Abstract
The Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) family has a key role in parasite survival, transmission, and virulence. PfEMP1 are exported to the erythrocyte membrane and mediate binding of infected erythrocytes to the endothelial lining of blood vessels. This process aids parasite survival by avoiding spleen-dependent killing mechanisms, but it is associated with adhesion-based disease complications. Switching between PfEMP1 proteins enables parasites to evade host immunity and modifies parasite tropism for different microvascular beds. The PfEMP1 protein family is one of the most diverse adhesion modules in nature. This review covers PfEMP1 adhesion domain classification and the significant role it is playing in deciphering and deconvoluting P. falciparum cytoadhesion and disease.
Keywords: malaria, Plasmodium falciparum, antigenic variation, var, pathogenesis
Introduction
Cytoadhesion of Plasmodium falciparum infected erythrocytes (IE) is a major virulence determinant associated with pathological complications from IE binding to the endothelial lining of blood vessels [1]. Although this deadly parasite adhesion trait has been recognized for over a century [2], the molecular interactions involved in parasite binding in brain and other microvasculature are only partially understood. This deficiency exists in part because of the complexity of the var gene/P. falciparum erythrocyte membrane protein 1 (PfEMP1) family that mediates endothelial binding [3]. Each parasite genotype encodes approximately 60 var gene copies and there is limited overlap of var repertoires between parasite genotypes [4–6]. Switching between var genes modifies the antigenic and binding properties of IEs, and orchestrates parasite binding tropism for placenta [7] and possibly other microvascular sites [8].
PfEMP1 proteins evolve under opposing binding and antibody selection pressures. This has resulted in extensive diversification of PfEMP1 adhesion domains. Within the protein family, some binding properties are common to many PfEMP1 [9], while others are rare or may have evolved to exploit specialized microvascular niches (e.g. placental binding) [7;10]. A major issue for pathogenesis research is whether specific PfEMP1-host receptor interactions are involved in severe malaria and, if so, whether there are common pathogenic mechanisms that could be targeted for intervention. This review covers the introduction of a system of PfEMP1 adhesion domain classification [11] and its application to malaria disease research.
PfEMP1 adhesion domain classification
At the time of their discovery [12–14], a significant clue into PfEMP1 binding function was that they encode a recognizable binding module from Plasmodium erythrocyte invasion ligands, called the Duffy binding-like (DBL) domain [15;16]. This homology showed the PfEMP1 ectodomain contained multiple DBL domains and a new domain termed the cysteine-rich interdomain region (CIDR) [14]. Early sequence comparisons indicated that individual PfEMP1 domains maintained less than 50% amino acid identity and were much more divergent than DBL domains in erythrocyte invasion ligands [12–14]. The variability in PfEMP1 size and sequence suggested a potential explanation for parasite binding differences [17], but given the massive sequence diversity in the PfEMP1 family it was unknown if PfEMP1 binding was predictable or if there would be any disease binding patterns.
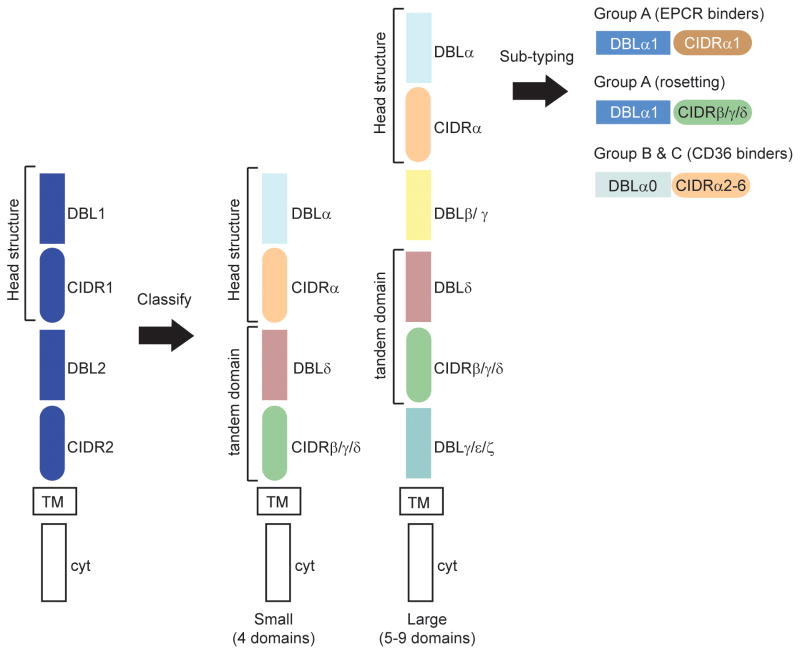
To investigate PfEMP1 structure and function, we used phylogenetic criteria to classify adhesion domains into different sequence types [11]. This analysis was performed on the first 20 var gene sequences in Genbank. It showed that DBL domains could be classified into four major types (α, β, γ, and δ) and CIDR domains into three major types (α, β, and γ). It also revealed higher domain organization in PfEMP1 proteins. Small PfEMP1 contained four extracellular domains; a DBLα-CIDRα tandem followed by a DBLδ-CIDRβ/γ tandem (Fig. 1). Large PfEMP1 proteins contained the same DBL-CIDR tandems, but had additional DBL domain types (β or γ) domains inserted before or after the C-terminal tandem (Fig. 1). The N-terminal DBL-CIDR tandem is the most conserved extracellular region and is referred to as the semi-conserved protein head structure [14]. Within a given var repertoire head structures maintain less than 50% amino acid identity highlighting the extensive diversification within the family [5].
Fig 1.
Adhesion domain classification of PfEMP1 proteins. The blue PfEMP1 shows a typical arrangement of PfEMP1 domains. The first arrow indicates how adhesion domain classification reveals higher domain organization in PfEMP1. Specific DBL and CIDR domain types form preferential tandem domain arrangements (DBLα-CIDRα and DBLδ-CIDRβ/γ/δ). The same tandem arrangements are present in small (4-domain) and large (5 or more domain) PfEMP1, but large proteins contain unique DBL subtypes (β or γ) that are not present in small proteins. The second arrow indicates that further sub-classification of adhesion domains (e.g. CIDRα1) distinguishes three different PfEMP1 head structure types and functional differences between group A, B, and C proteins. TM is transmembrane, cyt is cytoplasmic tail.
At the time of this adhesion domain classification, the CIDR domain in the semi-conserved head structure had already been shown to bind CD36 [18;19] and ICAM1 binding had been mapped to a DBLβ domain [20] (Fig. 2). However, it was not known what proportion of PfEMP1 variants encoded CD36 or ICAM1 binding activity or if binding was predictable. Notably, one of first twenty PfEMP1 proteins had a distinct protein head structure; a DBLα-CIDRγ tandem instead of the more characteristic DBLα-CIDRα tandem. This unusual DBLα-CIDRγ head structure was known to mediate “rosetting”, or the binding of IEs with uninfected red blood cells [21], but it was not known if it bound CD36. In addition, DBLβ domains were restricted to large PfEMP1 (Fig. 1). Together, these findings raised the questions whether small and large PfEMP1 encoded distinct binding properties and if adhesion domain classification could help predict PfEMP1 binding properties [11].
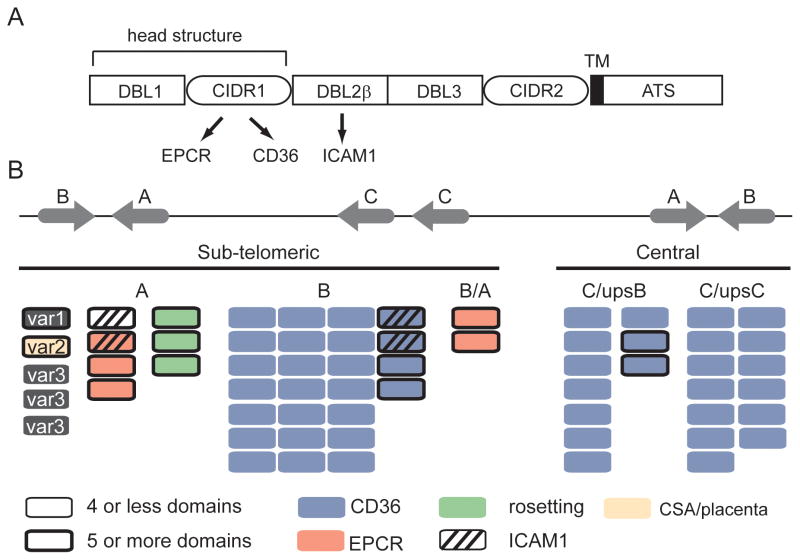
Fig 2.
The genome organization and predicted PfEMP1 binding properties of the 3D7 genome reference isolate. (A) Schematic representation of PfEMP1 domain architecture. The CIDR domain in the PfEMP1 head structure has diverged into CD36 (group B and C) and EPCR (group A) binding variants; binding is highly predictable by adhesion domain classification. ICAM1 binding has been mapped to specific DBLβ subtypes following the PfEMP1 head structure. (B) Group A and B var genes are located in sub-telomeric regions and group C var in central chromosome regions [4]. The predicted binding properties for the 3D7 genome reference isolate are shown based on adhesion domain classification and recombinant protein binding studies. Blue proteins encode CD36-binding CIDR domains [9], red proteins encode EPCR-binding CIDR domains [23], green proteins encode head structures associated with rosetting [27], striped proteins encode DBLβ3 or DBLβ5 domains associated with ICAM1 binding [37–39], and the beige protein (VAR2CSA) is linked to CSA binding in the placenta [7]. Large PfEMP1 proteins with 5 or more domains are distinguished by a thick border.
The initial observations on PfEMP1 architecture have largely held up [4–6]. While the number of DBL (α, β, γ, δ, ε and ζ) and CIDR (α, β, γ, δ, and PAM) sequence classes have slightly increased as more proteins have been analyzed, the same higher order domain organizations have been identified in geographically diverse parasites [6]. Although var repertoires are highly divergent, the majority of PfEMP1 are classified into three main groups on the basis of upstream sequence (ups) and chromosome location (Fig. 2) [22]. Group A (upsA) and group B (upsB) are present in subtelomeric regions and transcribed in opposite orientations. Group C (either upsC or upsB) are found in central chromosome regions. The var repertoire also contains three unusual strain transcendent variants (var1CSA, var2CSA, and type 3 var). Subsequent sequence comparisons have also led to further sub-classification of domains (e.g. CIDRα1.1) and the identification of PfEMP1 domain cassettes (DC), or tandem arrangements of two or more domains of particular subclasses (e.g. DC8) (Fig. 3) [6]. The fact that DCs are discernable despite extensive gene recombination in the var gene family suggests they may have important structural and functional significance.
Fig 3.
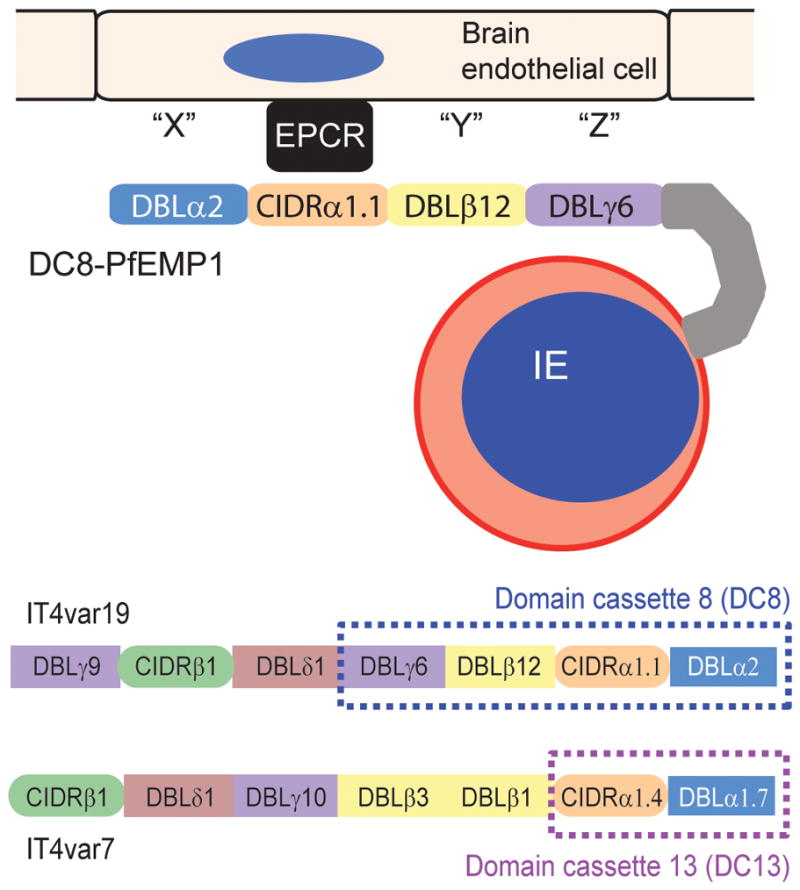
EPCR-binding PfEMP1 variants. The top shows a schematic of a DC8-expressing infected erythrocyte binding to brain endothelial cells. All four domains in the DC8 cassette encode binding activity for brain endothelial cells [44], EPCR binding maps to the CIDRα1.1 [23]. The bottom compares the protein architectures of a DC8 (IT4var19) and a DC13 (IT4var7) PfEMP1 variant, both of which have been shown to be selected on brain endothelial cells [42;44] and to encode EPCR binding activity in the orange colored CIDR domain [23]. Differences in domain composition between DC8, DC13, and other EPCR binding PfEMP1 variants have potential implications for the overall PfEMP1 binding specificities and parasite binding tropism for brain and other microvascular beds.
CIDR sequence classification and binding
To understand how the var gene repertoire has evolved under binding selection, it is valuable to understand one parasite genotype and determine if binding properties are predictable across parasites. The PfEMP1 head structure has an important role in IE binding and has provided proof of concept that CIDR sequence classification can predict binding differences.
The most common adhesion property of the CIDR domain is CD36 binding [18]. Based on repertoire-wide binding comparisons [9], over 80% of PfEMP1 proteins in the 3D7 genome reference isolate encode CD36 binding activity in the CIDR1 domain (Fig. 2). CD36 binding activity is limited to group B and C proteins [9;23] and associated with CIDRα2–6 sequence types. Remarkably, CD36 binding sequences only have ~40–50% amino acid identity [9] illustrating the predictive power of adhesion domain classification.
In contrast, Group A proteins have two distinct protein head structures; DBLα1-CIDRα1 and DBLα1-CIDRβ/γ/δ (Fig. 1) [6]. Group A proteins are of significant interest because they are commonly expressed in malaria naïve hosts and severe malaria [24–26], and recent analysis suggests the two group A head structures have diverged in binding properties. While CIDRα1 domains (subtypes 1.1, 1.4, 1.5, and 1.7) encode binding activity for endothelial protein C receptor (EPCR) [23], the DBLα1-CIDRβ/γ/δ head structure is linked to rosetting [27]. Both EPCR-binding and rosetting parasites have been associated with severe malaria [23;28]. Of interest, the CIDRβ/γ/δ domain type present in rosetting head structures are more typically found in the C-terminal tandem domain position (Fig. 1). The domain type did not bind CD36 or EPCR [9;23] and there has been no systematic analysis to determine its adhesion properties.
Together, these findings lead to the hypothesis that PfEMP1 head structures have diverged under selection for CD36, EPCR, and rosetting (Fig. 2B). Remarkably, there are essentially stable relationships between CIDR sequence classification and parasite binding properties over vast geographic distances, suggesting that adhesion domain classification reflects the evolutionary origin of ancient adhesion traits that spread throughout the parasite population. These findings raise questions, too, about the origins of CD36 and EPCR-binding parasites and whether related CIDR domains in the chimpanzee malaria P. reichenowii [6], or potentially other Laverania clade Plasmodium species [29], encode similar binding properties. Significantly, adhesion domain classification suggests that group A var genes have diverged from groups B and C in binding properties, and group A may have further diversified into EPCR binders and rosetting variants (Fig. 2B). The functional specialization of PfEMP1 subfamilies may be reinforced by gene recombination hierarchies [5;30], and this, in turn, may shape parasite binding tropism for different microvascular niches. In addition, it may not be coincidental that CD36 and EPCR are also expressed on many cells in the immune system. This may provide a mechanism for the parasite to modulate the innate or adaptive immune responses via these adhesive interactions [31].
Although CIDR sequence classification is predictive of domain binding, what we ultimately want to know is how accurately it predicts parasite binding. Using highly monoclonal parasite lines, there is a spectrum of CD36 binding from none (VAR2CSA), low (UpsA rosetting variants and EPCR binders), to moderate-high (group B and C) [32;33]. These findings suggest the CIDR domain makes a significant contribution to parasite-CD36 binding activity, but may not be the only binding partner. By comparison to group B and C expressing parasite lines, only a small number of group A-expressing parasite lines have been studied, and thus it will be important to validate how well EPCR-binding and IE rosetting are predicted by adhesion domain classification.
Adhesion domain classification and binding differences between small and large PfEMP1
Small and large PfEMP1 differ in domain composition (Fig. 1), suggesting the possibility that large proteins have unique binding properties. One example is the interaction between DBLβ domains and intercellular adhesion molecule 1 (ICAM1). ICAM1 can act in cooperation with CD36 to mediate binding of IEs under flow-based adhesion conditions [34], and has been proposed to be a candidate receptor for cerebral malaria [35;36]. ICAM1 binding is linked to the DBLβ domain immediately following the PfEMP1 head structure (Fig. 2), but not all DBLβ-containing PfEMP1 bind ICAM1 [20;37–39]. ICAM1 binding has been associated with DBLβ5 in group B and C proteins and DBLβ3 in group A proteins [20;37–39]. Whereas ICAM1 is a long molecule (>18nm) that has a bent rod shape and sticks out above the cellular glycocalyx, CD36 is a smaller plasma membrane protein with N- and C-terminal transmembrane anchors [40;41]. These physical features may explain the order and arrangement of CD36 and ICAM-1 binding modules in PfEMP1 proteins. As ICAM1 assists in capturing IEs from blood flow, other host receptors may serve this function in small or large PfEMP1 lacking ICAM1 binding domains.
DC8 and DC13 PfEMP1 proteins and severe malaria
A major question in pathogenesis research is whether there are common disease mechanisms that can be targeted for intervention. Recent evidence shows that a restricted subset of DC8 and DC13-containing PfEMP1 is selected on brain endothelial cells and is associated with severe pediatric malaria [32;42;43]. DC8 is a rare and unusual chimeric gene resulting from a recombination event between a group B and a group A var gene (B/A), while DC13 is a group A variant. The DC8 cassette consists of 4 domains (DBLα2-CIDRα1.1-DBLβ12-DBLγ4/6) and DC13 consists of 2 domains (DBLα1.7-CIDRα1.4) (Fig. 3). DC8- and DC13-expressing parasite lines have avid binding activity for diverse brain and non-brain endothelial cells [32;44]. However, as predicted from CIDR1 domain classification, they do not rely on CD36 for endothelial binding [32;42], indicating novel receptors are involved in cerebral binding. Work on CD36 binding parasites indicates that CD36 receptor clustering and recruitment of additional receptors is important to ensure tight adhesion to endothelial cells [45]. Notably, all four DC8 domains encode endothelial binding activity [32;44]. The CIDRα1 domain in DC8, DC13 and a subpopulation of group A variants bind EPCR [23] (Fig. 3), but the binding partners for the other DC8 domains remain to be defined.
The finding that EPCR binding parasites are associated with severe malaria has important implications for malaria pathogenesis. EPCR is a receptor for the serum factor protein C and promotes its conversion to activated protein C (APC) by the thrombin-thrombomodulin complex [46]. The protein C-EPCR pathway plays a key role in regulating blood clotting and endothelial barrier properties [46]. Pediatric autopsies showed decreased EPCR staining and increased fibrin deposition where IEs had adhered to cerebral microvessels [47]. These findings suggest there may be a causal link between IE cytoadhesion and blood brain barrier dysfunction (Fig. 1). The brain may be especially vulnerable to organ-specific pathology because brain cells have lower EPCR and thrombomodulin levels than other microvessels, which together limits their capacity to generate APC in response to elevated thrombin levels [48]. Key questions for pathogenesis research are whether EPCR-binding parasites inhibit APC generation or trigger the loss of EPCR at sites of sequestration.
Whereas endothelial activation and dysfunction are associated with disease severity [49], sequestration location matters. We hypothesize that EPCR-binding is a risk factor for severe malaria, but disease outcome will probably depend on the constellation of adhesion traits encoded into a specific PfEMP1 variant. DC8, DC13 and other EPCR-binding group A variants have different domain compositions from each other (Fig. 3) and this will likely influence parasite microvascular tropism and organ-specific pathology. It is currently unknown whether all EPCR-binding parasites have equivalent disease potential. Besides improving our understanding of how EPCR-binding parasites may affect EPCR function, important issues for pathogenesis research are whether all DC8 cassettes encode the same binding properties and how the combinatorial binding specificity of different EPCR-binding PfEMP1 influences cerebral binding tropism, endothelial signaling/dysfunction, and clinical symptoms.
Conclusions and future perspectives
IE cytoadhesion is complex, but binding studies and adhesion domain classification have begun to decipher how it works. To understand the complete PfEMP1 interactome map, we will need to map PfEMP1 functions to specific adhesion domain types, assign functions to the different PfEMP1 groups, learn if PfEMP1 adhesion traits are predictable, and test if this leads to predictable disease outcomes in clinical cohorts. The ultimate goal would be to design anti-adhesion interventions or new treatment options to target adhesion-based host pathogenic mechanisms.
A detailed understanding of the PfEMP1 interactome is possible through a combination of repertoire-wide binding screens with PfEMP1 recombinant proteins and monoclonal parasite lines [9;33;38;39], coupled with clinical cohort studies. New P. falciparum-endothelial cell binding models [32;42;44] and large scale human plasma membrane protein arrays [23] are accelerating research discoveries and have great potential to decipher binding and disease mechanisms. This combination of approaches has distinguished different types of PfEMP1 head structures and binding differences between small and large PfEMP1 proteins. It has also revealed functional specialization of the A, B, and C var groups. More recently, adhesion domain classification was crucial in distinguishing domain cassettes [6] and identifying an important role of DC8 and other EPCR binding parasites in severe malaria [23;32;42;43]. This leads to testable hypotheses for PfEMP1 binding and disease mechanisms.
As more receptor adhesion traits are fully characterized, there are likely to be other predictable traits because adhesion domain classification probably reflects the ancestral relationship of binding properties that arose and spread throughout the parasite population. Binding exceptions can also be expected and sequence types should not be thought of as monolithic binding traits. The concept of adhesion domain classification has been extended to short sequence fragment homology blocks [6], which may offer further ability to discriminate binding properties. In this context, sub-classification of DBLβ domains lent greater predictive power to ICAM1 binding [33] and homology blocks may distinguish rosetting variants [50;51]. In conclusion, adhesion domain classification provides a conceptual framework to investigate parasite binding and gain molecular insight into malaria pathogenesis. Sequence classification permits repertoire-wide predictions of P. falciparum host receptor interactions and to make strong inferences about PfEMP1 binding properties. The ultimate test is how well binding predictions are generalizable across geographically diverse parasites and is able to provide mechanistic insight into disease.
The PfEMP1 protein family is a major virulence determinant for P. falciparum
PfEMP1 size and sequence polymorphism obscures binding patterns
Domain classification can predict binding and protein functional specialization
Classification provides a framework to gain molecular insight into severe malaria
Acknowledgments
This work was supported by RO1 AI47953 and R56A1104238-01A1. I apologize for not discussing and citing all relevant publications due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could a3ect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–9. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.White NJ, Turner GD, Day NP, Dondorp AM. Lethal malaria: Marchiafava and Bignami were right. J Infect Dis. 2013;208:192–8. doi: 10.1093/infdis/jit116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Current Opinion in Microbiology. 2006;9:374–80. doi: 10.1016/j.mib.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, Christodoulou Z, et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rask TS, Hansen DA, Theander TG, Gorm PA, Lavstsen T. Plasmodium falciparum Erythrocyte Membrane Protein 1 Diversity in Seven Genomes - Divide and Conquer. PLoS Comput Biol. 2010;6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–91. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 8.Montgomery J, Mphande FA, Berriman M, Pain A, Rogerson SJ, Taylor TE, et al. Differential var gene expression in the organs of patients dying of falciparum malaria. Mol Microbiol. 2007;65:959–67. doi: 10.1111/j.1365-2958.2007.05837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson BA, Welch TL, Smith JD. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol Microbiol. 2003;47:1265–78. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- 10.Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–4. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 11.Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol Biochem Parasitol. 2000;110:293–310. doi: 10.1016/s0166-6851(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 12.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 13.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–10. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 15.Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–4. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 17.Rowe JA, Claessens A, Corrigan RA, Arman M. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications. Expert Rev Mol Med. 2009;11:e16. doi: 10.1017/S1462399409001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baruch DI, Ma XC, Singh HB, Bi X, Pasloske BL, Howard RJ. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood. 1997;90:3766–75. [PubMed] [Google Scholar]
- 19.Smith JD, Kyes S, Craig AG, Fagan T, Hudson-Taylor D, Miller LH, et al. Analysis of adhesive domains from the A4VAR Plasmodium falciparum erythrocyte membrane protein-1 identifies a CD36 binding domain. Mol Biochem Parasitol. 1998;97:133–48. doi: 10.1016/s0166-6851(98)00145-5. [DOI] [PubMed] [Google Scholar]
- 20.Smith JD, Craig AG, Kriek N, Hudson-Taylor D, Kyes S, Fagen T, et al. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proc Natl Acad Sci U S A. 2000;97:1766–71. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388:292–5. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 22.Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–5. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004;199:1179–90. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, et al. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Molecular and Biochemical Parasitology. 2006;150:211–8. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warimwe GM, Keane TM, Fegan G, Musyoki JN, Newton CR, Pain A, et al. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci U S A. 2009;106:21801–6. doi: 10.1073/pnas.0907590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghumra A, Semblat JP, Ataide R, Kifude C, Adams Y, Claessens A, et al. Induction of strain-transcending antibodies against Group A PfEMP1 surface antigens from virulent malaria parasites. PLoS Pathog. 2012;8:e1002665. doi: 10.1371/journal.ppat.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson J, Helmby H, Hill AV, Brewster D, Greenwood BM, Wahlgren M. Human cerebral malaria: association with erythrocyte rosetting and lack of anti-rosetting antibodies. Lancet. 1990;336:1457–60. doi: 10.1016/0140-6736(90)93174-n. [DOI] [PubMed] [Google Scholar]
- 29.Prugnolle F, Durand P, Ollomo B, Duval L, Ariey F, Arnathau C, et al. A Fresh Look at the Origin of Plasmodium falciparum, the Most Malignant Malaria Agent. PLoS Pathog. 2011;7:e1001283. doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraemer SM, Smith JD. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol Microbiol. 2003;50:1527–38. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 31.Urban BC, Ferguson DJ, Pain A, Willcox N, Plebanski M, Austyn JM, et al. Plasmodium falciparum-infected erythrocytes modulate the maturation of dendritic cells. Nature. 1999;400:73–7. doi: 10.1038/21900. [DOI] [PubMed] [Google Scholar]
- 32.Avril M, Tripathi AK, Brazier AJ, Andisi C, Janes JH, Soma VL, et al. A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc Natl Acad Sci U S A. 2012;109:E1782–90. doi: 10.1073/pnas.1120534109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janes JH, Wang CP, Levin-Edens E, Vigan-Womas I, Guillotte M, Melcher M, et al. Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog. 2011;7:e1002032. doi: 10.1371/journal.ppat.1002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooke BM, Berendt AR, Craig AG, MacGregor J, Newbold CI, Nash GB. Rolling and stationary cytoadhesion of red blood cells parasitized by Plasmodium falciparum: separate roles for ICAM-1, CD36 and thrombospondin. Br J Haematol. 1994;87:162–70. doi: 10.1111/j.1365-2141.1994.tb04887.x. [DOI] [PubMed] [Google Scholar]
- 35.Ochola LB, Siddondo BR, Ocholla H, Nkya S, Kimani EN, Williams TN, et al. Specific Receptor Usage in Plasmodium falciparum Cytoadherence Is Associated with Disease Outcome. PLoS One. 2011;6:e14741. doi: 10.1371/journal.pone.0014741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner GD, Morrison H, Jones M, Davis TM, Looareesuwan S, Buley ID, et al. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am J Pathol. 1994;145:1057–69. [PMC free article] [PubMed] [Google Scholar]
- 37.Bengtsson A, Joergensen L, Rask TS, Olsen RW, Andersen MA, Turner L, et al. A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies. J Immunol. 2013;190:240–9. doi: 10.4049/jimmunol.1202578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howell DP, Levin EA, Springer AL, Kraemer SM, Phippard DJ, Schief WR, et al. Mapping a common interaction site used by Plasmodium falciparum Duffy binding-like domains to bind diverse host receptors. Mol Microbiol. 2008;67:78–87. doi: 10.1111/j.1365-2958.2007.06019.x. [DOI] [PubMed] [Google Scholar]
- 39.Oleinikov AV, Amos E, Frye IT, Rossnagle E, Mutabingwa TK, Fried M, et al. High throughput functional assays of the variant antigen PfEMP1 reveal a single domain in the 3D7 Plasmodium falciparum genome that binds ICAM1 with high affinity and is targeted by naturally acquired neutralizing antibodies. PLoS Pathog. 2009;5:e1000386. doi: 10.1371/journal.ppat.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jun CD, Carman CV, Redick SD, Shimaoka M, Erickson HP, Springer TA. Ultrastructure and function of dimeric, soluble intercellular adhesion molecule-1 (ICAM-1) J Biol Chem. 2001;276:29019–27. doi: 10.1074/jbc.M103394200. [DOI] [PubMed] [Google Scholar]
- 41.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claessens A, Adams Y, Ghumra A, Lindergard G, Buchan CC, Andisi C, et al. A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells. Proc Natl Acad Sci U S A. 2012;109:E1772–81. doi: 10.1073/pnas.1120461109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci U S A. 2012;109:E1791–1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avril M, Brazier AJ, Melcher M, Sampath S, Smith JD. DC8 and DC13 var Genes Associated with Severe Malaria Bind Avidly to Diverse Endothelial Cells. PLoS Pathog. 2013;9:e1003430. doi: 10.1371/journal.ppat.1003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis SP, Lee K, Gillrie MR, Roa L, Amrein M, Ho M. CD36 recruits alpha(5)beta(1) integrin to promote cytoadherence of P. falciparum-infected erythrocytes. PLoS Pathog. 2013;9:e1003590. doi: 10.1371/journal.ppat.1003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouwens EA, Stavenuiter F, Mosnier LO. Mechanisms of anticoagulant and cytoprotective actions of the protein C pathway. J Thromb Haemost. 2013;11 (Suppl 1):242–53. doi: 10.1111/jth.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moxon CA, Wassmer SC, Milner DA, Jr, Chisala NV, Taylor TE, Seydel KB, et al. Loss of endothelial protein C receptors links coagulation and inflammation to parasite sequestration in cerebral malaria in African children. Blood. 2013;122:842–51. doi: 10.1182/blood-2013-03-490219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laszik Z, Mitro A, Taylor FB, Jr, Ferrell G, Esmon CT. Human protein C receptor is present primarily on endothelium of large blood vessels: implications for the control of the protein C pathway. Circulation. 1997;96:3633–40. doi: 10.1161/01.cir.96.10.3633. [DOI] [PubMed] [Google Scholar]
- 49.Cunnington AJ, Walther M, Riley EM. Piecing together the puzzle of severe malaria. Sci Transl Med. 2013;5:211ps18. doi: 10.1126/scitranslmed.3007432. [DOI] [PubMed] [Google Scholar]
- 50.Rorick MM, Rask TS, Baskerville EB, Day KP, Pascual M. Homology blocks of Plasmodium falciparum var genes and clinically distinct forms of severe malaria in a local population. BMC Microbiol. 2013;13:244. doi: 10.1186/1471-2180-13-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warimwe GM, Fegan G, Musyoki JN, Newton CR, Opiyo M, Githinji G, et al. Prognostic indicators of life-threatening malaria are associated with distinct parasite variant antigen profiles. Sci Transl Med. 2012;4:129ra45. doi: 10.1126/scitranslmed.3003247. [DOI] [PMC free article] [PubMed] [Google Scholar]