Abstract
Pneumocystis jirovecii is a fungal parasite that colonizes specifically humans and turns into an opportunistic pathogen in immunodeficient individuals. The fungus is able to reproduce extracellularly in host lungs without eliciting massive cellular death. The molecular mechanisms that govern this process are poorly understood, in part because of the lack of an in vitro culture system for Pneumocystis spp. In this study, we explored the origin and evolution of the putative biotrophy of P. jirovecii through comparative genomics and reconstruction of ancestral gene repertoires. We used the maximum parsimony method and genomes of related fungi of the Taphrinomycotina subphylum. Our results suggest that the last common ancestor of Pneumocystis spp. lost 2,324 genes in relation to the acquisition of obligate biotrophy. These losses may result from neutral drift and affect the biosyntheses of amino acids and thiamine, the assimilation of inorganic nitrogen and sulfur, and the catabolism of purines. In addition, P. jirovecii shows a reduced panel of lytic proteases and has lost the RNA interference machinery, which might contribute to its genome plasticity. Together with other characteristics, that is, a sex life cycle within the host, the absence of massive destruction of host cells, difficult culturing, and the lack of virulence factors, these gene losses constitute a unique combination of characteristics which are hallmarks of both obligate biotrophs and animal parasites. These findings suggest that Pneumocystis spp. should be considered as the first described obligate biotrophs of animals, whose evolution has been marked by gene losses.
Keywords: obligate parasite, lifestyle, gene families, neutral drift, convergent evolution
Introduction
Pneumocystis jirovecii is a fungal parasite causing Pneumocystis pneumonia in human beings with deficient immune system, a disease which is often fatal if untreated. The Pneumocystis genus includes several species, each displaying strict host specificity for a given mammalian species, for example, P. jirovecii infects humans, P. carinii rats, and P. murina mice. These fungi are members of the Taphrinomycotina, a subphylum of Ascomycota (Redhead et al. 2006), which encompasses organisms with remarkably diverse lifestyles ranging from free-living saprophytes (e.g., Schizosaccharomyces spp.) to biotrophic plant pathogens (e.g., Taphrina spp.).
Two categories of fungal parasitism are recognized: Necrotrophy where food is obtained from killed host cells, and biotrophy where food is obtained from living host cells (Kemen and Jones 2012). In general, necrotrophs secrete many cell wall degrading enzymes as well as toxins to kill host cells. On the other hand, biotrophs secrete low amounts of lytic enzymes and cause little damage to their host. Biotrophic lifestyle includes a continuum from facultative to obligate states (Kemen and Jones 2012; Spanu 2012). Facultative biotrophs require a living host only for a specific stage of their life cycle and can usually be easily grown axenically in the laboratory. Obligate biotrophs present an absolute requirement for their host to survive and usually cannot be grown axenically in the laboratory. Evolution toward obligate biotrophy is generally associated with losses of pathways, whose end products can be scavenged from the host (Spanu et al. 2010; Duplessis et al. 2011). In the case of human parasites, true pathogens refer to those able to cause disease in healthy individuals, whereas opportunistic pathogens are those causing disease only in the case of immunosuppression. These definitions are used in this work.
Pneumocystis spp. dwell almost exclusively in mammalian lungs where they apparently do not trigger massive host cell death. They carry out their sex life cycle within their host, which is a hallmark of obligate parasitism and which has been observed among fungi only in plant biotrophs (Spanu 2012). Moreover, attempts to obtain axenic in vitro culture have failed so far. Based on these characteristics, a parallel between Pneumocystis spp. and plant obligate biotrophs has been suggested (Cushion and Stringer 2010). Subsequent findings reinforced this resemblance. Indeed, Pneumocystis spp. have been suggested to be obligate parasites because of their relatively small genome with a high Adenine and Thymine (AT) content and the loss of the capacity of synthesizing most amino acids (Hauser et al. 2010; Cisse et al. 2012). Moreover, Pneumocystis spp. lack virulence factors commonly found in pathogenic fungi, such as glyoxylate cycle, toxins, and secondary metabolism (Cushion et al. 2007; Cisse et al. 2012).
The origin and evolution of biotrophy in Pneumocystis spp. can be explored through the hypothetical reconstruction of ancestral gene repertoires. This reconstruction is based on the reliable identification of homologous gene families across related organisms. There are currently two methods for that purpose: The maximum parsimony (MP) and the maximum likelihood (ML) (Wolf and Koonin 2013). MP is designed to find the ancestral state that corresponds to the minimum number of character changes necessary to match the species phylogeny with the observed presence–absence patterns. In MP, different models of evolution can be applied, for example, the irreversible Dollo model (Farris 1977), which assumes that once an ancestral character is lost during the evolution of a particular organism, it cannot be regained. The MP method is based on a stochastic birth-and-death model of evolution. This model assumes that a new gene can appear only once in a phylogeny and that it can further evolve through duplication and loss in the descendant lineages. ML is based on the likelihood that ancestral states would evolve under a stochastic model of evolution to reproduce the observed presence–absence patterns (Cohen et al. 2008; Csuros 2010). Although ML has been suggested to be more robust than MP (Wolf and Koonin 2013), this method requires large amounts of data and is generally not applicable to undersampled clades.
In this study, we have determined ubiquitous and species-specific gene families in the Taphrinomycotina subphylum. We then applied a parsimonious scenario to reconstruct ancestry of several lineages, and inferred gene families’ losses and gains. Our results suggest that the acquisition of biotrophy in Pneumocystis spp. was marked by gene losses in biosynthetic pathways, whose products are likely to be scavenged from the lung environment.
These findings provide insights into the intimate relation of these microorganisms with their mammalian hosts.
Materials and Methods
Data Sources and Proteome Completion
Incomplete versions of the proteomes of P. jirovecii and T. deformans are present in UniProt (www.uniprot.org, last accessed November 11, 2013). Therefore, we completed the protein data sets with manually curated gene predictions (Cisse et al. 2012, 2013). These data sets as well as P. carinii predicted proteins and expressed sequence tags are available at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm (last accessed July 27, 2014). Pneumocystis carinii genomic data sets and predicted proteins correspond to data published by others (Cushion et al. 2007; Hauser et al. 2010; Cisse et al. 2012; Slaven et al. 2006). Pneumocystis jirovecii genome raw reads and transcriptome assembly, as well as T. deformans expressed sequence tags (ESTs) were downloaded from EBI (http://www.ebi.ac.uk, last accessed November 11, 2013). The transcriptome assemblies and ESTs were translated into the six open reading frames. The predicted proteomes of Saccharomyces cerevisiae, Ustilago maydis, Neurospora crassa, Yarrowia lipolytica, Ashbya gossypii, Rhizopus oryzae, Aspergillus fumigatus, and Debaryomyces hansenii were obtained from UniProt (www.uniprot.org, last accessed August 26, 2013). The proteomes of S. pombe, S. octosporus, S. japonicus, and S. cryptosporus were from the Broad Institute (http://www.broadinstitute.org, last accessed November 5, 2013), and correspond to data published by Rhind et al. (2011). The level of completion of each protein data set was estimated by scanning with 458 core conserved eukaryotic proteins from the CEGMA pipeline (Parra et al. 2007) using hmmer3 (Eddy 2011) with an e value of 10−5 as threshold.
Gene Families and Annotation
Gene families were built using OrthoMCL (Li et al. 2003) with a Markov inflation of 1.5 and a maximum e value of 10−5. Repetitive elements were identified using TransposonPSI (http://transposonpsi.sourceforge.net/, last accessed July 27, 2014) and TBLASTn searches with an e value of 10−10 (Altschul et al. 1997) in Repbase (Jurka et al. 2005). Enrichment in Gene Ontology (GO) terms between different groups was identified using Fisher’s exact test with a P value of 0.05 and corrected for multiple testing using BLAST2GO (Conesa et al. 2005). Enzyme prediction and mapping to KEGG biochemical pathways were performed using BLAST2GO and PRIAM (Claudel-Renard et al. 2003). Proteases were classified according to MEROPS database (Rawlings et al. 2012).
Ancestral Genome Reconstruction and Gene Gains and Losses
The species phylogeny was obtained from our previous study (Cisse et al. 2012) and edited using FigTree (http://tree.bio.ed.ac.uk/software/figtree/, last accessed July 27, 2014). The true phylogenetic tree along with bootstrap values and branch lengths is shown in supplementary figure S3, Supplementary Material online. Gene presence and absence patterns were extracted from OrthoMCL proteomes clustering using custom Perl scripts. This matrix was used to infer ancestral genomes using Count (Csuros 2010). Count uses phylogenetic birth-and-death models in a stochastic probability inference. The rate models were computed and optimized under the gain–loss–duplication model with the Poisson family size distribution. The rate of variation across families was set to 4:1:1:4 gamma categories for the edge length, the loss rate, gain rate, and the duplication rate, respectively. The convergence criteria applied were set to 100 rounds for the optimization rounds with a likelihood threshold of 0.1. The family history was determined under the Dollo parsimony assumption.
Specific Gene Searches
All searches were first performed in the proteomes. In case of absence, searches were systematically carried out on the translated transcriptome assemblies, genomes, and finally on raw sequences reads if necessary, to confirm the absence.
RNA Interference Machinery
The S. pombe Dicer and Argonaute proteins were used for screening using BLASTp (e value < 0.01) and TBLASTn (e value < 10−3), respectively. The hidden Markov models-based searches were conducted using hmmer3 (0.1) using following Pfam models (Punta et al. 2012): PAZ (PFAM no. accession PF02170), dicer dimerization (PF03368), helicase conserved c-terminal (PF00271), and ResIII (PF04851). The results were manually examined to remove false positives. The S. cerevisiae killer virus was used as template for mapping using P. jirovecii-unassembled genome reads and Newbler’s gsMapper. Homologs to ScV-A (Gag, major capside protein), ScV-M1 (K1 preprotoxin), ScV-M2 (K2 preprotoxin), ScV-M28 (K28 preprotoxin), and ZbV-M (Zygocin preprotoxin) were searched through the P. jirovecii genome and raw reads using TBLASTn (e value < 0.1). Sequence accession numbers and details are given in Schmitt and Breinig (2006).
Thiamine Biosynthesis Pathway
To search the hallmark genes of the thiamine biosynthesis, the S. pombe thiamine thiazole synthase (thi2), the thiamine pyrophosphokinase (tnr3), the phosphomethyl–pyrimidine kinase, and the thiamine biosynthetic bifunctional enzyme (thi4) were used for screening using BLASTp (e value < 0.01) and TBLASTn (e value < 10−3). We ran hmmer3 (e value < 0.1) using the different domains composing the S. pombe proteins, that is, phosphomethylpyrimidine kinase (PF08543), TENA/THI-4/PQQC family (PF03070), hydroxyethylthiazole kinase family (PF02110), thiamine monophosphate synthase (PF02581), and thiamine pyrophosphokinase (PF04265 and PF04263). For the thi2 gene, we searched for the InterPro domain thiazole biosynthetic enzyme family (IPR002922). The results were manually inspected to avoid false positives.
Nitrogen Metabolism
We used the T. deformans nitrite and nitrate reductases as query in order to screen using BLASTp (e value < 0.01) and TBLASTn (e value < 10−3). Domain-based searches were conducted with hmmer3 (0.1) with the following characteristic domains: FAD-binding (PF00970), oxidoreductase NAD-binding (PF00175), nitrite/sulfite reductase ferredoxin-like half (PF03460), and nitrite and sulphite reductase 4Fe–4S (PF01077). We retrieved all proteins containing at least one domain and aligned them using MAFTT (Katoh and Standley 2013) with E-INS-i option to take into account of the presence of multiple domains. As these proteins belong to large protein families, the results were manually examined to remove false positives (e.g., flavoproteins, sulfite reductases, or cytochrome reductases).
Sulfite Metabolism and Molybdopterin Biosynthesis
The S. pombe proteins were used for the screening using BLASTp (e value < 0.1) and TBLASTn (e value < 10−3). The molybdopterin biosynthesis genes were identified in T. deformans based on protein similarity with the genes of the obligate biotroph Albugo laibachii (Kemen et al. 2011). Domain-based searches were conducted using hmmer3 (e value < 0.1) with the following characteristic domains: FAD binding (PF000667), oxidoreductase NAD-binding (PF00175), Mo-co oxidoreductase dimerisation (PF03404), oxidoreductase molybdopterin-binding domain (PF00174), Molybdenum cofactor synthesis C (PF06463), Radical SAM superfamily (PF04055), MoaE (PF02391), molybdopterin binding (PF00994), MoeA C-terminal region (PF03454), and MoeA N-terminal region (PF03453).
Transporters
The transporters were identified using two complementary approaches. The proteomes were compared with the tcdb proteins collection (Saier et al. 2009) using ssearch (Pearson 1991). In parallel, putative amino acid transporters were identified using BLAST2GO with BLASTp (10−10) and InterProscan options. The sequences were then classified according to the tcdb families’ classification system to assess putative substrate. Statistical tests were performed using (R Development Core Team 2008).
Data Accessibility
Proteome clustering results and matrix, scripts, and sequences alignments are available from GitHub (https://github.com/ocisse/Pneumocystis_comparative, last accessed July 27, 2014).
Results
Identification of Orthologous Gene Families
To investigate the evolutionary dynamics of gene families in the Taphrinomycotina subphylum, we established the orthologous relationships among the proteomes of seven Taphrinomycotina, four Saccharomycotina, two Pezizomycotina, one Ustilaginomycotina, and one Mucormycotina. This corresponded to a set of 101,030 predicted proteins. Based on the presence of 458 core eukaryotic proteins (Parra et al. 2007), P. jirovecii predicted proteins and its de novo assembled transcripts showed a completeness of 89% and 70%, respectively. There was an overlap of 98% between these two latter sets, suggesting that most of the P. jirovecii genes were captured (supplementary table S1, Supplementary Material online). Except that of P. carinii with 79%, the fungal proteomes had a completeness of at least 97%. The proteomes clustering yielded 8,368 families and 2,143 single taxa clusters (i.e., containing at least two proteins from the same species), which included 81% of P. jirovecii predicted proteins.
P. jirovecii Proteome Content
Sixty-eight percent of the 3,898 P. jirovecii predicted proteins had at least one ortholog in the proteome of another fungus, including in a distantly related one, suggesting an ancient origin (supplementary fig. S1, green and blue slices, Supplementary Material online). These conserved genes were primarily devoted to the basal cellular and metabolic activities (supplementary table S1, Supplementary Material online). Four percent of the P. jirovecii proteins were genus specific and enriched for glycoprotein biosynthetic activity (supplementary fig. S1, red slices, Supplementary Material online). As a comparison, the Schizosaccharomyces genus-specific genes represented 10% (pink slices), suggesting a greater genetic innovation than in the Pneumocystis genus. Two percent of P. jirovecii proteins, which encode mostly hypothetical proteins, were included in gene families specific to this organism (light gray slices).
Gene Families’ Gain and Loss Patterns in Reconstructed Ancestors
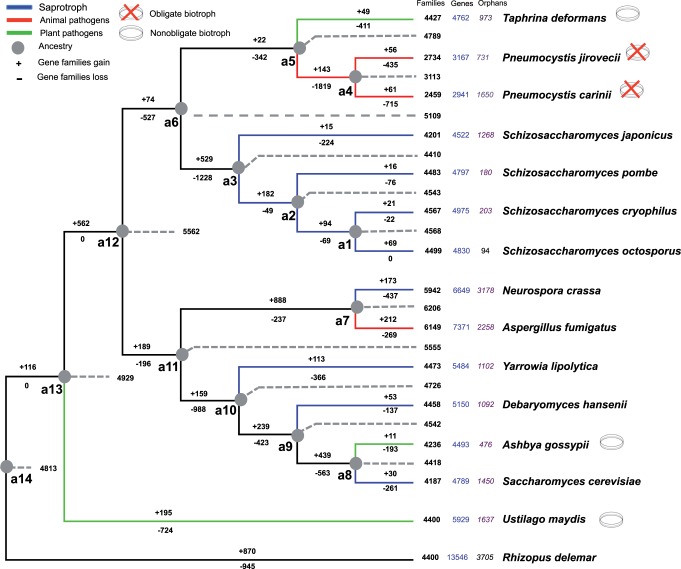
Biotrophy in Pneumocystis and Taphrina lineages would have appeared in ancestors as a consequence of a shift of host or environment. Thus, the gene families’ losses and gains in these ancestors should provide insights regarding the possible acquisition of biotrophy. To obtain a dynamic view of these events, we derived a gene presence–absence matrix from the gene clustering and projected these patterns onto a species phylogeny obtained from 458 shared single copy orthologs (Cisse et al. 2012). We then applied a parsimonious evolutionary scenario using the MP method based on the Dollo model to infer gene gain and loss events. The reconstructed ancestor of Pneumocystis spp. “a4” appeared to have had at least 3,113 gene families, which indicated a reduction of approximately 30% as compared with the ancestors of T. deformans “a5” and of Schizosaccharomyces spp. “a3” (fig. 1). There was a significant difference in the number of gene families’ gains between Taphrinomycotina members and their reconstructed ancestors (Wilcoxon rank sum test [one-side test], P = 0.02), with the highest number in the ancestor of Schizosaccharomyces spp. “a3” (n = 529). The difference in the number of gene losses among the Taphrinomycotina members was also significant (Wilcoxon rank sum test, P = 0.01), with the highest number in the last common ancestor of the Pneumocystis spp. “a4” (n = 1,819). These observations held true when the numbers of gene gains and losses were normalized by proteome size (data not shown).
Fig. 1.—
Lifestyle and projected gene families’ gain and loss patterns in representative fungal genomes. The gene families’ loss and gain patterns are projected onto each branch of the species phylogeny. The fungal species phylogeny corresponds to a cladogram derived from the ML tree of our previous study (Cisse et al. 2012; supplementary fig. S3, Supplementary Material online). The numbers on the branch of the phylogenetic tree corresponds the gene families’ gains (+) and losses (−). The lifestyle of the different species is indicated by the color of the branch (blue for saprotrophs, red for animal pathogens, and green for plant pathogens). Biotrophic organisms are indicated by Petri dishes, red crosses indicating the absence of a culture system. Ancestral gene families’ content was reconstructed using the Dollo parsimony assumption (represented by the gray doted lines and the letter “a” which stands for ancestor). For each species, the number of gene families, genes, and orphan genes is displayed. The number of gene families is also displayed for inferred ancestral taxa.
Gene Losses Associated with the Acquisition of Obligate Biotrophy in the Pneumocystis Genus
We focused on two time points in relation to the possible acquisition of obligate biotrophy in Pneumocystis ancestry: The common ancestor of the Taphrina and Pneumocystis genera “a5,” and the ancestor of the Pneumocystis genus “a4.” Notably, T. deformans and Pneumocystis spp. display distinct behaviors, that is, the former can be grown axenically in vitro (Fonseca and Rodrigues 2011), whereas the latter ones could not so far. Thus, we hypothesized that gene losses during the transition from the “a5” to “a4” occurred. We optimized rates for gene birth-and-models and applied Dollo parsimony to infer gene families’ gain and loss events. The investigation of the matrix of these events highlighted 2,324 ancestral genes that are present in “a5,” and subsequently lost in “a4.” In this analysis, we required that the inferred ancestral genes were also conserved in both Taphrina and Schizosaccharomyces lineages. These genes were significantly enriched for the following GO terms: Oxidation–reduction (Fisher’s exact test, P = 4.5 × 10−11), carbohydrate metabolism (P = 2.3 × 10−4), amino acid transport (P = 8.7 × 10−4), and sulfur compound metabolic process (P = 0.009) (supplementary table S3, Supplementary Material online).
To investigate whether gene losses could have occurred through massive pseudogenization, we conducted a comprehensive search of pseudogenes in P. jirovecii (supplementary material, Supplementary Material online; P. carinii could not be analyzed because of its high fragmentation). We found evidence of 75 pseudogenes (supplementary table S7, Supplementary Material online), which correspond to a ratio of 0.02 pseudogene per annotated protein-coding gene. This value is close to those of 0.03 and 0.01 reported, respectively, for S. cerevisiae and S. pombe (Echols et al. 2002; Wood et al. 2002).
Loss of Amino Acid and Purine Metabolisms
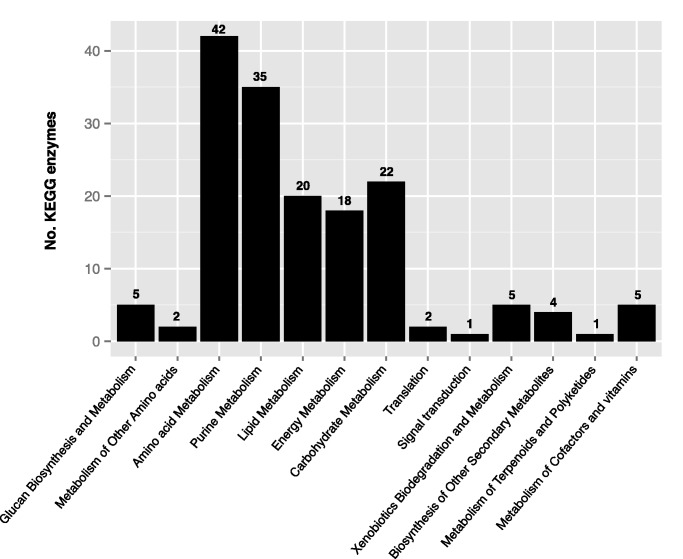
Among the 2,324 genes present in ancestor “a5” but lost in the Pneumocystis genus, 183 enzymes were mapped, and 42% of them turned out to be involved in the amino acid and purine metabolisms (fig. 2). The overrepresentation of the amino acid metabolism category is consistent with the previously reported loss of most enzymes dedicated to the biosynthesis of amino acids (Hauser et al. 2010; Cisse et al. 2012). These acquired auxotrophies imply scavenging of these compounds from the host or lung environment. Many fungi use various amino acid permeases and oligopeptide transporters to scavenge amino acids from their environment. For example, the biotroph rut fungus Uromyces fabae uses an AAT1p amino acid transporter for histidine uptake (Struck et al. 2002), and the necrotroph fungus Fusarium oxysporum uses a general permease for amino acids uptake (Divon et al. 2005). We screened the proteomes of all species used in this study to identify transporters. Pneumocystis jirovecii and P. carinii displayed a significant reduction in the amino acid/polyamine/organocation family as compared with other members of the Taphrinomycotina subphylum (Fisher’s exact unilateral test, P value = 1.3 × 10−7; supplementary table S3, Supplementary Material online). The major facilitator superfamily, which are versatile transporters that can carry amino acids, also showed a significant reduction in Pneumocystis spp. (Fisher’s exact unilateral test, P value = 4.08 × 10−15; supplementary table S3, Supplementary Material online).
Fig. 2.—
Functional categories of ancestral genes lost in the Pneumocystis genus. The 2,324 ancestral genes that appeared to be present in the last common ancestor “a5,” and subsequently lost in the Pneumocystis genus, were reconstructed using MP and mapped onto KEGG biochemical pathways atlas.
Loss of Purine Degradation Pathway
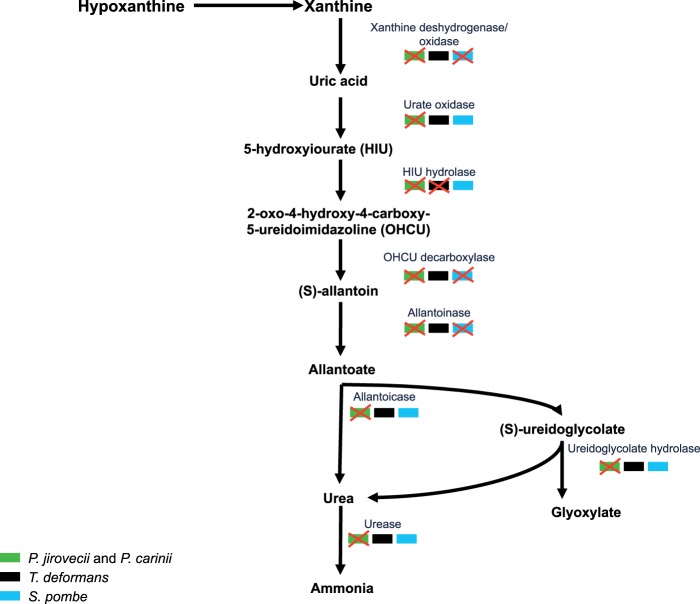
Nineteen percent of the 183 ancestral enzymes conserved in T. deformans but lost in the Pneumocystis genus are related to purine metabolism (fig. 2). This metabolism includes the synthesis and degradation of adenine and guanine, which are fundamental and well-conserved pathways across all life forms (Caetano-Anolles et al. 2007). The purine degradation allows the recycling of purines and their use as secondary source of nitrogen in case of limiting conditions. We detected the presence of the hallmark genes of the inosine 5′-phosphate and uridine 5′-phosphate biosynthesis pathways in P. jirovecii and P. carinii (supplementary table S5, Supplementary Material online). These compounds are the precursors of the synthesis of purines and pyrimidines, suggesting that the de novo purine biosynthesis takes place in Pneumocystis spp. However, no matches for all the catabolic enzymes involved in the degradation of purines were detected in P. jirovecii and P. carinii, whereas most of them were present in T. deformans and Schizosaccharomyces spp. (fig. 3 and supplementary table S4, Supplementary Material online).
Fig. 3.—
Purine degradation pathway in members of the Taphrinomycotina subphylum. The scheme indicates the presence or absence of purine metabolism genes in Pneumocystis jirovecii, P. carinii, Taphrina deformans, and Schizosaccharomyces pombe. The map diagram is based on the purine degradation pathway described by Lee et al. (2013). The colored rectangles represent the genes in the organisms. Schizosaccharomyces pombe is able to use different purines as starting point for purine degradation as described elsewhere (Kinghorn and Fluri 1984) (e.g., hypoxanthine, xanthine, uric acid, allantoin, and allantoic acid). The pathway has not been experimentally characterized in T. deformans.
Loss of Nitrogen and Sulfur Assimilation Pathways
Nitrogen and sulfur are essential for life. The key enzymes of the inorganic nitrogen and sulfur metabolisms, that is, the nitrate and nitrite reductases, as well as the sulfite reductase and oxidase, were found to be absent in the Pneumocystis and Schizosaccharomyces genera, while being preserved in T. deformans (supplementary table S4, Supplementary Material online). The nitrate reductase is a molybdenum-dependant enzyme, which is involved in the nitrate reduction during the nitrogen acquisition (Campbell 1999). Consistently, the complete molybdopterin biosynthesis pathway was lost in the Pneumocystis and Schizosaccharomyces genera (supplementary table S4, Supplementary Material online).
Loss of Thiamine Biosynthesis Pathway
Thiamine is an essential cofactor required by almost all living organisms. No matches for the key enzymes thiamine thiazole synthase and accessory enzyme kinases, except the thiamine pyrophosphokinase, were detected in P. jirovecii and P. carinii (supplementary table S4, Supplementary Material online). These enzymes appeared to be highly conserved in other members of the Taphrinomycotina subphylum, except the thiamine thiazole synthase in T. deformans.
Loss of RNAi Machinery
The RNAi machinery is involved in the genome defense by controlling the movement of repetitive mobile elements and the regulation of chromatin modification. No matches for the Argonaute and Dicer proteins were detected in P. jirovecii and P. carinii, whereas they were present in T. deformans and Schizosaccharomyces spp. (supplementary table S4, Supplementary Material online).
Virulence Associated Proteases
We analyzed several secreted peptidases of the MEROPS subfamilies, that is, A01, C13, G01, M35, S08, S09, S10, and S14. These are important fungal virulence factors (Monod et al. 2002), which might be involved in the degradation of the extracellular matrix in human lungs. The number of these proteases in Pneumocystis spp. is slightly lower (0.3–0.4% of their proteomes) than in most of the other Taphrinomycotina members (0.4–0.7%) (supplementary table S5, Supplementary Material online). Interestingly, P. jirovecii and P. carinii along with T. deformans harbor a specific ATP-dependent clp protease of the S14 family involved in the hydrolysis of proteins (UniProt accession: L0PC06), which is lacking in the Schizosaccharomyces spp. On the other hand, Pneumocystis spp. showed an underrepresentation of the A01 and S09 families which have been suggested to be crucial virulence factors in many fungi such as the human pathogen Candida albicans (Schaller et al. 2005).
Pneumocystis spp. Genomic Features
In order to investigate their evolution, several features of Pneumocystis spp. genomes were analyzed (supplementary material, Supplementary Material online). We estimated the genome-wide Ka/Ks ratio by the analysis of 898 single copy orthologous genes shared by Pneumocystis and Schizosaccharomyces spp. The mean Ka/Ks values of these genes were, respectively, 0.06 and 0.05 in the two genera (supplementary fig. S2A, Supplementary Material online). These values are smaller than one which suggests that these genes evolve under purifying selection.
The proportion of repetitive DNA in P. jirovecii genome, including transposons, retrotransposons, and low complexity DNA, but excluding the subtelomeric gene families, was estimated to 9.8% (supplementary table S6, Supplementary Material online; P. carinii could not be analyzed because of its high fragmentation). This value is higher than those of the plant pathogen T. deformans (1.5), of the free-living yeasts S. cerevisiae (4.8) and S. pombe (5.2), and of the extreme obligate parasite Encephalitozoon cuniculi (0.7).
Further analyses revealed intergenic region lengths in P. jirovecii which are similar to those observed in the free-living yeasts and T. deformans, but larger than those of E. cuniculi (supplementary fig. S2B, Supplementary Material online). The gene density observed in P. jirovecii (481 genes per Mb) was similar to those of S. cerevisiae (487), S. pombe (554), and T. deformans (431), but clearly lower than that observed in E. cuniculi (816; supplementary table S6, Supplementary Material online).
Discussion
Biotrophy is a character that has emerged independently in various fungal clades as well as in oomycetes (Kemen and Jones 2012; Spanu 2012), and which has been described exclusively in plant parasites so far. This study was undertaken to explore the signature of biotrophy in the human fungal parasite P. jirovecii. By reconstructing the ancestral genomes of the Taphrinomycotina subphylum, we discovered several gene losses in the Pneumocystis genus compatible with its obligate biotrophy. It is unlikely that we missed these genes because of the incompleteness of the genomes as 1) the losses were observed in both P. jirovecii and P. carinii, and 2) the searches were conducted on independent and partially overlapping genomic and transcriptomic data sets, which are expected to cover 100% of the genomes.
Because a large proportion of the genome was found to be under purifying selection, there is a high probability that the losses of genes we detected in Pneumocystis spp. have resulted from neutral genetic drift rather than from positive selection. Genetic drift might have been favored by the relative small effective population size of Pneumocystis spp., which is expected from the strict host specificity and absence of free-living forms of these fungi. The relatively large proportion of repetitive DNA, large intergenic region lengths, and low gene density in P. jirovecii are also more compatible with the genetic drift hypothesis than with positive selection. Furthermore, the ratio of pseudogenes in P. jirovecii was found to be small and similar to those of nonbiotrophic free-living organisms, suggesting that a massive appearance of such nonfunctional genes has not been a major mechanism of gene loss.
Most amino acids biosyntheses were lost in Pneumocystis spp. (Hauser et al. 2010; Cisse et al. 2012), a hallmark of obligate parasitism. Such loss was not observed in plant biotrophs so far (Spanu 2012), but occurs systematically in organisms feeding on other animals, such as protozoan parasites of humans, as well as, for example, in Homo sapiens (Payne and Loomis 2006). In fungal biotrophs, losses of biosynthetic pathways are generally compensated by expanded transporters families to scavenge the products of these pathways (Spanu et al. 2010; Duplessis et al. 2011). However, we observed a reduced panel of transporters susceptible to carry amino acids in Pneumocystis spp. This reduction may be linked to the loss of the amino acids biosynthesis pathways and suggests the use of an alternative strategy. Biochemical experiments showed that amino acids uptake in P. carinii may occur by a facilitated diffusion across the membrane (Basselin, Lipscomb, et al. 2001; Basselin, Qiu, 2001). Pneumocystis spp. have been previously suggested to scavenge other crucial compounds from their hosts, that is, sterols (Joffrion and Cushion 2010).
Nitrogen and sulfur assimilation pathways as well as the purine catabolism were lost in Pneumocystis spp. In the absence of assimilation from inorganic sources, nitrogen and sulfur are essential compounds which can be obtained from the degradation of purines, or from proteolysis. Purine catabolism is unlikely to be a source in P. jirovecii and P. carinii because we did not detect the key genes of this pathway. In the absence of the sulfite oxidase, which is required to metabolize cysteine and methionine, proteolysis does not seem to be the source of sulfur, source which thus remains to be characterized. On the other hand, proteolysis could be the source of nitrogen which would be consistent with the fact that animal tissues are rich in proteins. This would contrast from the saprotroph S. pombe, which uses the purine degradation for this purpose (Kinghorn and Fluri 1984). The loss of the purine degradation has not been observed in fungal biotrophs so far and thus represents a specificity of Pneumocystis spp., which might also be related to their adaptation to animal hosts. On the other hand, the loss of inorganic nitrogen and sulfur assimilation is frequently observed in fungal plant biotrophs (Spanu 2012), but this is not a hallmark of biotrophy as it is also observed in Schizosaccharomyces spp. that are saprotrophs. An interesting alternative to the possibility of acquiring essential compounds from the host is the Black Queen hypothesis (Morris et al. 2012). This hypothesis proposes that the loss of capability at the individual level can be compensated by the community. In the case of Pneumocystis spp., this community would be the other microorganisms sharing the same environment. This could be investigated through studies of the metabolic capacities of the lung microbiota associated with Pneumocystis pneumonia.
The key enzymes for thiamine biosynthesis were not detected in Pneumocystis spp. This loss is also a hallmark of biotrophy (Spanu 2012) and suggests that this vitamin is available from the mammalian host lungs. The missing thiamine thiazole synthase is a turnover suicide enzyme that needs to be replaced after each reaction (Chatterjee et al. 2011).
Our searches revealed that the RNAi machinery is lost in Pneumocystis spp. This loss would have occurred after the divergence of the Taphrina and Pneumocystis genera but before diversification in the Pneumocystis genus. The removal of RNAi has been suggested to confer a selective advantage to pathogens because it would enhance genome evolution by allowing the free movement of transposons and retrotransposons (Oliver and Greene 2009). The RNAi machinery is also absent in the fungi S. cerevisiae, U. maydis, and Cryptococcus gattii, as well as in the protozoan parasites Trypanosoma cruzi and Plasmodium falciparum (Nicolas et al. 2013). The significance of this loss remains to be elucidated.
Our findings suggest that obligate biotrophy arose in Pneumocystis spp. in conjunction with gene losses of multiple pathways. The vital compounds that can be scavenged from the host lungs probably allowed these losses. Together with other characteristics, that is, a sex life cycle within the host, the absence of massive destruction of host cells, the absence of an axenic in vitro culture system so far, and the lack of virulence factors, these gene losses strongly suggest that Pneumocystis spp. should be considered as obligate biotrophs. The reduction of the genome size of Pneumocystis spp. contrasts with the increase of this feature often observed in fungal plant obligate biotrophs (Spanu 2012). This increase corresponds to a proliferation of retrotransposons, which would create genetic variability and diversity including panels of effectors. As in oomycetes, the genome size reduction in Pneumocystis spp. might be compensated by the genetic diversity generated by sexuality, suggesting that they might be “obligate sexual organisms” (Spanu 2012). Another hypothesis is that the niches in animal hosts do not require a variety of effectors to sustain infection but rather an increased fitness. This increased fitness might be obtained by the reduction of genome size, a feature which is observed in most human parasites, eukaryotic as well as prokaryotic (Sakharkar 2004; Hauser et al. 2010).
In contrast to necrotrophy, biotrophy implies a long-lasting and tight relationship with the living host and its immune system. Such relationship requires mechanisms which are expected to be specific to each host. One mechanism might be the antigenic variation encoded by the contingency gene families present at the telomeres of Pneumocystis spp., which may help maintaining the infection despite the host immune system. Thus, one can speculate that the strict host specificity and the biotrophy of Pneumocystis spp. were acquired simultaneously. The obligate nature of biotrophy may have emerged later and implied coevolution with the host.
A limitation of our study was the use of the Dollo parsimony model, which assumes that lost ancestral traits cannot be reacquired. Indeed, the model is not appropriate to detect genes brought by horizontal gene transfer events, which are thought to be relatively frequent in lower eukaryotes such as fungi (Fitzpatrick 2012). The possible occurrence of such transfers, which may have brought important new traits within the Pneumocystis genus, deserves further studies.
In conclusion, Pneumocystis spp. harbor a unique combination of characteristics which are hallmarks of both obligate biotrophs and animal parasites. The evolutionary events underlying biotrophy in Pneumocystis spp and the closely related hemibiotroph T. deformans appear drastically different, which suggests a parallel rather than a convergent evolution. Pneumocystis spp. should be considered as the first described obligate biotrophs of animals, which can turn into opportunistic biotrophic pathogens upon immune deficiency of the host. The lifestyle of P. jirovecii differs from that of other human fungal parasites. Indeed, Malassezia and Candida spp. can be considered as obligate commensals and opportunistic necrotrophs, whereas Dermatophytes would include both facultative and obligate true necrotrophs, which, possibly, might be transiently commensals.
Supplementary Material
Supplementary material, tables S1–S7, and figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Swiss National Science Foundation grant 310030-124998 to P.M.H. and M.P. and Swiss National Science Foundation fellowship grant no. 151780 to O.H.C. The computations were performed at the Vital-IT Center for high-performance computing (http://www.vital-it.ch) of the SIB Swiss Institute of Bioinformatics. SIB receives financial supports from the Swiss Federal Government through the State Secretariat for Education and Research (SER). The authors thank Thomas Bernard, Sébastien Moretti, and Lorenzo Cerutti for help with sequence analyses. They also thank Pierre Bady for help with statistical tests as well as Ioannis Xenarios and Frederic Masclaux for discussions. They also thank the two anonymous reviewers for their constructive comments.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basselin M, Lipscomb KJ, Qiu YH, Kaneshiro ES. Transport of aspartic acid, arginine, and tyrosine by the opportunistic protist Pneumocystis carinii. Biochim Biophys Acta. 2001;1511:349–359. doi: 10.1016/s0005-2736(01)00293-0. [DOI] [PubMed] [Google Scholar]
- Basselin M, Qiu YH, Lipscomb KJ, Kaneshiro ES. Uptake of the neutral amino acids glutamine, leucine, and serine by Pneumocystis carinii. Arch Biochem Biophys. 2001;391:90–98. doi: 10.1006/abbi.2001.2389. [DOI] [PubMed] [Google Scholar]
- Caetano-Anolles G, Kim HS, Mittenthal JE. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci U S A. 2007;104:9358–9363. doi: 10.1073/pnas.0701214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell WH. Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:277–303. doi: 10.1146/annurev.arplant.50.1.277. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, et al. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature. 2011;478:542–546. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse OH, et al. Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. MBio. 2013;4:e00055–00013. doi: 10.1128/mBio.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse OH, Pagni M, Hauser PM. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. MBio. 2012;4:e00428–00412. doi: 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O, Rubinstein ND, Stern A, Gophna U, Pupko T. A likelihood framework to analyse phyletic patterns. Philos Trans R Soc Lond B Biol Sci. 2008;363:3903–3911. doi: 10.1098/rstb.2008.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- Cushion MT, et al. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS One. 2007;2:e423. doi: 10.1371/journal.pone.0000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushion MT, Stringer JR. Stealth and opportunism: alternative lifestyles of species in the fungal genus Pneumocystis. Annu Rev Microbiol. 2010;64:431–452. doi: 10.1146/annurev.micro.112408.134335. [DOI] [PubMed] [Google Scholar]
- Divon HH, Rothan-Denoyes B, Davydov O, Pietro DIA, Fluhr R. Nitrogen-responsive genes are differentially regulated in planta during Fusarium oxyspsorum f. sp. lycopersici infection. Mol Plant Pathol. 2005;6:459–470. doi: 10.1111/j.1364-3703.2005.00297.x. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S, et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A. 2011;108:9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols N, et al. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes comparing genes and pseudogenes. Nucleic Acids Res. 2002;30:2515–2523. doi: 10.1093/nar/30.11.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS. Phylogenetic analysis under Dollo’s law. Syst Zool. 1977;26:77–88. [Google Scholar]
- Fitzpatrick DA. Horizontal gene transfer in fungi. FEMS Microbiol Lett. 2012;329:1–8. doi: 10.1111/j.1574-6968.2011.02465.x. [DOI] [PubMed] [Google Scholar]
- Fonseca Á, Rodrigues MG. Taphrina fries. In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts, a taxonomic study, 5th ed. Vol. 2. Amsterdam (The Netherlands): Elsevier; 2011. pp. 823–858. [Google Scholar]
- Hauser PM, et al. Comparative genomics suggests that the fungal pathogen Pneumocystis is an obligate parasite scavenging amino acids from its host’s lungs. PLoS One. 2010;5:e15152. doi: 10.1371/journal.pone.0015152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffrion TM, Cushion MT. Sterol biosynthesis and sterol uptake in the fungal pathogen Pneumocystis carinii. FEMS Microbiol Lett. 2010;311:1–9. doi: 10.1111/j.1574-6968.2010.02007.x. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011;9:e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E, Jones JD. Obligate biotroph parasitism: can we link genomes to lifestyles? Trends Plant Sci. 2012;17:448–457. doi: 10.1016/j.tplants.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Kinghorn JR, Fluri R. Genetic studies of purine breakdown in the fission yeast Schizosaccharomyces pombe. Curr Genet. 1984;8:99–105. doi: 10.1007/BF00420225. [DOI] [PubMed] [Google Scholar]
- Lee IR, et al. Characterization of the complete uric acid degradation pathway in the fungal pathogen Cryptococcus neoformans. PLoS One. 2013;8:e64292. doi: 10.1371/journal.pone.0064292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of orthologgroups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin DJ, Hibbett DS, Lutzoni F, Spatafora JW, Vilgalys R. The search for the fungal tree of life. Trends Microbiol. 2009;17:488–497. doi: 10.1016/j.tim.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Monod M, et al. Secreted proteases from pathogenic fungi. Int J Med Microbiol. 2002;292:405–419. doi: 10.1078/1438-4221-00223. [DOI] [PubMed] [Google Scholar]
- Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio. 2012;3:e00036–12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas FE, Torres-Martinez S, Ruiz-Vazquez RM. Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 2013;9:e1003089. doi: 10.1371/journal.ppat.1003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. Bioessays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Payne SH, Loomis WF. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot Cell. 2006;5:272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redhead SA, Cushion MT, Frenkel JK, Stringer JR. Pneumocystis and Trypanosoma cruzi: nomenclature and typifications. J Eukaryot Microbiol. 2006;53:2–11. doi: 10.1111/j.1550-7408.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- Rhind N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team R: A language and environment forstatistical computing. Vienna (Austria): R Foundation for Statistical Computing. 2008 Available from: http://www.R-project.org. [Google Scholar]
- Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar KR, et al. Genome reduction in prokaryotic obligatory intracellular parasites of humans: a comparative analysis. Int J Syst Evol Microbiol. 2004;54:1937–1941. doi: 10.1099/ijs.0.63090-0. [DOI] [PubMed] [Google Scholar]
- Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005;48:365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- Schmitt MJ, Breinig F. Yeast viral killer toxins: lethality and self-protection. Nat Rev Microbiol. 2006;4:212–221. doi: 10.1038/nrmicro1347. [DOI] [PubMed] [Google Scholar]
- Slaven BE, et al. Draft assembly and annotation of the Pneumocystis carinii genome. J Eukaryot Microbiol. 2006;53(Suppl. 1): S89–S91. doi: 10.1111/j.1550-7408.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- Spanu PD. The genomics of obligate (and nonobligate) biotrophs. Annu Rev Phytopathol. 2012;50:91–109. doi: 10.1146/annurev-phyto-081211-173024. [DOI] [PubMed] [Google Scholar]
- Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- Struck C, Ernst M, Hahn M. Characterization of a developmentally regulated amino acid transporter (AAT1p) of the rust fungus Uromyces fabae. Mol Plant Pathol. 2002;3:23–30. doi: 10.1046/j.1464-6722.2001.00091.x. [DOI] [PubMed] [Google Scholar]
- Thines M, Kamoun S. Oomycete-plant coevolution: recent advances and future prospects. Curr Opin Plant Biol. 2010;13:427–433. doi: 10.1016/j.pbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV. Genome reduction as the dominant mode of evolution. Bioessays. 2013;35:829–837. doi: 10.1002/bies.201300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteome clustering results and matrix, scripts, and sequences alignments are available from GitHub (https://github.com/ocisse/Pneumocystis_comparative, last accessed July 27, 2014).