Abstract
Rapid divergence of gene copies after duplication is thought to determine the fate of the copies and evolution of novel protein functions. However, data on how long the gene copies continue to experience an elevated rate of evolution remain scarce. Standard theory of gene duplications based on some level of genetic redundancy of gene copies predicts that the period of accelerated evolution must end relatively quickly. Using a maximum-likelihood approach we estimate preduplication, initial postduplication, and recent postduplication rates of evolution that occurred in the mammalian lineage. We find that both gene copies experience a similar in magnitude acceleration in their rate of evolution. The copy located in the original genomic position typically returns to the preduplication rates of evolution in a short period of time. The burst of faster evolution of the copy that is located in a new genomic position typically lasts longer. Furthermore, the fast-evolving copies on average continue to evolve faster than the preduplication rates far longer than predicted by standard theory of gene duplications. We hypothesize that the prolonged elevated rates of evolution are determined by functional properties that were acquired during, or soon after, the gene duplication event.
Keywords: gene duplication, evolution, selection
Introduction
The study of gene duplications continues to be at the forefront of molecular evolution due to their likely role in the emergence of new functions (Bridges 1936; Ohno 1970; Conant and Wolfe 2008; Ponting 2008; Hahn 2009; Innan and Kondrashov 2010). New functions are thought to emerge in the process of sequence and regulatory divergence of the gene copies that result from a gene duplication event. Therefore, the efforts in theory and empirical observations of gene duplications remain focused on the rate of evolution after gene duplication. Models of gene duplication evolution suggest that either one (Ohno 1970; Hughes 1999) or both (Force et al. 1999) gene copies experience a brief period of accelerated evolution (Hahn 2009; Innan and Kondrashov 2010), an elevated ratio of nonsynonymous (dn) and synonymous (ds) rates, followed by a return to the preduplication levels (Walsh 1995; Stoltzfus 1999; Lynch and Conery 2003; Konrad et al. 2011; Proulx 2012). It is thought that within the initial period of evolution new functions are forged.
Several approaches have been used to detect the acceleration of evolution after a gene duplication. First, dn/ds ratios between duplicated pairs in the same genome were correlated with ds values, where ds is a proxy for time since duplication. The observation that for values of ds > 0.1 the dn/ds appear to be relatively constant was taken as evidence that the observed plateau must correspond to the preduplication levels (Lynch and Conery 2000; Vinogradov 2012). Second, the dn/ds ratios of duplicated and nonduplicated genes consistently show that recently duplicated genes evolve faster than that of nonduplicated genes (Kondrashov et al. 2002; Nembaware et al. 2002; Jordan et al. 2004; Yampolsky and Bouzinier 2014), although genes that duplicated a long time ago appear to be more conserved (Davis and Petrov 2004; Jordan et al. 2004). The observation that dn/ds ratios measured in paralogous comparisons were higher than when the dn/ds is measured between nonduplicated orthologs suggested an acceleration of evolution in gene copies.
These two approaches, however, cannot be used to determine at which time point after the gene duplication event the gene duplications return to their preduplicated rates of evolution. The first approach suffers from a lack of a comparison to other genes, as it is not clear whether or not the plateau of ds > 0.1 (Lynch and Conery 2000) corresponds to the preduplication levels. The second approach compares different genes and as some genes are more likely to be duplicated than others (Kondrashov et al. 2002; Jordan et al. 2004; Kondrashov 2012) it may create an inherent bias. Furthermore, under both approaches the dn/ds ratio is measured between two paralogous sequences. Therefore, the dn/ds estimated across the time since the gene duplication and is expected to be elevated for all comparisons due to the inclusion of the initial fast period of evolution right after the gene duplication (fig. 1A). Thus, neither of these two approaches makes it possible to reliably address the issue of the long-term impact of gene duplication on the rate of evolution.
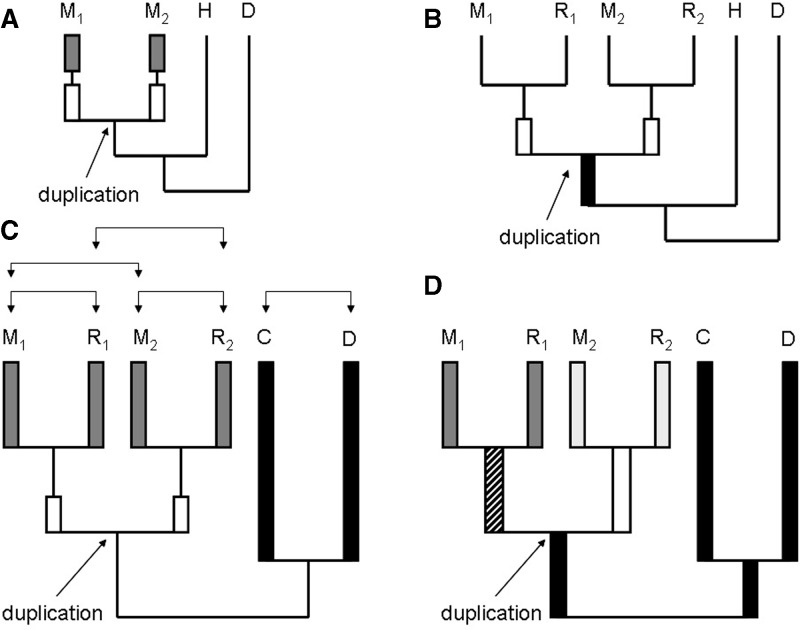
Fig. 1.—
Phylogenetic topology of different approaches that may be used to study the evolution of gene duplications. When considering paralogs in only one species it is not possible to distinguish between recent evolution (gray box on the phylogeny) and the initial evolutionary acceleration (A). This issue can be resolved when considering dn/ds between orthologs of two species that have split after the gene duplication (B). However, if the preduplication rate of evolution is determined from the internal branch (black box) it may be difficult to resolve given the proximity to the expected acceleration of evolution immediately after the gene duplication (white boxes). In this study we avoid both of the compounding issues by focusing on five pairwise sequence comparisons, shown with brackets in (C). We can distinguish recent evolution of the duplicated genes (M1–R1 and M2–R2, gray boxes) from the initial acceleration occurring immediately after the gene duplication event (white boxes). Similarly, we use the divergence between sequences from two species (C, D) that have diverged prior to the gene duplication event as a proxy for preduplication rates of evolution (black boxes). We date the gene duplication by estimating ds between the two paralogs in the two species (M1–M2 and R1–R2). (D) We estimated dn and ds on the phylogeny while restricting the model to estimating five different dn/ds values across the following branch segments indicated as follows: Black = preD, striped = ipostD1, white = ipostD2, dark gray = rpostD1, light gray = rpostD2.
To measure the long-term effects of gene duplication on the rate of evolution, it is necessary to estimate the preduplication rate of evolution and the recent rate of evolution of the paralogs. For very recent gene duplications measuring the rate of recent evolution is trivial as an estimate of dn/ds between them is sufficient. It is feasible to estimate recent dn/ds values from an older gene duplication by comparing orthologs of the gene copies of two species that separated after the emergence of the gene duplication in question (fig. 1B).
The preduplication rate of evolution can be estimated as the dn/ds observed on the internal branch leading up to the gene duplication event (fig. 1B). This approach has been utilized by Pegueroles et al. (2013) and Cusack and Wolfe (2007). Briefly, Pegueroles et al. (2013) identified instances of gene duplication that were broadly characterized by a phylogenetic relationship in figure 1B. They studied whether or not the rates of duplicated genes measured as dn/ds between the mouse and rat orthologs of the duplicated genes were significantly different from the rate of evolution on the internal branch leading up to the gene duplication. They found that the younger gene duplications indeed evolve faster than the preduplication rates. However, the older gene duplications, those that occurred between approximately 70 and approximately 43 Ma, were found to evolve at a rate indistinguishable from that in the preduplication branch. Cusack and Wolfe (2007) utilized a congruent approach to study the long-term impact on the rate of evolution stemming from the whole-genome duplication (WGD) in yeast. The rate of preduplication evolution was similarly inferred from an internal branch leading up to the duplication event.
These two studies presented with slightly contradictory results. Pegueroles et al. (2013) suggested that gene duplications return to their preduplication rates of evolution relatively quickly, on the order of old world and new world primate divergence (Chatterjee et al. 2009). In contrast, Cusack and Wolfe (2007) observed that in yeast the surviving duplications from the WGD event continue to evolve faster than those genes that have lost their extra copy. Unfortunately, the Cusack and Wolfe (2007) study was focused on the remnants of the WGD event and used very long distances between some of the genes in the phylogeny, with ds > 1, reducing the reliability of the dn/ds estimates. An earlier study suggested that dn/ds on internal branches just prior to duplication may be accelerated in vertebrates (Johnston et al. 2007).
Here, we utilize two different methods to study the persistence of the acceleration of the rate of evolution after gene duplication. First, we employ a similar method to that used by previous studies (Cusack and Wolfe 2007; Johnston et al. 2007; Pegueroles et al. 2013) estimating the rate of evolution across different segments of the phylogeny that includes a recent gene duplication. Second, we investigate the rates of evolution after gene duplications using data from gene duplications with tangible synonymous divergence distance and avoiding estimating the rate of evolution on internal phylogenetic branches.
Materials and Methods
We obtained protein-coding sequences for the mouse, rat, human, orangutan, dog, and cow from ENSEMBL (Vilella et al. 2009). We used the data in ENSEMBL to identify orthology relationships in these genomes. We selected the one-to-one dog–cow orthologs and for these orthologous pairs we identified instances of one-to-many orthology between dog and mouse (human) genes. For such mouse (human) genes, we selected those that showed a one-to-one orthology to the rat (orangutan) genes. This approach allowed us to identify a preliminary set of genes that were duplicated after the dog−human split but before the mouse–rat (human–orangutan) divergence, with the exception of those cases where a deletion of one copy occurred in the dog–cow lineage. Furthermore, these homologs included all cases when more than one duplication occurred along the aforementioned phylogenetic segment.
For all of these cases, we aligned all of the homologs using the protein sequence with MUSCLE (Edgar 2004) and reverse-translated to create a multiple nucleotide alignment. We then used the codeml package (Yang 2007) to estimate dn, ds, and dn/ds values across the phylogeny specifying five different areas of the phylogeny with independent dn/ds values with model = 2, clock = 0 and a user-defined tree that as shown in figure 1D. We also used the codeml program from the PAML package (Yang 2007) to estimate dn and ds between some pairwise comparisons of sequences from the sextuplet alignment.
For those homologous clusters that included more than one paralog in the mouse (human) genome, we estimated the order in which the duplications occurred. We obtained ds measurements for all pairwise mouse (human) paralogs. We then identified the pair with the smallest ds value as being the most recent gene duplication and created a set (((Mouse1–Rat1),(Mouse2–Rat2)),(Cow,Dog)) for that pair. We then removed at random one of the two paralogs and its rat (orangutan) ortholog and found the pair with the lowest ds among the remaining paralogs and the same set of six homologs was created. The procedure was then repeated recursively until only a single pair of mouse (human) paralogs was left. In the final data set, 25 out of 90 sets of six homologs originated from clusters representing more than one duplication between the dog–human common ancestor and the mouse–rat (human–orangutan) divergence.
To eliminate instances when the one-to-many orthology between dog and mouse (human) was caused by a gene loss in the dog–cow lineage, we eliminated all instances when the absolute ds values among the paralogous comparisons were higher than expected to have originated after the dog–human split. Specifically, we eliminated all instances when either of the pairwise comparisons shown in figure 1 had a ds > 1. This approach must have also concurrently eliminated most instances of misalignment or erroneous ortholog assignment. We estimated δ by subtracting dn/ds of the dog–cow orthologs from the dn/ds value of each of the two mouse–rat (human–orangutan) orthologous comparisons.
Synteny analysis was performed by corresponding in ENSEMBL the neighboring genes of each dog gene to the neighboring genes of the paralogs in the mouse (human) genome. A mouse (human) homolog was identified as the original gene copy if its gene neighbors were orthologs to the gene neighbors of the dog gene. We excluded tandem duplications from this analysis.
Results and Discussion
We used mammalian genomes due to the availability of sequence data and an established phylogeny among major groups. We searched for gene duplications that have occurred after the split of Laurasiatheria (the group includes cow and dog) and Euarchontoglires (human and mouse) lineages but before the divergence of mouse–rat or human–orangutan lineages. Thus, we identified genes that were found in single copy in dog and cow, but were duplicated in mouse (human) and rat (orangutan) genomes (see Materials and Methods). Overall, we found 90 cases of such sextuplet homologs, with a gene duplication common to the mouse–rat (human–orangutan) lineage and single copy orthologs present in both the dog and the cow. We use a shorthand notation to label the cow (C), dog (D), mouse copy 1 (M1), mouse copy 2 (M2), rat copy 1 (R1), rat copy 2 (R2), human copy 1 (H1), human copy 2 (H2), orangutan copy 1 (O1), and orangutan copy 2 (O2) where M1–R1 (H1–O1) and M2–R2 (H2–O2) are one-to-one orthologs.
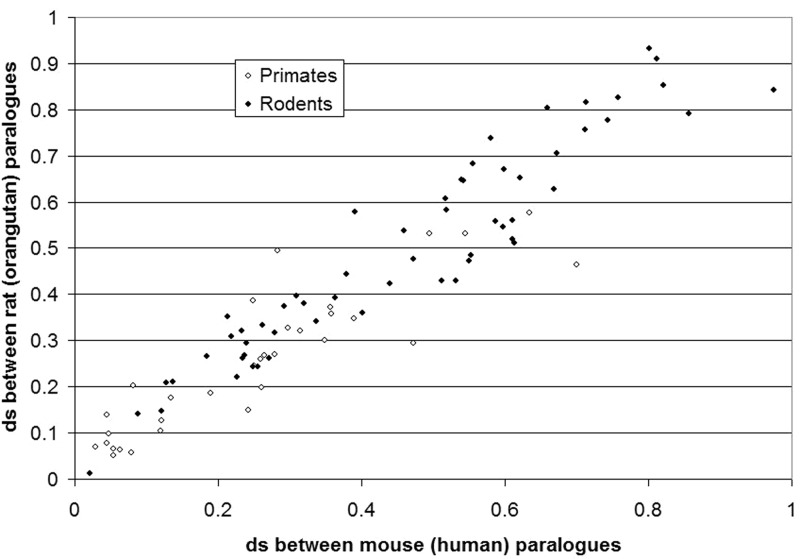
Due to the possible prevalence of gene conversion we first analyzed whether or not different rates of synonymous divergence are observed between gene copies different species. We correlated the estimated ds values obtained from M1–M2 (H1–H2) and R1–R2 (O1–O2) pairwise paralogous comparisons. We observe a very good correlation of the ds in paralogs of different species (fig. 2), indicating that gene conversion was not a large factor in the evolution of these genes. Of course, we cannot exclude the possibility of frequent gene conversion that occurred in both mouse (human) and rat (orangutan) lineages for a specific gene duplication. However, we are certain that this cannot affect much of our data as in that case we would have expected to see more paralogous sequence comparisons with a low ds.
Fig. 2.—
Relationship of synonymous divergence between gene duplications in sister species. The relationship between the synonymous divergence of paralogs in two species, M1–M2 (H1–H2) and R1–R2 (O1–O2) on the Y axis. The rodent and primate comparisons are shown with black and white circles, respectively.
Next, we introduce, δ, a measure of the difference between the preduplication and the recent postduplication rates of evolution. We measured these rates of evolution in two different ways. In a maximum-likelihood approach, we used PAML (see Materials and Methods) to estimate the preduplication (preD), two initial postduplication (ipostD1 and ipostD2), and two different recent postduplication (rpostD1 and rpostD2) rates of evolution or dn/ds (fig. 1D). We estimated the dn/ds separately in the two different copies (fig. 1) due to the possibility of asymmetrical evolution of the gene copies (Zhang et al. 2003; Cusack and Wolfe 2007; Han et al. 2009; Panchin et al. 2010; Pegueroles et al. 2013) and classify the gene copies into those with high and low dn/ds values, fast and slow rate evolution, respectively. We then measured δf = max(rpostD1, rpostD2) − preD and δs = min(rpostD1, rpostD2) − preD, which represent a difference in the pre- and postduplication rate of evolution for fast- and slow-evolving gene copies, respectively.
We used the average ds summed across the branches separating the paralogous gene copies M1–M2 and R1–R2 (H1–H2 and O1–O2; see fig. 1D), as a proxy for the length of time since the origin of the gene duplication. This allowed us to access how long after the gene duplication event the rate of evolution (dn/ds) remains elevated. We found three patterns when we compared δf and δs in duplications of different age (fig. 3A). First, the slower-evolving gene copies appear to evolve at the preduplication rates regardless of how long ago the duplication event occurred. This observation is consistent with our selection of gene duplications that are older than the mouse–rat divergence such that we were expected to miss the initial phase of acceleration that may be affecting the slow copy. Second, the rate of evolution in the younger fast-evolving duplications appears to be higher than in older duplications. Finally, the rate of evolution in fast-evolving gene duplications appears to be elevated even for the oldest duplications in our dataset with δf appearing to have reached a plateau at δf ∼ 0.15 for gene duplications older than ds ∼ 0.2 (fig. 3A).
Fig. 3.—
Difference in the rate of evolution in duplicated and nonduplicated orthologs. Average δf (white) and δs (gray) in bins. The number of duplications (average ds from the sum of branches connecting M1–M2 [H1–H2] and R1–R2 [O1–O2] nodes) is shown for each bin (A). The δ for internal branches (ipostDf and ipostDs) is shown in (B), branches of length 0 were removed from the analysis resulting in 20 (average ds = 0.17) and 21 (average ds = 0.18) duplications in one bin for ipostDf and ipostDs, respectively. The average δf (white) and δs (gray) excluding gene families are shown in (C). The average δf (white) and δs (gray) from pairwise comparisons are shown in (D), with average ds calculated similarly from pairwise comparisons of M1–M2 (H1–H2) and R1–R2 (O1–O2).
When considering perfectly symmetrical rates of evolution one copy is expected to evolve slower than the other due to the random variation of the dn/ds measurements. However, in a truly symmetrical case the increase in the rate of evolution in the fast-evolving copy will be of the same magnitude as the decrease in the rate of evolution of the slow copy. Our observation that δf > 0 cannot be explained by the stochastic segregation of slow and fast gene copies because the observed increase of dn/ds in the fast copy is of much higher than the slight, insignificant decrease in the dn/ds of the slow copy.
The observation that the slow copy is evolving at the preduplication rate may be explained in two ways. First, the slow copy may have never experienced an acceleration in the rate of evolution. Second, the acceleration of evolution occurred but was short-lived such that it returns to normal within a modest timeframe (ds < 0.2). We therefore compared the rate of evolution of the initial postduplication branches leading up to the mouse–rat divergence (fig. 1D). We calculated δif and δis for the internal branch leading up to the slow copy and fast copy, respectively. Specifically, δif = ipostDf − preD, where ipostDf is the dn/ds value from the branch leading up to the branch with δf. Conversely, δis = ipostDs − preD, where ipostDf is the dn/ds value from the branch leading up to the branch with δs. We found that δis and δif are not significantly different even for gene duplications that have emerged recently (fig. 3B), indicating that the slow copy evolves faster immediately after gene duplication and subsequently returns to the preduplication levels.
It has been suggested that the old copy, the one that is not relocated in a gene duplication event, typically evolves slower than the new copy after the gene duplication (Cusack and Wolfe 2007; Han et al. 2009). We see the same pattern in our data. Due to the prevalence of tandem duplications in mammals it was often not possible to distinguish between the old and the new copies. For 26 cases, however, we used data on synteny between the rodent (primate) genome and that of dog and cow genomes to discriminate between the original and the new copy. For 21 out of 26 duplications, the fast-evolving copy was the novel copy (Fisher’s exact test, P = 0.02). We hypothesize that the old copy is more likely to maintain the previous function and, therefore, more likely to have the preduplication levels of dn/ds.
Due to the decrease in selection pressure in primates caused by a smaller effective population size (Li 1997), it may be possible that the observed acceleration of the dn/ds in the duplicated genes is mostly influenced by the data from primate evolution. However, the fast-evolving gene copies are still significantly faster than the preduplication levels when we consider only rodent data, although the primate fast copies appear to have been accelerated to a greater degree (table 1). An increase in the strength of selection is anticipated in the rodent lineage relative to the cow–dog lineage (Li 1997), indicating that our observation of an increased δf is robust to lineage-specific changes in selection pressures.
Table 1.
Average dn/ds Values (Standard Error) for Slow and Fast-Evolving Gene Copies in Rodents and Primates
| Fast Copy | Slow Copy | |
|---|---|---|
| Rodents | 0.13 (±0.027) | −0.026 (±0.015) |
| Primates | 0.46 (±0.15) | 0.038 (±0.06) |
In our data set, we include genes that have duplicated more than once since the divergence of rodents (primates) and the dog–cow common ancestor. We employ a scheme to separate such instances into individual duplication events (see Materials and Methods); however, it may be possible that some of the gene copies in our data set are influenced by more than one recently emerged copy. We therefore performed the same analyses having retained only those genes that have experience only a single gene duplication event since the rodent (primate) and dog–cow divergence. We find the same patterns in the subset of genes with a single gene duplication (fig. 3C), indicating that multiple gene duplications do not substantially affect our results.
Finally, we have sought to replicate some of our results without attempting to reconstruct the rate of evolution in internal branches. We thus devised an approach that was based on five sequence comparisons among the orthologous and paralogous pairs of sequences estimating dn and ds (fig. 1C). We measured the rate of preduplication evolution O = dn/ds observed in the C–D comparison. We then measured P1 and P2 as dn/ds values in the M1–R1 (H1–O1) and M2–R2 (H2–O2) orthologous comparisons, respectively, which estimate the recent rate of postduplication evolution. We then measured δf = max(P1, P2) − O and δs = min(P1, P2) − O, which represent a difference in the pre- and postduplication rate of evolution for fast- and slow-evolving gene copies, respectively. The average ds of paralogous comparisons, M1–M2 and R1–R2 (H1–H2 and O1–O2) was used as a proxy for the length of time since the origin of the gene duplication (fig. 1C).
The data obtained through pairwise sequence comparisons confirm our conclusions that 1) the slower-evolving gene copies appear to evolve at the preduplication rates regardless of how long ago the duplication event occurred, 2) the rate of evolution is higher in younger fast-evolving duplications, and 3) the rate of evolution in fast-evolving gene duplications appears to be elevated even for the oldest duplications (fig. 3D). Due to the limitations of the pairwise comparisons using this approach we cannot confirm the observation of the initial acceleration in the rate of evolution of the slow-evolving gene copies.
Conclusions
Our results are consistent with the observations that asymmetrical evolution in gene duplication is common (Zhang et al. 2003; Cusack and Wolfe 2007; Han et al. 2009; Panchin et al. 2010; Pegueroles et al. 2013; Yampolsky and Bouzinier 2014) and with persistent, long-term faster evolution after a gene duplication in yeast (Scannell and Wolfe 2008). The observed persistence in the acceleration of evolution is surprising as it is not predicted by the more widespread theories of gene duplication. Most models of gene duplications that are based on a certain level of redundancy of gene duplications predict that the effect of the gene duplication event will be eliminated by accumulating mutations relatively quickly (Walsh 1995; Stoltzfus 1999; Lynch and Conery 2003; Konrad et al. 2011; Proulx 2012). Our data suggest that one of the gene copies seems to acquire some property other than redundancy, such as a new function (Notebaart et al. 2005; Assis and Bachtrog 2013), either at the duplication event or later, that results in a persistent faster rate of evolution than the ancestral single copy gene, or its ancestrally positioned paralog.
The following view on gene duplication evolution emerges from our data. First, immediately after gene duplication both gene copies experience an initial acceleration in their rate of evolution. Subsequently, one of the gene copies, which is typically the copy in the original genomic location, quickly returns to the preduplication levels in the timeframe of ds ∼ 0.15. The second copy in the novel location eventually (ds > 0.2) achieves a stationary evolutionary rate. However, it does not appear to reach the preduplication levels of evolution even after a substantial period of time (ds > 0.6). Furthermore, the novel copy takes longer to reach the new stationary rate of evolution than is necessary for the old copy to return to the preduplication rates. Unfortunately, due to the scarcity of our data we cannot address the issue of deviation of specific gene copies from the described course of evolution. We believe that the functional characterization of individual cases may reveal the properties of the new gene copies that result in their persistent accelerated evolution.
Acknowledgments
The work was supported by a grant of the HHMI International Early Career Scientist Program (55007424), the Spanish Ministry of Economy and Competitiveness (EUI-EURYIP-2011-4320) as part of the EMBO YIP program, two grants from the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013–2017 (Sev-2012-0208)” and (BFU2012-31329), the European Union and the European Research Council under grant agreement 335980_EinME.
Literature Cited
- Assis R, Bachtrog D. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A. 2013;110:43. doi: 10.1073/pnas.1313759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB. The bar “gene”—a duplication. Science. 1936;83:210–211. doi: 10.1126/science.83.2148.210. [DOI] [PubMed] [Google Scholar]
- Chatterjee HJ, Ho SY, Barnes I, Groves C. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol. 2009;9:259. doi: 10.1186/1471-2148-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH. Not born equal: increased rate asymmetry in relocated and retrotransposed rodent gene duplicates. Mol Biol Evol. 2007;24:679–686. doi: 10.1093/molbev/msl199. [DOI] [PubMed] [Google Scholar]
- Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:E55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Adaptive evolution of genes and genomes. New York: Oxford University Press; 1999. [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Johnston CR, O’Dushlaine C, Fitzpatrick DA, Edwards RJ, Shields DC. Evaluation of whether accelerated protein evolution in chordates has occurred before, after, or simultaneously with gene duplication. Mol Biol Evol. 2007;24:315–323. doi: 10.1093/molbev/msl162. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Wolf YI, Koonin EV. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002 doi: 10.1186/gb-2002-3-2-research0008. 3. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A, Teufel AI, Grahnen JA, Liberles DA. Toward a general model for the evolutionary dynamics of gene duplicates. Genome Biol Evol. 2011;3:1197–1209. doi: 10.1093/gbe/evr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Molecular evolution. Sunderland (MA): Sinauer; 1997. [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary demography of duplicate genes. J Struct Funct Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- Nembaware V, Crum K, Kelso J, Seoighe C. Impact of the presence of paralogs on sequence divergence in a set of mouse-human orthologs. Genome Res. 2002;12:1370–1376. doi: 10.1101/gr.270902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notebaart RA, Huynen MA, Teusink B, Siezen RJ, Snel B. Correlation between sequence conservation and the genomic context after gene duplication. Nucleic Acids Res. 2005;33:6164–6171. doi: 10.1093/nar/gki913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Panchin AY, Gelfand MS, Ramensky VE, Artamonova II. Asymmetric and non-uniform evolution of recently duplicated human genes. Biol Direct. 2010;5:54. doi: 10.1186/1745-6150-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Albà MM. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol. 2013;30:1830–1842. doi: 10.1093/molbev/mst083. [DOI] [PubMed] [Google Scholar]
- Ponting CP. The functional repertoires of metazoan genomes. Nat Rev Genet. 2008;9:689–698. doi: 10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- Proulx SR. Multiple routes to subfunctionalization and gene duplicate specialization. Genetics. 2012;190:737–751. doi: 10.1534/genetics.111.135590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Wolfe KH. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 2008;18:137–147. doi: 10.1101/gr.6341207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Large scale of human duplicate genes divergence. J Mol Evol. 2012;75:25–33. doi: 10.1007/s00239-012-9516-1. [DOI] [PubMed] [Google Scholar]
- Walsh JB. How often do duplicated genes evolve new functions? Genetics. 1995;139:421–428. doi: 10.1093/genetics/139.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky LY, Bouzinier MA. Faster evolving Drosophila paralogs lose expression rate and ubiquity and accumulate more non-synonymous SNPs. Biol Direct. 2014;9:2. doi: 10.1186/1745-6150-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang P, Gu Z, Li WH. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 2003;4:R56. doi: 10.1186/gb-2003-4-9-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]