Abstract
Mutations in GATA4 and TBX5 are associated with congenital heart defects in humans. Interaction between GATA4 and TBX5 is important for normal cardiac septation, but the underlying molecular mechanisms are not well understood. Here, we show that Gata4 and Tbx5 are co-expressed in the embryonic atria and ventricle, but after E15.5, ventricular expression of Tbx5 decreases. Co-localization and co-immunoprecipitation studies demonstrate an interaction of Gata4 and Tbx5 in the developing atria and ventricles, but the ventricular interaction declines after E14.5. Gata4+/−;Tbx5+/− mouse embryos display decreased atrial and ventricular myocardial thickness at E11.5, prior to cardiac septation. To determine the cell lineage in which the interaction was functionally significant in vivo, mice heterozygous for Gata4 in the myocardium or endocardium and heterozygous for Tbx5 (Gata4MyoDel/wt;Tbx5+/− and Gata4EndoDel/wt;Tbx5+/−, respectively) were generated. Gata4MyoDel/wt;Tbx5+/− mice displayed embryonic lethality, thin myocardium with reduced cell proliferation, and atrioventricular septation defects similar to Gata4;Tbx5 compound heterozygotes while Gata4EndoDel/wt;Tbx5+/− embryos were normal. Cdk4 and Cdk2, cyclin-dependent kinases required for myocardial development and septation were reduced in Gata4+/−;Tbx5+/− hearts. Cdk4 is a known direct target of Gata4 and the regulation of Cdk2 in the developing heart has not been studied. Chromatin immunoprecipitation and transactivation studies demonstrate that Gata4 and Tbx5 directly regulate Cdk4 while only Tbx5 activates Cdk2 expression. These findings highlight the mechanisms by which disruption of the Gata4 and Tbx5 interaction in the myocardium contributes to cardiac septation defects in humans.
INTRODUCTION
Congenital heart disease (CHD) is the most common type of human birth defect with an incidence of 8–9 per 1000 live births (1). CHD may involve a variety of cardiac structures, and cardiac septal defects are the most common type of CHD (2). CHD is the result of perturbation of well-conserved developmental pathways that regulate heart development (3,4). The precise etiologies for CHD remain unknown, but ∼80% of CHD is proposed to have a multifactorial etiology involving a combination of genetics and environmental factors (5).
Cardiac morphogenesis is a complex process regulated by numerous highly conserved transcription factors including Gata4 and Tbx5 (6,7). While mutations in these genes have been reported to cause a spectrum of heart malformations in humans, they are most commonly associated with defects of cardiac septation (8,9). Gata4 encodes for a zinc finger transcription factor while Tbx5 encodes a T-box transcription factor. Both are expressed in the developing heart and are critical for multiple events during heart development (10,11). We previously linked mutations in GATA4 to cardiac septation defects in humans by studying large families with autosomal-dominant disease (9). In one family with 17 affected family members, we identified a disease-causing point mutation in GATA4 that disrupted a novel interaction with TBX5, which is the etiology of Holt–Oram syndrome that is characterized by defects in cardiac septation (8). Of note, this disease-causing glycine to serine mutation at codon 296 (G296S) has been associated with a similar cardiac phenotype by other investigators (12,13). Furthermore, Gata4 and Tbx5 demonstrated a physical interaction in vitro, synergistically activated downstream target genes and exhibited a genetic interaction in vivo, suggesting that these transcription factors regulated a gene program critical for cardiac septation (9,14,15).
The molecular mechanisms underlying the cardiac septation defects that occur in the setting of these transcription factors mutations remain unclear. Mice harboring the highly penetrant Gata4 disease-causing G296S mutation demonstrated subtle defects in atrial septation and also exhibited abnormal cardiomyocyte proliferation. This phenotype was postulated to be secondary to the lack of interaction with Tbx5 (16). Mice which are compound heterozygotes for both Gata4 and Tbx5 display a complex phenotype of embryonic lethality secondary to myocardial hypoplasia and atrioventricular septal defects (AVSD) demonstrating the importance of dosage for these two transcription factors during heart development and suggesting that they regulate common molecular pathways critical for normal cardiac morphogenesis (14).
Here, we show that Gata4 and Tbx5 are co-expressed in the atria from embryonic day (E)11.5 to postnatal day (P)7, but ventricular expression of Tbx5 is reduced at late embryonic and postnatal stages. In vitro and in vivo assays demonstrate the interaction of Gata4 and Tbx5 in the atria at E11.5 and E14.5; however, this interaction is diminished in the left ventricle at E14.5. Analysis of Gata4;Tbx5 compound heterozygote embryos demonstrate a thin ventricular myocardium starting at E11.5 prior to atrioventricular septation. To define the cell lineage in which Gata4 and Tbx5 exhibit a functional interaction, we generated Gata4;Tbx5 compound mutant mice in which Gata4 was heterozygous in either the myocardium or endocardium and Tbx5 was heterozygous in all lineages. Heterozygous deletion of myocardial Gata4 in the setting of Tbx5 haploinsufficiency resulted in AVSD and a thin myocardium with associated cardiomyocyte proliferation deficits. Additional experiments find downregulation of Cdk2 and Cdk4 mRNA in Gata4+/−;Tbx5+/− embryonic hearts. Chromatin immunoprecipitation (ChIP) and transactivation assays demonstrate that both Gata4 and Tbx5 regulate Cdk4 expression while only Tbx5 activates Cdk2. These studies show the importance of Gata4 and Tbx5 for myocardial proliferation in the developing heart and suggest that reduced cardiomyocyte proliferation may contribute to abnormal atrioventricular septation.
RESULTS
Gata4 and Tbx5 expression during heart development
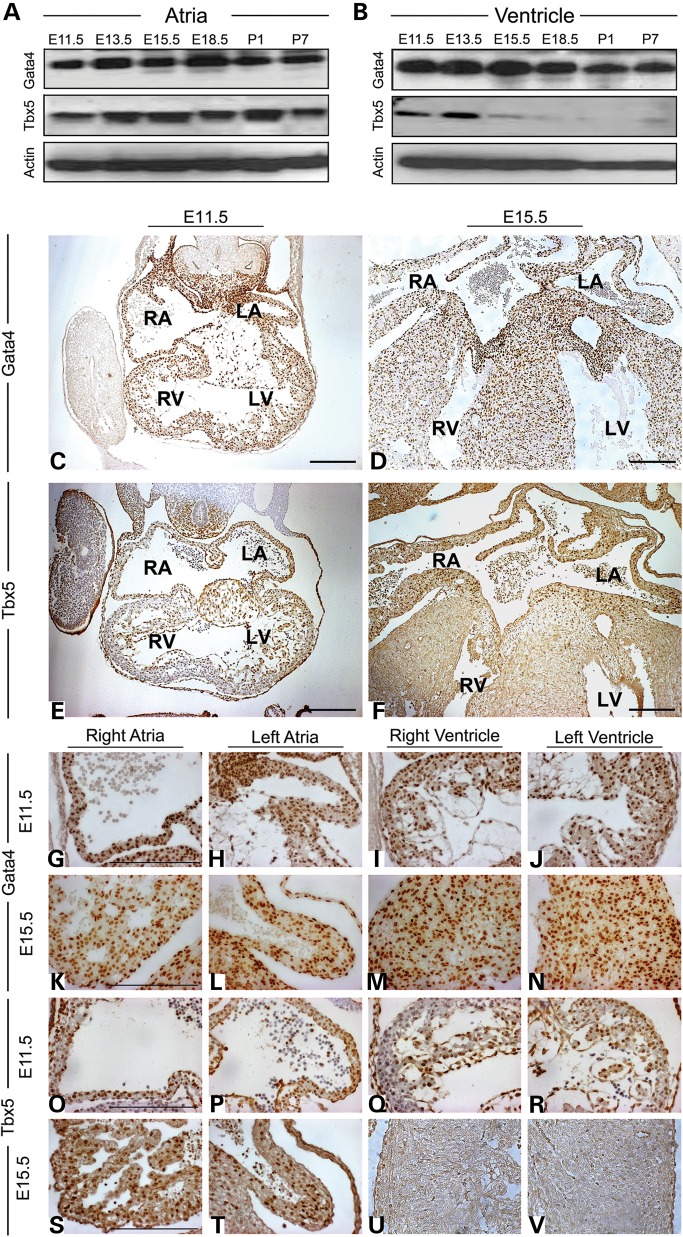
Gata4 and Tbx5 have been shown to play various roles at different stages of cardiac morphogenesis. Therefore, to determine the precise expression of Gata4 and Tbx5 in atria and ventricle of developing heart, we examined the protein expression of Gata4 and Tbx5 in the murine atria and ventricle by western blot from E11.5 to P7. Surprisingly, we found that Tbx5 expression level begins to diminish only in the ventricle at E15.5; however, the atrial expression is maintained in the postnatal heart (Fig. 1A and B). Gata4 was expressed in both the atria and ventricle during these timepoints (Fig. 1A and B). To determine the chamber-specific expression of these transcription factors, we assayed Gata4 and Tbx5 expression by immunohistochemistry at E11.5 and E15.5. Gata4 was expressed in both the atrial and ventricular myocardium at both timepoints (Fig. 1C, D and G–N). At E11.5, Tbx5 was predominantly expressed in both atria and left ventricle with lower levels of expression noted in the right ventricular myocardium (Fig. 1E and O–R). At E15.5, expression of Tbx5 was nearly absent in both ventricles (Fig. 1F and S–V). Of note, both Gata4 and Tbx5 were expressed in the developing atrioventricular cushions at E11.5, but at E15.5, only Gata4 was expressed (Fig. 1C–F). A similar expression pattern was noted in the endocardium where Gata4 and Tbx5 were both expressed at E11.5, but only Gata4 expression was noted at E15.5 (Fig. 1C–F).
Figure 1.
Cardiac expression of Gata4 and Tbx5 in the developing embryo and neonate. Protein expression of Gata4 and Tbx5 in the murine atria (A) and ventricle (B) during different embryonic and postnatal stages by immunoblot. Actin expression is shown as loading control. Immunohistochemistry for Gata4 (C, D and G–N) and Tbx5 (E, F and O–V) on coronal sections of the embryonic heart at E11.5 (C, E, G–J and O–R) and E15.5 (E, F, K–N, S–V). Expression of Gata4 is found in the atrial and ventricular endocardium and myocardium at E11.5 and E15.5, while Tbx5 is predominantly expressed in both atrial and left ventricular endocardium and myocardium at E11.5 and becomes restricted to the atria at E15.5. (G–J), (K–N), (O–R) and (S–V) represent high-magnification images of (C), (D), (E) and (F), respectively. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. Scale bars indicate 200 µm.
Co-localization and interaction of Gata4 and Tbx5 in the developing atria and ventricle
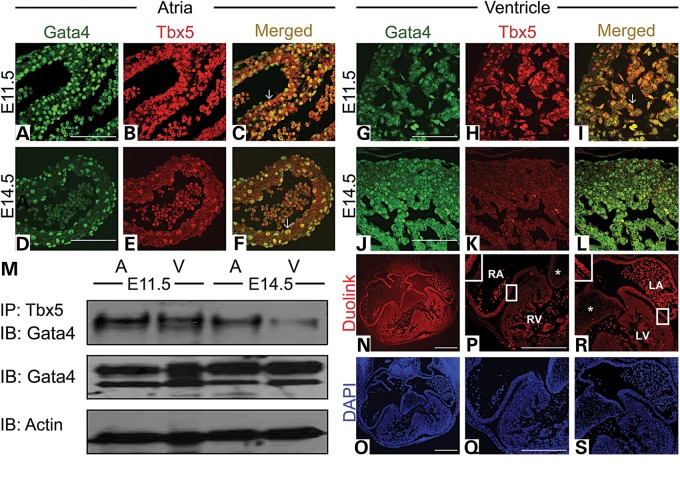
Our expression studies demonstrated that Gata4 and Tbx5 are both expressed in the atria throughout development; however, in the ventricle, they show similar expression patterns only in the left ventricle until E15.5. To determine whether Gata4 and Tbx5 were co-localized in the developing myocardium, we performed immunofluorescence staining of Gata4 and Tbx5 at E11.5 and E14.5. Gata4 and Tbx5 were found to co-localize in the nucleus of atrial (arrow in Fig. 2A–C) and ventricular cardiomyocytes (arrow in Fig. 2G–I) at E11.5, and in the atrial cardiomyocytes at E14.5 (arrow in Fig. 2D–F). However, the co-localization was not seen in the left ventricle at E14.5 (Fig. 2J–L). In the endocardium and endocardial cushions, we noted co-expression of Gata4 and Tbx5 at E11.5 but only in the atrial endocardium was this co-expression maintained at E14.5 (Supplementary Material, Fig. S1 and Fig. 2A–L). To determine whether Gata4 and Tbx5 physically interacted in the developing heart in vivo, we performed co-immunoprecipitation experiments. Immunoprecipitation was performed with Tbx5 followed by immunoblotting with Gata4 and demonstrated an in vivo Gata4 and Tbx5 interaction in the E11.5 and E14.5 atria and in the E11.5 ventricle (Fig. 2M). This interaction was significantly weakened in the E14.5 ventricle and was consistent with our immunofluorescence studies (Fig. 2M). A Duolink assay, which allows for visualization of protein interactions in tissue, was also performed to assess the chamber-specific interaction of Gata4 and Tbx5 in vivo (see Materials and Methods for additional details). The Duolink assay indicated an interaction of Gata4 and Tbx5 in vivo in the E11.5 right and left atria and to a lesser extent in the left ventricle (Fig. 2N–S). No evidence of interaction was noted in the right ventricle and atrioventricular cushions at E11.5 (asterisk in Fig. 2P and R).
Figure 2.
Gata4 and Tbx5 are co-expressed and physically interact during heart development. Immunofluorescent staining for Gata4 (A, D, G and J) and Tbx5 (B, E, H and K) in E11.5 (A–C and G–I) and E14.5 (D–F and J–L) atria and left ventricle demonstrates co-localization in the E11.5 atria and ventricle and E14.5 atria but not in the E14.5 left ventricle (L). Arrows highlight nuclear co-localization. (M) Immunoprecipitation (IP) with Tbx5 antibody demonstrates an interaction with Gata4 on immunoblot (IB) in E11.5 atria and ventricle and E14.5 atria. Interaction is decreased in the E14.5 ventricle. Actin expression is shown as loading control. (N, P and R) Duolink assay on histologic sections of E11.5 atria and ventricle demonstrates the decreased signal in the ventricle as compared to the atria. Corresponding DAPI staining is shown in blue (O, Q and S). *, highlights lack of interaction in atrioventricular cushions. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. Scale bars indicate 200 µm.
Development of myocardial hypoplasia in Gata4;Tbx5 compound heterozygote embryos occurs prior to defects in atrioventricular cushions
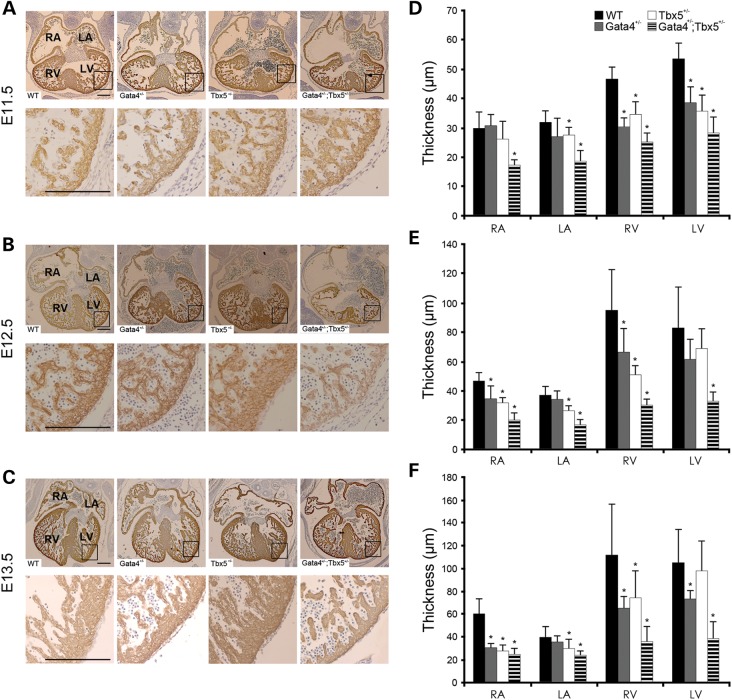
We previously reported that Gata4;Tbx5 compound heterozygote mice showed embryonic lethality starting at E15.5 and had myocardial hypoplasia with a complete AVSD (14). To define the underlying mechanisms for these cardiac phenotypes, we serially examined Gata4;Tbx5 compound heterozygote embryos at earlier timepoints. Quantitative measurement of the atrial and ventricular myocardial walls was performed using histologic sections from E11.5, E12.5 and E13.5 embryos that were immunostained with anti-myosin heavy-chain antibody to delineate the embryonic myocardium.
We found a significant decrease in the atrial and ventricular myocardial wall thickness of Gata4;Tbx5 compound heterozygotes starting at E11.5 when compared with wild-type as well as single heterozygote littermates (Fig. 3). Interestingly, the Gata4+/− and Tbx5+/− embryos also demonstrated modest levels of myocardial thinning at all three timepoints when compared with wild type (Fig. 3). Quantification of the endocardial cushions demonstrated a subtle but significant decrease in atrioventricular cushion cell number in Gata4;Tbx5 compound heterozygote embryos at E11.5 but this was not different from the Gata4+/− and Tbx5+/− single heterozygote embryos (Supplementary Material, Fig. S2A). After E12.5, the cushion hypoplasia was more severe in the Gata4;Tbx5 compound heterozygotes (Supplementary Material, Fig. S2B–C). The dorsal mesenchymal protrusion (DMP) has also been shown to be critical for proper atrioventricular septation (17–20). Histologic examination of Gata4;Tbx5 compound heterozygote E11.5 embryos demonstrated the absence of the DMP (n = 3/3, Supplementary Material, Fig. S3), but as described, the DMP was also not found in a subset of Tbx5 heterozygote littermates (n = 2/3), which do not display an AVSD phenotype (19). The DMP appeared normal in wild-type and Gata4 heterozygote littermates at E11.5 (Supplementary Material, Fig. S3). Based on these observations, the lack of DMP alone could not account for the AVSD phenotype found in Gata4;Tbx5 compound heterozygotes.
Figure 3.
Thin myocardium in Gata4;Tbx5 compound mutant embryos develops as early as E11.5. (A–C) Coronal sections through the embryonic heart at E11.5, E12.5 and E13.5 are shown. Sections are stained with α-MHC to label cardiomyocytes. Decreased atrial and ventricular myocardial wall thickness in Gata4;Tbx5 compound heterozygotes at E11.5 (A), E12.5 (B) and E13.5 (C) when compared with littermates. (D–F) Quantitative analyses of wall thickness in the atria and compact ventricular layer are shown. WT, wild type; RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. *P value <0.05. Scale bars indicate 200 µm.
In vivo analysis of cell lineage requirements for Gata4–Tbx5 interaction in cardiac morphogenesis
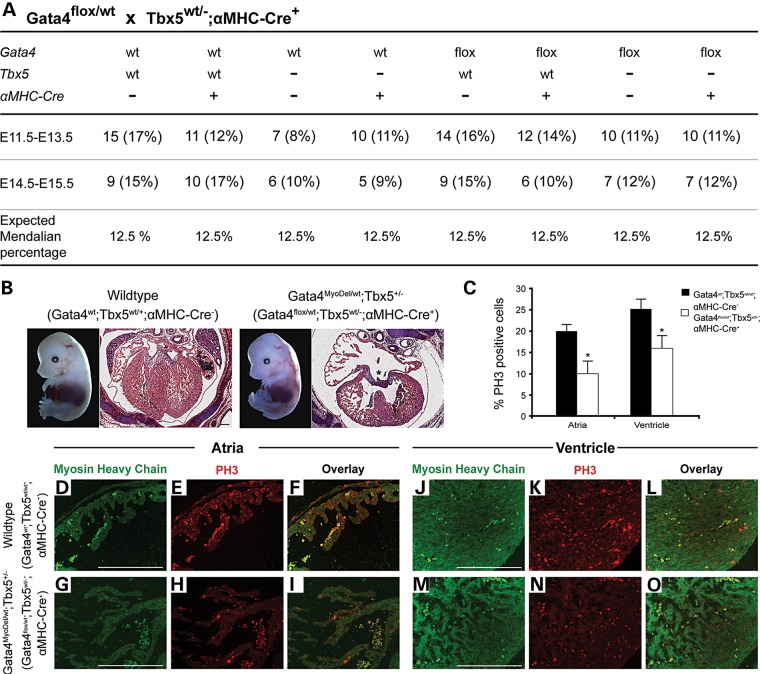
To determine the functional interaction between Gata4 and Tbx5 in different cell lineages in vivo, we generated mice that were compound heterozygotes for the Tbx5-null allele and a cell lineage-specific deletion of Gata4. To perform these studies, we first generated mice that were heterozygote for a Tbx5-null allele and a tissue-specific Cre. Lineage-specific deletion of the Gata4 was performed by crossing these mice (Tbx5+/−;Cre+) with mice harboring a floxed Gata4 allele (21). Tissue-specific deletion was obtained by using Cre under the regulation of Tie2, which is predominantly expressed in the endocardium and endothelium and alpha-myosin heavy-chain (α-MHC), which is specific for the late embryonic myocardium (starting after E9.5) (22,23). While expected Mendelian ratios were found for Gata4flox/wt;αMHC-Cre+;Tbx5+/−embryos, which are heterozygous for Gata4 and Tbx5 in cardiomyocytes (Gata4MyoDel/wt;Tbx5+/−) between E11.5–E15.5, the Gata4MyoDel/wt;Tbx5+/− were embryonic lethal (Fig. 4A and data not shown). Histologic examination demonstrated a thin myocardium and an AVSD at E14.5–E15.5, similar to the Gata4;Tbx5 compound heterozygotes (Fig. 4B). Immunohistochemistry for Gata4 demonstrated a reduction of Gata4 expression specifically within the atrial and ventricular myocardium in Gata4MyoDel/wt;Tbx5+/− embryos in comparison with wild-type littermates (Supplementary Material, Fig. S4). Immunofluorescence for phospho-histone 3 (PH3), a mitosis marker, demonstrated reduced myocardial proliferation both in atrial and ventricular myocardium in Gata4MyoDel/wt;Tbx5+/− at E14.5 (Fig. 4C–O). Analysis of the Gata4flox/wt;Tie2-Cre+;Tbx5+/− mice, which are heterozygous for Gata4 and Tbx5 in the endocardium (Gata4EndoDel/wt;Tbx5+/−) showed no obvious embryonic lethality, cardiac phenotype or myocardial proliferation deficits (Supplementary Material, Fig. S5). These studies suggested that heterozygous loss of Gata4 and Tbx5 in the myocardium results in myocardial proliferation deficits that contribute to a thin myocardial wall and also to defects in atrioventricular septation.
Figure 4.
Myocardial hypoplasia, atrioventricular septal defects, decreased cardiomyocyte proliferation in embryos with myocardial deletion of Gata4 and Tbx5 haploinsufficiency (Gata4MyoDel/wt;Tbx5+/−). (A) Frequency of genotypes obtained from intercrossing Gata4flox/wt mice with Tbx5wt/−;αMHC-Cre+ mice and demonstrates normal Mendelian ratios for Gata4flox/wt;Tbx5wt/−;αMHC-Cre+ (Gata4MyoDel/wt;Tbx5+/−) embryos from E11.5 to E15.5. (B) Gross images of E14.5 embryos and histologic sections through E14.5 heart are shown. Gata4MyoDel/wt;Tbx5+/− (Gata4flox/wt;Tbx5wt/−;αMHC-Cre+) mutant displays a thin myocardium and an atrioventricular septal defect (*) (n = 3). (C–O) Decreased cardiomyocyte proliferation in the E14.5 atria and ventricle as determined by phospho-histone H3 (PH3) staining in Gata4MyoDel/wt;Tbx5+/− mutants (D–O). Quantification is shown in (C). α-MHC staining (green) is shown to label cardiomyocytes. *P value < 0.05. Scale bars indicate 200 µm.
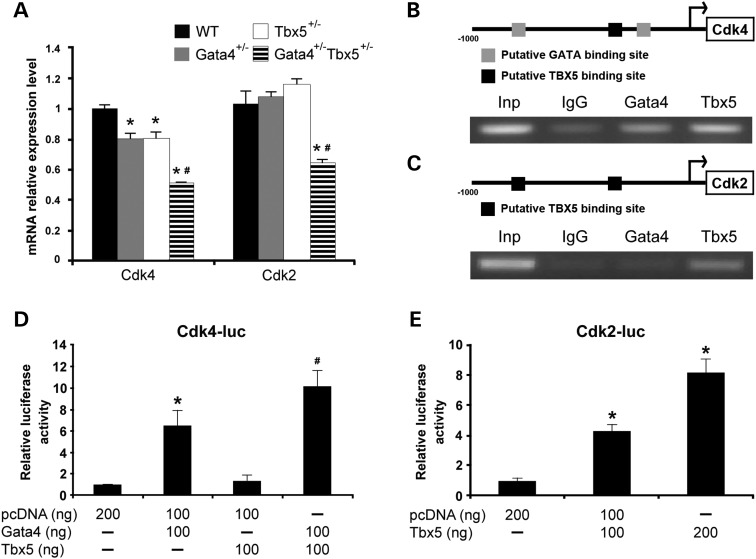
Gata4 and Tbx5 regulation of Cdk4 and Cdk2
Our data suggested that cardiomyocyte proliferation deficits may underlie the cardiac morphogenetic defects that are found in Gata4;Tbx5 compound heterozygote embryos and that myocardial Gata4 is required for both the proliferation deficits and AVSD phenotypes. Consistent with this, mice that are null for Cdk2 and Cdk4, cyclin-dependent kinases important for cell proliferation, display similar cardiac phenotypes (24,25). We examined the expression of Cdk2 and Cdk4 mRNA in Gata4;Tbx5 compound heterozygote hearts by qRT–PCR and found decreased levels of Cdk2 and Cdk4 when compared with wild-type and single heterozygote littermates (Fig. 5A). While it has been previously reported that Gata4 functions as a direct transcriptional activator of Cdk4, the role of Tbx5 in the regulation of Cdk4 or the regulation of Cdk2 by either Gata4 or Tbx5 has not been studied (26). We examined the proximal upstream sequences, 1 kb upstream of transcriptional start site (tss) of murine Cdk4 (NM_009870) and Cdk2 (NM_016756) for potential binding sites of Gata4 and Tbx5. We identified two potential GATA-binding sites (−180 and – 659 bp upstream of tss) in the Cdk4 upstream sequence, which have been previously described (Fig. 5B) (26). In addition, one putative TBX5-binding site (−280 bp upstream of tss) was also noted (Fig. 5B) (27). In contrast, Cdk2 has two potential TBX5-binding sequences (−340 and −610 bp upstream of tss) within the promoter proximal region (Fig. 5C). Therefore, we performed ChIP assays using E13.5 whole embryonic hearts to determine whether these two transcription factors directly bound to the proximal promoters of these cell-cycle genes. Gata4 and Tbx5 antibodies were used to pull down the associated genomic DNA followed by targeted amplification of Cdk4 and Cdk2 promoter regions containing putative GATA and TBX5-binding sites. Using ChIP, enrichment of the promoter sequence for Cdk4 was found when immunoprecipitation was performed using a Gata4 antibody, as previously described in P19CL6 cells (Fig. 5B) (26). Interestingly, we also detected a notable enrichment of Cdk4 promoter sequence when the chromatin was immunoprecipitated with the Tbx5 antibody suggesting a potential cooperative role for Gata4 and Tbx5 in the regulation of Cdk4 (Fig. 5B). The cooperativity of Gata4 and Tbx5 for Cdk4 activation was shown by performing transactivation assays using the previously published murine Cdk4 promoter in HeLa cells, which contains two GATA and one Tbx5-binding sites, as described above (26). Transfection of Gata4 increased luciferase activity by 6-fold compared with the cells transfected with empty vector, as previously published, and Tbx5 by itself had no increased transactivation ability (Fig. 5D). Interestingly, co-transfection of Gata4 and Tbx5 significantly increased activation when compared with Gata4 alone, suggesting the synergistic activation of Cdk4 by Gata4 and Tbx5, potentially by a direct interaction between Gata4 and Tbx5 (Fig. 5D). Further, knockdown of Gata4 and Tbx5 using siRNA in primary cardiomyocytes from E11.5 embryos resulted in decreased expression of Cdk4 and reduced proliferation (Supplementary Material, Fig. S6). Similar assays were performed for Cdk2 to investigate the possible role of Gata4 and Tbx5 on the activation of Cdk2. ChIP assays demonstrated enrichment of the promoter region of murine Cdk2 only with immunoprecipitation using Tbx5 antibody (Fig. 5C). Not surprisingly, no enrichment was found when immunoprecipitation was performed with the Gata4 antibody as there were no GATA-binding sites within the 1 kb upstream sequence of Cdk2. In luciferase reporter assays, we detected a significant activation of Cdk2 promoter when cells were transfected with Tbx5 in comparison with the empty vector (Fig. 5E). Examination of published ChIP-seq datasets for Gata4 and Tbx5 demonstrated only similar findings for Tbx5 regulation of Cdk4, but the majority of these studies have been performed in adult cardiomyocytes or HL-1 cells, potentially accounting for the differences (Supplementary Material, Fig. S7). Together, the combination of our ChIP and luciferase assays demonstrates that Gata4 and Tbx5 can activate the transcription of Cdk4 while only Tbx5 is a regulator of Cdk2.
Figure 5.
Regulation of Cdk4 and Cdk2 by Gata4 and Tbx5. (A) Decreased mRNA expression of Cdk4 and Cdk2 in E11.5 Gata4;Tbx5 mutant hearts as measured by qRT–PCR. *P value < 0.05 in comparison with wild type; #P value < 0.05 comparison between single heterozygotes and double heterozygotes. (B) Schematic representation of the sequence of upstream Cdk4 containing two putative GATA-binding sites and one putative TBX5-binding site. ChIP of E13.5 murine hearts demonstrated enrichment of Cdk4 promoter region with immunoprecipitation with Gata4 or Tbx5 when compared with IgG control. (C) Schematic representation of the sequence of upstream Cdk2 containing two putative TBX5-binding sites. ChIP of E13.5 murine hearts demonstrated enrichment of Cdk2 promoter region with immunoprecipitation with Tbx5 when compared with IgG control. Inp, input. (D) Luciferase reporter assay using the murine promoter of Cdk4 demonstrate that murine Gata4 protein activates the reporter but Tbx5 does not. In addition, synergistic activation is seen with transfection with both Gata4 and Tbx5. (E) Luciferase reporter assay demonstrates that Tbx5 activates Cdk2 promoter in a dose-dependent manner. Luciferase activity is normalized to β-galactosidase. *P value < 0.05 when compared with empty vector; #P value < 0.05 comparison between Gata4 alone and Gata4 with Tbx5.
DISCUSSION
Gata4 and Tbx5 play important roles during heart development and mutations in these genes predominantly cause congenital cardiac septation defects in humans (8–11). Here, we have performed detailed expression studies and demonstrated that Gata4 and Tbx5 are co-expressed in the developing atria and ventricle until E15.5, after which ventricular expression of Tbx5 decreases. Consistent with this, in vitro and in vivo analysis demonstrates physical interaction of Gata4 and Tbx5 throughout atrial development but a diminished ventricular interaction after E14.5. Staged analysis of Gata4;Tbx5 compound heterozygote embryos demonstrated that myocardial abnormalities starting at E11.5 occurred prior to the loss of atrioventricular cushion cells. In vivo studies demonstrate that heterozygous deletion of myocardial Gata4 in the setting of Tbx5 haploinsufficiency (Gata4MyoDel/wt;Tbx5+/−) leads to cardiomyocyte proliferation defects with associated AVSDs that are seen in Gata4;Tbx5 compound heterozygote mice. Furthermore, we found reduced expression of the cell proliferation genes, Cdk2 and Cdk4, in Gata4+/−;Tbx5+/− mutant hearts and demonstrated that Gata4 and Tbx5 regulate Cdk4 and Cdk2 expression. These studies demonstrate the importance of Gata4–Tbx5 in the cooperative regulation of cardiomyocyte proliferation in the developing embryo and that suggests a role for myocardial Gata4 and Tbx5 in atrioventricular septation.
Numerous transcriptional and signaling pathways have been described which direct discrete aspects of cardiac morphogenesis (3,6,7). The mechanisms by which these cardiac morphogenetic events are regulated appear to be via cardiac transcription factors including Gata4, Tbx5 and Nkx2.5, which are well described to function together during heart development and also during cardiac regeneration (28). These three transcription factors have been shown not only to regulate common downstream target genes but also physically interact (9,29–31). While these factors are arguably the best studied, additional transcription factors, such as Srf, Mef2a, Mef2c among others, also play critical roles (33,33). Our data further define how Gata4 and Tbx5 function together during heart development. While it has been shown that Gata4 and Tbx5 are required for cardiomyocyte differentiation, we specifically show that proper levels of Gata4 and Tbx5 in the developing myocardium are required for cardiomyocyte proliferation during later stages of heart development. In addition, our data suggest that DNA-binding by Tbx5 may not be required for the cooperative regulation of common downstream target genes by Gata4 and Tbx5. These studies only further demonstrate the complexity of cardiac transcriptional network during heart development and how its disruption may lead to cardiac malformations.
The development of a four-chambered mammalian heart requires contributions from several cell types, which undergo numerous cellular processes such as endothelial–mesenchymal transformation (4). Of these, the proliferation of embryonic cardiomyocytes is fundamental to forming a normally formed heart (34). Gata4 has roles in both embryonic and postnatal heart and numerous publications have demonstrated its role in early cardiogenesis and early cardiac morphogenetic events (10,35). As we and others have shown, Gata4 is expressed in the heart throughout development and its role in regulating cardiac proliferation during later stages of cardiac development is becoming increasingly recognized. Gata4 has been shown to regulate CyclinD2 and Cdk4 during cardiac development (26,36). In addition, we previously reported that a human disease-causing point mutation in GATA4 results in cardiomyocyte proliferation defects in vivo and likely contributes to the development of defects in atrial septation (16). While initial studies suggested that Tbx5 could inhibit cardiomyocyte proliferation when overexpressed in an embryonic chick heart, recent work has shown that a critical role in cell cycle progression and as a direct regulator of Cdk6, a cell cycle-dependent kinase (19,37,38). Our work demonstrates that Tbx5 promotes cardiomyocyte proliferation in cooperation with Gata4 in the regulation of Cdk4 and may also be involved in the regulation of Cdk2.
AVSDs, which account for about 5% of all CHD, were traditionally thought to be solely the result of improper endocardial cushion development, (39,40) and recent publications have highlighted the importance of the DMP, a component of the second heart field, in atrioventricular septation (18–20). Our findings show the importance of a normal ventricular myocardium during the remodeling of the endocardial cushions into portions of the tricuspid and mitral valves along with inferior and superior portions of the atrial and ventricular septae, respectively (19,20,41). Although not completely elucidated, proper development of the embryonic myocardium has previously been demonstrated to be critical for atrioventricular septation as myocardial deletion of several members of the Bmp-signaling pathway, including Bmp4, Alk3 and Smad4, results in cardiac septation defects along with cardiomyocyte proliferation defects (20,42,43). While our work suggests that the AVSD phenotype identified in Gata4;Tbx5 and Gata4MyoDel/wt;Tbx5+/− (Gata4flox/wt;αMHC-Cre+;Tbx5+/−) embryos is the result of perturbations in myocardial signaling to the adjacent atrioventricular cushion mesenchyme although we cannot exclude the possibility that loss of the DMP is also contributing to the AVSD phenotype. A defect in DMP development is found in a subset of Tbx5 heterozygote mice but they exhibit only ostium primum atrial septal defects (19,31). It remains a possibility that the AVSD phenotype in the Gata4;Tbx5 compound heterozygotes may be the result of both a perturbation of myocardial proliferation and abnormal DMP development. Of note, a subset of mice harboring a hypomorphic allele for Gata4 in a C57BL6/J background display AVSD (44). Gata4 has not been described to have a role in the development of the DMP, suggesting that loss of Gata4 alone potentially by affecting myocardial proliferation can lead to AVSD. Additional investigation is required to define the molecular pathways regulated by Gata4 and Tbx5 in the myocardium and DMP during atrioventricular septation. Ultimately, identification of these downstream targets will lead to an improved understanding of the molecular basis of AVSD and potential identify novel candidate genes for AVSD in humans.
MATERIALS AND METHODS
Ethics statement
Research was approved by the Research Institute at Nationwide Children's Hospital (Protocol No. AR09-00040) and conforms to the Guide for the Care and Use of Laboratory Animals.
Mouse strains and genotyping
Mice heterozygous for null alleles for Gata4 and Tbx5 were generated and genotyped as previously described (14). These lines have been maintained in mixed genetic backgrounds: Gata4 are in 129/C57BL6 while Tbx5 are in Black Swiss/129. To generate the double heterozygote mice used in this study, Gata4 heterozygote mice were mated to Tbx5 heterozygote mice. Mice harboring the Gata4-floxed allele, Tie2-Cre allele and α-MHC Cre allele have been previously generated and were genotyped as previously described (21–23).
Breeding and collection of mouse embryos
Mice were maintained on a 0600 to 1800 hours light–dark cycle, with noon of the day of observation of a vaginal plug defined as E0.5. Pregnant mothers were sacrificed at various embryonic timepoints. Littermates were used as controls for histologic sections, gene expression studies and quantitative reverse transcription-polymerase chain reaction (qRT–PCR).
Histologic section
For histological analysis, embryos hearts were fixed in 4% paraformaldehyde (PFA) and paraffin embedded. Hematoxylin and eosin (H&E) staining was carried out on heart sections using standard methodology.
Immunohistochemistry
Histologic sections were deparafinnized in xylene and rehydrated in phosphate-buffered saline (PBS). Antigen retrieval was performed on sections with sodium citrate buffer at 120°C for 30 min and 0.3% hydrogen peroxide at room temperature for 45 min. Sections were then blocked by 5% goat, rabbit or horse serum in PBS followed by incubation with mouse anti-α-MHC (1:400, ab15, Abcam, Cambridge, MA, USA), goat anti-Gata4 (1:500, sc-1237, Santa Cruz, Dallas, TX, USA) and rabbit anti-Tbx5 (1:500, 426500, Invitrogen, Grand Island, NY, USA) antibodies overnight at 4°C. Sections were then washed in PBS and stained with secondary antibody biotinylated goat anti-mouse IgG (1;500, BA-9200, Vector Labs, Burlingame, CA, USA), biotinylated rabbit anti-goat IgG (1:500, BA-5000, Vector Labs) and biotinylated horse anti-rabbit IgG (1:500, BA-5000v, Vector Labs) for 1 h at room temperature. Sections were washed in PBS and incubated in ABC solution (PK-6100, Vector Labs) at room temperature for 30 min followed by DAB solution (SK-4100, Vector Labs) for 2 min. The sections were then counterstained in hematoxylin (H-3404, Vector Labs) and dehydrated.
Myocardium wall thickness measurement
The myocardial wall thickness of atria and ventricle was determined by averaging three measurements of the width of atrial wall and the width from the subepicardial layer to the edge of the compact ventricular myocardium, respectively, in histological sections from each embryo examined. Minimum of n = 3 per group.
Isolation and culturing of embryonic cardiomyocytes
Cardiomyocytes were prepared from E11.5 mouse embryos by the conventional preplating method (45). Briefly, hearts were minced and digested with collagenase type II (C1889, Sigma-Aldrich, St. Louis, MO, USA) solution. To enrich for cardiomyocytes, the cells were preplated for 2 h to remove the nonmyocyte population. Cells were cultured in DMEM medium containing 10% FBS after transfection.
Immunofluorescence
Histologic sections were deparafinnized in xylene and rehydrated in PBS. Sections were fixed in 2.5% PFA at 4°C for 15 min and permeabilized in 0.1 Triton X-100 in PBS at room temperature for 3 min. Sections were then blocked by 1% BSA at room temperature for 30 min followed by incubation with mouse anti-α-MHC (1:250, ab15, Abcam), rabbit anti-PH3 (1:500, 06-570, Upstate, Billerica, MA, USA), goat anti-Gata4 (1:50, sc-1237, Santa Cruz) and rabbit anti-Tbx5 (1:25, 426500, Invitrogen) antibodies overnight at 4°C. Sections were then washed in PBS and stained with secondary antibodies Alexa Fluor 488 anti-rabbit (1:200, A-11008, Invitrogen), Alexa Fluor 488 anti-mouse (1:200, A-11001, Invitrogen) and Alexa Fluor 594 anti-goat (1:200, A-11055, Invitrogen) for 1 h at room temperature.
Immunoblotting
Murine atria and ventricles were isolated at different embryonic and postnatal timepoints and homogenized in RIPA buffer using a Dounce homogenizer. The cell lysate was obtained from tissue after incubation on ice for 30 min and centrifugation at 10 000g for 10 min at 4°C (repeated twice). Forty micrograms of cell lysate were loaded per lane and separated using 10% SDS–acrylamide gels, and transferred to immuneblot PVDF membranes (Bio-rad). After blocking with 5% nonfat milk in PBST, the membrane was probed with primary monoclonal mouse anti-Gata4 (1:200, sc-25310, Santa Cruz), rabbit anti-Tbx5 (1:200, 426500, Invitrogen) and rabbit anti-β-actin (1:5000, ab1801, Abcam) antibodies. The membranes were further probed with horseradish peroxidase-conjugated horse anti-mouse IgG (PI-2000), and goat anti-rabbit (PI-1000) (1:5000, Vector Labs). Cultured cardiomyocyte cells were also used for immunoblotting after lysing with RIPA lysis buffer and probed with rabbit anti-CDK4 (sc-260, Santa Cruz) primary monoclonal antibodies.
Co-immunoprecipitation
Atria and ventricles of frozen hearts at E11.5 and E14.5 were weighed and added to lysis buffer (20 mm Tris HCl pH 8.0, 137 mm NaCl, 10% glycerol, 1% Nonidet P-40, 2 mm EDTA) to a final volume of 200 mg/ml. Tissue was homogenized for 10 s and incubated at 4°C with rotation for 1 h before centrifugation at 10 000g for 20 min to remove insoluble debris. Following protein normalization, lysates were precleared using Dynabeadsprotein G (10003D, Invitrogen) for 30 min at 4°C with rotation. Beads were discarded, and 200 μg of precleared lysate was used per reaction. Lysates were incubated for overnight at 4°C with rotation with 2 μg of polyclonal rabbit anti-Tbx5 (426500, Invitrogen). Dynabeads protein G (20 μl) was added to each reaction, and tubes were rotated for a further 1 h at 4°C. Beads were washed four times for 5 min each time with 1 ml lysis buffer on ice using a Dynamag-2 magnet (12321D, Invitrogen). Proteins were eluted from beads in 10 μl 2× NuPAGE sample buffer and subjected to SDS–PAGE electrophoresis and western blotting with mouse anti-Gata4 (1:200, sc-25310, Santa Cruz) as described above.
Duolink assay
Interaction of Gata4 with Tbx5 was detected by the proximity ligation assay (PLA) kit Duolink (Olink Bioscience, Uppsala, Sweden), PLA probe anti-rabbit minus for the detection of the rabbit anti-Tbx5 antibody (426500, Invitrogen); PLA probe anti-goat PLUS for the detection of the mouse anti-Gata4 (sc-25310, Santa Cruz) antibodies. The PLA was performed according to the manufacturer's protocol; negative controls were included (44). Based on in situ PLA technology, Gata4 and Tbx5 antibodies were recognized by PLA probes, which contain unique DNA strands that will amplify when two PLA probes are in close proximity (<40 nm). The labeled complementary oligonucleotide probes will highlight the amplification product to indicate the interaction of Gata4 and Tbx5.
Gene expression analysis
RNA was purified from hearts at embryonic day 11.5 of mutant embryos and their respective wild-type littermates using Trizol (Invitrogen). One hundred nanograms of total RNA were used for reverse transcription using the SuperScript® VILO™cDNA Synthesis Kit (Invitrogen) and real-time qRT–PCR was performed using Applied Biosystems 7500 real-time PCR machine. Commercially available SYBR Green (Applied Biosystems, Carlsbad, CA, USA) PCR mix was utilized for the following genes: mouse Cdk2, Cdk4. The sequences for primers are shown Supplementary Material, Table S1. Mean relative gene expression was calculated after normalized to 18S ribosomal RNA using the ΔΔCt method. Three independent experiments were performed in triplicate.
Knockdown of Gata4 and Tbx5 using siRNA
Two pre-designed siRNAs targeting the coding region of the murine Gata4 (s201378, Life Technology, Grand Island, NY, USA) and murine Tbx5 (s74784, Life Technology) genes were used in this study. Negative control siRNA (4390843, Life Technology) was used as a control siRNA targeting no known gene. For transfection, electroporation reactions were conducted with Nucleofector® transfection system (Lonza, Basel, Switzerland) according to the manufacturer's protocol with the isolated mouse embryonic cardiomyocytes.
Luciferase assay
The mouse CDK4 pAUG-β-gal reporter was generously provided to us by Dr B.L. Black (26). A 827-bp fragment containing 771 bp upstream and 56 bp downstream of the transcriptional start site from the mouse Cdk4 promoter region was cloned into pCR™2.1-TOPO vector (Invitrogen) by using the following primers: 5′-CTTTTTAATATTCCGCGGGAGGTTTAC-3′ and 5′-GGGCAGCTGGATCCTTCGGGCCAGAC-3′. The Cdk4 promoter was then subcloned into pGL3-Basic Vector (Promega, Madison, WI, USA) by using KpnI and XhoI according to the manufacturer's protocol. A 1106-bp fragment containing 843 bp upstream and 263 bp downstream of the transcriptional start site from the mouse Cdk2 promoter region was cloned into pCR™2.1-TOPO vector (Invitrogen) by using the following primers: 5′-ATGAACCCCGAAACAGTGAG-3′ and 5′-TGCCCTCTCCAATCTTCTCCA-3′. The Cdk2 promoter was then subcloned into pGL3-Basic Vector (Promega) by using KpnI and XhoI according to the manufacturer's protocol. The directionality of all the resulting constructs were confirmed by sequencing using both vector- and insert-specific primers. HeLa cells were transiently transfected with 100 ng Cdk4-luciferase reporter or Cdk2-luciferase reporter plus 50 ng pCMV-lacZ plasmid, in combination with 100 ng of Gata4 myc-tagged or Tbx5 FLAG-tagged expression vectors using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Luciferase activity was measured 48 h after transfection using the Luciferase Reporter Assay (Promega) according to the manufacturer's protocol as previously described (46). Mean luciferase activity was calculated after normalization to β-galactosidase assay. Three independent experiments were performed in triplicate.
Chromatin immunoprecipitation
ChIP was performed from 60 pooled E13.5 embryonic hearts using EZ-ChIP™ (Millipore, Billerica, MA, USA) according to the manufacturer's protocol. Briefly, chromatin from fixed hearts was sonicated and immunoprecipitation was performed with primary monoclonal mouse anti-Gata4 (1:40, sc-25310, Santa Cruz) and polyclonal goat anti-Tbx5 (1:40, sc-17866, Santa Cruz) antibodies followed by reversal of cross-linking and DNA purification. Immunoenriched DNA targets were then amplified by PCR for the promoter region of the following genes: murine Cdk2 and Cdk4. The sequences for primers are shown in Supplementary Material, Table S1.
Statistical analysis
Statistical comparisons were performed using Student's t-test, and a P value of <0.05 was considered significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by funding from the National Institutes of Health (R01 HL088965) and American Heart Association to V.G.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Morphology Core at the Research Institute at Nationwide Children's Hospital for histology support, Dr W.T. Pu for providing us mice harboring the Gata4-floxed allele, Dr B.G. Bruneau for providing the Tbx5 mutant mice, Dr B.L. Black for providing us with the mouse CDK4 pAUG-β-gal reporter construct and Dr J. Lincoln for critical review of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Fahed A.C., Gelb B.D., Seidman J.G., Seidman C.E. Genetics of congenital heart disease: the glass half empty. Circ. Res. 2013;112:707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman J.I., Kaplan S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava D. Genetic regulation of cardiogenesis and congenital heart disease. Annu. Rev. Pathol. 2006;1:199–213. doi: 10.1146/annurev.pathol.1.110304.100039. [DOI] [PubMed] [Google Scholar]
- 4.Garg V. Insights into the genetic basis of congenital heart disease. Cell. Mol. Life. Sci. 2006;63:1141–1148. doi: 10.1007/s00018-005-5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards A.A., Garg V. Genetics of congenital heart disease. Curr. Cardiol. Rev. 2010;6:91–97. doi: 10.2174/157340310791162703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson E.N. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCulley D.J., Black B.L. Transcription factor pathways and congenital heart disease. Curr. Top. Dev. Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basson C.T., Bachinsky D.R., Lin R.C., Levi T., Elkins J.A., Soults J., Grayzel D., Kroumpouzou E., Traill T.A., Leblanc-Straceski J., et al. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 9.Garg V., Kathiriya I.S., Barnes R., Schluterman M.K., King I.N., Butler C.A., Rothrock C.R., Eapen R.S., Hirayama-Yamada K., Joo K., et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 10.Zhou P., He A., Pu W.T. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr. Top. Dev. Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
- 11.Greulich F., Rudat C., Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 2011;91:212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 12.Sarkozy A., Conti E., Neri C., D'Agostino R., Digilio M.C., Esposito G., Toscano A., Marino B., Pizzuti A., Dallapiccola B. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J. Med. Genet. 2005;42:e16. doi: 10.1136/jmg.2004.026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang R., Fan L.L., Huang H., Cao B.B., Li X.P., Peng D.Q., Xia K. A novel mutation of GATA4 (K319E) is responsible for familial atrial septal defect and pulmonary valve stenosis. Gene. 2014;534:320–323. [PubMed] [Google Scholar]
- 14.Maitra M., Schluterman M.K., Nichols H.A., Richardson J.A., Lo C.W., Srivastava D., Garg V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev. Biol. 2009;326:368–377. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Packham E.A., Brook J.D. Interaction makes the heart grow stronger. Trends. Mol. Med. 2003;9:407–409. doi: 10.1016/j.molmed.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Misra C., Sachan N., McNally C.R., Koenig S.N., Nichols H.A., Guggilam A., Lucchesi P.A., Pu W.T., Srivastava D., Garg V. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS. Genet. 2012;8:e1002690. doi: 10.1371/journal.pgen.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blom N.A., Ottenkamp J., Wenink A.G., Gittenberger-de Groot A.C. Deficiency of the vestibular spine in atrioventricular septal defects in human fetuses with down syndrome. Am J Cardiol. 2003;91:180–184. doi: 10.1016/s0002-9149(02)03106-5. [DOI] [PubMed] [Google Scholar]
- 18.Snarr B.S., Wirrig E.E., Phelps A.L., Trusk T.C., Wessels A. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev Dyn. 2007;236:1287–1294. doi: 10.1002/dvdy.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie L., Hoffmann A.D., Burnicka-Turek O., Friedland-Little J.M., Zhang K., Moskowitz I.P. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell. 2012;23:280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs L.E., Phelps A.L., Brown E., Kakarla J., Anderson R.H., van den Hoff M.J., Wessels A. Expression of the BMP receptor Alk3 in the second heart field is essential for development of the dorsal mesenchymal protrusion and atrioventricular septation. Circ. Res. 2013;112:1420–1432. doi: 10.1161/CIRCRESAHA.112.300821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pu W.T., Ishiwata T., Juraszek A.L., Ma Q., Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev. Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Kisanuki Y.Y., Hammer R.E., Miyazaki J., Williams S.C., Richardson J.A., Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 23.Gaussin V., Van de Putte T., Mishina Y., Hanks M.C., Zwijsen A., Huylebroeck D., Behringer R.R., Schneider M.D. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc. Natl Acad. Sci. USA. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthet C., Klarmann K.D., Hilton M.B., Suh H.C., Keller J.R., Kiyokawa H., Kaldis P. Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Dev. Cell. 2006;10:563–573. doi: 10.1016/j.devcel.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Barriere C., Santamaria D., Cerqueira A., Galan J., Martin A., Ortega S., Malumbres M., Dubus P., Barbacid M. Mice thrive without Cdk4 and Cdk2. Mol. Oncol. 2007;1:72–83. doi: 10.1016/j.molonc.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojas A., Kong S.W., Agarwal P., Gilliss B., Pu W.T., Black B.L. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol. Cell. Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh T.K., Packham E.A., Bonser A.J., Robinson T.E., Cross S.J., Brook J.D. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet. 2001;10:1983–1994. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- 28.Xin M., Olson E.N., Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell. Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durocher D., Charron F., Warren R., Schwartz R.J., Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO. J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiroi Y., Kudoh S., Monzen K., Ikeda Y., Yazaki Y., Nagai R., Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 31.Bruneau B.G., Nemer G., Schmitt J.P., Charron F., Robitaille L., Caron S., Conner D.A., Gessler M., Nemer M., Seidman C.E., et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 32.He A., Kong S.W., Ma Q., Pu W.T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl Acad. Sci. USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlesinger J., Schueler M., Grunert M., Fischer J.J., Zhang Q., Krueger T., Lange M., Tönjes M., Dunkel I., Sperling S.R. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS. Genet. 2011;7:e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahuja P., Sdek P., MacLellan W.R. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol. Rev. 2007;87:521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallagher J.M., Komati H., Roy E., Nemer M., Latinkic B.V. Dissociation of cardiogenic and postnatal myocardial activities of GATA4. Mol. Cell. Biol. 2012;32:2214–2223. doi: 10.1128/MCB.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamak A., Temsah R., Maharsy W., Caron S., Paradis P., Aries A., Nemer M. Cyclin D2 rescues size and function of GATA4 haplo-insufficient hearts. Am. J. Physiol. Heart. Circ. Physiol. 2012;303:H1057–H1066. doi: 10.1152/ajpheart.00250.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatcher C.J., Kim M.S., Mah C.S., Goldstein M.M., Wong B., Mikawa T., Basson C.T. TBX5 transcription factor regulates cell proliferation during cardiogenesis. Dev. Biol. 2001;230:177–188. doi: 10.1006/dbio.2000.0134. [DOI] [PubMed] [Google Scholar]
- 38.Goetz S.C., Brown D.D., Conlon F.L. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133:2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B., Weidenfeld J., Lu M.M., Maika S., Kuziel W.A., Morrisey E.E., Tucker P.W. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131:4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- 40.Wu B., Wang Y., Lui W., Langworthy M., Tompkins K.L., Hatzopoulos A.K., Baldwin H.S., Zhou B. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circ. Res. 2011;109:183–192. doi: 10.1161/CIRCRESAHA.111.245035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian Y., Yuan L., Goss A.M., Wang T., Yang J., Lepore J.J., Zhou D., Schwartz R.J., Patel V., Cohen E.D., et al. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev. Cell. 2010;18:275–287. doi: 10.1016/j.devcel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao K., Kulessa H., Tompkins K., Zhou Y., Batts L., Baldwin H.S., Hogan B.L. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes. Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi X., Yang G., Yang L., Lan Y., Weng T., Wang J., Wu Z., Xu J., Gao X., Yang X. Essential role of Smad4 in maintaining cardiomyocyte proliferation during murine embryonic heart development. Dev. Biol. 2007;311:136–146. doi: 10.1016/j.ydbio.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 44.Rajagopal S.K., Ma Q., Obler D., Shen J., Manichaikul A., Tomita-Mitchell A., Boardman K., Briggs C., Garg V., Srivastava D., et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J. Mol. Cell Cardiol. 2007;43:677–685. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ieda M., Kanazawa H., Kimura K., Hattori F., Ieda Y., Taniguchi M., Lee J.K., Matsumura K., Tomita Y., Miyoshi S., et al. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat. Med. 2007;13:604–612. doi: 10.1038/nm1570. [DOI] [PubMed] [Google Scholar]
- 46.Chang S-W., Mislankar M., Misra C., Huang N., DaJusta D., Harrison S.M., McBride K.L., Baker L.A., Garg V. Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat. 2013;34:1226–1230. doi: 10.1002/humu.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.