Abstract
Aims: High salinity stress impairs plant growth and development. Trehalose metabolism has been implicated in sugar signaling, and enhanced trehalose metabolism can positively regulate abiotic stress tolerance. However, the molecular mechanism(s) of the stress-related trehalose pathway and the role of individual trehalose biosynthetic enzymes for stress tolerance remain unclear. Results: Trehalose-6-phosphate phosphatase (TPP) catalyzes the final step of trehalose metabolism. Investigating the subcellular localization of the Arabidopsis thaliana TPP family members, we identified AtTPPD as a chloroplast-localized enzyme. Plants deficient in AtTPPD were hypersensitive, whereas plants overexpressing AtTPPD were more tolerant to high salinity stress. Elevated stress tolerance of AtTPPD overexpressors correlated with high starch levels and increased accumulation of soluble sugars, suggesting a role for AtTPPD in regulating sugar metabolism under salinity conditions. Biochemical analyses indicate that AtTPPD is a target of post-translational redox regulation and can be reversibly inactivated by oxidizing conditions. Two cysteine residues were identified as the redox-sensitive sites. Structural and mutation analyses suggest that the formation of an intramolecular disulfide bridge regulates AtTPPD activity. Innovation: The activity of different AtTPP isoforms, located in the cytosol, nucleus, and chloroplasts, can be redox regulated, suggesting that the trehalose metabolism might relay the redox status of different cellular compartments to regulate diverse biological processes such as stress responses. Conclusion: The evolutionary conservation of the two redox regulatory cysteine residues of TPPs in spermatophytes indicates that redox regulation of TPPs might be a common mechanism enabling plants to rapidly adjust trehalose metabolism to the prevailing environmental and developmental conditions. Antioxid. Redox Signal. 21, 1289–1304.
Introduction
High soil salinity is a major environmental constraint for plant growth and development and a worldwide phenomenon that negatively affects agricultural productivity. Salt stress can disturb numerous cellular and physiological processes, including chloroplast integrity and photosynthesis (41). It leads to metabolic imbalances and production of excess levels of reactive oxygen species (ROS) (32, 51). Plants have evolved various strategies to cope with salinity, including recovery of ion homeostasis and accumulation of osmoprotectants (53).
Carbohydrates are thought to play a crucial role in acclimation to osmotic stress conditions such as drought, cold, and salt stress. Soluble sugars, such as mono- and disaccharides as well as raffinose family oligosaccharides, can function as osmolytes to maintain cell turgor, to protect membranes and proteins, and to act as radical scavengers upon abiotic stress conditions (40).
Innovation.
A regulatory role of trehalose metabolism in diverse biological processes, including stress tolerance, has emerged. We provide evidence that the trehalose-6-phosphate (T6P) metabolizing enzymes (trehalose-6-phosphate phosphatases [TPPs]) are present in the cytosol, nucleus, and chloroplasts. Genetic data indicate a role for the chloroplast-localized AtTPPD in modulating carbohydrate metabolism and salt stress tolerance. Biochemical data show that AtTPPD is a direct target for redox-dependent activity modulation. Based on the evolutionary conservation of the redox-sensitive cysteine residues, we propose a new mechanism regulating the TPP activity by post-translational redox modification, thereby linking trehalose metabolism to the cellular redox state.
Trehalose, a nonreducing disaccharide, is widely distributed in nature. In bacteria, fungi, and invertebrates, it serves as reserve carbohydrate, compatible solute, and protectant against environmental stress (22). In plants, only certain desiccation-tolerant species, for example, Myrothamnus flabellifolius (9, 20), accumulate trehalose to sufficiently high levels to function as an osmolyte and to stabilize proteins and membranes. In most angiosperms, however, trehalose is present in trace amounts and abiotic stress increases the levels of trehalose only moderately (24, 25, 27, 31, 34, 36, 60, 64). Nevertheless, different studies in these plants showed that trehalose metabolism can contribute to the acquisition of tolerance to environmental stresses via not yet understood mechanism(s) (23).
In addition to the ancestral connection relating trehalose metabolism with stress, the trehalose pathway plays an important role in regulating plant growth and development under nonstress conditions (12, 21, 29, 30, 65, 78, 79, 85). Trehalose-6-phosphate (T6P) has been suggested to function as a signaling metabolite communicating the plant's carbohydrate status to other pathways involved in growth, development, and responses to the environment (46). Thus, the multiple functions of trehalose metabolism in plants are likely caused by its role in sugar signaling.
In plants, trehalose is synthesized in a two-step process (56). Trehalose-6-phosphate synthase (TPS; EC2.4.1.15) catalyzes the formation of T6P from UDP-glucose and glucose-6-phosphate. T6P is dephosphorylated by trehalose-6-phosphate phosphatase (TPP; EC3.1.3.12) to generate trehalose. Trehalose can be hydrolyzed by trehalase (TRE; EC3.2.1.28) to glucose. In Arabidopsis thaliana, there are 11 genes coding for TPSs (class I and II trehalose biosynthesis genes), 10 genes encoding for TPPs (class III trehalose biosynthesis genes), and 1 gene coding for TRE.
There has been great interest in trehalose metabolism as a means of engineering environmental stress tolerance in crops. In numerous studies, heterologous expression of microbial trehalose biosynthesis genes in different plant species conferred protection to abiotic stresses (24, 33), but often led to growth aberrations; this is likely due to the regulatory role of trehalose metabolism in plant development. Overexpression of different isoforms of TPS from rice conferred enhanced resistance of rice plants to salinity, cold, and drought (43). In response to cold stress, the TPP activity and trehalose levels have been reported to transiently increase in rice (60). Overexpression of OsTPP1 rendered rice more tolerant to salinity and cold, although no increase in the trehalose content could be observed (28). In Arabidopsis, constitutive overexpression of AtTPS1 slightly increased T6P and trehalose levels and enhanced drought tolerance (4), and plants deficient in AtTPS5 function showed a reduced basal thermotolerance (77). Interestingly, overexpression of AtTRE decreased trehalose levels and enhanced drought resistance, whereas loss of AtTRE1 elevated trehalose contents and decreased drought tolerance (80). These data illustrate that intact trehalose metabolism is important in plants to cope with environmental stress and that genetic engineering of the trehalose pathway can enhance stress tolerance. Yet, the precise function of trehalose metabolism and its metabolites, T6P and trehalose, under environmental stress conditions is not yet clear. Moreover, the specific roles of the various different isoforms of the enzymes in the trehalose biosynthesis pathway remain to be further elucidated.
The Arabidopsis genome contains 10 TPP genes (AtTPPA-J). Functional complementation of yeast Δtps2 mutants indicated that all genes code for active TPP enzymes (81, 84). AtTPP promoter reporter lines and transcriptional analyses showed that specific AtTPPs have distinct tissue-specific expression patterns during development and in response to different environmental stimuli, possibly indicating a functional diversification (44, 71, 79, 81). In line with this view, an increased shoot size and an increased number of epidermal cells were observed for tppb plants, while tppg knockout plants showed an abscisic acid (ABA)-related stomatal phenotype, and tppa tppg double mutants displayed a hairy root phenotype (79, 81).
Chloroplasts are of endosymbiotic origin and have essential roles in photosynthesis and associated metabolic pathways. They also function as important environmental sensors and communicate changes to adjust metabolism and modulate gene expression (11, 26, 58, 66). Feeding of T6P to isolated chloroplasts induced accumulation of starch (38), suggesting that at least part of trehalose metabolism is present in chloroplasts. However, it remains elusive whether any TPP, the enzyme that uses T6P to catalyze the formation of trehalose, is present in chloroplasts. Furthermore, it remains unknown whether an innate trehalose metabolism in chloroplasts might be important for plant stress tolerance. In this work, we have systematically analyzed the subcellular localization of the Arabidopsis TPPs. We identified AtTPPD and AtTPPE as chloroplast-localized enzymes and investigated the functional role of AtTPPD in salt stress tolerance. Furthermore, we provide biochemical evidence for a redox-dependent enzymatic regulation of TPPs.
Results
Subcellular localization of the Arabidopsis TPP gene family
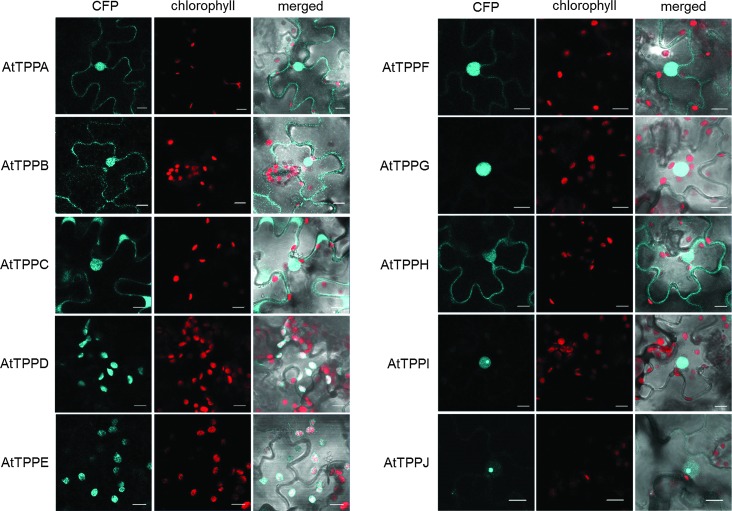
We explored the subcellular localization of the 10 Arabidopsis TPP family members in a systematic approach. Bioinformatics-based predictions (http://suba.plantenergy.uwa.edu.au) suggested that several AtTPPs might be present in the chloroplasts. To experimentally verify the subcellular localization of the 10 AtTPPs, the full-length coding sequences of AtTPPA-J were cloned and their localization as cyan fluorescent protein (CFP) fusion proteins was investigated in transiently transformed tobacco leaves. Interestingly, distinct AtTPPs showed different subcellular localization (Fig. 1). Chloroplast localization, indicated by an overlap of the CFP signal and the chlorophyll autofluorescence, was observed for two AtTPPs: AtTPPD and AtTPPE. AtTPPA, AtTPPB, AtTPPC, AtTPPF, and AtTPPH fused to the CFP showed fluorescence in the cytosol and nucleus. AtTPPG, AtTPPI, and AtTPPJ CFP fusion proteins predominantly showed fluorescence in the nucleus. For AtTPPI and AtTPPJ, a strong signal was found in the nucleolus. The differential subcellular localization of the diverse AtTPP family members indicates a role for trehalose metabolism in different cellular compartments.
FIG. 1.
Subcellular localization of AtTPPA-J-CFP fusion proteins in Nicotiana tabacum. Tobacco leaves were infiltrated with Agrobacterium tumefaciens containing the constructs of interest (35S::AtTPPA-J-CFP), and expression of AtTPP fluorescent proteins was visualized by confocal laser scanning microscopy 4 days after infiltration. The CFP signal (blue) is shown in the first channel and chlorophyll autofluorescence (red) in the second. The third channel shows the merged image. The scale bars represent 10 μm. CFP, cyan fluorescent protein.
AtTPPD is a positive regulator of salt stress tolerance
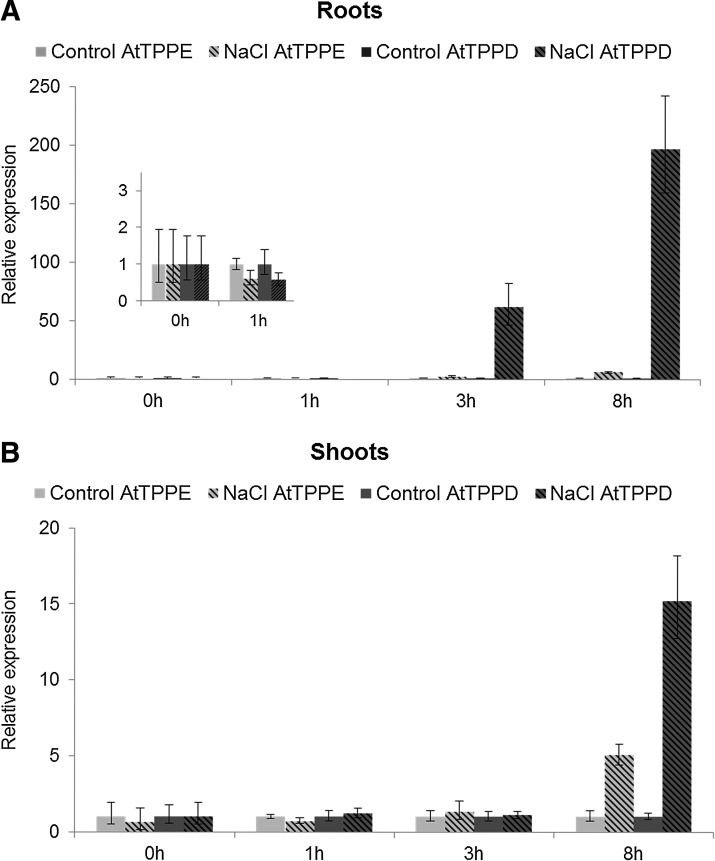
To investigate whether the two chloroplast-localized AtTPPs, AtTPPD and AtTPPE, might be involved in the Arabidopsis salt stress response, we first analyzed the expression of AtTPPD and AtTPPE in plants exposed to high salinity. Real-time polymerase chain reaction (qRT-PCR) analysis showed that the AtTPPD transcripts were strongly upregulated by salt stress in shoots and in roots, whereas AtTPPE showed only a minor transcriptional induction in shoots and no induction in roots (Fig. 2).
FIG. 2.
Transcriptional induction of AtTPPD by salt stress. AtTPPD and AtTPPE expression in roots (A) and shoots (B) of plants under control and salt stress conditions was analyzed by qRT-PCR. Ten-day-old seedlings were transferred to plates without (control) or with supplementation of 200 mM NaCl. Expression was normalized to PP2A and Ubi1 and is represented as fold change of expression relative to the respective controls. Data are mean±SD of three independent experiments. qRT-PCR, real-time polymerase chain reaction.
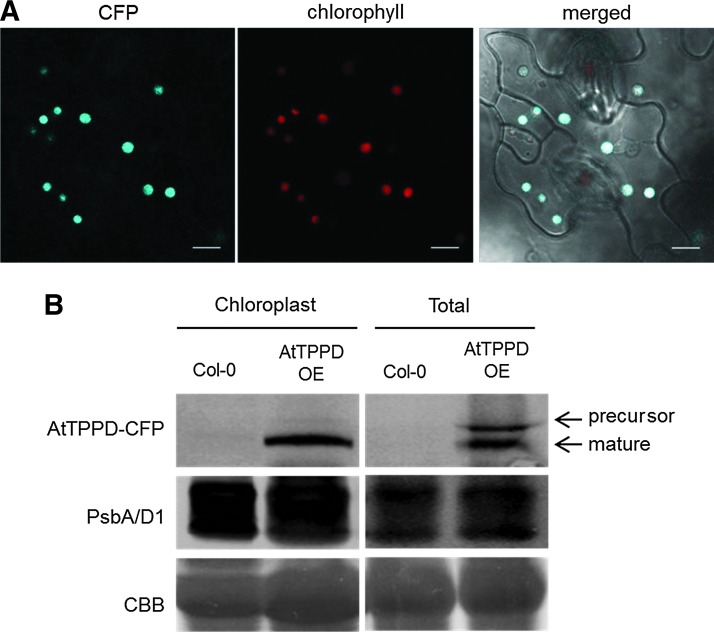
Based on the chloroplast-localization and gene expression data, we further focused our studies on AtTPPD. To analyze the biological function of AtTPPD, we obtained tppd T-DNA insertion knockout plants (Supplementary Fig. S1A, B; Supplementary Data are available online at www.liebertpub.com/ars) and plants overexpressing AtTPPD tagged with CFP from the 35S promoter (Supplementary Fig. S1B, C). AtTPPD mutant plants did not show obvious growth perturbations (Supplementary Fig. S1D–F). Consistent with the transient expression data, AtTPPD localized to the chloroplasts in leaves of Arabidopsis plants expressing AtTPPD-CFP (Fig. 3A). To confirm the subcellular localization, we performed Western blot analysis on total protein extracts and on protein extracts from purified chloroplasts. The photosystem II protein D1 (PsbA) was used as a chloroplast marker protein. In line with the microscopic analyses, the AtTPPD protein was detected in the chloroplast, while both the likely precursor and the mature AtTPPD were detected in total protein extracts (Fig. 3B).
FIG. 3.
Chloroplast localization of AtTPPD. (A) Subcellular localization of AtTPPD protein in leaves of Arabidopsis thaliana plants stably transformed with 35S::AtTPPD-CFP. Ten-day-old seedlings were analyzed by confocal microscopy. The CFP signal (blue) is shown in the first channel and chlorophyll autofluorescence (red) in the second. The third channel shows the merged image. The scale bars represent 10 μm. (B) Western blot analysis of A. thaliana leaf total protein and purified chloroplast extracts from 4-week-old wild-type (Col-0) and AtTPPD overexpressor line (AtTPPD OE). AtTPPD-CFP proteins were detected with an anti-GFP antibody. The photosystem II protein D1 antibody (PsbA/D1) was used as a chloroplast marker. CBB, Coomassie Brilliant Blue.
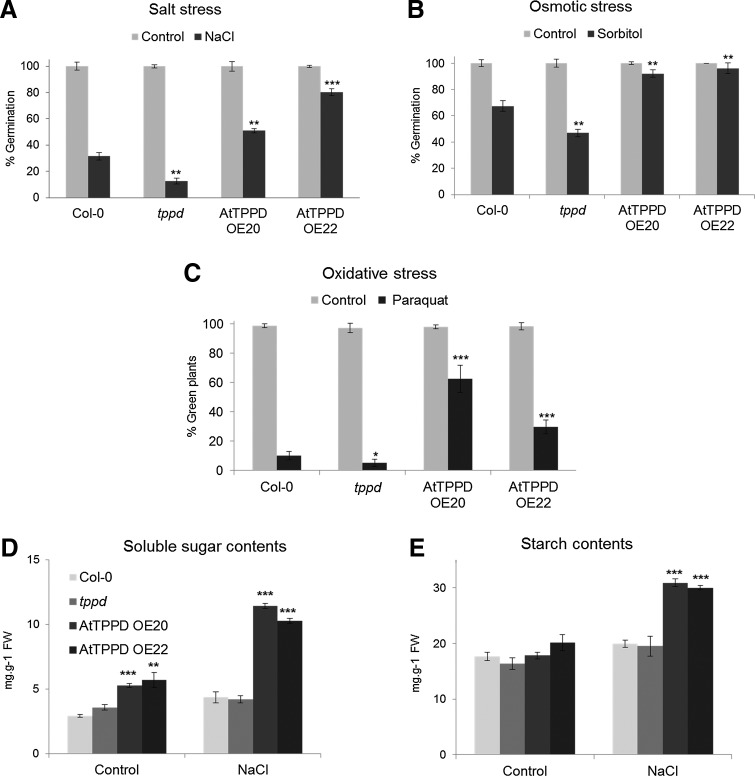
We next analyzed whether AtTPPD is important for tolerance to salt stress. On a medium supplemented with NaCl, germination efficiency was significantly reduced in tppd mutants compared with wild-type Col-0 plants grown under the same high salt stress conditions (Fig. 4A). In contrast, plants overexpressing AtTPPD germinated significantly better under high salinity conditions as compared with wild-type plants. Exposing soil-grown plants to high salinity led to chlorosis of wild-type and tppd plants, while plants overexpressing AtTPPD remained green (Supplementary Fig. S1G, H), further supporting a positive role of AtTPPD in salt stress tolerance. High salt conditions induce osmotic and ionic stress. Under high osmotic conditions, germination efficiency of plants deficient in AtTPPD was significantly reduced, whereas that of AtTPPD overexpressors was significantly enhanced (Fig. 4B), indicating a positive function of AtTPPD in osmotic stress tolerance. Salinity leads to enhanced production of ROS. Exposure of plants to paraquat, a herbicide that generates ROS within chloroplasts (6), showed that tppd mutants were significantly more sensitive and TPPD-overexpressing plants more tolerant (Fig. 4C), indicating that AtTPPD modifies tolerance to oxidative stress.
FIG. 4.
AtTPPD regulates stress tolerance. (A, B) Germination efficiency of wild-type Col-0, tppd, and two AtTPPD-CFP overexpressor lines (AtTPPD OE20 and AtTPPD OE22) on half-strength MS plates supplemented with 200 mM NaCl (A) and 360 mM sorbitol (B) is represented as the percentage relative to normal growth plates. Data are mean±relative SD of three independent biological experiments with n=70 for each genotype. Asterisks indicate a significant difference (**p≤0.01, ***p≤0.001) using Student's t-test for pairwise comparison to Col-0 under stress conditions. (C) Oxidative stress tolerance. Wild-type Col-0, tppd, and two AtTPPD-CFP overexpressor lines (AtTPPD OE20 and AtTPPD OE22) were grown on half-strength MS plates supplemented with 1 μM paraquat. The percentage of green seedlings on paraquat-containing plates relative to normal growth plates is represented. Data are mean±relative SD of three independent biological experiments with n=50 for each genotype. Asterisks indicate a significant difference (*p≤0.05, ***p≤0.001) using Student's t-test for pairwise comparison to Col-0 under stress conditions. (D, E) Soluble sugar (D) and starch (E) contents of wild-type Col-0, tppd, and two AtTPPD-CFP overexpressor lines (AtTPPD OE20 and AtTPPD OE22). Three-week-old soil-grown plants were watered without (control) or with supplementation of 200 mM NaCl. Data are mean±relative SD of four independent technical experiments. Asterisks indicate a significant difference (**p≤0.01, ***p≤0.001) using Student's t-test for pairwise comparison to Col-0 within the same condition.
Accumulation of soluble sugars contributes to the osmotic adjustment under salinity conditions (40). To investigate whether the positive role of AtTPPD in stress tolerance might involve this physiological acclimation response, we determined soluble sugar levels of wild-type and of plants deficient in or overexpressing AtTPPD under nonstress and high soil salinity conditions (Fig. 4D). Under normal growth conditions, the soluble sugar content was similar in wild-type and tppd plants, but was increased in AtTPPD-overexpressing plants. Salinity conditions moderately increased soluble sugar contents in wild-type and tppd plants. However, consistent with increased stress tolerance, AtTPPD overexpressors showed a significantly enhanced stress-induced increase in the soluble sugar content as compared with wild-type plants.
Trehalose metabolism has been shown to influence starch accumulation (29, 38, 46, 47). Thus, we also analyzed starch levels (Fig. 4E). Under normal growth conditions, AtTPPD-deficient and -overexpressing plants had starch levels similar to wild type. However, under salt stress conditions, AtTPPD overexpressors accumulated starch, pointing to a possible role of AtTPPD in modulating starch levels under stress conditions. Altogether, the data show that AtTPPD is involved in regulating sugar metabolism and stress tolerance.
AtTPPD activity is redox sensitive
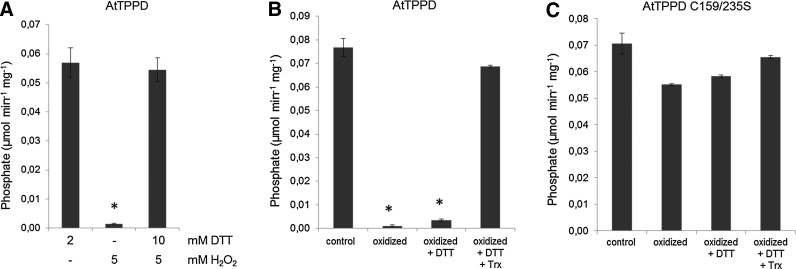
The modified tolerance of AtTPPD mutants to enhanced ROS production and the presence of cysteine residues in the amino acid sequence of AtTPPD prompted us to analyze whether AtTPPD might be redox regulated. Unless reduced dithiothreitol (DTT) was added to the reaction mixture, purified recombinant AtTPPD protein had only little catalytic activity toward T6P. Oxidation of AtTPPD led to inactivation of the enzyme, which was fully reversible by the subsequent addition of reduced DTT (Fig. 5A and Supplementary Fig. S2), suggesting that inactivation was not due to ROS damage and that AtTPPD activity can be regulated by the redox state.
FIG. 5.
Redox regulation of AtTPPD activity. (A) Redox sensitivity of AtTPPD activity. The activity of pre-reduced AtTPPD (2 mM) was analyzed after oxidation with 5 mM H2O2 followed by the addition of 10 mM DTT for reactivation. (B, C) Trx-mediated reduction of oxidized AtTTPD. Oxidized AtTPPD (B) and AtTPPD C159/235S (C) double mutants were dialyzed and subsequently treated with 0.05 mM DTT in the absence or presence of 5 μM Escherichia coli Trx. Averages and standard deviations were calculated from three independent enzymatic assays. Asterisks indicate the significant difference (*p<0.001) as determined by Student's t-test for pairwise comparison to the respective control condition. DTT, dithiothreitol; H2O2, hydrogen peroxide; Trx, thioredoxin.
In plants, the redox transmitter thioredoxin (Trx) directly reduces redox-sensitive thiols, thereby regulating a number of important plastidic target proteins (68). Thus, we investigated whether oxidized AtTPPD can be reactivated by Trx. Coincubation of fully oxidized and inactive AtTPPD with Trx and low concentration of DTT in the reactivation treatment resulted in a drastic stimulation of AtTPPD activity compared with DTT treatment alone (Fig. 5B), indicating that AtTPPD can be efficiently reduced by Trxs.
Mechanism of redox-dependent TPPD regulation
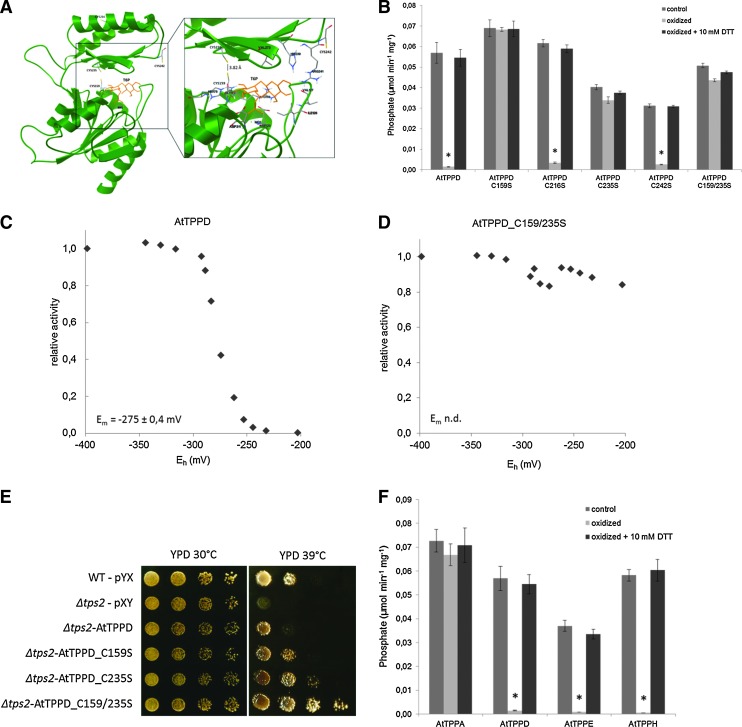
To understand the molecular effects of redox regulation of AtTPPD, we analyzed the spatial distribution of the four cysteine residues C159, C216, C235, and C242 in a structural model of AtTPPD, which was generated on the basis of the crystal structure of Thermoplasma acidophilum TPP (TaTPP) (62). TPPs belong to the magnesium-dependent haloacid-dehalogenase (HAD) superfamily of phosphatases/phosphotranferases. The three HAD-specific motifs are highly conserved between AtTPPD and TaTPP (Supplementary Fig. S3). TPP proteins consist of a core and a cap domain. The substrate T6P binds to the interface, forming hydrogen bonds with amino acid residues of both domains. C159 of AtTPPD is located in the core domain; the other three cysteine residues are part of the cap domain (Fig. 6A). Interestingly, the distance between the sulfur atoms of C159 and C235 is 3.82 Å, which would allow a disulfide bridge formation under oxidizing conditions.
FIG. 6.
Redox-sensitive cysteine residues regulate AtTPP activity. (A) Overall representation of the AtTPPD protein model (green) with the ligand T6P (orange) modeled into the active site. The detailed representation on the right shows T6P in the binding cavity of AtTPPD; the interacting residues as well as C159, C216, C235, and C242 are represented as balls and sticks. The AtTPPD homology model structure was created according to TaTPP [(2); PDB-ID: 1U02] using SWISS-MODEL (1). The ligand T6P (PDB Ligand-ID:T6P) was placed into the active site using AutoDock 4 and AutoDockTools (http://autodock.scripps.edu). (B) Cys159 and Cys235 mediate TPPD redox sensitivity. Activities of wild-type AtTPPD and of the AtTPPD variants C159S, C216S, C235S, and C242S (single mutants) and C159/235S (double mutant) were analyzed after oxidation (5 mM H2O2) and subsequent reactivation with 10 mM DTT as compared with the pre-reduced control (2 mM DTT). Averages and standard deviations were calculated from three independent enzymatic reactions. Asterisks indicate the significant difference (*p<0.001) as determined by Student's t-test for pairwise comparison to the pre-reduced control. (C, D) Redox titrations of AtTPPD (C) and AtTPPD C159/235S (D). Recombinant proteins were incubated with 20 mM DTT in different dithiol/disulfide ratios at pH 7 and 25°C. The data were fitted to the Nernst equation with n=2 (using XLFit curve fitting software for Excel). The midpoint redox potential Em (mean±SD of three experiments) for AtTPPD is indicated. (E) Complementation of the heat-sensitive yeast Δtps2. The Saccharomyces cerevisiae knockout strain Δtps2 was transformed with wild-type AtTPPD and AtTPPD variants C159S, C235S, and C159/235S. Wild-type yeast and Δtps2 cells were also transformed with the empty vector pYX424 as control. Transformed cells were spotted on YPD plates and their growth at 30°C and 39°C analyzed after 4 days. (F) Redox regulation of different AtTPP isoforms. Activities of AtTPPA, AtTPPD, AtTPPE, and AtTPPH were analyzed after oxidation (5 mM H2O2) and subsequent reactivation with 10 mM DTT as compared with the pre-reduced control (2 mM DTT). Averages and standard deviations were calculated from three independent enzymatic reactions. Asterisks indicate the significant difference (*p<0.001) as determined by Student's t-test for pairwise comparison to the pre-reduced control. TaTPP, Thermoplasma acidophilum TPP; T6P, trehalose-6-phosphate; YPD, yeast peptone dextrose.
To experimentally explore the significance of C159 and C235 for redox regulation of AtTPPD, each cysteine residue was mutated to serine, and the TPP activity of single- and C159/235S double-mutant proteins was investigated (Fig. 6B and Supplementary Table S1). Oxidizing conditions inactivated wild-type AtTPPD, AtTPPD C216S, and AtTPPD C242S. However, single C159S and C235S mutant and C159/235S double-mutant proteins maintained a high phosphatase activity under oxidizing conditions, providing biochemical evidence that C159 and C235 convey post-translational redox-dependent regulation of AtTPPD. Consistently, oxidation and subsequent incubation with Trx had no significant impact on AtTPPD C159/235S activity (Fig. 5C).
To investigate whether AtTPPD responds to physiologically meaningful redox potentials, a redox titration analysis with defined mixtures of reduced and oxidized DTT was performed (Fig. 6C, D). At pH 7.0, AtTPPD shifted from an active form at −350 mV to an inactive form at −200 mV, whereas the AtTPPD C159/235S activity was only reduced by 15%, fitting a Nernst equation for a two-electron oxidoreduction of a disulfide with a midpoint redox potential (Em) of −275 mV. This result is within the range of values for other redox-regulated chloroplastic enzymes involved in the starch metabolism of the Calvin cycle (49, 70, 74, 75).
To further analyze the significance of the two redox-sensitive cysteine residues, we made use of the Saccharomyces cerevisiae TPP knockout strain Δtps2 (Fig. 6E). Under control conditions (30°C), wild-type cells and Δtps2 cells expressing AtTPPD, AtTPPD C159S, AtTPPD C235S, or AtTPPD C159/235S grew equally well. At 39°C, wild-type AtTPPD (partially) complemented the heat-sensitive phenotype of Δtps2. Interestingly, Δtps2 cells expressing the C/S mutated versions of AtTPPD grew better at high-temperature conditions than cells expressing wild-type AtTPPD, providing evidence for a regulatory role of C159 and C235 on AtTPPD activity in vivo.
Evolutionary conservation of TPP redox sensitivity
To gain insight into whether redox regulation might be a general mechanism to modulate TPP activity, we aligned the protein sequence of the 10 Arabidopsis TPPs (Supplementary Fig. S4). The cysteine residue corresponding to AtTPPD C235 is conserved in all AtTPPs, while the cysteine residue corresponding to C159 is only present in AtTPPD, AtTPPE, AtTPPH, AtTPPI, and AtTPPJ.
Next, we tested the notion whether conservation of these two cysteine residues might predict TPP redox sensitivity and assessed the activity of purified recombinant AtTPPE, AtTPPH, and AtTPPA under oxidizing and reducing conditions as examples (Fig. 6F). Indeed, similar to AtTPPD, the activity of AtTPPE and AtTPPH, which contain the two cysteine residues corresponding to the ones conveying redox sensitivity to AtTPPD, was highly sensitive to the redox state. On the other hand, the activity of AtTPPA, which has five cysteine residues, including the cysteine corresponding to AtTPPD C235 (but none at the position equivalent to C159), remained high under oxidative conditions. These results provide further evidence that the AtTPPD activity is regulated by disulfide bridge formation between C159 and C235.
Archaea, bacteria, protists, invertebrates, and plants use class III TPP enzymes to synthesize trehalose (5). To investigate whether the mechanism of cysteine-based redox regulation of TPP proteins is evolutionarily conserved, we examined the amino acid sequence of TPP enzymes of diverse species from different kingdoms for the presence of cysteine residues corresponding to C159 and C235 in AtTPPD (Table 1). The two redox-sensitive cysteine residues are not present in archaea, bacteria, protists, nematodes, insects, and red algae. In the class of Viridiplantae (green plants), comprising green algae and land plants, the cysteine corresponding to C235 is conserved in all the TPP genes analyzed. Interestingly, the cysteine corresponding to C159 is only present in gymnosperm and angiosperm plants and, as in Arabidopsis, several but not all TPP isoforms contain the two redox-sensitive cysteine residues. It is worth noting that acquisition of the redox-sensitive domain in spermatophytes (seed plants) appears to have occurred in parallel to TPP gene expansion. In evolutionary terms, the introduction of new regulatory cysteine residues to a subclass of TPPs may have allowed a rapid response of TPP activity to cellular redox changes, enabling plants to readily adjust trehalose metabolism to developmental and environmental signals.
Table 1.
Evolutionary Conservation of Redox-Sensitive Cysteine Residues
| Species | Prot_ID/Locus | AtTPPD_C159 | AtTPPD_C235 | |
|---|---|---|---|---|
| Prokaryotes | ||||
| Archaea | Themoplasma acidophilum | Q9HIW7 | No | No |
| Bacteria | Escherichia coli | P31678 | No | No |
| Eukaryotes | ||||
| Protists | Phytophtora infestans | DONMM9 | No | No |
| Animals | ||||
| Nematodes | Caenorhabditis elegans | Q9XTQ5 | No | No |
| Caenorhabditis briggsae | A8X485 | No | No | |
| Brugia malayi | A8NS89 | No | No | |
| Insects | Drosophila melanogaster | C9QPE7 | No | No |
| Q9VM18 | No | No | ||
| Anopheles gambiae | Q7PJ67 | No | No | |
| Plants | ||||
| Red algae | Cyanidioschyzon merolae | CMQ148C | No | No |
| Galdieria sulphuraria | M2XOW6 | No | No | |
| Green algae | Ostreococcus tauri | QOOWH8 | No | Yes |
| Chlamydomonas reinhardtii | A8JGL2 | No | Yes | |
| Mosses | Physcomitrella patens | A9RWB0 | No | Yes |
| A9S4E1 | No | Yes | ||
| A9SZE3 | No | Yes | ||
| A9TV90 | No | Yes | ||
| A9TC22 | No | Yes | ||
| Ferns | Selaginella mollendorffii | D8SOQ9 | No | Yes |
| D8SBD6 | No | Yes | ||
| D8SUZ5 | No | Yes | ||
| Gymnosperms | Picea glauca | est_Pagl_157592577 | Yes | Yes |
| UniGene_1928047 | Yes | Yes | ||
| UniGene_5419283 | Yes | Yes | ||
| UniGene_2303759 | n.d. | Yes | ||
| Picea abies | MA_9055473g | Yes | Yes | |
| MA_9368348g | n.d. | Yes | ||
| MA_331962g | Yes | n.d | ||
| Pinus taeda | isotig18254 | Yes | Yes | |
| scaffold172504.1 | Yes | Yes | ||
| tcaffold5357 | Yes | Yes | ||
| Angiosperm monocots | Oryza sativa | Q75WV3 | No | Yes |
| QODDI1 | Yes | Yes | ||
| Q9FWQ2 | No | Yes | ||
| Q10KF5 | Yes | Yes | ||
| Q6ZAL2 | Yes | Yes | ||
| Q6ZGP8 | Yes | Yes | ||
| Q7XI41 | Yes | Yes | ||
| Q7XT34 | No | Yes | ||
| QOD6F4 | No | Yes | ||
| Q6H5L4 | Yes | Yes | ||
| Zea mays | Q1W5S8 | Yes | Yes | |
| B6STK8 | Yes | Yes | ||
| Q1W5S7 | Yes | Yes | ||
| B4FVF6 | No | Yes | ||
| B4FPT4 | Yes | Yes | ||
| B4FST3 | Yes | Yes | ||
| COHHB8 | Yes | Yes | ||
| Angiosperm dicots | Arabidopsis thaliana | At5g51460 | No | Yes |
| At1g78090 | No | Yes | ||
| At1g22210 | No | Yes | ||
| At1g35910 | Yes | Yes | ||
| At2g22190 | Yes | Yes | ||
| At4g12430 | No | Yes | ||
| At4g22590 | No | Yes | ||
| At4g39770 | Yes | Yes | ||
| At5g10100 | Yes | Yes | ||
| At5g65140 | Yes | Yes | ||
| Medicago truncatula | G7IYG6 | No | Yes | |
| G7J6S8 | No | Yes | ||
| G7JK40 | No | Yes | ||
| G7KAB1 | Yes | Yes | ||
| Vitis vinifera | F6HP93 | No | Yes | |
| LOC100248172 | No | Yes | ||
| LOC100251837 | No | Yes | ||
| LOC100258645 | No | Yes | ||
| EOCRI8 | Yes | Yes | ||
| F6HCY3 | No | Yes | ||
| LOC100258773 | Yes | Yes | ||
| Populus trichocarpa | B9GVI1 | No | Yes | |
| B9N3Q1 | No | Yes | ||
| B9I4N1 | No | Yes | ||
| B9H748 | Yes | Yes | ||
| A9PCZ6 | Yes | Yes | ||
| B9GUL5 | Yes | Yes | ||
| B9I555 | No | Yes | ||
| B9HF10 | Yes | Yes | ||
| B9ICS4 | No | Yes | ||
Presence of cysteine residues corresponding to C159 and C235 of TPPD in several species of archaea, bacteria, protists, invertebrates, lower and higher plants. Due to the lack of complete gene annotation in gymnosperm genomes, some TBLASTN searches retrieved incomplete transcripts or genomic scaffolds, which only allowed determination of the presence of one of the two redox-sensitive cysteine residues.
n.d., not determined.
Discussion
Salt stress induces a complex response, including a change in the cellular redox balance and metabolic adjustments. In response to environmental stress, carbon use needs to be tightly controlled. Trehalose levels rise upon high salinity, and trehalose metabolism has been implicated in regulating stress tolerance. T6P is considered as a signaling metabolite indicating the sugar status and regulating carbohydrate use. TPPs catabolize T6P to generate trehalose and thus regulate T6P and trehalose levels. In this work, we provide genetic and biochemical evidence that redox-sensitive AtTPPD localizes to chloroplasts and is involved in regulating sugar levels and salt stress tolerance.
AtTPPD is a novel component promoting salt stress tolerance. High salinity conditions strongly induced AtTPPD transcript levels in roots and shoots, and plants deficient in AtTPPD were hypersensitive to salinity and osmotic stress in germination assays. In line with a positive role of AtTPPD during stress response, plants overexpressing AtTPPD showed an enhanced tolerance to high salinity and high osmotic conditions. In rice, overexpression of OsTPP1 increased tolerance to salt stress (28). However, OsTPP1 does not contain the two redox-active cysteine residues and OsTPP1 and AtTPPD reside in different phylogenetic clades, suggesting that they are not orthologous. In Arabidopsis, salt-triggered transcriptional regulation of several AtTPP family members (44) points toward an involvement of AtTPPs, located in different subcellular compartments, in adjusting trehalose metabolism to salinity conditions and underlines the importance of trehalose metabolism for Arabidopsis salt stress tolerance.
Trehalose metabolism is linked to sugar and starch metabolism. Internal sucrose levels were shown to strongly correlate with T6P levels (46, 54, 76, 83, 87). Sucrose feeding enhanced T6P levels and overexpression of Escherichia coli TPP led to increased sucrose levels under normal growth conditions, indicating a bidirectional regulation of sucrose and T6P (89). In line with this study, plants overexpressing AtTPPD showed elevated levels of soluble sugars. Sugar accumulation is a well-known adaptive response to osmotic stress (40). The elevated soluble sugar levels in AtTPPD-overexpressing lines might be vital at the onset of stress. Under prolonged salt stress, AtTPPD-overexpressing plants accumulated even higher levels of soluble sugars, which can serve for osmoregulation under salt stress conditions and thereby contribute to the improved stress tolerance in these lines. Additionally, increased starch levels in AtTPPD-overexpressing plants under high soil salinity underline the importance of AtTPPD in regulating carbohydrate metabolism under stress conditions. Previous studies indicated a role for trehalose metabolism in regulating starch accumulation and degradation in unstressed plants. tps1 mutants accumulate starch during seed development (29). Feeding of trehalose to Arabidopsis plants led to carbon reallocation and starch accumulation (7, 19, 38, 61, 88), and plants overexpressing E. coli or yeast TPS or TPP showed altered starch levels (16, 18, 38, 39, 47, 67, 73, 87). Interestingly, unstressed plants with elevated AtTPPD showed starch levels similar to wild type, whereas under high soil salinity conditions, they accumulated starch indicating that AtTPPD is able to specifically regulate the accumulation of transitory starch under stress conditions. While the mechanism(s) of how AtTPPD regulates carbohydrate metabolism are likely complex and should be addressed in future studies, our data already provide strong evidence for a role of AtTPPD in adjusting carbohydrate utilization to the physiological requirements under stress conditions. The concomitant accumulation of transitory starch and the increased amounts of soluble sugars in AtTPPD overexpressors under stress, together with their improved stress tolerance, might point toward an enhanced carbohydrate metabolism that is advantageous under salt stress.
tppd plants were hypersensitive to salinity and osmotic stress during early development, whereas adult plants deficient in AtTPPD did not show an altered sensitivity to high salinity, and levels of soluble sugars and starch were comparable to wild type. Functional redundancy in adult plants might account for this observation. Alternatively, AtTPPD might have different roles in the stress response depending on the developmental stage. In this regard, it is interesting that T6P inhibits the activity of SnRK1, a key component in energy signaling, in seedlings but not in mature leaves (93).
It is assumed that T6P and trehalose are mainly produced in the cytosol (38, 47). Indeed, our data indicate a cytoplasmic localization of several AtTPPs (AtTPPA, AtTPPB, AtTPPC, AtTPPF, and AtTPPH). Similarly, AtTPS1, AtTPS2, and AtTPS4 GFP fusion proteins were reported to be predominantly cytoplasmic (82). On the other hand, we found AtTPPA, AtTPPB, AtTPPC, AtTPPF, AtTPPG, AtTPPH, AtTPPI, and AtTPPJ fusion proteins in the nucleus. In line with a possible nuclear trehalose biosynthesis pathway, AtTPS1 has also been detected in the nucleus (1, 82), suggesting that trehalose metabolism is also involved in directly regulating nuclear processes.
Notably, we detected AtTPPD and AtTPPE fusion proteins in chloroplasts. Together with recent analyses indicating a minor fraction of T6P in chloroplasts (47) and immunolocalization studies indicating that a fraction of AtTPS1 expressed in tobacco leaves is in the chloroplast (1), our data suggest the presence of trehalose biosynthesis in chloroplasts.
Chloroplasts are sensors of environmental information and play a vital, although not fully understood role in acquiring stress tolerance. Targeting heterologous expression of yeast TPS1 to chloroplasts led to increased levels of trehalose and enhanced osmotic stress tolerance of tobacco and Arabidopsis plants without perturbing plant development as observed when microbial trehalose biosynthesis genes were expressed in the cytosol (35, 42). The modified stress tolerance of plants deficient or overexpressing AtTPPD now provides evidence for an innate chloroplast trehalose metabolism-based mechanism for acquiring stress tolerance.
Prolonged stress can damage chloroplasts and thereby interfere with its various vital functions. Based on the analysis of tobacco plants overexpressing yeast TPS1 in chloroplasts, it has been suggested that trehalose production in chloroplasts might have a direct protective function on thylakoid membranes and thereby promote stress tolerance (42). Salt stress increases trehalose levels (25, 27, 36); yet, the amounts of trehalose determined from total cell extracts are too low for a direct protective function. A future challenge will be to determine whether trehalose might accumulate locally in chloroplasts to levels sufficient to contribute to osmoprotection, or whether, the more likely case, the modified stress tolerance observed in AtTPPD mutants is due to its influence on the flux through the trehalose pathway thus modulating sugar sensing/signaling and carbohydrate use.
Changes in environmental conditions alter the redox state of chloroplasts (50, 52, 72). The modified redox status of the chloroplasts is instrumental in regulating the activity of numerous metabolic enzymes accordingly (14, 68). Interestingly, this work now provides evidence for a post-translational, redox-dependent regulation of AtTPPD. AtTPPD was inactivated by oxidizing conditions and the activity was restored in a reducing environment. C159 and C235 were identified to convey redox regulation and mutation of these cysteine residues rendered AtTPPD redox insensitive. The redox control of TPPD activity appears to involve conformational changes. Structural analysis indicated that under oxidizing conditions, C159 and C235 can form an intramolecular disulfide bridge, which could modify the substrate binding cavity located between the cap and the core domain and, thus, alter the catalytic activity of AtTPPD.
AtTPPD expression rescued the growth arrest of S. cerevesiae TPP knockout strain Δtps2 under restrictive temperatures consistent with previous data (81, 84). Interestingly, mutating C159 and C235 further enhanced the growth of AtTPPD expressing Δtps2 cells at high temperatures. This result confirms the regulatory role of these cysteine residues and supports the biochemical analyses of purified AtTPPD proteins.
Environmental stress enhances chloroplast ROS production (50, 58). Plants have evolved sophisticated means to counterbalance the oxidative inactivation of redox-sensitive proteins under stress conditions. For example, the ferredoxin-Trx system was found to restore the activity of enzymes following their oxidative deactivation by ROS (8, 91). We observed that oxidized inactive AtTPPD was efficiently reduced by E. coli Trx. In chloroplasts, several Trx family oxidoreductases, which regulate the protein redox state, have been implicated in the response to oxidative stress: the ferredoxin-linked Trx system (8, 15, 37); the C-type NADPH-linked thioredoxin reductase (NTRC, which possesses a Trx domain in addition to the NADPH-Trx reductase domain) (57, 69); the glutathione/glutaredoxin system (13, 92); and the stress-induced Trx CDSP32 (63). Considering that AtTPPD enhances stress tolerance and Trx family oxidoreductases play a fundamental role in oxidative stress response in chloroplasts, it is interesting to speculate that targeted reduction of AtTPPD under stress conditions might counterbalance its oxidation triggered inactivation. It will thus be interesting in future studies to investigate by which redox regulatory network(s) AtTPPD is targeted in planta and how this regulation influences signaling by trehalose metabolism (under environmental stress conditions).
The redox modulation of TPP activity appears to be a common regulatory mechanism. Notably, the two redox-sensitive cysteine residues are also present in AtTPPE, AtTPPH, AtTPPI, and AtTPPJ proteins. Post-translational modification of cysteine residues plays a fundamental regulatory role in a wide variety of cellular processes in the cytosol, nucleus, and chloroplasts (48). We detected AtTPPD-CFP and AtTPPE-CFP in chloroplasts, AtTPPH-CFP in the cytosol and nucleus, and AtTPPI-CFP and AtTPPJ-CFP in the nucleus, indicating that redox modulation of distinct AtTPPs regulates trehalose metabolism in different subcellular compartments. From an evolutionary point of view, the conservation of the two redox-sensitive cysteine residues in spermatophytes supports the notion that TPP redox regulation is a basic mechanism to modulate TPP activity in planta, thus enabling rapid adjustment of trehalose metabolism to environmental variations.
Materials and Methods
Plant growth and stress treatments
A. thaliana ecotype Columbia (Col-0) was used in all experiments. Stratified seeds were either transferred to a half-strength Murashige and Skoog medium supplemented with 1% sucrose (½ MS; Duchefa) or to the soil and cultivated in a 16-h light/8-h dark photoperiod at 130 μmol·m−2·s−1 light intensity and 60% relative humidity. For in-soil high salinity stress, plants were watered with a 200 mM NaCl solution. For germination assay under high salt and osmotic stress conditions, seeds were germinated on ½ MS or ½ MS supplemented with 200 mM NaCl and 360 mM sorbitol, respectively, in a 16-h light/8-h dark regime. For oxidative stress, seedlings were grown on ½ MS or ½ MS supplemented with 1 μM paraquat (Sigma). Seeds of different genotypes used for one experiment were propagated in the same growth chamber at the same time.
Plant material and plasmid constructs
The tppd mutant (SALK_013114C) was genotyped using AtTPPD_ex1 and AtTPPD_ex6 primers (Supplementary Table S2). The coding sequence of all AtTPPs was amplified from A. thaliana ecotype Col-0 cDNA by a Pfu-based PCR using gene-specific oligonucleotides (Supplementary Table S2) and cloned with the In-fusion™ Advantage PCR Cloning Kit (Clontech) into the expression vector pGreenII0029 under the control of the CaMV 35S promoter. The CFP tag was cloned as NotI fragments after the coding sequences.
Protein subcellular localization
Agrobacterium tumefaciens (GV3101) transformed with the constructs 35S::AtTPPA-J-CFP were resuspended in a 5% sucrose solution containing 100 μM acetosyringone at an OD600 0.8 and infiltrated in Nicotiana tabacum leaves. Subcellular localization was examined 4 days after transformation by confocal laser scanning microscopy (LSM 710 Zeiss Spectral Confocal). CFP emission was excited at 458 nm (455–533 nm) and chlorophyll fluorescence between 657 and 726 nm. The subcellular localization of Arabidopsis plants stably transformed with the 35S::AtTPPD-CFP was analyzed in 10-day-old seedlings by confocal laser scanning microscopy using the settings described above.
Gene expression analysis
Arabidopsis Col-0 seedlings grown on a mesh in liquid ½ MS were transferred to either ½ MS or ½ MS supplemented with 200 mM NaCl for 0, 1, 3, and 8 h. Total RNA was isolated by phenol/chloroform extraction using the TRIzol® Reagent (Sigma), followed by sodium chloride/sodium citrate-isopropanol precipitation to improve RNA purity and treated with DNase using the RNeasy plant mini kit (Qiagen). For real-time-PCR, cDNA was synthesized from 1 μg of total RNA using the qScript™cDNA Synthesis kit (Quanta). The resulting single-stranded cDNA was diluted fivefold. Three microliters of the diluted samples were used for real-time PCR with the SYBR® green Supermix (Bio-Rad) on an IQ5 multicolor real-time PCR detection system (Bio-Rad). The thermal cycling conditions were composed of 95°C for 30 s followed by 40 cycles at 95°C for 5 s and 58°C for 15 s. The experiments were carried out in triplicate for each data point. The relative quantification in gene expression was determined using the 2−ΔΔCt method (45). Primer efficiencies were calculated by relative standard curves. PP2A and Ubi1 were used as internal control genes, as described by Czechowski et al. (17). Normalized gene expression was represented relative either to wild type or to control condition. The oligonucleotide primers used for analyzing AtTPPD transcripts spanned the exons 5 and 6 and for AtTPPE exons 9 and 10 (Supplementary Table S1).
Chlorophyll contents
Chlorophyll contents were measured from 4-week-old, soil-grown plants in a 16-h light/8-h dark photoperiod at 130 μmol·m−2·s−1 light intensity and 60% relative humidity. Rosette leaves were harvested and snap frozen in liquid nitrogen. Chlorophyll of four replicates per genotype was extracted overnight at 4°C with 1 ml 95% ethanol. Chlorophyll contents in mg·g−1 FW (Chl a+b=5.24OD664+22.24OD649) were calculated according to (59).
Chloroplast isolation
Plants used for isolation of chloroplasts were grown in short day conditions (8-h/16-h photoperiod). Intact chloroplasts were isolated from 4-week-old Arabidopsis plants. Leaves were harvested in the morning and immediately put on ice before homogenization in the SlnA buffer (450 mM sorbitol, 10 mM Tricin/KOH, pH 8.4, 10 mM EDTA, 5 mM NaHCO3, 1 g/L bovine serum albumin) using a Waring blender. The homogenized plant material was filtered through two layers of Miracloth (Calbiochem) and centrifuged for 5 min at 1590 g at 4°C without brake. The pellet was recovered in 1×SlnB buffer (2×SlnB buffer: 600 mM sorbitol, 40 mM Tricin/KOH, pH 7.6, 10 mM MgCl2, and 5 mM EDTA), loaded on a 50% (v/v) Percoll™ (GE Healthcare)/2×SlnB buffer gradient, and centrifuged for 6 min at 2500 g at 4°C in a swing-out rotor. Intact chloroplasts were removed from the 50% Percoll interphase, washed in 1×SlnB buffer, and used for immunoblot analysis.
Protein extraction and Western blot analysis
Total proteins from A. thaliana leaves and chloroplast extracts were separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and blotted on a PVDF (Immobilon®-FL; Millipore) membrane. Membranes were probed with a 1:1000 dilution of anti-GFP (11814460001; Roche) or with a 1:25,000 dilution of anti-Psba/D1 (AS05084-10; Agrisera) antibodies. Goat anti-mouse IgG conjugated to IR Dye® 800CW (926-32210; LI-COR Biosciences) and goat anti-rabbit IgG conjugated to ID Dye® 800CW (926-32211; LI-COR Biosciences) were used (1:20,000) as secondary antibodies against anti-GFP and anti-PsbA/D1 antibodies, respectively. Membranes were scanned in the 800 channel using the Odyssey IR Imager (LI-COR Biosciences).
Sugar and starch measurements
Plants were grown in soil in a 16-h light/8-h dark photoperiod at 130 μmol·m−2·s−1 light intensity and 60% relative humidity for 2 weeks. Subsequently, plants were either watered with a 150 mM NaCl or normally watered (control) for 8 days. Rosette leaves were harvested at the end of the photoperiod and snap frozen in liquid nitrogen. Sugar and starch extractions were done in quadruplates from ground material. Soluble carbohydrates were extracted with 1 ml of 80% ethanol for 1 h at 4°C and after several washings of the pellet with the same solvent, starch was extracted with 1.5% HCl for 1 h at 80°C. Soluble sugars and starch were quantified by the anthrone-sulfuric acid assay (Anthrone ACS reagent, sc-206057; Santa Cruz) (90).
Protein modeling
The AtTPPD model was created using swissmodel (http://swissmodel.expasy.org) (3) according to TPP from T. acidophilum TaTPP (PDB-ID: 1u02) (62). The ligand T6P (PDB Ligand-ID: T6P) was placed into the active site of AtTPPD using AutoDock 4 and AutoDockTools (http://autodock.scripps.edu).
Recombinant protein expression
AtTPP was cloned into the pGEX6 expression vector, expressed in E. coli BL21, and purified according to the manufacturer's instructions (GE Healthcare). In brief, an overnight culture was used to inoculate the expression culture at an OD600 0.2. The expression culture was induced at OD600 0.6 with 1 mM IPTG and grown for 4 h at 28°C. Cells were harvested and proteins extracted in the TBS buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl) after lysis with 1 mg/ml lysozyme in the presence of 2 mM PMSF. The protein lysate was column purified and the tag was cut with PreScission protease [Turbo3c (HRV3C) Protease, purchased from A.G. Scientific] and successful expression verified by SDS-PAGE.
AtTPP activity measurements
For analyzing AtTPP activity and its sensitivity to oxidants and antioxidants, purified proteins were incubated with DTT/GSH and H2O2/GSSG in a total volume of 10 μl for 30 min at room temperature in a reaction buffer (50 mM Tris/HCl pH 7.5, 150 mM MgCl2, 200 mM NaCl). Subsequently, the substrate (T6P, 0.01 mM) was added to the reaction (final volume of 50 μl) and incubated at 37°C for 30 min. The reaction was stopped by adding 100 μl BIOMOL Green reagent (purchased at Enzo Life Sciences) and the absorption at 620 nm read after 20 min using a microtiter plate reader (71). For Trx treatment, oxidized proteins were dialyzed using Amicon Ultra-0.5 ml Centrifugal Filters 10 kDa MWCO (MerckMillipore) according to the manufacturer's instructions, and subsequently incubated with reduced DTT with or without Trx (E. coli Trx; Sigma). Thiol titration was performed at 25°C and pH 7.0 using defined ratios of reduced and oxidized DTT at a total concentration of 20 mM. The experimental data were fitted to the Nernst equation (using XLFit curve fitting software for Excel) with n=2 and a midpoint redox potential of −330 mV for DTT at pH 7.0. All measurements were performed in triplicates and repeated with proteins from independent expressions.
Yeast complementation assay
For the yeast complementation assay, the S. cerevisiae BY4741 wild-type strain (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the Δtps2 deletion strain YDR074W were used. The coding sequences of wild-type AtTPPD and of AtTPPD C159S, AtTPPD C235S and AtTPPD C159/235S were cloned into the yeast multicopy plasmid pYX424 with a TPI promotor and a LEU2 marker. As control, the wild-type and Δtps2 strains were transformed with the empty vector. Yeast was transformed by adding 3 μg plasmid DNA together with 100 μg single-stranded carrier DNA to cells from an overnight culture resuspended in 200 μl of transformation buffer (200 mM LiAc, 100 mM DTT, 40% PEG 4000 in TE pH 8.0), followed by a 15-min incubation at 30°C and a 45-min incubation at 45°C. Transformed cells were grown on a synthetic defined medium without leucine (SD-leu) containing 2% glucose. Drop assays were performed by diluting an overnight culture to an OD600 of 0.1 and sequential 1:5 dilutions were spotted on a yeast peptone dextrose medium containing 2% glucose. Plates were incubated at 30°C and 39°C and analyzed after 4 days.
Protein sequence alignment
Protein sequences of Class III trehalose biosynthesis enzymes from prokaryotes, protists, animals, red and green algae, moss, fern, and angiosperms were retrieved from KEGG (www.genome.jp/kegg/catalog/org_list.html) and aligned using CLC Main workbench.
TPP homologous sequences in gymnosperms were identified through TBLASTN (2) searches using the AtTPPD protein sequence as a query. For Picea abies, a database (61,393 CDS) of high confidence (predicted protein coding loci with >70% coverage by transcripts) and medium confidence (30%–70% coverage) gene predictions (55) was used. In the case of Picea glauca, Transcriptome Assembly v1.0 (68,297 transcripts) from the SMarTForests Project and Unigene set #15 from NCBI (27,848 transcripts) databases were analyzed (10). For Pinus taeda, both transcriptome and genomic databases from the PineRefSeq Project (86) were used, such as TreeGenesTranscriptome assembly v1.0 (77,249 transcripts) and Pinus taeda Genomic Assembly Contigs V1.0 (∼14 million scaffolds), respectively.
Supplementary Material
Abbreviations Used
- ABA
abscisic acid
- CBB
Coomassie Brilliant Blue
- CFP
cyan fluorescent protein
- DTT
dithiothreitol
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- HAD
haloacid dehalogenase
- H2O2
hydrogen peroxide
- NTRC
NADPH-linked thioredoxin reductase
- PCR
polymerase chain reaction
- qRT-PCR
real-time polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TaTPP
Thermoplasma acidophilum TPP
- T6P
trehalose-6-phosphate
- TPP
trehalose-6-phosphate phosphatase
- TPS
trehalose-6-phosphate synthase
- TRE
trehalase
- ROS
reactive oxygen species
- Trx
thioredoxin
- YPD
yeast peptone dextrose
Acknowledgments
The authors thank A. Auer and B. Dekrout for technical assistance and the Austrian Science Foundation for supporting the work with the grant P20375-B03.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Almeida AM, Santos M, Villalobos E, Araujo SS, van Dijck P, Leyman B, Cardoso LA, Santos D, Fevereiro PS, and Torne JM. Immunogold localization of trehalose-6-phosphate synthase in leaf segments of wild-type and transgenic tobacco plants expressing the AtTPS1 gene from Arabidopsis thaliana. Protoplasma 230: 41–49, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold K, Bordoli L, Kopp J, and Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, and Iturriaga G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136: 3649–3659, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avonce N, Mendoza-Vargas A, Morett E, and Iturriaga G. Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6: 109, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babbs CF, Pham JA, and Coolbaugh RC. Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol 90: 1267–1270, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae H, Herman E, and Sicher R. Exogenous trehalose promotes non-structural carbohydrate accumulation and induces chemical detoxification and stress response proteins in Arabidopsis thaliana grown in liquid culture. Plant Sci 168: 1293–1301, 2005 [Google Scholar]
- 8.Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, and Buchanan BB. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci U S A 100: 370–375, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi G, Gamba A, Limiroli R, Pozzi N, Elster R, Salamini F, and Bartels D. The unusual sugar composition in leaves of the resurrection plant Myrothamnus flabellifolia. Physiol Plant 87: 223–226, 1993 [Google Scholar]
- 10.Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Yuen MM, Keeling CI, Brand D, Vandervalk BP, Kirk H, Pandoh P, Moore RA, Zhao Y, Mungall AJ, Jaquish B, Yanchuk A, Ritland C, Boyle B, Bousquet J, Ritland K, Mackay J, Bohlmann J, and Jones SJ. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29: 1492–1497, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldana C, Fernie AR, Willmitzer L, and Steinhauser D. Unraveling retrograde signaling pathways: finding candidate signaling molecules via metabolomics and systems biology driven approaches. Front Plant Sci 3: 267, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chary SN, Hicks GR, Choi YG, Carter D, and Raikhel NV. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol 146: 97–107, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng NH, Liu JZ, Brock A, Nelson RS, and Hirschi KD. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem 281: 26280–26288, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Chibani K, Couturier J, Selles B, Jacquot JP, and Rouhier N. The chloroplastic thiol reducing systems: dual functions in the regulation of carbohydrate metabolism and regeneration of antioxidant enzymes, emphasis on the poplar redoxin equipment. Photosynth Res 104: 75–99, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, and Miginiac-Maslow M. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278: 23747–23752, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Cortina C. and Culiáñez-Macià FA. Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci 169: 75–82, 2005 [Google Scholar]
- 17.Czechowski T, Stitt M, Altmann T, Udvardi MK, and Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debast S, Nunes-Nesi A, Hajirezaei MR, Hofmann J, Sonnewald U, Fernie AR, and Bornke F. Altering trehalose-6-phosphate content in transgenic potato tubers affects tuber growth and alters responsiveness to hormones during sprouting. Plant Physiol 156: 1754–1771, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delatte TL, Sedijani P, Kondou Y, Matsui M, de Jong GJ, Somsen GW, Wiese-Klinkenberg A, Primavesi LF, Paul MJ, and Schluepmann H. Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol 157: 160–174, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drennan PM, Smith MT, Goldsworthy D, and van Staden J. The occurence of trehalose in the leaves of the desiccation-tolerant angiosperm Myrothamnus flabellifolius welw. J Plant Physiol 142: 493, 1993 [Google Scholar]
- 21.Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, and Graham IA. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235, 2002 [DOI] [PubMed] [Google Scholar]
- 22.Elbein AD, Pan YT, Pastuszak I, and Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology 13: 17R–27R, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Fernandez O, Bethencourt L, Quero A, Sangwan RS, and Clement C. Trehalose and plant stress responses: friend or foe? Trends Plant Sci 15: 409–417, 2010 [DOI] [PubMed] [Google Scholar]
- 24.Fernandez O, Vandesteene L, Feil R, Baillieul F, Lunn J, and Clément C. Trehalose metabolism is activated upon chilling in grapevine and might participate in Burkholderia phytofirmans induced chilling tolerance. Planta 236: 355–369, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Fougere F, Le Rudulier D, and Streeter JG. Effects of salt stress on amino acid, organic acid, and carbohydrate composition of roots, bacteroids, and cytosol of Alfalfa (Medicago sativa L.). Plant Physiol 96: 1228–1236, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foyer CH, Neukermans J, Queval G, Noctor G, and Harbinson J. Photosynthetic control of electron transport and the regulation of gene expression. J Exp Bot 63: 1637–1661, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, and Wu RJ. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci U S A 99: 15898–15903, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge LF, Chao DY, Shi M, Zhu MZ, Gao JP, and Lin HX. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228: 191–201, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Gomez LD, Baud S, Gilday A, Li Y, and Graham IA. Delayed embryo development in the Arabidopsis trehalose-6-phosphate synthase 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 46: 69–84, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Gomez LD, Gilday A, Feil R, Lunn JE, and Graham IA. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64: 1–13, 2010 [DOI] [PubMed] [Google Scholar]
- 31.Guy C, Kaplan F, Kopka J, Selbig J, and Hincha DK. Metabolomics of temperature stress. Physiol Plant 132: 220–235, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Hasegawa P, Bressan R, Zhu JK, and Bohnert H. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499, 2000 [DOI] [PubMed] [Google Scholar]
- 33.Iordachescu M. and Imai R. Trehalose biosynthesis in response to abiotic stresses. J Integr Plant Biol 50: 1223–1229, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, and Guy CL. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karim S, Aronsson H, Ericson H, Pirhonen M, Leyman B, Welin B, Mantyla E, Palva ET, Van Dijck P, and Holmstrom KO. Improved drought tolerance without undesired side effects in transgenic plants producing trehalose. Plant Mol Biol 64: 371–386, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Kempa S, Krasensky J, Dal Santo S, Kopka J, and Jonak C. A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS One 3: e3935, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keryer E, Collin V, Lavergne D, Lemaire S, and Issakidis-Bourguet E. Characterization of Arabidopsis mutants for the variable subunit of ferredoxin:thioredoxin reductase. Photosynth Res 79: 265–274, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, and Geigenberger P. Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci U S A 102: 11118–11123, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondrák M, Marincs F, Kalapos B, Juhász Z, and Bánfalvi Z. Transcriptome Analysis of potato leaves expressing the trehalose-6-phosphate synthase 1 gene of yeast. PLoS One 6: e23466, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krasensky J. and Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63: 1593–1608, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larcher W. Responses of Plants to Environmental Stresses. San Diego, CA: Academic Press, Inc., II, 2nd Ed., 1980 [Google Scholar]
- 42.Lee S-B, Kwon H-B, Kwon S-J, Park S-C, Jeong M-J, Han S-E, Byun M-O, and Daniell H. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed 11: 1–13, 2003 [Google Scholar]
- 43.Li H-W, Zang B-S, Deng X-W, Wang X-P. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234: 1007–1018, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Li P, Ma S, and Bohnert HJ. Coexpression characteristics of trehalose-6-phosphate phosphatase subfamily genes reveal different functions in a network context. Physiol Plant 133: 544–556, 2008 [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ. and Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Lunn JE, Feil R, Hendriks JH, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, and Stitt M. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martins MC, Hejazi M, Fettke J, Steup M, Feil R, Krause U, Arrivault S, Vosloh D, Figueroa CM, Ivakov A, Yadav UP, Piques M, Metzner D, Stitt M, and Lunn JE. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol 163: 1142–1163, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer Y, Buchanan BB, Vignols F, and Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 43: 335–367, 2009 [DOI] [PubMed] [Google Scholar]
- 49.Mikkelsen R, Suszkiewicz K, and Blennow A. A novel type carbohydrate-binding module identified in alpha-glucan, water dikinases is specific for regulated plastidial starch metabolism. Biochemistry 45: 4674–4682, 2006 [DOI] [PubMed] [Google Scholar]
- 50.Miller G, Suzuki N, Ciftci-Yilmaz S, and Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467, 2010 [DOI] [PubMed] [Google Scholar]
- 51.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410, 2002 [DOI] [PubMed] [Google Scholar]
- 52.Munne-Bosch S, Queval G, and Foyer CH. The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol 161: 5–19, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munns R. and Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681, 2008 [DOI] [PubMed] [Google Scholar]
- 54.Nunes C, O'Hara LE, Primavesi LF, Delatte TL, Schluepmann H, Somsen GW, Silva AB, Fevereiro PS, Wingler A, and Paul MJ. The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol 162: 1720–1732, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hallman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Kaller M, Luthman J, Lysholm F, Niittyla T, Olson A, Rilakovic N, Ritland C, Rossello JA, Sena J, Svensson T, Talavera-Lopez C, Theissen G, Tuominen H, Vanneste K, Wu ZQ, Zhang B, Zerbe P, Arvestad L, Bhalerao R, Bohlmann J, Bousquet J, Garcia Gil R, Hvidsten TR, de Jong P, MacKay J, Morgante M, Ritland K, Sundberg B, Thompson SL, Van de Peer Y, Andersson B, Nilsson O, Ingvarsson PK, Lundeberg J, and Jansson S. The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584, 2013 [DOI] [PubMed] [Google Scholar]
- 56.Paul MJ, Primavesi LF, Jhurreea D, and Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol 59: 417–441, 2008 [DOI] [PubMed] [Google Scholar]
- 57.Perez-Ruiz JM, Spinola MC, Kirchsteiger K, Moreno J, Sahrawy M, and Cejudo FJ. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pogson BJ, Woo NS, Forster B, and Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609, 2008 [DOI] [PubMed] [Google Scholar]
- 59.Porra RJ, Thompson WA, and Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394, 1989 [Google Scholar]
- 60.Pramanik MH. and Imai R. Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol Biol 58: 751–762, 2005 [DOI] [PubMed] [Google Scholar]
- 61.Ramon M, Rolland F, Thevelein JM, Van Dijck P, and Leyman B. ABI4 mediates the effects of exogenous trehalose on Arabidopsis growth and starch breakdown. Plant Mol Biol 63: 195–206, 2007 [DOI] [PubMed] [Google Scholar]
- 62.Rao KN, Kumaran D, Seetharaman J, Bonanno JB, Burley SK, and Swaminathan S. Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications. Protein Sci 15: 1735–1744, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rey P, Cuine S, Eymery F, Garin J, Court M, Jacquot JP, Rouhier N, and Broin M. Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J 41: 31–42, 2005 [DOI] [PubMed] [Google Scholar]
- 64.Rizhsky L, Liang HJ, Shuman J, Shulaev V, Davletova S, and Mittler R. When Defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, and Jackson D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230, 2006 [DOI] [PubMed] [Google Scholar]
- 66.Scheibe R. and Dietz KJ. Reduction-oxidation network for flexible adjustment of cellular metabolism in photoautotrophic cells. Plant Cell Environ 35: 202–216, 2012 [DOI] [PubMed] [Google Scholar]
- 67.Schluepmann H, Pellny T, van Dijken A, Smeekens S, and Paul M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci U S A 100: 6849–6854, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schurmann P. and Buchanan BB. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274, 2008 [DOI] [PubMed] [Google Scholar]
- 69.Serrato AJ, Perez-Ruiz JM, Spinola MC, and Cejudo FJ. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279: 43821–43827, 2004 [DOI] [PubMed] [Google Scholar]
- 70.Seung D, Thalmann M, Sparla F, Abou Hachem M, Lee SK, Issakidis-Bourguet E, Svensson B, Zeeman SC, and Santelia D. Arabidopsis thaliana AMY3 is a unique redox-regulated chloroplastic alpha-amylase. J Biol Chem 288: 33620–33633, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shima S, Matsui H, Tahara S, and Imai R. Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J 274: 1192–1201, 2007 [DOI] [PubMed] [Google Scholar]
- 72.Sierla M, Rahikainen M, Salojarvi J, Kangasjarvi J, and Kangasjarvi S. Apoplastic and chloroplastic redox signaling networks in plant stress responses. Antioxid Redox Signal 18: 2220–2239, 2013 [DOI] [PubMed] [Google Scholar]
- 73.Singh V, Louis J, Ayre BG, Reese JC, Pegadaraju V, and Shah J. Trehalose phosphate synthase11-dependent trehalose metabolism promotes Arabidopsis thaliana defense against the phloem-feeding insect Myzus persicae. Plant J 67: 94–104, 2011 [DOI] [PubMed] [Google Scholar]
- 74.Sparla F, Costa A, Lo Schiavo F, Pupillo P, and Trost P. Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis. Plant Physiol 141: 840–850, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sparla F, Pupillo P, and Trost P. The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. J Biol Chem 277: 44946–44952, 2002 [DOI] [PubMed] [Google Scholar]
- 76.Sulpice R, Flis A, Ivakov AA, Apelt F, Krohn N, Encke B, Abel C, Feil R, Lunn JE, and Stitt M. Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol Plant 7: 137–155, 2014 [DOI] [PubMed] [Google Scholar]
- 77.Suzuki N, Bajad S, Shuman J, Shulaev V, and Mittler R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 283: 9269–9275, 2008 [DOI] [PubMed] [Google Scholar]
- 78.van Dijken AJ, Schluepmann H, and Smeekens SC. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 135: 969–977, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Houtte H, Lopez-Galvis L, Vandesteene L, Beeckman T, and Van Dijck P. Redundant and non-redundant roles of the trehalose-6-phosphate phosphatases in leave growth, root hair specification and energy-responses in Arabidopsis. Plant Signal Behav 8, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Houtte H, Vandesteene L, Lopez-Galvis L, Lemmens L, Kissel E, Carpentier S, Feil R, Avonce N, Beeckman T, Lunn JE, and Van Dijck P. Overexpression of the trehalase gene AtTRE1 leads to increased drought stress tolerance in Arabidopsis and is involved in abscisic acid-induced stomatal closure. Plant Physiol 161: 1158–1171, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vandesteene L, Lopez-Galvis L, Vanneste K, Feil R, Maere S, Lammens W, Rolland F, Lunn JE, Avonce N, Beeckman T, and Van Dijck P. Expansive evolution of the trehalose-6-phosphate phosphatase gene family in Arabidopsis. Plant Physiol 160: 884–896, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vandesteene L, Ramon M, Le Roy K, Van Dijck P, and Rolland F. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol Plant 3: 406–419, 2010 [DOI] [PubMed] [Google Scholar]
- 83.Veyres N, Danon A, Aono M, Galliot S, Karibasappa YB, Diet A, Grandmottet F, Tamaoki M, Lesur D, Pilard S, Boitel-Conti M, Sangwan-Norreel BS, and Sangwan RS. The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence. Plant J 55: 665–686, 2008 [DOI] [PubMed] [Google Scholar]
- 84.Vogel G, Aeschbacher RA, Muller J, Boller T, and Wiemken A. Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J 13: 673–683, 1998 [DOI] [PubMed] [Google Scholar]
- 85.Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, and Schmid M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707, 2013 [DOI] [PubMed] [Google Scholar]
- 86.Wegrzyn JL, Lee JM, Tearse BR, and Neale DB. TreeGenes: a forest tree genome database. Int J Plant Genomics 2008: 412875, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wingler A, Delatte TL, O'Hara LE, Primavesi LF, Jhurreea D, Paul MJ, and Schluepmann H. Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol 158: 1241–1251, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wingler A, Fritzius T, Wiemken A, Boller T, and Aeschbacher RA. Trehalose induces the ADP-glucose pyrophosphorylase gene, ApL3, and starch synthesis in Arabidopsis. Plant Physiol 124: 105–114, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yadav UP, Ivakov A, Feil R, Duan GY, Walther D, Giavalisco P, Piques M, Carillo P, Hubberten HM, Stitt M, and Lunn JE. The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J Exp Bot 65: 1051–1068, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yemm EW. and Willis AJ. The estimation of carbohydrates in plant extracts by anthrone. Biochem J 57: 508–510, 1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zaffagnini M, Bedhomme M, Groni H, Marchand CH, Puppo C, Gontero B, Cassier-Chauvat C, Decottignies P, and Lemaire SD. Glutathionylation in the photosynthetic model organism Chlamydomonas reinhardtii: a proteomic survey. Mol Cell Proteomics 11: M111.014142, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zaffagnini M, Bedhomme M, Marchand CH, Couturier JR, Gao XH, Rouhier N, Trost P, and Lemaire SP. Glutaredoxin s12: unique properties for redox signaling. Antioxid Redox Signal 16: 17–32, 2012 [DOI] [PubMed] [Google Scholar]
- 93.Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, and Paul MJ. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.