aged children. Pediatrics. 2010;125(3):554–565. doi: 10.1542/peds.2009-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade C, Roy-Engel AM, Deininger PL. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol. 2013;3(6):639–645. doi: 10.1016/j.coviro.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akchiche N, Bossenmeyer-Pourie C, Kerek R, Martin N, Pourie G, Koziel V, Daval JL. Homocysteinylation of neuronal proteins contributes to folate deficiency-associated alterations of differentiation, vesicular transport, and plasticity in hippocampal neuronal cells. FASEB J. 2012;26(10):3980–3992. doi: 10.1096/fj.12-205757. [DOI] [PubMed] [Google Scholar]
- Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147(12):5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arria AM, Derauf C, Lagasse LL, Grant P, Shah R, Smith L, Lester B. Methamphetamine and other substance use during pregnancy: preliminary estimates from the Infant Development, Environment, and Lifestyle (IDEAL) study. Matern Child Health J. 2006;10(3):293–302. doi: 10.1007/s10995-005-0052-0. [DOI] [PubMed] [Google Scholar]
- Bae S, Zhang L. Prenatal cocaine exposure increases apoptosis of neonatal rat heart and heart susceptibility to ischemia-reperfusion injury in 1-month-old rat. Br J Pharmacol. 2005;144(7):900–907. doi: 10.1038/sj.bjp.0706129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Zhu X, Zhang Y, Zhang S, Zhang L, Xue L, Zhang X. Abnormal hippocampal BDNF and miR-16 expression is associated with depression-like behaviors induced by stress during early life. PLoS One. 2012;7(10):e46921. doi: 10.1371/journal.pone.0046921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Tingling JD, Tsai PC, Miranda RC. Dysregulation of microRNA expression and function contributes to the etiology of fetal alcohol spectrum disorders. Alcohol Res. 2013;35(1):18–24. doi: 10.35946/arcr.v35.1.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer NV, Weng MK, Zimmer B, Ivanova VN, Chambers SM, Nikolaeva E, Leist M. Epigenetic changes and disturbed neural development in a human embryonic stem cell-based model relating to the fetal valproate syndrome. Hum Mol Genet. 2012;21(18):4104–4114. doi: 10.1093/hmg/dds239. [DOI] [PubMed] [Google Scholar]
- Bellinger DC. Teratogen update: lead and pregnancy. Birth Defects Res A Clin Mol Teratol. 2005;73(6):409–420. doi: 10.1002/bdra.20127. [DOI] [PubMed] [Google Scholar]
- Berry RJ, Li Z, Erickson JD, Li S, Moore CA, Wang H, Correa A. Prevention of neural-tube defects with folic acid in China. China-U.S. Collaborative Project for Neural Tube Defect Prevention. N Engl J Med. 1999;341(20):1485–1490. doi: 10.1056/NEJM199911113412001. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146(6):866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res. 2002;26(3):347–351. [PubMed] [Google Scholar]
- Bihaqi SW, Huang H, Wu J, Zawia NH. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer’s disease. J Alzheimers Dis. 2011;27(4):819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Pantazis NJ, Karacay B, Bonthius NE, Taggard Da, Lothman EW. Alcohol exposure during the brain growth spurt promotes hippocampal seizures, rapid kindling, and spreading depression. Alcohol Clin Exp Res. 2001;25(5):734–745. [PubMed] [Google Scholar]
- Boscarino JA. Posttraumatic stress disorder, exposure to combat, and lower plasma cortisol among Vietnam veterans: findings and clinical implications. J Consult Clin Psychol. 1996;64(1):191–201. doi: 10.1037//0022-006x.64.1.191. [DOI] [PubMed] [Google Scholar]
- Bose R, Onishchenko N, Edoff K, Janson Lang AM, Ceccatelli S. Inherited effects of low-dose exposure to methylmercury in neural stem cells. Toxicol Sci. 2012;130(2):383–390. doi: 10.1093/toxsci/kfs257. [DOI] [PubMed] [Google Scholar]
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180(5):462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne ML. Maternal exposure to caffeine and risk of congenital anomalies: a systematic review. Epidemiology. 2006;17(3):324–331. doi: 10.1097/01.ede.0000208476.36988.44. [DOI] [PubMed] [Google Scholar]
- Buonocore G, Bracci R, Weindling AM. Neonatology: a practical approach to neonatal diseases. Milan; New York: Springer; 2010. [Google Scholar]
- Burd L, Klug MG, Martsolf JT, Kerbeshian J. Fetal alcohol syndrome: neuropsychiatric phenomics. Neurotoxicol Teratol. 2003;25(6):697–705. doi: 10.1016/j.ntt.2003.07.014. [DOI] [PubMed] [Google Scholar]
- Buscariollo DL, Fang X, Greenwood V, Xue H, Rivkees SA, Wendler CC. Embryonic Caffeine Exposure Acts via A1 Adenosine Receptors to Alter Adult Cardiac Function and DNA Methylation in Mice. PLoS One. 2014;9(1):e87547. doi: 10.1371/journal.pone.0087547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo P, Ibanez F, Guajardo A, Llanos MN, Ronco AM. Impact of cadmium exposure during pregnancy on hepatic glucocorticoid receptor methylation and expression in rat fetus. PLoS One. 2012;7(9):e44139. doi: 10.1371/journal.pone.0044139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik I, Gallicchio L, Boyd K, Lam TK, Matanoski G, Tao X, Alberg AJ. Arsenic in drinking water and lung cancer: a systematic review. Environ Res. 2008;108(1):48–55. doi: 10.1016/j.envres.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Centers for Disease, C. Use of folic acid for prevention of spina bifida and other neural tube defects--1983–1991. MMWR Morb Mortal Wkly Rep. 1991;40(30):513–516. [PubMed] [Google Scholar]
- Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33(4):593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Bhaumik S, Nag Chaudhury A, Das Gupta S. Arsenic induced changes in growth development and apoptosis in neonatal and adult brain cells in vivo and in tissue culture. Toxicol Lett. 2002;128(1–3):73–84. doi: 10.1016/s0378-4274(01)00535-5. [DOI] [PubMed] [Google Scholar]
- Chefer VI, Denoroy L, Zapata A, Shippenberg TS. Mu opioid receptor modulation of somatodendritic dopamine overflow: GABAergic and glutamatergic mechanisms. European Journal of Neuroscience. 2009;30(2):272–278. doi: 10.1111/j.1460-9568.2009.06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Damayanti NP, Irudayaraj J, Dunn K, Zhou FC. Diversity of two forms of DNA methylation in the brain. Front Genet. 2014;5:46. doi: 10.3389/fgene.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ozturk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS One. 2013;8(3):e60503. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shin BC, Thamotharan S, Devaskar SU. Differential methylation of the micro-RNA 7b gene targets postnatal maturation of murine neuronal Mecp2 gene expression. Dev Neurobiol. 2014;74(4):407–425. doi: 10.1002/dneu.22126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenais B, Caruso A, Hiard S, Casse N. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene. 2012;509(1):7–15. doi: 10.1016/j.gene.2012.07.042. [DOI] [PubMed] [Google Scholar]
- Chorbov VM, Todorov AA, Lynskey MT, Cicero TJ. Elevated levels of DNA methylation at the OPRM1 promoter in blood and sperm from male opioid addicts. J Opioid Manag. 2011;7(4):258–264. doi: 10.5055/jom.2011.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103(7):2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronican AA, Fitz NF, Carter A, Saleem M, Shiva S, Barchowsky A, Lefterov I. Genome-wide alteration of histone H3K9 acetylation pattern in mouse offspring prenatally exposed to arsenic. PLoS One. 2013;8(2):e53478. doi: 10.1371/journal.pone.0053478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Addario C, Caputi FF, Ekstrom TJ, Di Benedetto M, Maccarrone M, Romualdi P, Candeletti S. Ethanol induces epigenetic modulation of prodynorphin and pronociceptin gene expression in the rat amygdala complex. J Mol Neurosci. 2013;49(2):312–319. doi: 10.1007/s12031-012-9829-y. [DOI] [PubMed] [Google Scholar]
- Dalton VS, Kolshus E, McLoughlin DM. Epigenetics and depression: return of the repressed. J Affect Disord. 2014;155:1–12. doi: 10.1016/j.jad.2013.10.028. [DOI] [PubMed] [Google Scholar]
- Darnaudery M, Maccari S. Epigenetic programming of the stress response in male and female rats by prenatal restraint stress. Brain Res Rev. 2008;57(2):571–585. doi: 10.1016/j.brainresrev.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Das A, Poole WK, Bada HS. A repeated measures approach for simultaneous modeling of multiple neurobehavioral outcomes in newborns exposed to cocaine in utero. Am J Epidemiol. 2004;159(9):891–899. doi: 10.1093/aje/kwh114. [DOI] [PubMed] [Google Scholar]
- Day NL, Richardson GA. Prenatal marijuana use: epidemiology, methodologic issues, and infant outcome. Clin Perinatol. 1991;18(1):77–91. [PubMed] [Google Scholar]
- De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20(10):609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNieri JA, Wang XY, Szutorisz H, Spano SM, Kaur J, Casaccia P, Hurd YL. Maternal Cannabis Use Alters Ventral Striatal Dopamine D2 Gene Regulation in the Offspring. Biol Psychiatry. 2011;70(8):763–769. doi: 10.1016/J.Biopsych.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z, Waters SB, Devesa V, Harmon AW, Thomas DJ, Styblo M. Metabolism and toxicity of arsenic in human urothelial cells expressing rat arsenic (+3 oxidation state)-methyltransferase. Toxicol Appl Pharmacol. 2005;207(2):147–159. doi: 10.1016/j.taap.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8(1):59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Ebrahim SHGJ. Pregnancy-related substance use in the United States during 1996–1998. Obstet Gynecol. 2003;101(2):374–349. doi: 10.1016/s0029-7844(02)02588-7. [DOI] [PubMed] [Google Scholar]
- El Marroun H, Tiemeier H, Steegers EAP, Jaddoe VWV, Hofman A, Verhulst FC, Huizink AC. Intrauterine Cannabis Exposure Affects Fetal Growth Trajectories: The Generation R Study. Journal of the American Academy of Child and Adolescent Psychiatry. 2009;48(12):1173–1181. doi: 10.1097/Chi.0b013e3181bfa8ee. [DOI] [PubMed] [Google Scholar]
- Famy C, Streissguth AP, Unis AS. Mental illness in adults with fetal alcohol syndrome or fetal alcohol effects. Am J Psychiatry. 1998;155(4):552–554. doi: 10.1176/ajp.155.4.552. [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64(3):303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, Mansuy IM. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68(5):408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Frary CD, Johnson RK, Wang MQ. Food sources and intakes of caffeine in the diets of persons in the United States. J Am Diet Assoc. 2005;105(1):110–113. doi: 10.1016/j.jada.2004.10.027. [DOI] [PubMed] [Google Scholar]
- Frassica JJ, Orav EJ, Walsh EP, Lipshultz SE. Arrhythmias in children prenatally exposed to cocaine. Arch Pediatr Adolesc Med. 1994;148(11):1163–1169. doi: 10.1001/archpedi.1994.02170110049008. [DOI] [PubMed] [Google Scholar]
- Friedman EH. Neurobiology of arrhythmias in children prenatally exposed to cocaine. Arch Pediatr Adolesc Med. 1995;149(10):1178–1179. doi: 10.1001/archpedi.1995.02170230132028. [DOI] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15(3):395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20(24):6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry. 2012;72(5):378–388. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueant JL, Namour F, Gueant-Rodriguez RM, Daval JL. Folate and fetal programming: a play in epigenomics? Trends Endocrinol Metab. 2013;24(6):279–289. doi: 10.1016/j.tem.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Crossey EL, Zhang L, Zucca S, George OL, Valenzuela CF, Zhao X. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PLoS One. 2011;6(5):e19351. doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvich N, Berman MG, Wittner BS, Gentleman RC, Klein PS, Green JB. Association of valproate-induced teratogenesis with histone deacetylase inhibition in vivo. FASEB J. 2005;19(9):1166–1168. doi: 10.1096/fj.04-3425fje. [DOI] [PubMed] [Google Scholar]
- Hackshaw A, Rodeck C, Boniface S. Maternal smoking in pregnancy and birth defects: a systematic review based on 173 687 malformed cases and 11.7 million controls. Hum Reprod Update. 2011;17(5):589–604. doi: 10.1093/humupd/dmr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hairston IS, Waxler E, Seng JS, Fezzey AG, Rosenblum KL, Muzik M. The role of infant sleep in intergenerational transmission of trauma. Sleep. 2011;34(10):1373–1383. doi: 10.5665/SLEEP.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsted CH, Villanueva J, Chandler CJ, Stabler SP, Allen RH, Muskhelishvili L, Poirier L. Ethanol feeding of micropigs alters methionine metabolism and increases hepatocellular apoptosis and proliferation. Hepatology. 1996;23(3):497–505. doi: 10.1002/hep.510230314. [DOI] [PubMed] [Google Scholar]
- Hanson ML, Brundage KM, Schafer R, Tou JC, Barnett JB. Prenatal cadmium exposure dysregulates sonic hedgehog and Wnt/beta-catenin signaling in the thymus resulting in altered thymocyte development. Toxicol Appl Pharmacol. 2010;242(2):136–145. doi: 10.1016/j.taap.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M. Minamata disease: methylmercury poisoning in Japan caused by environmental pollution. Crit Rev Toxicol. 1995;25(1):1–24. doi: 10.3109/10408449509089885. [DOI] [PubMed] [Google Scholar]
- Haycock PC. Fetal Alcohol Spectrum Disorders: The Epigenetic Perspective. Biol Reprod. 2009 doi: 10.1095/biolreprod.108.074690. biolreprod.108.074690 [pii] [DOI] [PubMed] [Google Scholar]
- Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the h19 imprinting control region. Biol Reprod. 2009;81(4):618–627. doi: 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- He F, Lidow IA, Lidow MS. Consequences of paternal cocaine exposure in mice. Neurotoxicol Teratol. 2006;28(2):198–209. doi: 10.1016/j.ntt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol. 2012;233(1):102–111. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Holaskova I, Elliott M, Hanson ML, Schafer R, Barnett JB. Prenatal cadmium exposure produces persistent changes to thymus and spleen cell phenotypic repertoire as well as the acquired immune response. Toxicol Appl Pharmacol. 2012;265(2):181–189. doi: 10.1016/j.taap.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19(8):1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexsters A, Claes S. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J Psychiatr Res. 2013;47(7):880–891. doi: 10.1016/j.jpsychires.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Hoyo C, Murtha AP, Schildkraut JM, Forman MR, Calingaert B, Demark-Wahnefried W, Murphy SK. Folic acid supplementation before and during pregnancy in the Newborn Epigenetics STudy (NEST) Bmc Public Health. 2011;11:Artn 46. doi: 10.1186/1471-2458-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizink AC. Prenatal cannabis exposure and infant outcomes: Overview of studies. Prog Neuropsychopharmacol Biol Psychiatry. 2013 doi: 10.1016/j.pnpbp.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Intarasunanont P, Navasumrit P, Waraprasit S, Chaisatra K, Suk WA, Mahidol C, Ruchirawat M. Effects of arsenic exposure on DNA methylation in cord blood samples from newborn babies and in a human lymphoblast cell line. Environ Health. 2012;11:31. doi: 10.1186/1476-069X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak Y, Ergui I, Young JI. Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.7. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jeng W, Wong AW, Ting AKR, Wells PG. Methamphetamine-enhanced embryonic oxidative DNA damage and neurodevelopmental deficits. Free Radic Biol Med. 2005;39(3):317–326. doi: 10.1016/j.freeradbiomed.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Jensen Pena C, Monk C, Champagne FA. Epigenetic effects of prenatal stress on 11beta-hydroxysteroid dehydrogenase-2 in the placenta and fetal brain. PLoS One. 2012;7(6):e39791. doi: 10.1371/journal.pone.0039791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara Y, Yasutake A, Adachi T, Hirayama K. Methylmercury transport across the placenta via neutral amino acid carrier. Arch Toxicol. 1996;70(5):310–314. doi: 10.1007/s002040050279. [DOI] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox TC, Chong S. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010;6(1):e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerek R, Geoffroy A, Bison A, Martin N, Akchiche N, Pourie G, Daval JL. Early methyl donor deficiency may induce persistent brain defects by reducing Stat3 signaling targeted by miR-124. Cell Death Dis. 2013;4:e755. doi: 10.1038/cddis.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, Christiani DC. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect. 2012;120(7):1061–1066. doi: 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Sartor MA, Rozek LS, Faulk C, Anderson OS, Jones TR, Dolinoy DC. Perinatal bisphenol A exposure promotes dose-dependent alterations of the mouse methylome. BMC Genomics. 2014;15:30. doi: 10.1186/1471-2164-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41(2):126–132. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- Kim KC, Kim P, Go HS, Choi CS, Yang SI, Cheong JH, Ko KH. The critical period of valproate exposure to induce autistic symptoms in Sprague-Dawley rats. Toxicol Lett. 2011;201(2):137–142. doi: 10.1016/j.toxlet.2010.12.018. [DOI] [PubMed] [Google Scholar]
- Kippler M, Engstrom K, Mlakar SJ, Bottai M, Ahmed S, Hossain MB, Broberg K. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics. 2013;8(5):494–503. doi: 10.4161/epi.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkbride JB, Errazuriz A, Croudace TJ, Morgan C, Jackson D, Boydell J, Jones PB. Incidence of schizophrenia and other psychoses in England, 1950–2009: a systematic review and meta-analyses. PLoS ONE. 2012;7(3):e31660. doi: 10.1371/journal.pone.0031660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Kemmel V, Taleb O, Aunis D, Maitre M. Pharmacological doses of gamma-hydroxybutyrate (GHB) potentiate histone acetylation in the rat brain by histone deacetylase inhibition. Neuropharmacology. 2009;57(2):137–147. doi: 10.1016/j.neuropharm.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10(12):1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Knezovich JG, Ramsay M. The effect of preconception paternal alcohol exposure on epigenetic remodeling of the h19 and rasgrf1 imprinting control regions in mouse offspring. Frontiers in genetics. 2012;3:10. doi: 10.3389/fgene.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect. 2013;121(8):971–977. doi: 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Batzer MA. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 2010;20(4):211–221. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczkowski KM. Caffeine in pregnancy. Arch Gynecol Obstet. 2009;280(5):695–698. doi: 10.1007/s00404-009-0991-6. [DOI] [PubMed] [Google Scholar]
- Kuehn BM. Panel advises tougher limits on lead exposure. JAMA. 2012;307(5):445. doi: 10.1001/jama.2012.50. [DOI] [PubMed] [Google Scholar]
- Kutay H, Klepper C, Wang B, Hsu SH, Datta J, Yu L, Ghoshal K. Reduced susceptibility of DNA methyltransferase 1 hypomorphic (Dnmt1N/+) mice to hepatic steatosis upon feeding liquid alcohol diet. PLoS One. 2012;7(8):e41949. doi: 10.1371/journal.pone.0041949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski MA, Roos A, Stein DJ, Thomas KG, Donald K. Effects of prenatal methamphetamine exposure: a review of cognitive and neuroimaging studies. Metab Brain Dis. 2013 doi: 10.1007/s11011-013-9470-7. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J International Human Genome Sequencing, C. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laufer BI, Mantha K, Kleiber ML, Diehl EJ, Addison SM, Singh SM. Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Dis Model Mech. 2013;6(4):977–992. doi: 10.1242/dmm.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester BM, ElSohly M, Wright LL, Smeriglio VL, Verter J, Bauer CR, Maza PL. The Maternal Lifestyle Study: drug use by meconium toxicology and maternal self-report. Pediatrics. 2001;107(2):309–317. doi: 10.1542/peds.107.2.309. [DOI] [PubMed] [Google Scholar]
- Levine A, Huang Y, Drisaldi B, Griffin EA, Jr, Pollak DD, Xu S, Kandel ER. Molecular mechanism for a gateway drug: epigenetic changes initiated by nicotine prime gene expression by cocaine. Sci Transl Med. 2011;3(107):107ra109. doi: 10.1126/scitranslmed.3003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CR, Staudinger K, Scheck L, Olive MF. The Effects of Maternal Separation on Adult Methamphetamine Self-Administration, Extinction, Reinstatement, and MeCP2 Immunoreactivity in the Nucleus Accumbens. Front Psychiatry. 2013;4:55. doi: 10.3389/fpsyt.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Li Y, Li Z, Liu Z, Zhang Z, Chang S, Wu J. Mechanism of folate deficiency-induced apoptosis in mouse embryonic stem cells: Cell cycle arrest/apoptosis in G1/G0 mediated by microRNA-302a and tumor suppressor gene Lats2. Int J Biochem Cell Biol. 2012;44(11):1750–1760. doi: 10.1016/j.biocel.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4(7):500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ramirez MA, Nicoli S. Role of miRNAs and epigenetics in neural stem cell fate determination. Epigenetics. 2013;9(1) doi: 10.4161/epi.27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W. Basic pharmacology of valproate: a review after 35 years of clinical use for the treatment of epilepsy. CNS Drugs. 2002;16(10):669–694. doi: 10.2165/00023210-200216100-00003. [DOI] [PubMed] [Google Scholar]
- Lu Q, Yang YT, Chen CS, Davis M, Byrd JC, Etherton MR, Chen CS. Zn2+-chelating motif-tethered short-chain fatty acids as a novel class of histone deacetylase inhibitors. J Med Chem. 2004;47(2):467–474. doi: 10.1021/jm0303655. [DOI] [PubMed] [Google Scholar]
- Lu SC, Huang ZZ, Yang H, Mato JM, Avila MA, Tsukamoto H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;279(1):G178–185. doi: 10.1152/ajpgi.2000.279.1.G178. [DOI] [PubMed] [Google Scholar]
- Maccani JZ, Koestler DC, Houseman EA, Marsit CJ, Kelsey KT. Placental DNA methylation alterations associated with maternal tobacco smoking at the RUNX3 gene are also associated with gestational age. Epigenomics. 2013;5(6):619–630. doi: 10.2217/epi.13.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, Marsit CJ. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics. 2010;5(7):583–589. doi: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier SE, Cramer JA, West JR, Sohrabji F. Alcohol exposure during the first two trimesters equivalent alters granule cell number and neurotrophin expression in the developing rat olfactory bulb. J Neurobiol. 1999;41(3):414–423. doi: 10.1002/(sici)1097-4695(19991115)41:3<414::aid-neu9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Mamelak M, Scharf MB, Woods M. Treatment of narcolepsy with gamma-hydroxybutyrate. A review of clinical and sleep laboratory findings. Sleep. 1986;9(1 Pt 2):285–289. doi: 10.1093/sleep/9.1.285. [DOI] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One. 2013;8(1):e55387. doi: 10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh DO, Myers GJ, Clarkson TW, Amin-Zaki L, Tikriti S, Majeed MA. Fetal methylmercury poisoning: clinical and toxicological data on 29 cases. Ann Neurol. 1980;7(4):348–353. doi: 10.1002/ana.410070412. [DOI] [PubMed] [Google Scholar]
- Mason JW, Giller EL, Kosten TR, Ostroff RB, Podd L. Urinary free-cortisol levels in posttraumatic stress disorder patients. J Nerv Ment Dis. 1986;174(3):145–149. doi: 10.1097/00005053-198603000-00003. [DOI] [PubMed] [Google Scholar]
- Matthews SG. Early programming of the hypothalamo-pituitary-adrenal axis. Trends Endocrinol Metab. 2002;13(9):373–380. doi: 10.1016/s1043-2760(02)00690-2. [DOI] [PubMed] [Google Scholar]
- McDowell MA, Dillon CF, Osterloh J, Bolger PM, Pellizzari E, Fernando R, Mahaffey KR. Hair mercury levels in U.S. children and women of childbearing age: reference range data from NHANES 1999–2000. Environ Health Perspect. 2004;112(11):1165–1171. doi: 10.1289/ehp.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO. Epigenomic Mechanisms of Early Adversity and HPA Dysfunction: Considerations for PTSD Research. Front Psychiatry. 2013;4:110. doi: 10.3389/fpsyt.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharg A. Venomous earth: how arsenic caused the world’s worst mass poisoning. Choice: Current Reviews for Academic Libraries. 2005;43(4):693–693. [Google Scholar]
- Mergler D, Anderson HA, Chan LH, Mahaffey KR, Murray M, Sakamoto M Toxicological Effects of, M. Methylmercury exposure and health effects in humans: a worldwide concern. Ambio. 2007;36(1):3–11. doi: 10.1579/0044-7447(2007)36[3:meahei]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Meyer K, Zhang H, Zhang L. Direct effect of cocaine on epigenetic regulation of PKCepsilon gene repression in the fetal rat heart. J Mol Cell Cardiol. 2009;47(4):504–511. doi: 10.1016/j.yjmcc.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Zhang H, Zhang L. Prenatal cocaine exposure abolished ischemic preconditioning-induced protection in adult male rat hearts: role of PKCepsilon. Am J Physiol Heart Circ Physiol. 2009;296(5):H1566–1576. doi: 10.1152/ajpheart.00898.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC. MicroRNAs and Fetal Brain Development: Implications for Ethanol Teratology during the Second Trimester Period of Neurogenesis. Front Genet. 2012;3:77. doi: 10.3389/fgene.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti B, Polazzi E, Contestabile A. Biochemical, molecular and epigenetic mechanisms of valproic acid neuroprotection. Curr Mol Pharmacol. 2009;2(1):95–109. doi: 10.2174/1874467210902010095. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Murugan S, Zhang C, Mojtahedzadeh S, Sarkar DK. Alcohol exposure in utero increases susceptibility to prostate tumorigenesis in rat offspring. Alcohol Clin Exp Res. 2013;37(11):1901–1909. doi: 10.1111/acer.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzik M, Marcus SM, Flynn HA. Psychotherapeutic treatment options for perinatal depression: emphasis on maternal-infant dyadic outcomes. J Clin Psychiatry. 2009;70(9):1318–1319. doi: 10.4088/JCP.09com05451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers GJ, Davidson PW. Prenatal methylmercury exposure and children: neurologic, developmental, and behavioral research. Environ Health Perspect. 1998;106(Suppl 3):841–847. doi: 10.1289/ehp.98106841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Itoh K, Yaoi T, Fujiwara Y, Sugimoto T, Fushiki S. Murine neocortical histogenesis is perturbed by prenatal exposure to low doses of Bisphenol A. J Neurosci Res. 2006;84(6):1197–1205. doi: 10.1002/jnr.21020. [DOI] [PubMed] [Google Scholar]
- National Institute on Drug Abuse. Prescription drug abuse and addiction. NIH Publication Number 11-4881. 2011:2. Printed July 2001, Reveised October 2011. [Google Scholar]
- Nakashima K. Mechanism of neural stem cell fate specification. Tanpakushitsu Kakusan Koso. 2004;49(6):718–726. [PubMed] [Google Scholar]
- Nehlig A, Debry G. Potential teratogenic and neurodevelopmental consequences of coffee and caffeine exposure: a review on human and animal data. Neurotoxicol Teratol. 1994;16(6):531–543. doi: 10.1016/0892-0362(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Nelson T, Kaufman E, Kline J, Sokoloff L. The extraneural distribution of gamma-hydroxybutyrate. J Neurochem. 1981;37(5):1345–1348. doi: 10.1111/j.1471-4159.1981.tb04689.x. [DOI] [PubMed] [Google Scholar]
- Novikova SI, He F, Bai J, Cutrufello NJ, Lidow MS, Undieh AS. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS One. 2008;3(4):e1919. doi: 10.1371/journal.pone.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell T, Kaye L, Plosay JJ., 3rd Gamma-hydroxybutyrate (GHB): a newer drug of abuse. Am Fam Physician. 2000;62(11):2478–2483. [PubMed] [Google Scholar]
- O’Connor MJ, Shah B, Whaley S, Cronin P, Gunderson B, Graham J. Psychiatric illness in a clinical sample of children with prenatal alcohol exposure. Am J Drug Alcohol Abuse. 2002;28(4):743–754. doi: 10.1081/ada-120015880. [DOI] [PubMed] [Google Scholar]
- O’Malley KD, Barr H. Fetal alcohol syndrome and seizure disorder. Can J Psychiatry. 1998;43(10):1051. [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Olsson IM, Bensryd I, Lundh T, Ottosson H, Skerfving S, Oskarsson A. Cadmium in blood and urine--impact of sex, age, dietary intake, iron status, and former smoking--association of renal effects. Environ Health Perspect. 2002;110(12):1185–1190. doi: 10.1289/ehp.021101185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishchenko N, Karpova N, Sabri F, Castren E, Ceccatelli S. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem. 2008;106(3):1378–1387. doi: 10.1111/j.1471-4159.2008.05484.x. [DOI] [PubMed] [Google Scholar]
- Otero NK, Thomas JD, Saski CA, Xia X, Kelly SJ. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcohol Clin Exp Res. 2012;36(10):1701–1709. doi: 10.1111/j.1530-0277.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2009;33(9):1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sci. 2007;81(12):979–987. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanza P, Gioiosa L, vom Saal FS, Parmigiani S. Effects of developmental exposure to bisphenol A on brain and behavior in mice. Environ Res. 2008;108(2):150–157. doi: 10.1016/j.envres.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. nature10102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkinsa A, Lehmanna C, Lawrencea RC, Kelly SJ. Alcohol exposure during development: Impact on the epigenome. International Journal of Developmental Neuroscience. 2013;31(6):391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Plume JM, Gibbons FX, Brody GH, Beach SR. The impact of recent alcohol use on genome wide DNA methylation signatures. Front Genet. 2012;3:54. doi: 10.3389/fgene.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Hall MN, Liu X, Ilievski V, Slavkovich V, Levy D, Gamble MV. Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PLoS One. 2012;7(5):e37147. doi: 10.1371/journal.pone.0037147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, Hernandez-Avila M. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117(9):1466–1471. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrone N, Mahaffey KR. Dynamics of mercury pollution on regional and global scales: atmospheric processes and human exposures around the world. New York: Springer; 2005. [Google Scholar]
- Polanco TA, Crismale-Gann C, Reuhl KR, Sarkar DK, Cohick WS. Fetal alcohol exposure increases mammary tumor susceptibility and alters tumor phenotype in rats. Alcohol Clin Exp Res. 2010;34(11):1879–1887. doi: 10.1111/j.1530-0277.2010.01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Aktas O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10(4):385–394. doi: 10.1038/ncb1700. ncb1700 [pii] [DOI] [PubMed] [Google Scholar]
- Qiang M, Denny A, Lieu M, Carreon S, Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics. 2011;6(9):1095–1104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, Fry RC. Prenatal arsenic exposure and the epigenome: Altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ Mol Mutagen. 2013 doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RV. Post-traumatic stress disorder and birthweight: methodological challenges. BJOG. 2012;119(1):110. doi: 10.1111/j.1471-0528.2011.03199.x. author reply 111. [DOI] [PubMed] [Google Scholar]
- Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010;2(1):87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss D, Zhang Y, Rouhi A, Reuter M, Mager DL. Variable DNA methylation of transposable elements: the case study of mouse Early Transposons. Epigenetics. 2010;5(1):68–79. doi: 10.4161/epi.5.1.10631. [DOI] [PubMed] [Google Scholar]
- Resendiz M, Chen Y, Ozturk NC, Zhou FC. Epigenetic medicine and fetal alcohol spectrum disorders. Epigenomics. 2013;5(1):73–86. doi: 10.2217/epi.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus GZ, Abelaira HM, dos Santos MA, Carlessi AS, Tomaz DB, Neotti MV, Quevedo J. Ketamine and imipramine in the nucleus accumbens regulate histone deacetylation induced by maternal deprivation and are critical for associated behaviors. Behav Brain Res. 2013;256:451–456. doi: 10.1016/j.bbr.2013.08.041. [DOI] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci. 2013;33(21):9003–9012. doi: 10.1523/JNEUROSCI.0914-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65(9):760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotta NT, Cunha GB. Prenatal exposure to cocaine: review of the neurobehavioral effects. J Pediatr (Rio J) 2000;76(3):179–184. doi: 10.2223/jped.122. [DOI] [PubMed] [Google Scholar]
- Rowold DJ, Herrera RJ. Alu elements and the human genome. Genetica. 2000;108(1):57–72. doi: 10.1023/a:1004099605261. [DOI] [PubMed] [Google Scholar]
- Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, Gen REDC. Genome-wide DNA methylation scan in major depressive disorder. PLoS ONE. 2012;7(4):e34451. doi: 10.1371/journal.pone.0034451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Vakili G, Kumaresan V, Schmidt HD, Famous KR, Chawla P, Vassoler FM, Cha JH. Cocaine-induced chromatin remodeling increases brain-derived neurotrophic factor transcription in the rat medial prefrontal cortex, which alters the reinforcing efficacy of cocaine. J Neurosci. 2010;30(35):11735–11744. doi: 10.1523/JNEUROSCI.2328-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139(1):15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Sanders AP, Smeester L, Rojas D, Debussycher T, Wu MC, Wright FA, Fry RC. Cadmium exposure and the epigenome: Exposure-associated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics. 2013;9(2) doi: 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders T, Liu Y, Buchner V, Tchounwou PB. Neurotoxic effects and biomarkers of lead exposure: a review. Rev Environ Health. 2009;24(1):15–45. doi: 10.1515/reveh.2009.24.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27(32):8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Kidd SK, Anderson DW. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett. 2013;217(1):75–81. doi: 10.1016/j.toxlet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut MC, Cingolani P, Sen A, Kruger A, Shaik A, Hirsch H, Ruden D. Epigenetics of early-life lead exposure and effects on brain development. Epigenomics. 2012;4(6):665–674. doi: 10.2217/epi.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Aroor AR. Epigenetic effects of ethanol on liver and gastrointestinal injury. World J Gastroenterol. 2006;12(33):5265–5271. doi: 10.3748/wjg.v12.i33.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Lee YJ, Park PH, Aroor AR. Acetaldehyde alters MAP kinase signalling and epigenetic histone modifications in hepatocytes. Novartis Found Symp. 2007;285:217–224. doi: 10.1002/9780470511848.ch16. discussion 224–218. [DOI] [PubMed] [Google Scholar]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging Role of Epigenetics in the Actions of Alcohol. Alcohol Clin Exp Res. 2008 doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS ONE. 2008;3(11):e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield KH, Pilsner JR, Lu Q, Wright RO, Guilarte TR. Dysregulation of BDNF-TrkB signaling in developing hippocampal neurons by Pb(2+): implications for an environmental basis of neurodevelopmental disorders. Toxicol Sci. 2012;127(1):277–295. doi: 10.1093/toxsci/kfs090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straif K, Benbrahim-Tallaa L, Baan R, Grosse Y, Secretan B, El Ghissassi F W. H. O. I. A. f. R. o. C. M. W Group. A review of human carcinogens--part C: metals, arsenic, dusts, and fibres. Lancet Oncol. 2009;10(5):453–454. doi: 10.1016/s1470-2045(09)70134-2. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Shivakumar M, Umapathy NS, Saito M, Mohan PS, Kumar A, Basavarajappa BS. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiol Dis. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, Abramovici A, Showalter L, Hu M, Shope CD, Varner M, Aagaard-Tillery K. In utero tobacco exposure epigenetically modifies placental CYP1A1 expression. Metabolism. 2010;59(10):1481–1490. doi: 10.1016/j.metabol.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, Ma J, Harris A, Patterson L, Brown KA, Shope C, Aagaard-Tillery KM. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6(11):1284–1294. doi: 10.4161/epi.6.11.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011 doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. The genome- and system-wide response of DNA methylation to early life adversity and its implication on mental health. Can J Psychiatry. 2013;58(12):697–704. doi: 10.1177/070674371305801208. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos PK, Michaelides M, Umegaki H, Volkow ND. D2R DNA transfer into the nucleus accumbens attenuates cocaine self-administration in rats. Synapse. 2008;62(7):481–486. doi: 10.1002/syn.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Rodriguez M, Lotfipour S, Leonard G, Perron M, Richer L, Veillette S, Paus T. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(7):1350–1354. doi: 10.1002/ajmg.b.31109. [DOI] [PubMed] [Google Scholar]
- Tong VTD, PM, Morrow B, D’Angelo DV, Farr SL, Rockhill KM, England LJ Centers for Disease Control and Prevention (CDC) Trends in smoking before, during, and after pregnancy--Pregnancy Risk Assessment Monitoring System, United States, 40 sites, 2000–2010. MMWR Surveill Summ. 2013;8(62):1–19. [PubMed] [Google Scholar]
- Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48(6):479–487. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PLoS One. 2012;7(1):e30148. doi: 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Watanabe Y. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69(2):359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Ungerer M, Knezovich J, Ramsay M. In utero alcohol exposure, epigenetic changes, and their consequences. Alcohol Res. 2013;35(1):37–46. doi: 10.35946/arcr.v35.1.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, Coombes RC. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res. 2001;7(4):971–976. [PubMed] [Google Scholar]
- vom Saal FS, Akingbemi BT, Belcher SM, Birnbaum LS, Crain DA, Eriksen M, Zoeller RT. Chapel Hill bisphenol A expert panel consensus statement: integration of mechanisms, effects in animals and potential to impact human health at current levels of exposure. Reprod Toxicol. 2007;24(2):131–138. doi: 10.1016/j.reprotox.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrba L, Munoz-Rodriguez JL, Stampfer MR, Futscher BW. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS One. 2013;8(1):e54398. doi: 10.1371/journal.pone.0054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Li Y. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod. 2009;24(3):562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- White LD, Cory-Slechta DA, Gilbert ME, Tiffany-Castiglioni E, Zawia NH, Virgolini M, Basha MR. New and evolving concepts in the neurotoxicology of lead. Toxicol Appl Pharmacol. 2007;225(1):1–27. doi: 10.1016/j.taap.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Wilhelm-Benartzi CS, Houseman EA, Maccani MA, Poage GM, Koestler DC, Langevin SM, Marsit CJ. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect. 2012;120(2):296–302. doi: 10.1289/ehp.1103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011 doi: 10.1038/nature10066. nature10066 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12(11):949–957. [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25(7):679–684. doi: 10.1101/gad.2036011. 25/7/679 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski DF, Nambisan M, Surve T, Alsdorf RM, Smith CR, Holmes LB Antiepileptic Drug Pregnancy, R. Increased rate of major malformations in offspring exposed to valproate during pregnancy. Neurology. 2005;64(6):961–965. doi: 10.1212/01.WNL.0000154516.43630.C5. [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Robson SC, Kouzarides T. Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell. 2012;151(2):278–288. doi: 10.1016/j.cell.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu J, Benbrahim-Tallaa L, Ward JM, Logsdon D, Diwan BA, Waalkes MP. Aberrant DNA methylation and gene expression in livers of newborn mice transplacentally exposed to a hepatocarcinogenic dose of inorganic arsenic. Toxicology. 2007;236(1–2):7–15. doi: 10.1016/j.tox.2007.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhang B, Liang G, Ping J, Kou H, Li X, Wang H. Caffeine-induced activated glucocorticoid metabolism in the hippocampus causes hypothalamic-pituitary-adrenal axis inhibition in fetal rats. PLoS One. 2012;7(9):e44497. doi: 10.1371/journal.pone.0044497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Shi YG. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. S1097-2765(11)00283-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8(5):712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376(3):563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- Yasutake AMM, Yamaguchi M, Hachiya N. Current Hair Mercury Levels in Japanese for Estimation of Methylmercury Exposure. Journal of Health Science. 2004;50(2):120–125. [Google Scholar]
- Yehuda R, Bierer LM. Transgenerational transmission of cortisol and PTSD risk. Prog Brain Res. 2008;167:121–135. doi: 10.1016/S0079-6123(07)67009-5. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Schmeidler J, Wainberg M, Binder-Brynes K, Duvdevani T. Vulnerability to posttraumatic stress disorder in adult offspring of Holocaust survivors. Am J Psychiatry. 1998;155(9):1163–1171. doi: 10.1176/ajp.155.9.1163. [DOI] [PubMed] [Google Scholar]
- Yeo M, Berglund K, Hanna M, Guo JU, Kittur J, Torres MD, Liedtke WB. Bisphenol A delays the perinatal chloride shift in cortical neurons by epigenetic effects on the Kcc2 promoter. Proc Natl Acad Sci U S A. 2013;110(11):4315–4320. doi: 10.1073/pnas.1300959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147(7):1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochum C, Doherty-Lyon S, Hoffman C, Hossain MM, Zelikoff JT, Richardson JR. Prenatal cigarette smoke exposure causes hyperactivity and aggressive behavior: Role of altered catecholamines and BDNF. Exp Neurol. 2014;254C:145–152. doi: 10.1016/j.expneurol.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Darwanto A, Linkhart TA, Sowers LC, Zhang L. Maternal cocaine administration causes an epigenetic modification of protein kinase Cepsilon gene expression in fetal rat heart. Mol Pharmacol. 2007;71(5):1319–1328. doi: 10.1124/mol.106.032011. [DOI] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013;37(Suppl 1):E108–115. doi: 10.1111/j.1530-0277.2012.01928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15(18):2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zhou FC. DNA methylation program during development. Front Biol (Beijing) 2012;7(6):485–494. doi: 10.1007/s11515-012-9246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Balaraman Y, Teng M, Liu Y, Singh RP, Nephew KP. Alcohol Alters DNA Methylation Patterns and Inhibits Neural Stem Cell Differentiation. Alcohol Clin Exp Res. 2011a;w35(4):735–746. doi: 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Chen Y, Love A. Cellular DNA methylation program during neurulation and its alteration by alcohol exposure. Birth Defects Res A Clin Mol Teratol. 2011;91(8):703–715. doi: 10.1002/bdra.20820. [DOI] [PubMed] [Google Scholar]
- Zhou X, Li Q, Arita A, Sun H, Costa M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol Appl Pharmacol. 2009;236(1):78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann U, Spring K, Wittchen HU, Himmerich H, Landgraf R, Uhr M, Holsboer F. Arginine vasopressin and adrenocorticotropin secretion in response to psychosocial stress is attenuated by ethanol in sons of alcohol-dependent fathers. Journal of Psychiatric Research. 2004;38(4):385–393. doi: 10.1016/j.jpsychires.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Zoroddu MA, Peana M, Medici S, Casella L, Monzani E, Costa M. Nickel binding to histone H4. Dalton Trans. 2010;39(3):787–793. doi: 10.1039/b916019c. [DOI] [PubMed] [Google Scholar]