Abstract
Complex I of the mitochondrial respiratory chain is a large multisubunit enzyme that assembles from nuclear and mtDNA-encoded components. Several complex I assembly factors have been identified, but their precise functions are not well understood. Here, we have investigated the function of one of these, NDUFAF7, a soluble matrix protein comprised of a DUF185 domain that harbors a methyltransferase motif. Knockdown of NDUFAF7 by siRNA in human fibroblasts produced a specific complex I assembly defect, as did morpholino-mediated knockdown of the zebrafish ortholog. Germline disruption of the murine ortholog was an early embryonic lethal. The complex I assembly defect was characterized by rapid, AFG3L2-dependent, turnover of newly synthesized ND1, the subunit that seeds the assembly pathway, and by decreased steady-state levels of several other structural subunits including NDUFS2, NDUFS1 and NDUFA9. Expression of an NDUFAF7 mutant (G124V), predicted to disrupt methyltransferase activity, impaired complex I assembly, suggesting an assembly factor or structural subunit as a substrate for methylation. To identify the NDUFAF7 substrate, we used an anti-ND1 antibody to immunoprecipitate complex I and its associated assembly factors, followed by mass spectrometry to detect posttranslational protein modifications. Analysis of an NDUFAF7 methyltransferase mutant showed a 10-fold reduction in an NDUFS2 peptide containing dimethylated Arg85, but a 5-fold reduction in three other NDUFS2 peptides. These results show that NDUFAF7 functions to methylate NDUFS2 after it assembles into a complex I, stabilizing an early intermediate in the assembly pathway, and that this function is essential for normal vertebrate development.
INTRODUCTION
NADH ubiquinone oxidoreductase (complex I) is the first complex of the oxidative phosphorylation system (OXPHOS). It catalyzes the oxidation of NADH and couples this to the translocation of protons from the mitochondrial matrix to the mitochondrial intermembrane space, contributing to the generation of a proton gradient that drives ATP production by complex V. Mammalian complex I is composed of 44 structural subunits, seven of which are encoded by the mitochondrial genome. It harbors the electron acceptor flavin mononucleotide and seven-iron sulfur clusters. Complex I has an L-shaped structure with a hydrophobic domain embedded in the inner mitochondrial membrane and a hydrophilic arm protruding towards the mitochondrial matrix. Deficiencies in complex I account for ∼40% of patients with OXPHOS disorders, whose incidence is ∼1:5000 live births (1, 2). These disorders are generally of early onset with severe phenotypes that result in early fatality (3).
The assembly of complex I initiates with the formation of a membrane intermediate containing the mitochondrial ND1subunit and a soluble subassembly of 200 kDa containing the nuclear subunits NDUFS2, NDUFS3, NDUFS7, NDUFS8 and NDUFA9. These two early assembly intermediates form a membrane-anchored subcomplex of ∼315 kDa (4) that nucleates the assembly of the holo-complex in a step-wise manner (5, 6). This process is orchestrated by nuclear-encoded assembly factors that include NDUFAF1-6, ACAD9, FOXRED1, NUBPL, ECSIT, AIF and TMEM126B; however, the fact that 40% of complex I deficient patients have no mutations in the structural subunits, makes it likely that additional assembly factors remained to be identified.
NDUFAF7 was identified as a nuclear-encoded mitochondrial protein with a putative role in complex I biogenesis using a genome subtraction strategy (7) and phylogenetic profiling (8, 9). It is a protein of broad taxonomic distribution, conserved from bacteria to mammals, that contains a PFAM domain of unknown function, DUF185 (10). In silico analysis using the COMPASS (Comparison of Multiple Protein Alignments with Assessment of Statistical Significance) platform identified a shared conserved region named motif I, which is present in RNA methylases with an S-adenosyl-l-methionine (SAM)-dependent methyltransferase fold (11). Additionally, structural modeling and biochemical characterization of NDUFAF7 homolog MidA of the slime mold Dictyostelium discoideum suggests that human NDUFAF7 methyltransferase domain is functionally relevant for complex I biogenesis (12).
Here, we have investigated the function of NDUFAF7 and show that it is an arginine methyltransferase essential for complex I biogenesis and embryonic development.
RESULTS
NDUFAF7 is a soluble mitochondrial matrix protein
We initially identified NDUFAF7 using a yeast genome subtraction strategy to identify putative complex I assembly factors (7), a result confirmed by phylogenetic profiling (9, 13). NDUFAF7 has 10 exons and 13 predicted transcript variants; however, only variants 1 and 4 are predicted to be translated into polypeptides of 49 and 38 kDa. An RT-PCR experiment in human fibroblasts identified two transcripts of 1.3 and 1.0 kb (Fig. 1A). A schematic of the structure of the two isoforms is shown in Supplementary Material, Figure S1. Both isoforms are also detected by immunoblot analysis (Supplementary Material, Fig. S2); however, we focused our studies on the long isoform. The long NDUFAF7 isoform encodes a 441 amino acid protein with a predicted mitochondrial targeting sequence. Immunofluorescence experiments on fibroblasts expressing NDUFAF7-HA showed that the protein was exclusively localized to the mitochondrial compartment (Fig. 1B), as had been previously reported (14), and this was confirmed by subcellular fractionation experiments (Fig. 1C). To determine whether NDUFAF7 was associated with mitochondrial membranes, purified mitochondria were extracted with alkaline carbonate. NDUFAF7 behaved like SDHA, a subunit of complex II, a peripherally associated mitochondrial protein, and not like the mitochondrial AAA+ protease AFG3L2 or the structural subunit of complex IV, COX2, both of which are integral inner mitochondrial membrane proteins (Fig. 1D).
Figure 1.
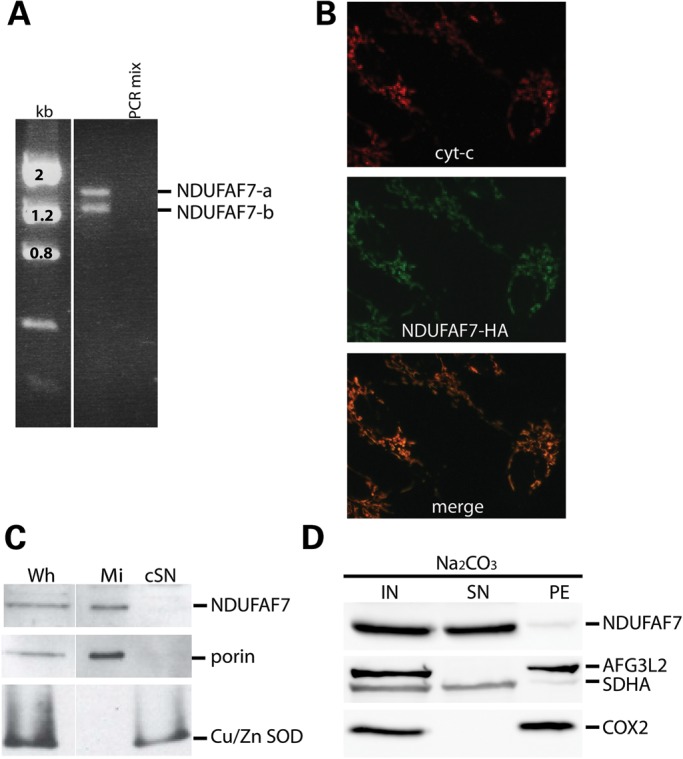
NDUFAF7 is a soluble mitochondrial protein. (A) Analysis of NDUFAF7 RT-PCR products showing two transcript variants in human fibroblasts. (B) Immunocytochemistry of control cells expressing NDUFAF7-HA. Antibodies to hemagglutinin (HA) and cytochrome c were used as indicated (C) SDS–PAGE immunoblot analysis of native NDUFAF7, porin and Cu/Zn SOD in whole-cell extracts (Wh), mitochondria (Mi) and cytosolic supernatants (cSN) (D) SDS–PAGE immunoblot analysis of alkaline carbonate extracted mitochondrial fractions: Input (IN), supernatant (SN) and pellet (PE). This analysis was performed in cells expressing an untagged version of the long isoform of NDUFAF7.
NDUFAF7 is essential for complex I assembly
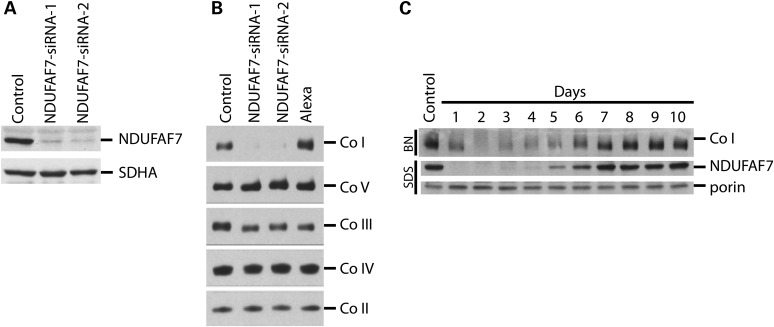
To investigate the function of NDUFAF7, we transiently knocked it down for 6 days using siRNA duplexes specifically targeted to the NDUFAF7 mRNA (NDUFAF7-siRNA-1 and siRNA-2). Immunoblot analysis showed decreased levels of NDUFAF7 protein using both siRNAs (Fig. 2A). To assess the steady-state levels of each of the five complexes of the OXPHOS, we performed a Blue Native PAGE (BN–PAGE) analysis on the knockdown cell lines (Fig. 2B). Fully assembled complex I was severely depleted in both cell lines, whereas the other OXPHOS complexes were unaffected. The NDUFAF7-siRNA-2 cell line was assayed by BN–PAGE and SDS–PAGE every day for 10 days to investigate the relationship between complex I assembly deficiency and the steady-state level of NDUFAF7 (Fig. 2C). This analysis showed that complex I re-assembled shortly after the recovery of NDUFAF7 at Day 5, demonstrating that its assembly depends on NDUFAF7.
Figure 2.
NDUFAF7 knockdown by siRNA. (A) Immunoblot analysis of NDUFAF7 and SDHA in control and two knockdown cell lines: NDUFAF7-siRNA-1 and NDUFAF7-siRNA-2. (B) BN–PAGE analysis of OXPHOS complexes (Co I–Co V) detected with subunit-specific antibodies in control, Alexa (siRNA transfection control) and knockdown cell lines: NDUFAF7-siRNA-1 and NDUFAF7-siRNA-2 (C) NDUFAF7 knockdown recovery analysis of Complex I (Co I) and NDUFAF7 by BN–PAGE (BN) and SDS–PAGE (SDS), respectively. Porin was used as a loading control.
NDUFAF7 is an early complex I assembly factor
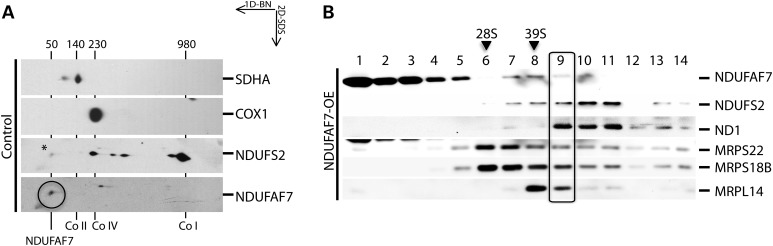
To investigate whether NDUFAF7 is organized as a higher molecular-weight complex, we performed 2D-SDS–PAGE experiments in control fibroblasts (Fig. 3A). NDUFAF7 was detected at ∼50 kDa, corresponding to its monomeric molecular weight, indicating that the majority of NDUFAF7 does not associate with a complex I subassembly. NDUFS2, a nuclear-encoded structural subunit of complex I that is required in the first steps of complex I assembly pathway was immunodetected in ∼200, 315 and 370 kDa complex I subassemblies (re-estimates of the sizes of these subcomplexes in (4) and in the fully assembled 980 KDa complex). To further explore a possible relationship between NDUFAF7 and complex I assembly intermediates, we analyzed a sucrose density gradient using an NDUFAF7 overexpressing cell line (Fig. 3B). Immunoblot analysis detected NDUFAF7 in fractions 1–5 and 7–10. Two complex I subunits, NDUFS2 and ND1, showed a peak in fractions 10 and 11, suggesting that some fraction of NDUFAF7 might interact with a complex I subassembly. Immunoprecipitation of the ND1 subunit from isolated mitochondria, followed by mass spectrometry analysis of chymotryptic peptides, identified NDUFAF7 (in addition to the majority of structural subunits and other assembly factors), further supporting a stable association with a subcomplex containing ND1 (data not shown).
Figure 3.
NDUFAF7 complex association analysis. (A) 2D-SDS–PAGE analysis of NDUFAF7 in a control cell line. NDUFAF7, complex I (Co I), complex IV (Co IV) and complex II (Co II) were identified by immunodetection of NDUFAF7, NDUFS2, COX1 and SDHA with specific antibodies. Numbers indicate the molecular weight of NDUFAF7 and fully assembled OXPHOS complexes. *Residual signal from NDUFAF7 antibody. (B) Sucrose density gradient analysis of NDUFAF7 overexpressing cell line (NDUFAF7-OE) followed by immunodetection of NDUFAF7 and complex I structural subunits ND1 and NDUFS2. Immunoblots of MRPS22, MRPS18B and MRPL14 indicate the peaks for the small (28S) and large (39S) mitochondrial ribosomal subunits.
NDUFAF7 knockdown decreases ND1 pulse labeling
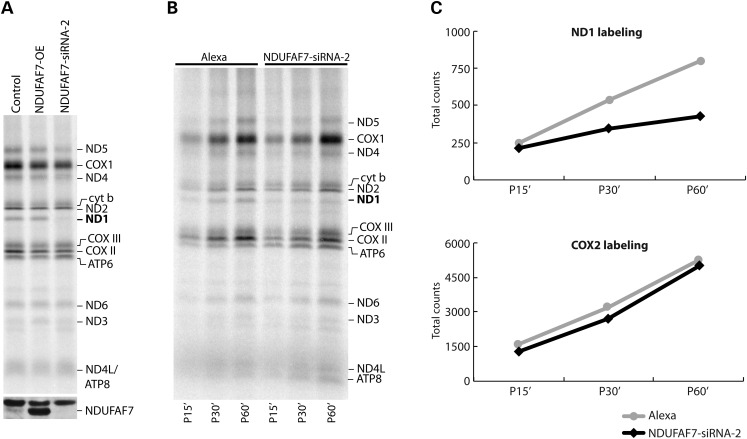
To test whether NDUFAF7 has a role in the translation of mt-DNA encoded polypeptides, control, NDUFAF7 over expressing (NDUFAF7-OE) and NDUFAF7-siRNA-2 cell lines were pulse-labeled labeled with [35S]methionine/cysteine in the presence of emetine to inhibit cytosolic translation (Fig. 4A). The knockdown of NDUFAF7 consistently resulted in an 80% decrease in the labeling of the ND1 subunit of complex I, whereas the labeling of all other complex I subunits (ND subunits) as well as subunits of complexes III (cyt b), IV (COX subunits) and V (ATP subunits) appeared normal. Northern blot analysis showed that the pulse translation phenotype observed in the NDUFAF7 knockdown was not due to an alteration in the steady-state levels of the ND1 transcript. In fact, ND1 mRNA levels were slightly increased compared with controls, and none of the other mitochondrial mRNAs tested were significantly different than in controls (Supplementary Material, Fig. S3).
Figure 4.
Translation phenotype in NDUFAF7 knockdown cells. (A) The mtDNA-encoded polypeptides were pulse labeled with [35S] methionine/cysteine for 60 min in control, NDUFAF7 overexpressing (OE) and NDUFAF7-siRNA-2 cell lines. The positions of the ND subunits of complex I; COX, subunits of complex IV, cyt b, subunit of complex III; ATP, subunits of complex V are indicated on the right. Immunoblot analysis (bottom) shows levels of NDUFAF7 in all cell lines (B) Pulse labeling for 15, 30 and 60 min of the 13 mtDNA-encoded polypeptides with [35S] methionine/cysteine in alexa (siRNA transfection control) and NDUFAF7-siRNA-2 cell lines. The positions of the ND subunits of complex I; COX, subunits of complex IV, cyt b, subunit of complex III; ATP, subunits of complex V are indicated on the right. (C) Label incorporation into the ND1 and COX2 polypeptides (expressed as total counts) was quantified using ImageQuant 5.2 software and plotted to show the rate of incorporation in both Alexa control and NDUFAF7-siRNA-2 cell lines.
To further investigate the apparent translation phenotype observed in the NDUFAF7-siRNA-2 knockdown, cells were pulse labeled with a [35S]methionine/cysteine mix for 15, 30 and 60 min, and then chased for 10 min (Fig. 4B). The NDUFAF7 knockdown caused a very severe ND1-labeling defect at early time points (15 min) compared with control; however, the labeling rate of the other 12 mtDNA-encoded polypeptides remained unaffected, indicating that the effect is specific to ND1 (Fig. 4C).
ND1 is rapidly degraded in NDUFAF7 knockdown
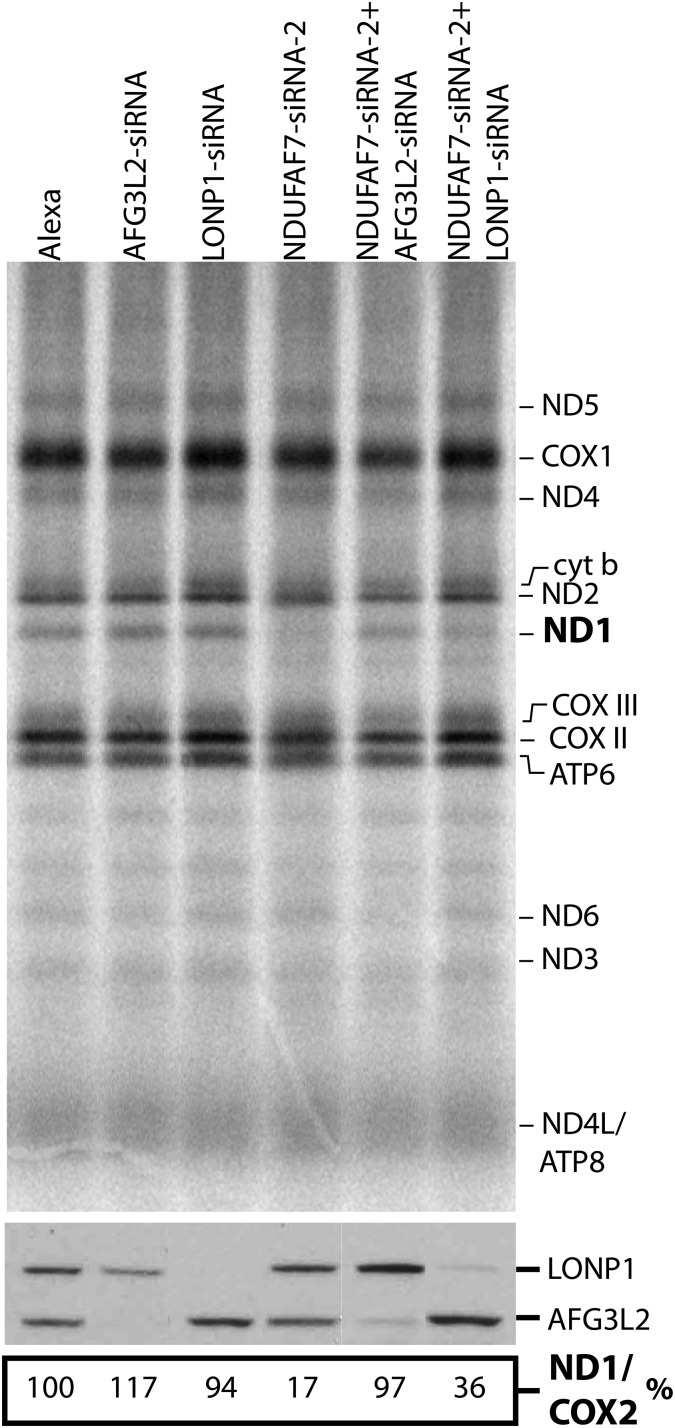
To directly test whether the pulse-labeling phenotype observed in NDUFAF7-siRNA-2 was due to decreased synthesis or to rapid degradation of the ND1 polypeptide, we depleted the mitochondrial inner membrane protease AFG3L2 and the mitochondrial soluble protease LONP1 by siRNA in the background of the NDUFAF7 knockdown. The proteases and NDUFAF7 were knocked down using siRNAs for 2 and 6 days, respectively, followed by a pulse translation experiment (Fig. 5). Depletion of AFG3L2 (NDUFAF7-siRNA-2+AFG3L2-siRNA) completely rescued ND1-labeling defect, whereas depletion of LONP1 showed only a partial rescue. These results demonstrate that the decrease in ND1 labeling in the pulse translation experiment is the result of rapid proteolysis of the ND1 subunit, largely mediated by AFG3L2.
Figure 5.
Knockdown of AFG3L2 rescues the ND1-labeling phenotype. Pulse labeling of mtDNA-encoded polypeptides with [35S] methionine/cysteine for 60 min in control (Alexa), AFG3L2-siRNA, LONP1-siRNA, NDUFAF7-siRNA-2 and double siRNA cell lines. The positions of the ND subunits of complex I; COX, subunits of complex IV, cyt b, subunit of complex III; ATP, subunits of complex V are indicated on the right. Immunoblot analysis (bottom) shows levels of LONP1 and AFG3L2 in all experiments. Numbers indicate incorporation of label in ND1 normalized to COX2.
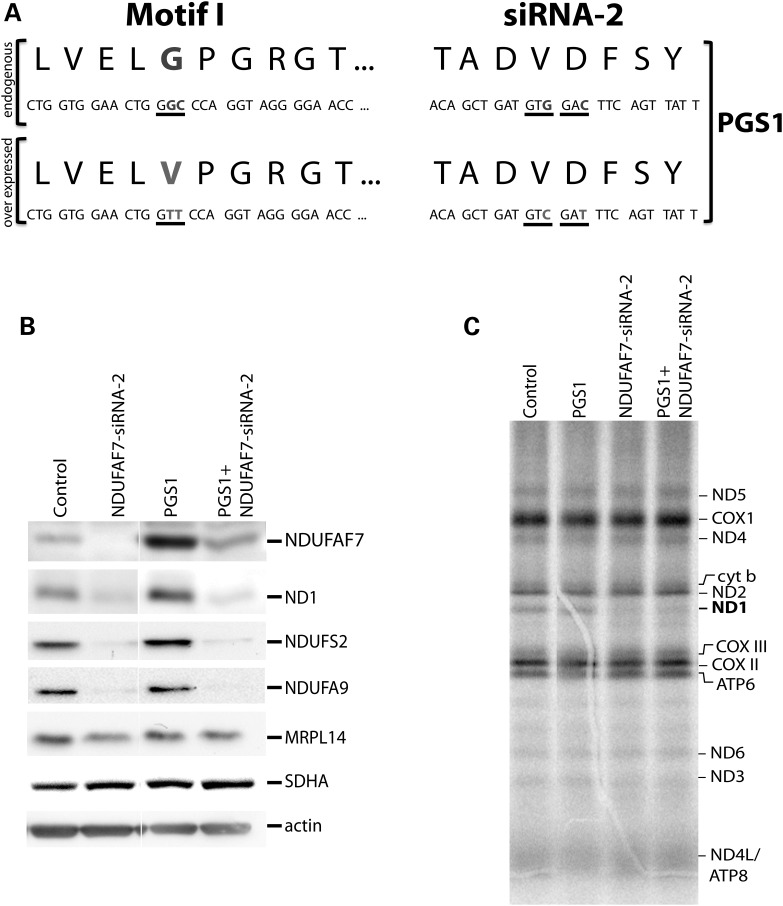
The methyltransferase motif I is necessary for NDUFAF7 function
Modeling and biochemical analysis performed on a mutant (G170V) in the DUF185 domain of MidA, the D. discoideum NDUFAF7 ortholog, suggested that the protein is an SAM-binding methyltransferase (12). To test this hypothesis in human cells, we generated a fibroblast cell line (PGS1) in which we expressed a construct containing a G124V mutation corresponding to the first glycine on motif I. Site-directed mutagenesis was used to generate two synonymous mutations in the vector expressing the G124V mutant, rendering it insensitive to NDUFAF7-siRNA-2. This strategy allowed us to selectively knockdown endogenous, wild-type NDUFAF7, while expressing the mutant (Fig. 6A). Immunoblot analysis of the NDUFAF7-siRNA-2 and PGS1-siRNA-2 cell lines showed nearly identical phenotypes: the steady-state levels of subunits ND1, NDUFS2 and NDUFA9 were severely reduced in both, whereas the large ribosomal subunit MRPL14 remained unaffected (Fig. 6B). A mitochondrial pulse translation experiment showed that ND1-labeling defect was reproducible in the PGS1-siRNA-2 cell line (Fig. 6C).
Figure 6.
Characterization of the methyltransferase motif I mutant cell line. (A) Base changes in motif 1 used to generate the G124V methyltransferase mutant, and the synonymous mutations incorporated into the same construct to render it insensitive to the siRNA-2 targeted to wild-type NDUFAF7. (B) Immunoblot analysis of complex I structural subunits ND1, NDUFS2 and NDUFA9 in control, NDUFAF7-siRNA-2 and PGS1 + NDUFAF7-siRNA-2 cell lines. The mitochondrial large ribosomal subunit MRPL14, SDHA and actin were used as loading controls (C) Pulse labeling of mtDNA-encoded polypeptides with [35S] methionine/cysteine for 60 min in control, PGS1, NDUFAF7-siRNA-2 and PGS1 + NDUFAF7-siRNA-2 cell lines. The positions of the ND subunits of complex I; COX, subunits of complex IV, cyt b, subunit of complex III; ATP, subunits of complex V are indicated on the right.
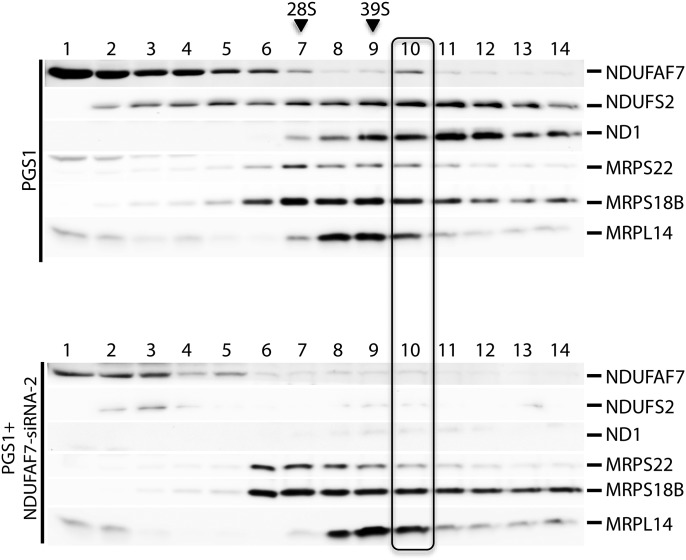
To further characterize the PGS1-siRNA-2 cell line, we performed a sucrose density gradient analysis (Fig. 7). While the majority of NDUFAF7 fractionated near the top of the gradient, a small amount was detectable in fraction 10 together with NDUFS2 and ND1, supporting the idea that some NDUFAF7 associates with a complex I intermediate.
Figure 7.
Sucrose gradient sedimentation analysis of cell lines expressing G124V NDUFAF7. Sucrose density gradient sedimentation analysis of PGS1 and PGS1 + NDUFAF7-siRNA-2 cell lines followed by immunodetection of NDUFAF7 and the complex I structural subunits NDUFS2 and ND1. Immunoblots of MRPS22, MRPS18B and MRPL14 indicate the peaks for the small (28S) and large (39S) mitochondrial ribosomal subunits.
Mass spectrometry analysis of PGS1 knockdown cell line
Taken together, the above data suggested that human NDUFAF7 acts as a protein methyltransferase whose substrate is a structural subunit or an assembly factor of complex I. In silico prediction (http://www.bioinfo.bio.cuhk.edu.hk/bpbppms/intro.jsp) together with pull-down experiments in Dictyostelium using the N-terminal fragment of NDUFS2-GST and cell extracts from MidA-GFP demonstrated that these two proteins interact, suggesting NDUFS2 as a putative NDUFAF7 substrate (12). Moreover, recent mass spectrometry data using 143B-cell line reported that NDUFS2 is dimethylated on R85, mediated by NDUFAF7 (14).
To identify NDUFAF7 substrates, we assessed the methylation profile of the PGS1-siRNA-2 fibroblast cell line following immunoprecipitation with an anti-ND1 antibody. This strategy allowed us to immunoprecipitate holo-complex I and several associated assembly factors (Supplementary Material, Table S1). Mass spectrometry analysis of the tryptic peptide KCDPHIGLLHRGTEK confirmed that Arg85 (numbered from the first amino acid in the mature, processed peptide) of NDUFS2 is symmetrically dimethylated in the control PGS1, but 10.6-fold less in the PGS1 knockdown cell line (Supplementary Material, Fig. S4; Table 1). As expected, all complex I structural subunits identified in the mass spectrometry analysis, including NDUFS2, were markedly reduced in the PGS1-siRNA-2 cell line. However, the relative abundance of two peptides in NDUFS1, one peptide of ND1, and three other peptides in NDUFS2 was reduced only ∼5-fold compared with control (Table 1). From these data, we conclude that a fraction of the NDUFS2 subunit is unmethylated in assembled complex I (or more likely in an assembly intermediate), in cells expressing the G124V NDUFAF7 mutant protein.
Table 1.
Mass spectrometry analysis of NDUFS2
| OXPHOS subunits | MS peptide | Relative abundance |
||
|---|---|---|---|---|
| PGS1 (control) | PGS1 + NDUFAF7-siRNA-2 (methyl transferase mutant) | Ratio (control/methyl transferase mutant) | ||
| NDUFS2 | (KCDPHIGLLHsmeRGTEK) | 5.79E + 07 | 5.46E + 06 | 10.6 |
| (NITLNFGPQHPAAHGVLR) | 1.88E + 08 | 3.90E + 07 | 5.7 | |
| (TQPYDVYDQVEFDVPVGSR) | 6.69E + 07 | 1.06E + 07 | ||
| (IIAQCLNK) | 9.99E + 07 | 1.70E + 07 | ||
| ND1 | (KGPNVVGPYGLLQPFADAMK) | 1.53E + 07 | 3.44E + 06 | 4.4 |
| NDUFS1 | (FASEIAGVDDLGTTGR) | 6.10E + 07 | 1.07E + 07 | 5.7 |
| (VAVTPPGLAR) | 7.91E + 07 | 1.40E + 07 | ||
| UQCRC2 | (TIAQGNLSNTDVQAAK) | 5.86E + 07 | 4.96E + 07 | 1.2 |
| (NALANPLYCPDYR) | 5.93E + 07 | 4.99E + 07 | ||
| COX2 | (LLDVDNR) | 8.56E + 07 | 4.73E + 07 | 1.6 |
| (VVLPIEAPIR) | 1.73E + 08 | 1.24E + 08 | ||
ND1 subunit immunoprecipitation in PGS1 and PGS1 knockdown (PGS1 + NDUFAF7-siRNA-2) cell lines followed by mass spectrometry analysis. The relative abundance of indicated tryptic peptides was obtained from the areas of Gaussian ion chromatograms. Where more than one peptide was identified, the mean ratio is shown.
Early defects in complex I assembly results in depletion of NDUFS2
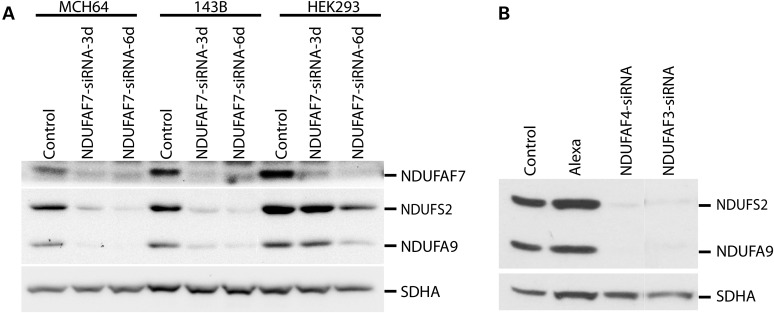
In the earliest stages of complex I assembly, the soluble subunit NDUFS2 and the membrane subunit ND1 form an ∼200 kDa subcomplex in the inner mitochondrial membrane. Early assembly defects result in the rapid degradation of the ND1 subunit and, as a concomitant effect, rapid turnover of many of the other complex I structural subunits (15–17). To confirm that the early complex I assembly defect resulting from NDUFAF7 depletion is a general phenomenon, we transiently depleted it in human fibroblasts (MCH64), 143B and HEK293 cell lines for 3 or 6 days, and examined the steady-state levels of different complex I subunits by immunoblot analysis (Fig. 8A). The levels of the NDUFS2 and NDUFA9 subunits were severely depleted in all three of these cell lines after 6 days of knockdown. Knockdown of two additional early assembly factors, NDUFAF3 or NDUFAF4, for 6 days similarly showed markedly reduced steady-state levels of both structural subunits (Fig. 8B).
Figure 8.
NDUFAF7 knockdown in different human cell lines. (A) Immunoblot analysis of NDUFAF7, NDUFS2 and NDUFA9 in mitochondria from MCH64, 143B and HEK293 cells after siRNA-mediated knockdown of NDUFAF7 for 3 or 6 days. SDHA was used as a loading control. (B) Same analysis as in (A) following knockdown of two different complex I assembly factors, NDUFAF3 and NDUFAF4, for 6 days.
Knockout of NDUFAF7 in mice is embryonically lethal
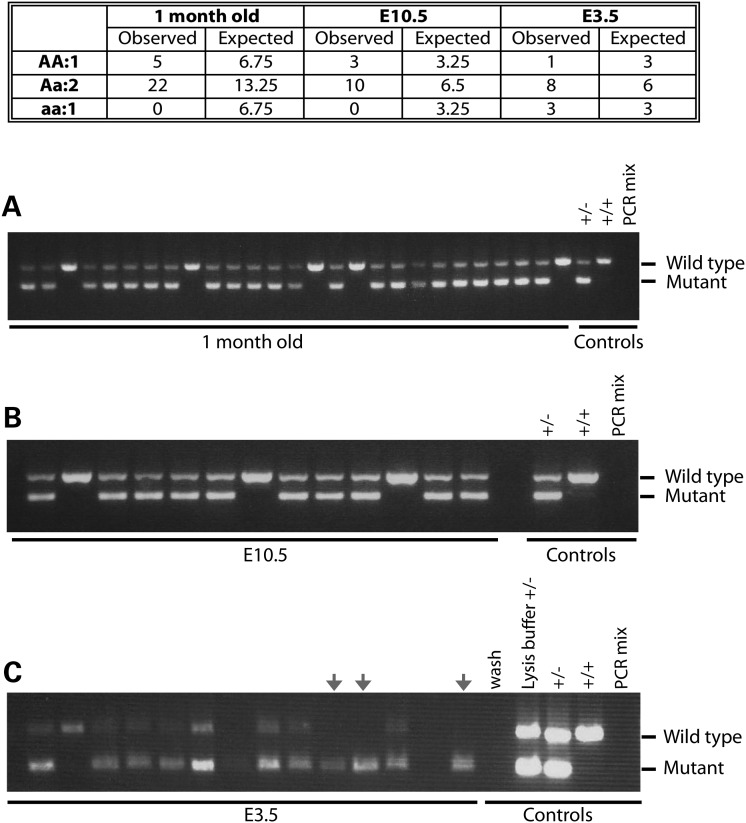
An NDUFAF7 germline knockout mice was generated by the gene trapping method. Heterozygous animals were crossed and progeny were genotyped by multiplex PCR (Fig. 9). One month after birth, no homozygous knockout animals were detected. To further investigate the NDUFAF7 knockout phenotype, embryos were harvested at embryonic Day 10.5. All genotyped embryos were either heterozygous or homozygous wild type. Finally, the NDUFAF7 knockout lethality phenotype was assessed at the blastocyst stage. Out of the 12 embryos analyzed, three were homozygous recessive, corresponding to the expected Mendelian segregation ratio (Fig. 9C).
Figure 9.
Embryonic lethality in mice with a germline disruption of NDUFAF7. Multiplex PCR products show NFUFAF7 wild-type (815 bp) and mutant (438 bp) bands. (A) One-month-old animals (n = 27) were genotyped using three different controls: heterozygous DNA (+/−), homozygous DNA (+/+) and negative control (no DNA). (B) Thirteen animals genotyped at embryonic Day 10.5, controls are the same as in (A). (C) Twelve animals genotyped at embryonic Day 3.5 using five different controls: M2 buffer in which embryos were collected and washed (wash), proteinase-K lysis buffer + heterozygote DNA (lysis buffer +/−), heterozygous DNA (+/−), homozygous DNA (+/+) and negative control (no DNA, PCR mix).
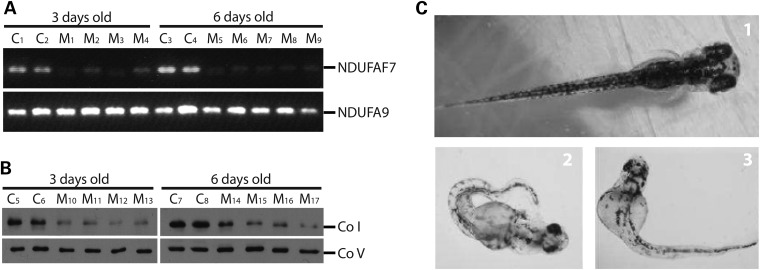
NDUFAF7 knockdown in zebrafish impairs complex I assembly
Considering that complex I is highly conserved in vertebrates, we decided to knockdown NDUFAF7 in the fish Danio rerio (zebrafish) using the morpholino (MO) microinjection technique. To modify NDUFAF7 mRNA splicing, we used two MO targeted to exon 3 and 6. RT-PCR analysis was performed to assess the mRNA levels of NDUFAF7 in nine MO-treated animals (Fig. 10A). At 3 days of knockdown, some variability was observed in NDUFAF7 transcripts levels but by 6 days of knockdown all tested animals were homogenously depleted of NDUFAF7 mRNA. As a control for RNA quality, we measured the mRNA levels of the nuclear-encoded subunit of complex I, NDUFA9, which were unchanged. BN–PAGE analysis showed that NDUFAF7 knockdown specifically affects the steady-state levels of complex I (Fig. 10B). Hatching was delayed in the majority of the MO-treated fish, and most showed morphological abnormalities compared with control animals (Fig. 10C).
Figure 10.
NDUFAF7 knockdown in zebrafish. (A) RT-PCR analysis of controls (C1–4) and MO knockdown fish (M1–9) at 3 and 6 days old using specific primers for amplification of NDUFAF7 and the complex I subunit, NDUFA9. (B) Controls (C5–8) and MO knockdown fish (M10–17) were analyzed at 3 and 6 days old by BN–PAGE to detect complex I (Co I) and V (Co V) using subunit-specific antibodies, NDUFAF9 and ATPα, respectively. (C) Control (1) and MO knockdown (2–3) fish phenotype at 4 days old.
DISCUSSION
The results presented here clearly establish NDUFAF7 as an evolutionarily conserved complex I assembly factor that is necessary for normal vertebrate development. Disruption of the zebrafish ortholog resulted in delayed hatching times and morphologically abnormal fish; a germline disruption of the murine gene was embryonic lethal. Although we were able to detect blastocysts with a homozygously disrupted gene in mice, we found no evidence for embryos with this genotype at E10.5, suggesting that it may be essential for implantation or very early embryogenesis.
Previous studies of NDUFAF7 and of MidA, the Dictyostelium ortholog (12, 14) implicated the human protein in complex I assembly, as had been suggested from studies using phylogenetic profiling to identify factors associated with the expression of complex I (9, 13). The siRNA-mediated knockdown of NDUFAF7 to nearly undetectable levels in human fibroblasts completely abrogated complex I assembly, and the re-appearance of the protein was associated with re-assembly of the complex, confirming it is an essential assembly factor. All of our experiments focused on the long isoform of NDUFAF7, and it is possible that the short isoform, which was also targeted by the siRNAs we used, interacts or modifies the function of the long isoform in some way, or even has a different substrate specificity, but this will require further investigation.
NDUFAF7 has a DUF185 domain containing a small region of identity with motif I of SAM-dependent methyltransferases. Motif I is one of the four loops that form the catalytic core of SAM-binding methyltransferases and it has a nine residue consensus sequence: hh (D/E) hGxGxG (18). In this study, we used site-directed mutagenesis to generate a G124V point mutation in the glycine-rich ‘GxGxG’ signature sequence. Biochemical characterization of this mutant cell line (PGS1 knockdown) demonstrated that G124 is required for NDUFAF7 function, indicating that the methyltransferase activity of NDUFAF7 is required for its function in complex I assembly.
SAM-dependent methyltransferases have a wide range of substrates including DNA, RNA and proteins; however, there is no consensus sequence/structure in SAM-methyltransferases that might permit accurate prediction of their substrates (19). We saw no evidence for impaired mitochondrial transcription or translation after knockdown of NDUFAF7, making it unlikely that NDUFAF7 was responsible for methylating a nucleic acid target in mitochondria, but rather suggesting a protein target in complex I or its associated assembly factors. The reported interaction of MidA with NDUFS2 in a yeast two-hybrid assay (12), and the identification of a symmetrically dimethylated arginine residue at residue 85 in mature NDUFS2 (20) clearly suggested this structural subunit as a potential substrate. We observed a nearly 11-fold reduction in the abundance of an NDUFS2 peptide dimethylated at Arg85 in the PGS1 knockdown cell line; however, three other peptides in NDUFS2 were only reduced ∼5.6-fold. As this analysis was carried out on an anti-ND1 immunoprecipitate, the NDUFS2 peptides that we detected must have been incorporated into the residual holoenzyme complex I (or a subassembly containing ND1). From this, we conclude that NDUFS2 is methylated in an NDUFAF7-dependent fashion after it is incorporated into an early assembly intermediate, and that this modification is necessary for the stability or further assembly of this subcomplex. Consistent with this we detected NDUFAF7 by mass spectrometric analysis of ND1 immunoprecipitates from isolated mitochondria, and we observed that a small fraction of NDUFAF7 co-fractioned on sucrose gradients with subcomplexes containing ND1 and NDUFS2. Studies of MidA also showed that while the majority of MidA forms a soluble dimer, a small amount associates with higher molecular-weight subcomplexes of complex I (12).
A recent study in human osteosarcoma (143B) cells similarly identified NDUFS2 as an NDUFAF7 substrate (14), but did not determine whether the methylation occurred before assembly into the 315 kDa subassembly nucleated by ND1. Additionally, they reported a progressive accumulation of a non-methylated NDUFS2 peptide during a time course knockdown of NDUFAF7 (14). Their results showed that the steady-state levels of NDUFS2 on immunoblot were not affected by NDUFAF7 knockdown, even though a BN–PAGE analysis showed a marked impairment of the assembly of holocomplex I that was revealed with an anti-NDUFS2 antibody. The results we present here show that knockdown of NDUFAF7 in three different human cell lines, including 143B, resulted in minimal immunodetectable levels of NDUFS2 (Fig. 9), consistent with our immunoprecipitation results, and with previous experiments in which early complex I assembly factors were depleted (17). Early complex I assembly defects result in rapid AFGL32-dependent proteolysis of newly synthesized ND1, the subunit that seeds the assembly process, and because other nuclear encoded structural subunits are not stabilized in a membrane bound subcomplex, they are likewise turned over by the quality control proteases. Contrary to results reported in (14), we were not able to find any evidence for the accumulation of non-methylated NDUFS2.
Mitochondrial protein methylation has only been found in bovine NDUFB3, another structural subunit of complex I, but neither the methyltransferase responsible for this methylation, or the functional relevance of it, have been described (20). Methylation of mitochondrial rRNA and tRNA has been more extensively characterized (21, 22). In mammals, the mitochondrial transcription factor B1 (TFB1M) and the NOL1/NOP2/Sun domain family, member 4 (NSUN4) are both rRNA methyltransferases essential for assembly of the mitochondrial ribosome (23, 24). The recent study by (14) identified 35 putative or known mitochondrial methyltransferases, the majority of which are uncharacterized and have no known function.
While this and a previous study implicate NDUFS2 as the substrate for NDUFAF7, it is entirely possible that it has additional complex I targets. In fact, in silico methylation analysis (25) predicts that the late complex I assembly factors Ecsit and FOXRED1 are potential arginine-methylated substrates; however, our preliminary mass spectrometry analysis of ND1 immunoprecipitates indicate that Ecsit is not methylated. Additional work will be required to assess whether methylation plays an important regulatory role in other factors involved in the complex I assembly pathway. As assembly defects account for nearly half of all cases of autosomal recessive complex I deficiencies, it is likely that patients with mutations in NDUFAF7 will eventually be identified.
MATERIALS AND METHODS
Cell culture
Primary cell lines were established from subject skin fibroblasts and immortalized by transduction with a retroviral vector expressing the HPV-16 E7 gene and the catalytic component of human telomerase (26). The fibroblasts were grown at 37°C in an atmosphere of 5% CO2 in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum.
NDUFAF7 antibody
An antibody against NDUFAF7 was generated by injecting rabbits with a synthetic peptide specific to human NDUFAF7 (GGRYQRNARQSKPF). The peptide was conjugated with the immunogenic carrier keyhole limpet hemocyanin via a reactive sulfhydryl group, and emulsified in Titermax (Pierce Biotechnology Inc.).
Mitochondrial isolation and localization experiments
Fibroblasts were resuspended in ice-cold SET buffer: 250 mm sucrose/10 mm Tris–HCl/1 mm EDTA (pH 7.4) supplemented with complete protease inhibitors (Roche) and homogenized with 10 passes through a prechilled, zero clearance homogenizer (Kimble/Kontes). The homogenized cellular extract was then centrifuged twice for 10 min at 600g to obtain a postnuclear supernatant. Mitochondria were pelleted by centrifugation for 10 min at 10 000g and washed once in the same buffer.
Immunofluorescence analysis was performed using a control cell line overexpressing NDUFAF7-HA. Glass slides were fixed in 4% paraformaldehyde at 50% confluence followed by 1 h incubation with HA and cytochrome c antibodies, which preceded the addition of secondary antibodies coupled with Alexa fluorochromes. Images were obtained with an inverted fluorescent microscope. SET homogenized fibroblasts (Wh) were centrifuged at 600g to separate soluble fraction (cSN) from nuclei/membranes. A second centrifugation at 10 000g was used to pellet the mitochondrial fraction (Mi). Fractions were analyzed by SDS–PAGE.
Purified mitochondria from control fibroblasts were extracted with 100 mm Na2CO3 (pH 11.5) at 4°C for 30 min and centrifuged at 184,000g to separate soluble and membrane fractions. Fractions were analyzed by SDS–PAGE.
NDUFAF7 constructs
cDNA from NDUFAF7 was amplified by One-Step RT-PCR (QIAGEN). The longest transcript of NDUFAF7 (CCDS1788) was cloned into pBABE Puro (27) using the Gateway Cloning Technology (Invitrogen). The accuracy of the clones was verified by DNA sequencing. Direct mutagenesis of NDUFAF7 was done using the site-directed mutagenesis lightning kit (Stratagene). Retroviral constructs were transiently transfected into the Phoenix packaging cell line using the HBS/Ca3(PO4)2 method. Control fibroblasts were infected 48 h later by exposure to virus-containing medium in the presence of 4 g/ml of polybrene (http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html).
RNAi transfection
For the NDUFAF7 knockdown experiments, two different Stealth RNA interference (RNAi) duplex constructs (Invitrogen, Carlsbad, CA, USA) were used; NDUFAF7-siRNA-1: UCAGAGACUUCAAGGUGGAAGAUAU and NDUFAF7-siRNA-2: ACAGCUGAUGUGGACUUCAGUUAUU. Cell lines were transfected twice, at Days 1 and 3 and analyzed at Day 6. Mitochondrial proteases were knocked down for 2 days. The constructs were designed using the BLOCK-iT™ RNAi Express website (http://rnaidesigner.invitrogen.com/rnaiexpress/rnaiExpress.jsp?CID=FL-RNAIEXPRESS). Fibroblasts were transfected using the LipofectamineTM RNAiMAX protocol in Opti-MEM® Reduced Serum Medium. BLOCK-iT™Alexa Fluor® Red Fluorescent Oligo was used to assess the transfection efficiency (transfection performed in parallel with the other Stealth constructs) and as a mock oligo transfection control (all reagents from Invitrogen).
SDS–PAGE immunoblot analysis
Whole-cell extracts were prepared by solubilization of pelleted cells at a 1:5 ratio with extraction buffer (1.5% dodecyl maltoside) for 30 min on ice. The soluble fraction was obtained by centrifugation at 20 000 g for 20 min at 4°C. Samples were mixed at a 1:1 ratio with 2× Laemmli sample buffer and solubilized proteins were loaded onto a 12% acrylamide:bisacrylamide (29:1) gel. Subsequent immunoblotting was done with the appropriate antibodies. Antibodies were obtained as follows: MitoSciences: NDUFS2, actin. Abcam: NDUFA9, ATPα, COX2, COX4, COX1, core I, SDHA. Sigma: porin and MRPL14. Proteintech Group: MRPS22 and MRPS18B. Stressgen: cu/ZnSOD. AFG3L2 (kindly provided by T. Langer), LONP1 (kindly provided by C. Suzuki) and ND1 (kindly provided by A. Lombes)
Sucrose density gradients
Mitochondria (500 μg) were lysed in buffer containing 260 mm sucrose, 100 mm KCl, 20 mm MgCl2, 10 mm Tris–Cl, pH 7.5, 1% Triton X-100, 5 mm β-mercaptoethanol, supplemented with complete protease inhibitors without EDTA, on ice for 20 min. Lysates were centrifuged for 45 min at 9400g at 4°C before loading them on 1 ml of a 10–30% discontinuous sucrose gradient (50 mm Tris–Cl, 100 mm KCl, 10 mm MgCl2). The loaded gradients were centrifuged at 32 000 rpm for 130 min in a Beckman SW60-Ti rotor (Beckman Coulter, Brea, CA, USA). After centrifugation, 14 fractions were collected and analyzed by SDS–PAGE.
BN–PAGE and 2D-SDS immunoblot analysis
Mitoplasts were prepared from fibroblasts, as described previously (28) by treating the cells with 0.8 mg digitonin/mg of protein. Mitoplasts were solubilized with 1% lauryl maltoside, and solubilized protein was used for electrophoresis. BN–PAGE (29) was used to separate the samples in the first dimension on 6–15% polyacrylamide gradients. For analysis of the second dimension, strips of the first-dimension gel were incubated for 30 min in 1% SDS and 1% β-mercaptoethanol. These strips were then run on a 10% tricine/SDS–PAGE to separate the proteins in the second dimension (29). Individual structural subunits of complexes I, II, III, IV and V were detected by immunoblot analysis using commercially available monoclonal antibodies (MitoSciences and Abcam), except for complex I, where a polyclonal antibody against subunit ND1 was used.
Pulse labeling of mitochondrial translation products
Labeling of the mitochondrial translation products was performed as previously described (30). Briefly, cells were labeled for the indicated times at 37°C in methionine and cysteine-free DMEM containing 200 µCi/ml [35S]methionine/cysteine and 100 µg/ml emetine, and chased for 10 min in regular DMEM. Protein extraction was done in labeled cells (50 μg) by resuspension in loading buffer containing 93 mm Tris–HCl, pH 6.7, 7.5% glycerol, 1% SDS, 0.25 mg bromophenol blue/ml and 3% mercaptoethanol, sonicated for 5 s, loaded and run on 15–20% polyacrylamide gradient gels. The labeled mitochondrial translation products were detected by direct autoradiography on a Phosphorimager. Quantitative analysis was done using the ImageQuant 5.2 software.
RNA blot analysis
RNA was isolated from NDUFAF7-siRNA-2 and control fibroblasts using the RNeasy Kit (Qiagen). Ten micrograms of total RNA were separated on a denaturing MOPS/formaldehyde agarose gel and transferred to a nylon membrane. ND1, ND5, ND6, COX1, ATP6/8 and 16S probes were labeled with [-32P]-dCTP (GE Healthcare) using the MegaPrime DNA-labeling kit (GE Healthcare). Hybridization was conducted according to the manufacturer's protocol using ExpressHyb Hybridization Solution (Clontech), and the radioactive signal was detected using the Phosphorimager system.
Immunoprecipitation experiments and mass spectrometry analysis
Purified mitochondria were cross linked with DSG (Disuccinimidyl glutarate) for 1.5 h on ice prior to protein extraction. Two milligrams of mitochondrial protein were extracted on ice from control (PGS1) and knockdown (PGS1 + NDUFAF7-siRNA-2) cell lines in 500 μl of extraction buffer (50 mm HEPES buffer, pH 7.6, 150 mm NaCl, 1% taurodeoxycholate), supplemented with complete protease inhibitors (Roche), for 45 min. The protein extract was centrifuged at 25 000g at 4°C, for 40 min, and the supernatant was used to immunoprecipitate ND1. Immunoprecipitation was performed on magnetic Dynabeads (Life Technologies) according to the manufacturer's instructions. The incubation of the protein extract with the beads was carried out overnight at 4°C. Bound protein was eluted with 400 μL of 0.1 m glycine (pH 2.5) with 0.5% DDM at 45°C followed by TCA precipitation. Elutions were analyzed by mass spectrometry (LTQ Orbitrap Velos (ThermoFisher Scientific, Bremen, Germany) after tryptic digestion. Proteins were identified from peptide mass and fragmentation data by interrogation of NCBInr protein database (version July 18, 2012) using Mascot 2.3 (Matrix Science). Mono, di- and tri-methylation of arginine were allowed as variable modifications, while carbamidomethylation was set as a fixed modification. The relative abundance of indicated peptides was estimated from the area of reconstructed ion chromatograms obtained with Qual Browser (Xcalibur, Thermo Fisher Scientific) using an m/z tolerance of 7 ppm.
NDUFAF7 knockout mice generation and genotyping
A NDUFAF7 germline knockout mice was generated by the gene trapping method with the auspices of The International Gene Trap Consortium (IGTC). DNA was purified from mice tails tissue using lysis buffer containing 100 mm Tris–Cl (pH 8.5), 5 mm EDTA, 0.2% SDS, 200 mm NaCl and 0.4 μg/μl of proteinase K. Tails were incubated at 55°C in lysis buffer. Fifty microlitres of 5 M potassium acetate (pH 4.8) was added 24 h after and solubilized tissue was spun at 4°C for 5 min at 14 000 rpm. Supernatants were decanted and mixed with 100% isopropyl alcohol. Samples were spun for 10 min at 14 000 rpm, pellets were washed with 70% ethanol followed by a final centrifugation for 10 min at 10 000 rpm. Pellets were air dried and resolubilized in water. Genotyping was done by multiplex PCR. Three primers were designed: WT-F: CACTCTCTGCCTCCAACTTATG, WT-R: CCCATCTACGATCACTTTCTTCAGC and MT-F: TTCCATCTGTTCCTGACCTTG. The wild-type allele was detected as a band of 815 bp, while the mutant allele generates a fragment of 428 bp.
Blastocyts were flushed from oviducts at embryonic Day 3.5 using M2 (EMD Millipore) buffer as described (31). For genotyping purposes, blastocysts were solubilized in lysis buffer containing: 20 mm Tris (pH 8), 100 mm KCl, 4 mm MgCl2, 0.9% Nonidet P-40, 0.9% Triton X-100 and 300 ng/μL of proteinase K.
Embryos were chirurgically removed from oviducts at embryonic Day 10.5 as described elsewhere (32). DNA was extracted from mouse tails as described above.
Zebrafish maintenance
Wild-type adult zebrafish were maintained on a 14/10 h light–dark cycle at 28.5°C under standard laboratory conditions (33). Embryos were allowed to develop in an incubator at 28.5°C for 72 or 144 h (34) and were then frozen in liquid nitrogen.
Morpholino knockdown
A MO targeting the zebrafish NDUFAF7 gene (ATTATTTGGATTACTTACCCTCAGG) was designed by Gene Tools (http://www.gene-tools.com/). The MO was resuspended in sterile water, and 0.04% xylene cyanol was added for injection visualization purposes. Approximately 10 ηl of a 1 μM solution were injected in zebrafish embryos at the 1- or 2-cell stage. After being injected, the eggs were kept in embryo water (5.03 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2·2H2O, 0.33 mm MgSO4·7H2O and 0.1% methylene blue.
Zebrafish RNA extraction, DNA amplification and protein extraction
RNA was extracted from injected and non-injected single fish at 72 h postfertilization using the RNeasy Plus Kit (Qiagen). One-Step RT-PCR kit (Qiagen) was used to generate cDNAs in order to test for the gene knockdown, with primers designed specifically for NDUFAF7 and for complex I NDUFA9 subunit, as a control. Individual fish were extracted in 1.5% dodecylmaltoside in MB2 buffer (0.75 m amino caproic acid, 1.075 m bis–tris, 2 mm EDTA) for BN–PAGE analysis as described above.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a CIHR grant (MT 15460) to E.A.S. O.Z.R. was supported by fellowships from CONACyT (Consejo Nacional de Ciencia y Tecnologia) (209378) and PBEEE (Fonds quebecois de la recherché sur la nature et les technologies) (140802).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Denis Faubert of the Institut de Recherches Clinique, Montreal for help with the mass spectrometry experiments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Schaefer A.M., McFarland R., Blakely E.L., He L., Whittaker R.G., Taylor R.W., Chinnery P.F., Turnbull D.M. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 2.Skladal D., Halliday J., Thorburn D.R. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain. 2003;126:1905–1912. doi: 10.1093/brain/awg170. [DOI] [PubMed] [Google Scholar]
- 3.Smeitink J.A., Zeviani M., Turnbull D.M., Jacobs H.T. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Andrews B., Carroll J., Ding S., Fearnley I.M., Walker J.E. Assembly factors for the membrane arm of human complex I. Proc. Natl. Acad. Sci. USA. 2013;110:18934–18939. doi: 10.1073/pnas.1319247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimaki M., Wang X., McKenzie M., Thorburn D.R., Ryan M.T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta. 2011;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Vogel R.O., Dieteren C.E., van den Heuvel L.P., Willems P.H., Smeitink J.A., Koopman W.J., Nijtmans L.G. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 2007;282:7582–7590. doi: 10.1074/jbc.M609410200. [DOI] [PubMed] [Google Scholar]
- 7.Ogilvie I., Kennaway N.G., Shoubridge E.A. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 2005;115:2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynen M.A., Duarte I., Chrzanowska-Lightowlers Z.M., Nabuurs S.B. Structure based hypothesis of a mitochondrial ribosome rescue mechanism. Biol. Direct. 2012;7:14–10. doi: 10.1186/1745-6150-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchler-Bauer A., Anderson J.B., Derbyshire M.K., DeWeese-Scott C., Gonzales N.R., Gwadz M., Hao L., He S., Hurwitz D.I., Jackson J.D., et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadreyev R., Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326:317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 12.Carilla-Latorre S., Gallardo M.E., Annesley S.J., Calvo-Garrido J., Grana O., Accari S.L., Smith P.K., Valencia A., Garesse R., Fisher P.R., et al. MidA is a putative methyltransferase that is required for mitochondrial complex I function. J. Cell. Sci. 2010;123:1674–1683. doi: 10.1242/jcs.066076. [DOI] [PubMed] [Google Scholar]
- 13.Gabaldon T., Rainey D., Huynen M.A. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I) J. Mol. Biol. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 14.Rhein V.F., Carroll J., Ding S., Fearnley I.M., Walker J.E. NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I. J. Biol. Chem. 2013;288:33016–33026. doi: 10.1074/jbc.M113.518803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saada A., Vogel R.O., Hoefs S.J., van den Brand M.A., Wessels H.J., Willems P.H., Venselaar H., Shaag A., Barghuti F., Reish O., et al. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am. J. Hum. Genet. 2009;84:718–727. doi: 10.1016/j.ajhg.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugiana C., Pagliarini D.J., McKenzie M., Kirby D.M., Salemi R., Abu-Amero K.K., Dahl H.H., Hutchison W.M., Vascotto K.A., Smith S.M., et al. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 2008;83:468–478. doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zurita Rendon O., Shoubridge E.A. Early complex I assembly defects result in rapid turnover of the ND1 subunit. Hum. Mol. Genet. 2012;21:3815–3824. doi: 10.1093/hmg/dds209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niewmierzycka A., Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 19.Liscombe David K., Louie Gordon V., Noel Joseph P. Architectures, mechanisms and molecular evolution of natural product Methyltransferases. Nat. Prod. Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 20.Carroll J., Fearnley I.M., Skehel J.M., Runswick M.J., Shannon R.J., Hirst J., Walker J.E. The post-translational modifications of the nuclear encoded subunits of complex I from bovine heart mitochondria. Mol. Cell. Proteomics. 2005;4:693–699. doi: 10.1074/mcp.M500014-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Helm M., Brule H., Degoul F., Cepanec C., Leroux J.P., Giege R., Florentz C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pintard L., Bujnicki J.M., Lapeyre B., Bonnerot C. MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 2002;21:1139–1147. doi: 10.1093/emboj/21.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camara Y., Asin-Cayuela J., Park C.B., Metodiev M.D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K., et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Cotney J., McKay S.E., Shadel G.S. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum. Mol. Genet. 2009;18:2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao J., Xu D., Tsai S.N., Wang Y., Ngai S.M. Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PLoS ONE. 2009;4:e4920. doi: 10.1371/journal.pone.0004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lochmuller H., Johns T., Shoubridge E.A. Expression of the E6 and E7 genes of human papillomavirus (HPV16) extends the life span of human myoblasts. Exp. Cell. Res. 1999;248:186–193. doi: 10.1006/excr.1999.4407. [DOI] [PubMed] [Google Scholar]
- 27.Morgenstern J.P., Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klement P., Nijtmans L.G., Van den Bogert C., Houstek J. Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal. Biochem. 1995;231:218–224. doi: 10.1006/abio.1995.1523. [DOI] [PubMed] [Google Scholar]
- 29.Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 30.Sasarman F., Shoubridge E.A. Radioactive labeling of mitochondrial translation products in cultured cells. Methods Mol. Biol. 2012;837:207–217. doi: 10.1007/978-1-61779-504-6_14. [DOI] [PubMed] [Google Scholar]
- 31.Nagy A., Gertsenstein M., Vintersten K., Behringer R. Manipulating the Mouse Embryo. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 32.Moreno-Ortiz H., Esteban-Perez C., Badran W., Kent-First M. Isolation and derivation of mouse embryonic germinal cells. J. Vis. Exp. 2009;32:e1635. doi: 10.3791/1635. doi:10.3791/1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: University of Oregon Press; 2000. [Google Scholar]
- 34.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.