Abstract
The Hippo/YAP pathway is an emerging signaling cascade involved in the regulation of stem cell activity and organ size. Alterations in Hippo signaling are also a common feature of human epithelial malignancies, although the molecular bases for this misregulation are unclear. As most of the current knowledge has been derived from work in the fruit fly, our understanding of mammalian Hippo/YAP signaling is still incomplete. To identify novel components of this pathway, we performed an RNAi-based kinome screen in human cells. Our screen identified several kinases not previously associated with Hippo signaling that strongly regulate the activity of the Hippo transducer YAP. Some of these kinases control processes such as response to stress, boundary formation, cell cycle and adhesion, and reflect novel inputs that may impinge on Hippo signaling and growth control. One of the hits, LKB1 (also known as Stk11), is a common tumor suppressor whose mechanism of action is only partially understood. We demonstrate that LKB1 acts through its substrates of the PAR-1 family (MARK1-4) to regulate the localization of the baso-lateral polarity complex and the activity of the core Hippo kinases. Murine and human LKB1-deficient tumors exhibit mislocalization of the basolateral determinant Scribble, reduced Hippo kinase activity, and enhanced YAP-driven transcription. Using xenograft assays and genetic analysis, we demonstrate that YAP is functionally important for the tumor suppressive effects of LKB1. Our results identify an important signaling axis that links YAP activation with LKB1 mutations, and have significant implications for the treatment of LKB1-mutant human malignancies. Additionally, our findings provide novel insight into the nature of inputs that speak to the Hippo/YAP signaling cascade.
Our understanding of human disease has benefited greatly from the study of developmental pathways in model organisms. Characterization of signaling cascades such as Wnt, Hedgehog, and Notch has particularly contributed to the understanding and treatment of cancer1. A more recently discovered signaling cascade is the Hippo pathway, originally described in Drosophila, and proposed to be a means by which organ size can be regulated. This pathway is highly conserved in mammals, where the mammalian hippo orthologues, MST1/2, phosphorylate the large tumor suppressor (LATS1/2) kinases, which in turn phosphorylate the transcriptional co-activator YAP, restricting its activity and stability2–4. In the absence of phosphorylation, YAP translocates to the nucleus where it binds to the TEA-domain transcription factors (TEAD1-4)5,6. Activation of YAP, or loss of upstream negative regulators leads to striking overgrowth and tumor phenotypes in epithelial tissues, in many cases driven by the expansion of tissue-resident stem cells3,4. Additionally, studies of human samples have demonstrated widespread Hippo pathway inactivation and nuclear YAP localization in multiple epithelial malignancies7–9. However, genomic analyses of common epithelial cancers have not revealed a significant rate of mutations in the known components of the pathway10. Recent data also suggest the presence of alternative kinases that might be responsible for YAP regulation9,11. Thus, common alterations of Hippo signaling in human cancer might be caused by mutations in genes currently not associated with the pathway.
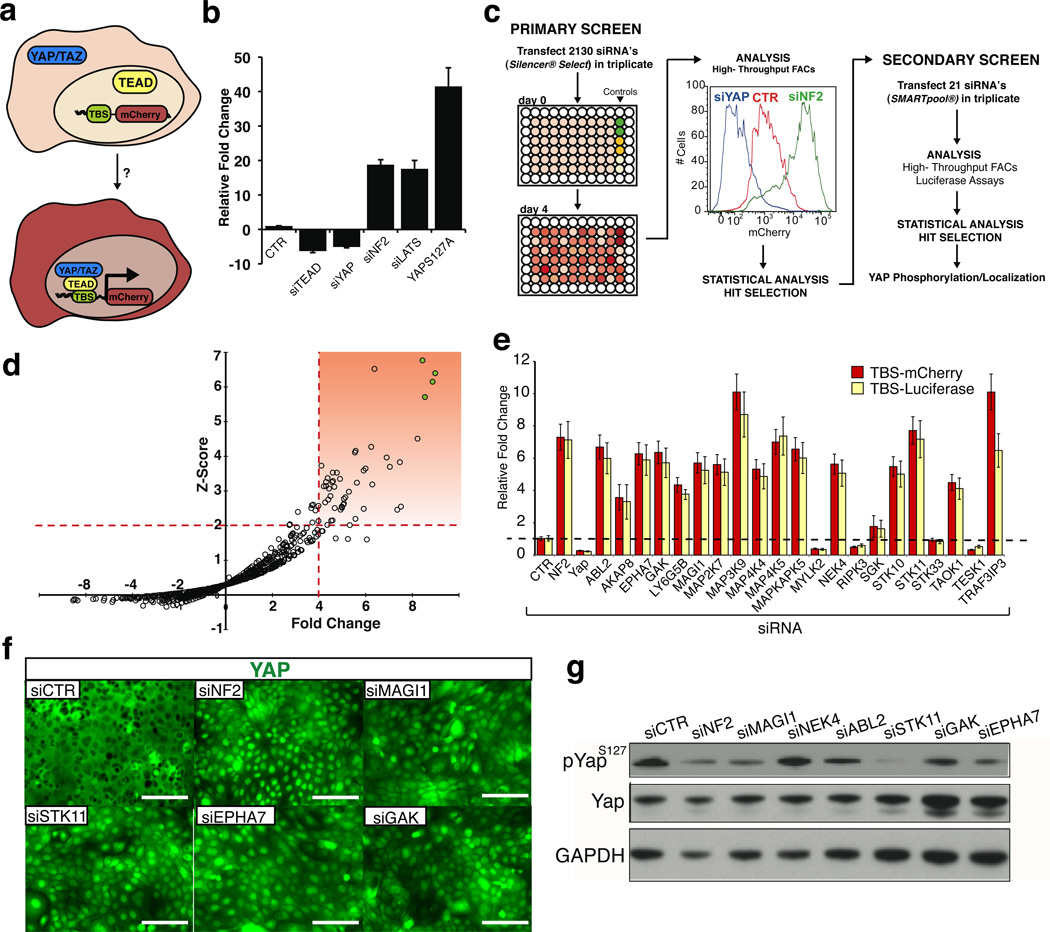
To identify potential kinases that can repress YAP/TEAD activity, we developed an improved transcriptional reporter containing 14 copies of the known TEAD DNA-binding sequence (Super TBS reporter) (Fig 1A)11. Functional assays revealed that this reporter faithfully recapitulated YAP/TEAD transcriptional activity, and was highly responsive to perturbations of endogenous upstream Hippo components such as LATS2 and the cytoskeleton-associated protein NF212,13 (Fig. 1B). Armed with a robust reporter for Hippo/YAP activity, we interrogated the effects of a human kinome siRNA library containing 2130 unique siRNA oligos for 710 kinase genes in a 293T cell line stably carrying the reporter (Fig 1C). Initial hits were identified by a statistical Z-score cutoff of 2 in addition to a > 4 fold-change of mean fluorescence intensity compared to scrambled siRNA controls (Fig 1D). Our high stringency statistical analysis revealed 21 kinases whose silencing resulted in enhanced STBS reporter activity (Fig 1D, Table S1). Through a secondary screen using a different commercial source of siRNAs to control for off-target effects, we confirmed that knockdown of 16 of these kinases robustly induced STBS-reporter activity (Fig 1E). Loss of 13 of these kinases also led to YAP nuclear accumulation even in high-density conditions where Hippo signaling is typically activated (Fig 1F, S1A). To further characterize these hits, we evaluated their effects on YAP phosphorylation at Serine 127 (S127), as this is a highly-conserved direct-substrate site for LATS1/2 and is one of the best characterized biochemical markers for Hippo-mediated YAP inactivation14. Silencing of eight of the 16 kinases resulted in decreases in YAPS127 phosphorylation (Fig 1G, S1B), indicating that some of these molecules regulate YAP activity independently of Hippo.
Figure 1. Kinome RNAi screen identifies novel regulators of Hippo/YAP signaling.
A) Graphical representation of Yap/Taz mediated STBS reporter activation in vitro. B) Validation of STBS reporter sensitivity using siRNA knockdown of known components of Hippo signaling. CTR=Scrambled negative control. C) Schematic of RNAi screening strategy. The RNAi screen was performed in 96 well plates using a stably expressing 293T-STBS-mCherry reporter cell line. Activation of the STBS-mCherry reporter was visualized 4 days following siRNA transfection. Fluorescence intensity was captured by flow cytometry. Statistical analysis was performed to identify genes for secondary screening and final selection of hits. D) Mean Z-score and mCherry reporter fold change (vs. scrambled controls) values for each triplicate siRNA oligo were plotted to identify hits that statistical thresholds of Z-score >2 and fold change greater than 4. Highlighted box represents hits satisfying these thresholds. Green filled circles represent siRNA knockdown of LATS2 as a positive control. E) A secondary siRNA screen identifies kinases that reproducibly raise STBS-mcherry reporter activity, performed using an alternative siRNA oligo source using two reporter systems. F) YAP immunolocalization in HaCaT cells following siRNA knockdown of kinases that regulate STBS reporter activity. G) Immunoblot for S127 Yap phosphorylation following siRNA knockdown of kinases from secondary screen. CTR represents Scr siRNA and NF2 siRNA is used as a positive control. Error bars represent ± SD from triplicate samples. Scale bars, 200 µm
Interestingly, four of the validated kinase hits (MAP2K7, MAP3K9, MAP4K4, MAP4K5) are part of an activating network of the c-Jun N-terminal kinase (JNK) branch of the mitogen activated kinase (MAP) pathway, a stress-activated cascade implicated in compensatory growth and tumorigenesis15. Silencing of these kinases does not lead to a reduction in YAP S127 phosphorylation, indicating a potentially new mode of YAP regulation (Fig S1B). A targeted analysis using RNA-interference and small molecule manipulation confirmed that only the JNK arm of the MAP kinase pathway controlled YAP/TEAD reporter activity (Fig S2A, S2B). While the role of JNK signaling in cancer is complex, our data support emerging findings suggesting that JNK activators are tumor suppressors, and implicate Hippo/YAP signaling as a downstream mechanism16,17. The ephrin receptor EPHA7 (Fig 1D-G), implicated in providing cell-positioning cues during development and mutated in lung cancer and lymphomas18,19, also regulates YAP activity. Intriguingly, other Ephrin-type A receptors (EPHA4, EPHA5, and EPHA8; Table S2) are also found to enhance STBS activity, indicating an important crosstalk between Ephrin signaling and Hippo. We also identify MAGI1 (Fig 1E-G, Table S1), a growth suppressive kinase also mutated in multiple human cancers10,20. GAK, a protein involved in clathrin-mediated endocytosis is also a hit21, as are the microtubule regulating kinases NEK4 and TESK122. Among the other regulators, a recently described Hippo-regulating kinase, TAOK1, was also identified (Fig 1, S1A,B, Table S1) 23.
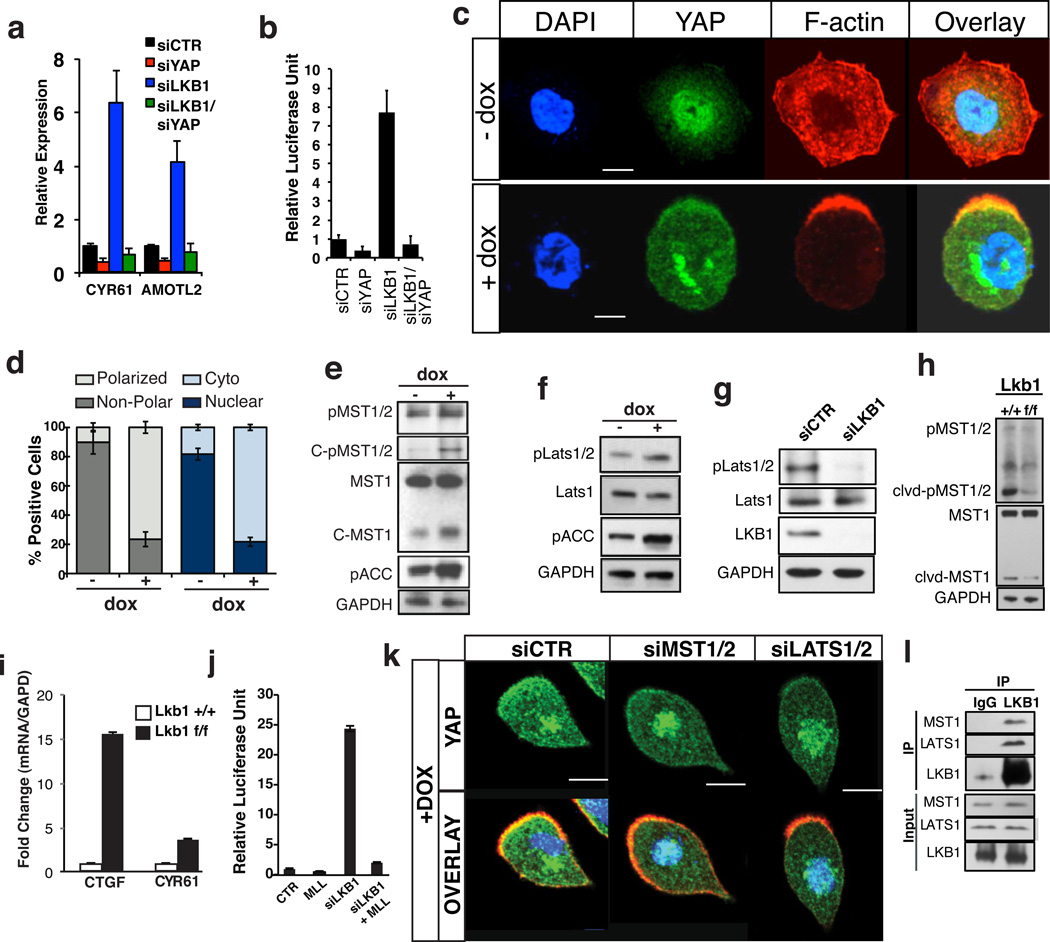
We were particularly interested by the fact that YAP phosphorylation was significantly repressed by STK11 knockdown (Fig 1E-G). STK11, also known as LKB1, is a well-established human tumor suppressor that controls, among other things, cellular metabolism, proliferation, and polarity24. The effect of LKB1 knockdown on YAP phosphorylation and localization was reproduced with multiple oligos and cell lines (Fig S3A-D). LKB1 knockdown also resulted in the upregulation of known YAP target genes, such as AMOTL2 and CYR616 (Fig 2A). This transcriptional response was entirely YAP-dependent, as endogenous target gene and reporter responses were suppressed in YAP/LKB1 double knockdown cells (Fig 2A, B, S3E).To further demonstrate a regulatory role of LKB1 upstream of YAP we utilized an engineered intestinal epithelial cell line (W4) in which LKB1-activity could be induced following treatment with Doxycycline (Dox)25. Dox-dependant LKB1-activity is evidenced by polarization and actin cytoskeleton rearrangements (Fig 2C, D). While YAP is predominantly nuclear at low cell densities, stimulation of LKB1 activity induced a striking and significant shift of YAP localization into the cytoplasm and actin cap of polarized cells (Fig 2C, D). Consistent with this, we observed significant reduction of YAP/TEAD transcriptional activity in Dox-treated cells (Fig S3F). Our results are consistent with a recent report indicating YAP activation in LKB1-mutant cell lines26.
Figure 2. LKB1 regulates YAP activity through the Hippo kinases.
A, B) LKB1 knockdown induces YAP target gene expression of AMOTL2and Cyr61 and TEAD reporter activation and is dependent on Yap expression. C) Immunofluorescence for F-actin (red), Yap (green) and nuclei (blue) in LS174T (W4) cells. Dox-inducible LKB1 activation after 24 hours results in LKB1-dependent cell polarization and Yap nuclear to cytoplasmic translocation. D) Quantification of cell polarization and Yap subcellular localization following Dox administration. E) MST1 activity in W4 cells is induced upon LKB1 activation (+Dox). Note increased MST1/2 phosphorylation in the full length and cleaved forms of MST1 and increase in levels of cleaved active MST1 peptide. F) Activity of Lats1/2 is increased upon LKB1 activation as measured by phosphorylation at Thr1079 LATS1/2. G) LATS1/2 phosphorylation at Thr1079 is abolished upon siRNA knockdown of LKB1 in MCF7 cells. H, I) Western blot analysis and qPCR of Yap target gene performed on liver lysates derived from Adeno-cre infected Lkb1 +/+ or Lkb1f/f mice 3months post infection. Both lines of mice also carried a p53 homozygous floxed allele. LKB1 deficiency leads to an overall decrease in cleaved activated MST1 and Thr183/Thr180 MST1/2 phosphorylation and increase in CTGF and CYR61 expression. J) Overexpression of LATS1, LATS2 and MOB1 in LKB1 knockdown 293T cells can restore STBS reporter activity. K) Knockdown of MST1/2 and LATS1/2 in Dox treated W4 cells suppresses LKB1-driven cytoplasmic translocation of YAP (green) when compared to the scrambled negative control (siCTR). L) Endogenous co-immunoprecipitation experiments using 293T cells expressing demonstrate physical association of Lkb1 with LATS1 and MST1. Error bars represent ± SD from triplicate samples. **, P≤0.01. Scale bars, 20 µm
We next determined whether LKB1 acts through the canonical Hippo kinases to regulate YAP. We observed increased MST1 activity, as measured by phosphorylation and the presence of a cleaved MST1 catalytic fragment following LKB1 activation in W4 cells (Fig 2E). Similarly, LKB1 activation led to a marked increase in phosphorylation of Threonine 1079 in LATS1/2 (Fig 2F). This residue marks LATS1/2 activation by MST1/2 and its co-activator Salvador14. Correspondingly, LKB1 silencing led to loss of LATS1/2 T1079 phosphorylation (Fig 2G). To confirm that LKB1 is important for MST1/2 activation, we utilized a mouse model in which Lkb1 was deleted in the liver via adeno-Cre. In agreement with MST1/2 loss-of-function phenotypes9, LKB1 deletion resulted in hepatomegaly and increased hepatocyte proliferation (Fig S4). As predicted, we also observed a significant decrease in the amount of cleaved and phosphorylated MST1 peptide in Lkb1-deficient livers and upregulation of YAP target genes (Fig 2H, I S4B). Supporting our findings that LKB1 acts upstream of the Hippo kinases, we find that expression of LATS1/2 and its co-activator Mob1 rescues the increase in YAP/TEAD transcriptional activity following knockdown of LKB1 (Fig 2J, S4E). Furthermore, knockdown of MST1/2 or LATS1/2 in Dox-treated W4 cells significantly suppress the LKB1-mediated shift in YAP subcellular localization (Fig 2K, Fig S5A-E). Supporting a regulatory role, we find that endogenous and overexpressed LKB1 can strongly interact with both LATS1 and MST1 in co-immunoprecipitation experiments (Fig 2L, S5F-G). support
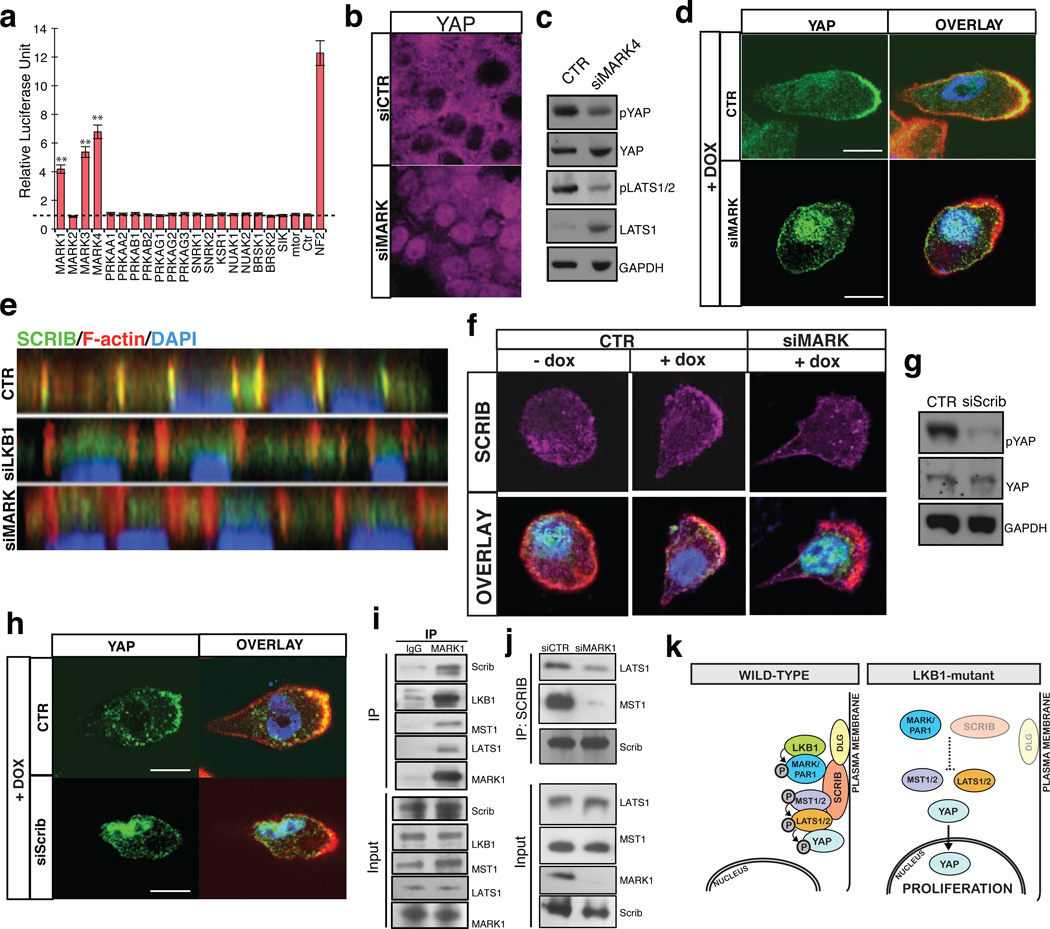
To shed light on a possible mechanism for regulation, we performed in vitro kinase assays and mass spectrometry analyses to determine whether MST1 or LATS2 could be direct targets of LKB1. Our results found no evidence for LKB1-mediated phosphorylation at potential consensus sites in either MST1 or LATS2 (data not shown), thus suggesting that the LKB1 effect on these kinases was indirect. We then performed a siRNA mini-screen evaluating most known downstream targets of LKB127, including AMPK and mTOR, commonly implicated in growth suppression by LKB1, for their ability to regulate the STBS reporter. This screen revealed that three members of the microtubule associated regulatory kinase (MARK) family (MARK1, 3 and 4; hereafter referred as MARKs) were able to robustly modulate TEAD-reporter activity (Fig 3A). These kinases are also hits in our primary kinome screen if lower hit-thresholds are selected (Table S2). The effect of MARKs knockdown was reproduced across several cell-types and with multiple oligos (Fig. S6A, B), and its effect on TEAD-reporter activity was also suppressed with concomitant knockdown of YAP (Fig S6C). Loss of MARK4 also results in enhanced YAP nuclear localization (Fig 3B) and decrease in LATS and YAP phosphorylation (Fig 3C). Suggesting that MARKs also act upstream of the Hippo kinases, overexpression of LATS and Mob1 can fully suppress the MARK4 knockdown effect on TEAD-reporter activity (Fig S6D).To ascertain whether MARKs were functionally downstream of LKB1, we knocked down MARKs in LKB1-induced W4 cells. Dox addition to W4 cells leads to MARK1 activation27(Fig S6E) and silencing of MARKs in this context resulted in a significant loss of cytoplasmic YAP translocation (Fig 3D, Fig S6F-H). Combined, these data demonstrate that LKB1 is exerting its effects on the Hippo pathway through its direct substrate, the MARKs.
Figure 3. MARKs/SCRIB act downstream of LKB1 to regulate Hippo/YAP.
A) Small scale RNAi screen on downstream substrates of LKB1 in 293T STBS-luc cells. Error bars represent ± SD from triplicate samples. B, C) Nuclear Yap accumulation and decreases in Lats and Yap phosphorylation following knockdown of MARKs. D) Suppression of LKB1-driven cytoplasmic translocation of YAP following MARK4 knockdown in Dox-treated W4 cells. Scale bars, 20 µm. E) Confocal immunofluorescent and Z-stack analysis for Scribble (SCRIB, green), F-actin (red) and nuclei (blue) in LKB1 and MARKs knockdown in MCF7 cells. Note mislocalization of SCRIB following LKB1 or MARKs silencing. F) Immunofluorescence in W4 cells demonstrates that LKB1 activation leads to SCRIB re-localization to the cell membrane and actin cap and require MARK expression. G) Knockdown of SCRIB in 293T cells reduces S127 Yap phosphorylation. H) Knockdown of SCRIB in Dox-induced LKB1 activated W4 cells suppressed YAP re-localization to the cytoplasm/actin cap. Scale bars, 20 µm. I) Endogenous co-immunoprecipitation of MARK1 demonstrates biochemical interactions with LKB1, SCRIB, MST1 and LATS1. J) Immunoblot for MST1 and LATS1, in SCRIB immunopreciptates derived from 293T cells with concomitant knockdown of MARK1. Adjusted lysate amounts were used for the IP to obtain equal levels of immunoprecipitated SCRIB. K) Diagram showing LKB1/MARK signaling axis controlling SCRIB localization and MST/LATS activation.
MARKs are also known as the PAR-1 family of proteins and have been implicated in the regulation of cell polarity and microtubule dynamics through different mechanisms28. In Drosophila, the PAR-1orthologue has been shown to phosphorylate and regulate localization of Discs large (Dlg)29, a member of the basolateral polarity complex also consisting of Lethal giant larvae (LGL) and Scribble (SCRIB)30,31. Proper localization of the basolateral polarity complex is required for Hippo pathway activity in both Drosophila and mammalian cells32–34. Thus, we posited that LKB1 could be regulating Hippo/YAP activity through regulation of the basolateral polarity complex via the MARKs. Indeed, we find that human MARK1 can also phosphorylate DLG (S7A), and that LKB1 or MARKs knockdown results in mislocalization of SCRIB (Fig 3E, S7B), and reduction of both SCRIB and DLG protein levels (Fig S7B-E). Demonstrating a direct role for LKB1 and MARKs in the localization of SCRIB, Dox-mediated activation of LKB1 in W4 cells results in SCRIB recruitment to the cellular membrane and the actin cap (Fig 3F). Knockdown of MARKs in this context reduces the sub-cellular localization shift of SCRIB (Fig 3F). As predicted, SCRIB knockdown also leads to an increase in TEAD-reporter activity and a decrease in YAP phosphorylation (Fig 3G, S8A, B). Importantly, knockdown of SCRIB in LKB1-activated W4 cells significantly rescues the shift of YAP localization to the cytoplasm and actin cap (Fig 3H, S8C-E), indicating that SCRIB is critical for LKB1-mediated regulation of YAP. Moreover, co-immunoprecipitation experiments demonstrate that endogenous MARK1 or expressed MARK4 physically associates with LKB1, MST1, LATS1 and SCRIB (Fig 3I, S8F), indicating the existence of a Hippo regulatory protein complex. It has been proposed that association of SCRIB with MST1/2 is important for the activation of the Hippo cascade34. We find that this association is highly dependent on MARKs (Fig 3J), as their loss impairs the interaction of both MST1/2 and LATS1/2 with SCRIB. Our data provide insight into a novel signaling axis downstream of LKB1 and PAR-1 kinases that regulates the interaction of the Hippo kinases with components of the basolateral polarity complex (Fig 3K). MARKs can also lead to changes in polarity by antagonizing the Par-3/Par-6 polarity complex35. Whether this and other substrates of the MARKs are also involved in controlling SCRIB remains to be investigated. A connection between Hippo/YAP signaling and the actin cytoskeleton has been recently demonstrated36. Considering that LKB1 and SCRIB have effects on the actin cytoskeleton 37,38,it is possible that actin fiber regulation could be an additional mechanism by which LKB1 modulates YAP activity.
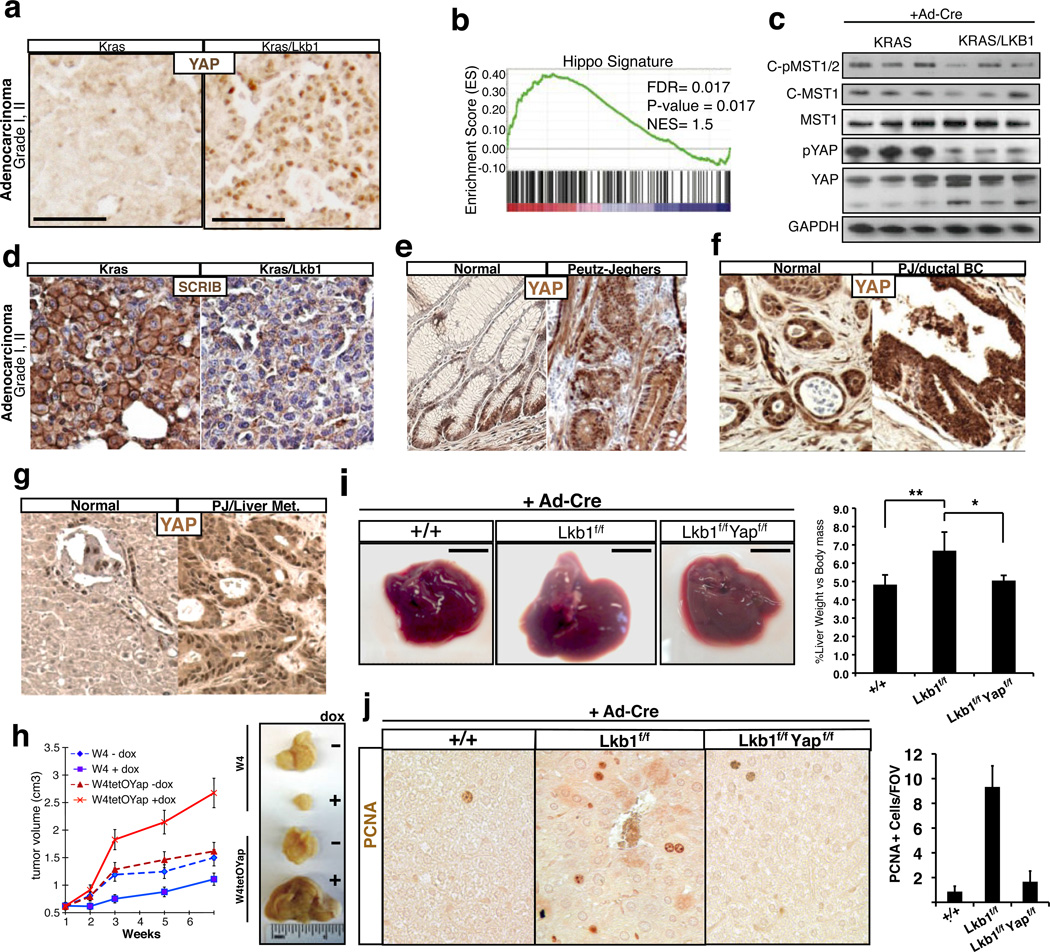
Lkb1 germline mutations are associated with Peutz-Jeghers Syndrome (PJS), an inherited disorder in which patients develop intestinal polyps and are at higher risk for developing multiple malignancies39. Lkb1 alterations are also present in many types of sporadic epithelial cancers, particularly lung and pancreatic carcinomas39. Loss of Lkb1 in mice is associated with more aggressive and metastatic potential of lung tumors 40. To corroborate our in vitro observations, we evaluated the status of Hippo signaling in lung tumors derived from mice carrying an activating K-Ras mutation (K) or the K-Ras transgene and concomitant LKB1 deletion (KL). Strikingly, we find that stage-matched KL adenocarcinomas were strongly positive for nuclear YAP in contrast to K tumors, which display predominantly cytoplasmic and diffuse YAP localization (Fig 4A). To further assess the extent of YAP transcriptional activity in Lkb1-null tumors, we carried out Gene Set Enrichment Analysis (GSEA) to examine the enrichment of a YAP transcriptional signature derived in our laboratory (Fig. S9). GSEA demonstrates a highly significant enrichment of this YAP signature in KL tumors (Fig 4B). Furthermore, biochemical analyses of tumor nodules also demonstrate decreased MST1 and YAP S127 phosphorylation in the KL genotype (Fig 4C). Furthermore, as predicted from our model, SCRIB localization is drastically altered and its expression reduced in KL tumors (Fig 4D). We also evaluated YAP status in a model of pancreatic neoplasia derived from tissue-specific deletion of Lkb1. Consistent with the lung tumor data, Lkb1-null pancreatic ductal adenocarcinomas exhibit robust YAP nuclear localization compared to control tissue (Fig S10A). Moreover, we find that gastrointestinal polyps of human PJS patients display an increase in nuclear YAP localization in both epithelial and smooth muscle cells compared to normal colon or juvenile polyposis polyps carrying SMAD4 mutations (Fig 4E, S10B). Examination of a malignant ductal breast adenocarcinoma and metastatic liver disease that developed in a PJS patient further revealed strong YAP nuclear accumulation in the tumor (Fig 4F, G). Taken together, these data show that genetic deletion of Lkb1 in both murine and human tissue leads to enhanced nuclear YAP activity.
Figure 4. YAP activity in LKB1 deficient tumors.
A) Immunohistochemistry for YAP on Ad-cre administered KrasG12D mutant (K) and KrasG12D/LKB1fl/fl (KL) on Grade I, II lung adenocarcinoma. Representative picture shown, n = 5 for each genotype. B) Gene set enrichment using a Hippo signature on K versus KL murine lung tumors. C) Western blot analysis for activated, phosphorylated and cleaved, MST1/2 and S127 Yap phosphorylation on K and KL murine lung nodules (n=3 mice). D) Immunohistochemistry for Scribble localization in K and KL lung adenocarcinomas, (n=5 mice). E) YAP localization assessed by immunohistochemistry in human intestinal tissue and Peutz-Jeghers (PJ) intestinal polyps. Representative data, n=3. F) Immunohistochemistry for YAP localization and expression in normal ductal tissue compared with ductal breast adenocarcinoma, and in normal human liver compared with metastatic liver adenocarcinoma derived from a Peutz-Jeghers syndrome patient (G). H) Subcutaneous xenograft assay using W4 cells, and W4 cells that co-express doxycycline inducible YapS127A. Tumor volumes for non-induced and induced tumors are shown. Representative tumors from non-induced and induced W4 and W4TetOYap xenografts are displayed. I-J) Genetic deletion of LKB1 and YAP1 in the liver following Ad-cre display restoration of organ size and hepatocyte proliferation. Animals received Ad-Cre at 1 month of age and tissues were collected 2.5 months later. Error bars represent ± SD from n=5 mice.
We next investigated functionally whether YAP acted downstream of LKB1 in tumor suppression. Using W4 cells, we found that inducible LKB1 activation has a powerful growth suppressive function in vitro (Fig S11A, B), and in xenografts (Fig 4H, S11C). However, expression of a YAPS217A mutant protein is able to significantly overcome all of LKB1 tumor suppressive effects (Fig 4H, S11). Silencing of either LATS2 or SCRIB also rescues growth suppression by LKB1 activation (Fig S12). To test if we could reverse the effects of Lkb1-loss by manipulating YAP-expression levels, we developed a doxycycline inducible-YAP shRNA A549 cell line (Fig S13A). A549 is a lung cancer cell line mutant for Lkb1 widely used in tumor growth and metastasis assays40. In both a soft-agar colony formation assay, and in vivo metastatic assays, we find that Yap-loss following Dox-treatment reduces the number and/or size of colonies and tumors (Fig S13A-E). Lung adenocarcinoma cell lines that are wild type for LKB1 and expressed lower levels of YAP1 were insensitive to YAP modulation (S13F-I). Finally, we utilized Ad-Cre infection in mice to demonstrate that conditional deletion of Yap1 suppresses the liver overgrowth phenotype (Fig 4I, S13J) and hepatocyte hyperplasia observed following acute deletion of Lkb1 (Fig 4J).Together these data provide multiple lines of evidence that Hippo/YAP is a functionally critical pathway downstream of of LKB1.
One important question in the Hippo/YAP field relates to the upstream signals that regulate the Hippo kinases. Our studies here have identified many novel molecules and pathways that might impinge on Hippo activity and growth control. As many of these kinases are also mutated in human cancer, their identification as regulators of YAP might provide a molecular explanation for the observations that YAP is highly active in numerous epithelial tumors, where mutations in the canonical Hippo components are not found. The tumor suppressive function of LKB1 has primarily been linked to its ability to regulate cellular metabolism through AMPK activation41. We demonstrate that LKB1 can also exert its tumor suppressive effects through activation of a PAR-1 mediated polarity axis that controls the Hippo signaling pathway. Manipulation of this signaling axis should be now evaluated for the treatment of LKB1 mutant tumors.
METHODS
RNAi screen
A human RNAi library against all known kinases in the genome was utilized (AMBION, silencer select). Reverse transfection (RNAi max, Invitrogen) of a final concentration of 5 nM siRNA was performed on approximately 10,000 HEK-293T-TBSmCherry cells per well of a 96-well plate. We estimate that close to 98% of 293T cells are transfected using this method. The screen was performed in triplicate, with 3 oligos for each gene. To ensure limited edge effects, outer rows and columns were not used. At four days post-transfection, cells in control wells are confluent. Plates are subsequently trypsinized and analyzed by flow cytometry (BD LSRII) to obtain the mean fluorescent intensity of each cell.
Hit Selection
Positive hits for each gene were identified as followed. Z-scores and fold changes were calculated for each oligo when compared to the negative control for each individual plate. Rigorous hit selection was performed by eliminating data that did not reproduce in at least two out of the three experiments for each oligo. Subsequently, these data were further filtered to identify oligos that reproducibly have a Z-score > 2 and fold change > 4. The final hit selection is based on the mean Z-score values and mean fold changes of each oligo for each gene, and if two or more oligo’s for each gene met these thresholds.
Immunofluorescence
Briefly, cells are plated on glass coverslips and then fixed in 4%PFA for 10 minutes at room temperature, followed by 3 washes in PBS. Cells are permeabilized in 0.01% Triton for 1 minute, followed by 3 washes in PBS. Cells are incubated in blocking buffer (0.5% FBS/PBS/0.01%Tween) for 1 hour, and then incubated overnight at 4°C in primary antibody (YAP- 1:1000, Cell Signaling, 1:400, Avruch, Scribble- 1:1000, Santa Cruz, GFP-1:500, Abcam) in blocking buffer. Coverslips are washed 3 times in PBS/0.01% Tween and then incubated in secondary antibody (1:5000, Molecular Probes) for 1 hour at room temperature. Coverslips are washed 3 times in PBS/0.01% tween and once in PBS prior to mounting on slides.
Immunohistochemistry
p53, p53/Lkb1 fl/fl mouse livers and Kras, Kras/Lkb1fl/fl mouse lungs were harvested and fixed in buffered formaldehyde (4%; pH 7.0; 16–24 h, 20°C), and then switch to 70% ethanol. Tissues were embedded in paraffin and 5 µm sections were mounted on positively charged slides. Tissues were deparaffinized and antigen retrieval was performed in pH 6.0 Citrate buffer for 30 minutes at 95°C. Endogenous peroxidase was blocked by incubation in 0.3% hydrogen peroxide. Sections were blocked in rabbit sera followed by primary incubation overnight at 4°C. Sections were washed and then incubated with HRP conjugated secondary antibody (30 min; 20°C), and development with 3,3′-diaminobenzidine tetrahydrochloride (DAB)/H2O2.Counterstaining was done with haematoxylin and samples were washed, dehydrated, and mounted with vectamount.
Immunoblotting
Cell lines and tissues were harvested, and processed for Western blotting by solubilizing extracts in lysis buffer (50mM Tris, 100mM NaCl, protease inhibitor cocktail, phosSTOP) and then resolved by PAGE under reducing conditions (4–12% SDS-PAGE Bis-Tris gels; MOPS buffer system) (Invitrogen; NuPAGE-MOPS system). The gels were blotted onto PVDF or nitrocellulose papers blocked (phospho-antibodies: pYap, pMST1/2, pLats1/2 PBS; 5% w/v BSA; 0.1% Tween-20, standard antibodies, TBS-T, 5% w/v milk), and the transferred proteins probed with the primary antibodies diluted in TBS-T, 5% w/v milk. After washing in TBS-T, antigens were detected using HRP-conjugated secondary antibodies (1:20,000 in TBS-T: Thermo), and visualized using enhanced chemoluminescence.
Microarray and Gene Set Enrichment Analysis
Reverse transfection of RNAi oligos for NF2, LATS2, YAP1, TAZ, and Scrambled RNAi was performed in HEK293T, HaCaT, and DLD1 cells with RNAiMAX (Invitrogen) according to manufacture’s instruction. Duplicate samples were prepared for each condition. Four days after transfection, confluent cell culture was harvested for RNA extraction with the Trizol reagent (Invitrogen). RNA quality assessment, cDNA synthesis, probe generation, array hybridization and scanning were carried out by Children’s Hospital Boston Molecular Genetics Microarray Core Facility. Datasets were analyzed with the on-line microarray analysis software GenePattern (Broad Institute) with default settings. Differentially expressed genes were defined as those with at least 2 fold change in NF2/LATS2 double knockdown cells and a p value smaller than 0.05. To generate a generic Hippo target gene signature, genes upregulated in all three NF2/LATS2 double knockdown cell lines were combined, and those without a gene symbol were eliminated from the list. For gene set enrichment analysis, we assembled a dataset from published gene expression profiles of lung adenocarcinomas developed in KrasG12D and KrasG12D LKBf/- or KrasG12D LKBf/f mouse models (supplemental table). The enrichment analysis was performed in the GSEA software available from the Broad Institute with default setting.
Cell Lines
LS174T, HEK293T, DLD1, MCF7, HaCaT cells were cultured in DMEM+10%FBS. A549 cells were cultured in RPMI supplemented with L-Glutamine and 10% FBS. The LS174T –W4 clone cell line was a generous gift from Dr. Hans Clevers (Utrecht Institute, Netherlands). Other cell lines were acquired from the American Type Culture Collection (Manassas, VA, USA).
Soft Agar Colony Formation Assay
Base agar consisted of low melt point 0.6% agar dissolved in RPMI or DMEM, 10% FBS, 1X Penicillin/Streptomycin. Top agar consisted of approximately 250 cells resuspended in 0.3% low melt point agar dissolved in RPMI or DMEM, 10% FBS, 1X Penicillin/Streptomycin in a well of a 6-well plate. Samples are incubated for 4 weeks following seeding and then stained with 0.1% crystal violet in 10% ethanol for 20 minutes.
In vitro Proliferation Assay
An MTS assay (Promega, Madison, WI USA) was used to determine the proliferation rate for the cell lines. For each cell line, 1000 cells per well were cultured in triplicate into 96-well plates and incubated for 0-7 days. At the time of culture and each day for a total of 7 days, a plate was analyzed by MTS assay. Absorption of light at 450 nm was measured as an assay of cell proliferation.
Mouse Models
P53flox/flox, LKB1flox/flox, LSL-KRASG12D, PDX-Cre have been previously described. Xenograft assays were performed in 5 week old male Nu/J mice (JAX labs, Bar Harbor, ME, USA), using 1x106 cells/100ul volume of matrigel (BD Biosciences, New Jersey, USA). Metastasis assays were performed in 5 week old male NOD/SCID mice (JAX labs, Bar Harbor, ME, USA) using 1x106 cells/100ul volume of PBS injected intravenously.
Tissue Samples
We studied formalin-fixed paraffin-embedded intestinal polyp sections of diagnosed Peutz-Jegher and Juvenille Polyposis patients with confirmed mutations in LKB1 and SMAD4, respectively.
Small Molecule Inhibitors
Metformin, 10 mM (EMD Millipore, Inc). AICAR, 2 mM (TOCRIS Biosciences, Inc.). Rapamycin, 1 mM (TOCRIS Bioscience, Inc.). MAP Kinase Signaling Pathway Inhibitor Panel was purchased from EMD Millipore (#444189) and incubated according to manufacturers recommended concentrations (FR180204: IC50 = 510 nM, JNK II: IC50 = 40 nM for JNK-1 and JNK-2 and 90 nM for JNK-3, JNK IX: pIC50 = 6.5, MEK1/2: IC50 = 220 nM, MK2a: IC50 = 60 nM, Inhibitor 5: IC50 = 40 nM, PD98059: IC50 = 10 uM, Inhibitor IV: IC50 = 10 nM, SB203580: IC50 = 600 nM, TPL2 Inhibitor: IC50 = 50 nM, ZM336372: IC50 = 70 nM).
Luciferase Assays
For stable cell lines expressing STBS-Luciferase, cell lysates were analyzed using Dual-glo luciferase assay (Promega, Inc.). Alternatively, cell lines not containing the reporter were transiently transfected with the STBS-firefly-Luciferase and firefly-Renilla constructs using Lipofectamine 2000 (Life Technologies, Inc.) and assayed 48 hours following plasmid transfection (Berthold Technologies, USA).
Supplementary Material
REFERENCES
- 1.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 2.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Current opinion in cell biology. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramos A, Camargo FD. The Hippo signaling pathway and stem cell biology. Trends in cell biology. 2012;22:339–346. doi: 10.1016/j.tcb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 6.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinhardt AA, et al. Expression of Yes-associated protein in common solid tumors. Human pathology. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, et al. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–2822. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 9.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catalogue Of Somatic Mutations In Cancer [Google Scholar]
- 11.Schlegelmilch K, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 14.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F. JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer research. 2012;72:379–386. doi: 10.1158/0008-5472.CAN-11-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nature genetics. 2012;44:165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schramek D, et al. The stress kinase MKK7 couples oncogenic stress to p53 stability and tumor suppression. Nature genetics. 2011;43:212–219. doi: 10.1038/ng.767. [DOI] [PubMed] [Google Scholar]
- 18.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oricchio E, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 2011;147:554–564. doi: 10.1016/j.cell.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaric J, et al. Identification of MAGI1 as a tumor-suppressor protein induced by cyclooxygenase-2 inhibitors in colorectal cancer cells. Oncogene. 2012;31:48–59. doi: 10.1038/onc.2011.218. [DOI] [PubMed] [Google Scholar]
- 21.Lee DW, Zhao X, Yim YI, Eisenberg E, Greene LE. Essential role of cyclin-G-associated kinase (Auxilin-2) in developing and mature mice. Molecular biology of the cell. 2008;19:2766–2776. doi: 10.1091/mbc.E07-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doles J, Hemann MT. Nek4 status differentially alters sensitivity to distinct microtubule poisons. Cancer research. 2010;70:1033–1041. doi: 10.1158/0008-5472.CAN-09-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boggiano JC, Vanderzalm PJ, Fehon RG. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev Cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baas AF, Smit L, Clevers H. LKB1 tumor suppressor protein: PARtaker in cell polarity. Trends Cell Biol. 2004;14:312–319. doi: 10.1016/j.tcb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Baas AF, et al. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell. 2004;116:457–466. doi: 10.1016/s0092-8674(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen HB, Babcock JT, Wells CD, Quilliam LA. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogene. 2012 doi: 10.1038/onc.2012.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lizcano JM, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. The EMBO journal. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurov J, Piwnica-Worms H. The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell cycle. 2007;6:1966–1969. doi: 10.4161/cc.6.16.4576. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. PAR-1 kinase phosphorylates Dlg and regulates its postsynaptic targeting at the Drosophila neuromuscular junction. Neuron. 2007;53:201–215. doi: 10.1016/j.neuron.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 31.Yamanaka T, Ohno S. Role of Lgl/Dlg/Scribble in the regulation of epithelial junction, polarity and growth. Front Biosci. 2008;13:6693–6707. doi: 10.2741/3182. [DOI] [PubMed] [Google Scholar]
- 32.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Current biology : CB. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 33.Parsons LM, Grzeschik NA, Allott ML, Richardson HE. Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly (Austin) 2010;4:288–293. doi: 10.4161/fly.4.4.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 35.Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- 36.Sansores-Garcia L, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. The EMBO journal. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Omelchenko T, Hall A. LKB1 tumor suppressor protein regulates actin filament assembly through Rho and its exchange factor Dbl independently of kinase activity. BMC cell biology. 2010;11:77. doi: 10.1186/1471-2121-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osmani N, Vitale N, Borg JP, Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Current biology : CB. 2006;16:2395–2405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26:7825–7832. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- 40.Ji H, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 41.Katajisto P, et al. The LKB1 tumor suppressor kinase in human disease. Biochimica et biophysica acta. 2007;1775:63–75. doi: 10.1016/j.bbcan.2006.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.