Abstract
Background
This study aimed to investigate the relationships of chitinase 3-like 1 (CHI3L1) single nucleotide polymorphisms (SNPs) and haplotypes with the development of uterine cervical cancer in Taiwanese women. The SNPs frequencies and haplotypes were also correlated with the clinicopathologic variables of cervical cancer, cancer recurrence, and patient survival.
Methodology and Principal Findings
Ninety-nine patients with invasive cancer and 61 with pre-cancerous lesions of the uterine cervix were compared to 310 healthy control subjects. Three SNPs rs6691378 (−1371, G/A), rs10399805 (−247, G/A) and rs4950928 (−131, C/G) in the promoter region, and one SNP rs880633 (+2950, T/C) in exon 5 were analyzed by real time polymerase chain reaction and genotyping. The results showed that the mutant homozygous genotype AA of CHI3L1 SNP rs6691378 and AA of rs10399805, and haplotypes AACC and AACT increased the risk of developing pre-cancerous lesions and invasive cancer. The patients with these risk haplotypes had higher than stage I tumors, larger tumors, and vaginal invasion. In logistic regression model, they also tended to have poor survival event [p = 0.078; odds ratio (OR): 2.99, 95% confidence interval (CI): 0.89–10.08] and a higher probability of recurrence event (p = 0.081; OR: 3.07, 95% CI: 0.87–10.81). There was a significant association between the CHI3L1 risk haplotypes and probability of recurrence (p = 0.002; hazard ratio: 6.21, 95% CI: 1.90–20.41), and a marginal association between the risk haplotypes and overall survival (p = 0.051; hazard ratio: 3.76, 95% CI: 0.99–14.29) in the patients with SCC, using Cox proportional hazard model.
Conclusion
The CHI3L1 SNPs rs6691378 and rs10399805 and CHI3L1 haplotypes all correlated with the development of cervical pre-cancerous lesions and invasive cancer. The cervical cancer patients with the CHI3L1 haplotypes AACC or AACT had poor clinicopathologic characteristics and poor recurrence and survival events. These risk haplotypes were associated with higher recurrence, especially in the patients with SCC.
Introduction
Chitinase3-like1 (CHI3L1) is a glycoprotein encoded by the chitinase 3-like 1 (CHI3L1) gene located on human chromosome 1q32.1 [1]. This glycoprotein is often referred to as YKL-40 or human cartilage glycoprotein-39 (HC gp-39) and is known to be a pro-inflammatory cytokine of chitinase [2], [3]. It is a secreted protein with a molecular weight of 40 kD and is identified by the N-terminal sequencing to be tyrosine (abbreviated as Y), lysine (K), and leucine (L) [4]. It is recognized as a growth factor for connective cells and a migration-promoting factor for endothelial cells, and is produced by a variety of cells including cancer cells, activated macrophages, and neutrophils. It is also known to play a role in inflammation, cell proliferation, anti-apoptosis, stimulation of angiogenesis, and regulation of extracellular tissue remodeling [2], [5]–[7].
Kjaergaard et al. used a population-based prospective study of the Danish general population to investigate the genetic variants of CHI3L1 that influence YKL-40 levels, and found that eight single nucleotide polymorphisms (SNPs) of the CHI3L1 gene are associated with plasma YKL-40 levels in the general population [8]. These CHI3L1 SNPs included rs10399805 (−247, G/A) and rs4950928 (−131, C/G) in the promoter region, and rs880633 (+2950, T/C) in exon 5 etc. However, Thomsen et al., also using the Danish population in the international MONICA (MONItoring trends and determinants of CArdiovascular disease) project, demonstrated that 12SNPs were associated with serum YKL-40 levels [9]. These CHI3L1 SNPs included rs6691378 (−1371, G/A), rs4950928, and rs880633 etc.
Uterine cervical cancer is the fifth most common type of malignancy among women in Taiwan, with an age-standardized incidence rate in 2009 of 11.86 per 100000 women according to the Health Promotion Administration of the Ministry of Health and Welfare. Its age-standardized mortality rate was 3.72 per 100000 women in 2011, making it the seventh top cause of cancer death in Taiwanese women.
Single nucleotide polymorphisms may affect promoter activity, gene expression, messenger RNA conformation (stability), and sub-cellular localization of mRNAs and/or proteins, and probably cause diseases [10]. Several recent studies have shown that genetic variations of CHI3L1 SNPs have an impact on inter-individual serum YKL-40 levels as well as susceptibilities to atopy, sarcoidosis, asthma, and lung function [11]–[14]. Pre-treatment serum levels of YKL-40 have also been reported to be elevated in cervical cancer [15]. To date, no study has correlated CHI3L1 SNPs with cervical cancer in Taiwanese women. Under the hypothesis that gene polymorphisms or haplotypes of the CHI3L1 gene have an impact on YKL-40 expression in cervical cancer, this study investigated the distribution of CHI3L1 gene polymorphisms and haplotypes among patients with cervical cancer and pre-cancerous lesions and normal controls to define their roles in cervical carcinogenesis in Taiwanese women. The SNP frequencies or haplotypes of CHI3L1 were further associated with clinicopathologic variables of cervical cancer, cancer recurrence, and patient survival. This study demonstrated significant associations of CHI3L1 SNPs and haplotypes with the development of pre-cancerous lesions and invasive cancer of the uterine cervix, and revealed that CHI3L1 haplotypes were related to the prognosis of cervical cancer patients.
Materials and Methods
Population
Four hundred and seventy women, including 99 patients with invasive cancer, 61 patients with pre-cancerous lesions of the uterine cervix, and 310 normal controls, were recruited consecutively into this study. The patients with invasive cervical cancer were clinically staged based on the 2009 International Federation of Gynecology and Obstetrics Classification and received routine treatment protocols at the Department of Obstetrics and Gynecology in Chung Shan Medical University Hospital, Taiwan, from March 1999 to October 2012. The patients with pre-cancerous lesions, which only comprised cervical high-grade dysplasia (high-grade squamous intraepithelial lesions) in this study and included moderate and severe dysplasia as well as carcinoma in situ, underwent colposcopy-directed cervical punch biopsy, large loop excision of the transformation zone, total abdominal hysterectomy, or total vaginal hysterectomy. The 310 controls with normal Papanicolaou smears were further verified using colposcopy during general examinations at the outpatient department of our hospital. The ages of the women with cervical invasive cancer, pre-cancerous lesions, and normal controls were 53.6±12.0, 42.7±12.7, and 44.7±9.8(mean ± SD) years, respectively. All of them were Taiwanese women who resided in central Taiwan. The Chung Shan Medical University Hospital Institutional Review Board approved this study (CSMUH IRB: CS12218, CS12219, CS14014), and informed written consent was obtained from each individual.
Selection of chitinase 3-like 1 gene polymorphisms
Three SNPs rs6691378 (−1371, G/A), rs10399805 (−247, G/A), and rs4950928 (−131, C/G) in the promoter region and one SNP rs880633 (+2950, T/C) in exon 5 were selected based on the Chinese HapMap (Han Chinese in Beijing, China) data and the studies of Thomsen et al. and Kjaergaard et al. [12], [16]. The minor allele frequencies (MAFs) of these SNPs were ≥5%.
Blood sample collection and genomic DNA extraction
In total, 160 blood specimens were collected from the patients with cervical invasive cancer and those with pre-cancerous lesions, and 310 blood specimens were obtained from the controls. Genomic DNA was extracted from EDTA anti-coagulated venous blood using a QIAamp DNA blood mini kit (Qiagen,Valencia, CA, USA) based on the manufacturer's protocol. The DNA was dissolved in Tris ethylene buffer (10 mmol/L Tris and 1 mmol/L EDTA; pH 7.8) and then quantified by a measurement of OD260. The final preparation was stored at −20°C and applied as templates in polymerase chain reaction (PCR).
Single nucleotide polymorphisms by real time-PCR and genotyping
Allelic discrimination of the rs6691378 (−1371, G/A), rs10399805 (−247, G/A), rs4950928 (−131, C/G), and rs880633 (+2950, T/C) polymorphisms was assessed using an ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and analyzed by SDS version 3.0 software (Applied Biosystems) using the TaqMan assay. The 10 µL final volume for each reaction contained 5 µL TaqMan Genotyping Master Mix, 0.25 µL TaqMan probe mix, and 10 ng genomic DNA. Real-time PCR included an initial denaturation step at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and then 60°C for one minute.
Statistical analysis
Analysis of variance (ANOVA) was used to analyze the age distribution of the study population and control subjects. Scheffe's test was used for post hoc analysis.
Hardy-Weinberg equilibrium was used to analyze the genotype distributions of rs6691378, rs10399805, rs4950928, and rs880633 in the normal controls (degree of freedom = 2). Chi-square or Fisher exact tests were used to examine the relationships among frequencies of CHI3L1 gene SNPs, alleles, and haplotypes, and the incidence of cervical neoplasia (including invasive cancer and pre-cancerous lesions), as well as various clinicopathologic parameters, including clinical stage (I or ≥II), histopathologic types such as squamous cell carcinoma (SCC) or adenocarcinoma, cell grading (well, or moderate and poor differentiation), invasion depth of cervical stroma (≤10 mm or >10 mm of stromal invasion depth), tumor diameter (≤4 or >4 cm), parametrium and vaginal invasion, and pelvic lymph node metastasis found during surgery.
Logistic regression model or multiple logistic regression were used to analyze multiple comparisons of SNP genotypes of the CHI3L1 gene polymorphisms before and after controlling for age between the patients with cervical neoplasia and the controls, or among patients with invasive cancer or pre-cancerous lesions and the controls. No age distribution was adjusted for allele comparisons (the allele number of each allele in each subgroup equaled two-fold of the number of homozygous genotypes plus one-fold of the number of heterozygous genotypes, therefore we could not analyze the age distribution among the subjects subgroups; for example the number of G alleles = 2 * the number of GG alleles+1 * the number of GA alleles) or haplotype distribution (arbitrary reclassification). The sample size was estimated with power (1-β), 0.8 and α, 0.05, and the powers were calculated when the comparisons among the subgroups reached statistical significances (p<0.05) with an adequate sample size using WinPepi software, version 10.0.
A logistic regression model was also used to correlate the various clinicopathologic parameters and CHI3L1 polymorphisms with cancer recurrence event or patient survival event. A Cox proportional hazard model was used to evaluate the effects of CHI3L1 haplotypes on the probability of recurrence or overall survival after adjusting for various clinicopathologic parameters in multivariate analysis relative to recurrence or survival time. When the follow-up period was included into survival or recurrence analysis, the patients were enrolled for overall survival analysis including 5-year survival rate or probability of recurrence between primary surgery and death or recurrence or the end of the study (October, 2013) using the Kaplan-Meier model and multivariate and univariate Cox regression models. A significant difference was set at p<0.05. Statistical analyses including odds ratio (OR) and adjusted odds ratio (AOR; controlling for age) and their 95% confidence interval (CI) were calculated by the SPSS, version 12.0 and WinPepi Software, version 10.0.
Results
The age distribution of the study subjects was significantly different between the patients with cervical cancer and those with pre-cancerous lesions (53.6±12.0 vs. 42.7±12.7 years, p<0.001) and between those with cervical cancer and the controls (53.6±12.0 vs. 44.7±9.8 years, p<0.001), but not between those with pre-cancerous lesions and the controls (42.7±12.7 vs. 44.7±9.8 years, p = 0.407). The genotype distributions of rs6691378, rs10399805, rs4950928, and rs880633 met the Hardy-Weinberg equilibrium, which was applied to the normal controls.
Association of chitinase 3-like 1 gene polymorphisms with cervical neoplasia
There were significant differences in the distributions of SNPs rs6691378 and rs10399805 of the CHI3L1 gene between the women with cervical neoplasia and the normal controls (p<0.001 and p<0.001, respectively) (Table 1). However, no such differences were observed in rs4950928 and rs880633. The mutant homozygous genotype AA of SNP rs6691378 was differently distributed between women with cervical neoplasia and the controls compared to the wild homozygous genotype GG and heterozygous genotype GA (p<0.001). Further controlling for age, the women with the mutant homozygous AA carried a higher risk of developing cervical neoplasia compared to those with the wild genotype GG (AOR: 3.53, 95% CI: 1.71–7.35) or GG/GA (AOR: 3.40, 95% CI: 1.69–6.85) (Table 1). The mutant homozygous genotype AA of SNP rs10399805 was also differently distributed between those with cervical neoplasia and the controls compared to the wild GG and heterozygous GA (p<0.001). After controlling for age, the women with the mutant homozygous AA had a higher risk of developing cervical neoplasia compared to those with the wild genotype GG (AOR: 3.68, 95% CI: 1.71–7.87) or GG/GA (AOR: 3.60, 95% CI: 1.82–7.52).
Table 1. Genotype distributions of the single nucleotide polymorphisms of the chitinase 3-like 1 gene in patients with neoplasia of the uterine cervix and normal controls.
| Variables | Normal controls (n = 310) | Cervical neoplasiab (n = 160) | P (power)a | Odds ratio (95% confidence interval) | Adjusted odds ratio (95% confidence interval)c |
| rs6691378 | |||||
| GGd | 156(50.3%) | 67 (41.9%) | <0.001a | 1.00 | 1.00 |
| GA | 138 (44.5%) | 67 (41.9%) | (1.0) | 1.13 (0.75–1.70) | 1.08 (0.71–1.64) |
| AA | 16 (5.2%) | 26 (16.2%) | 3.79 (1.90–7.52) | 3.53 (1.71–7.35) | |
| GGd | 156 (50.3%) | 67 (41.9%) | 0.082 | 1.00 | 1.00 |
| GA/AA | 154 (49.7%) | 93 (58.1%) | 1.41(0.96–2.07) | 1.32 (0.89–1.96) | |
| GG/GAd | 294 (94.8%) | 134 (83.2%) | <0.001a | 1.00 | 1.00 |
| AA | 16 (5.2%) | 26 (16.3%) | (1.0) | 3.57 (1.85–6.85) | 3.40 (1.69–6.85) |
| rs10399805 | |||||
| GGd | 166 (53.5%) | 73 (45.6%) | <0.001a | 1.00 | 1.00 |
| GA | 130 (42.0%) | 63 (39.4%) | (1.0) | 1.10 (0.73–1.66) | 1.05 (0.69–1.59) |
| AA | 14 (4.5%) | 24 (15.0%) | 3.89 (1.91–7.94) | 3.68 (1.71–7.87) | |
| GGd | 166(53.5%) | 73 (45.6%) | 0.103 | 1.00 | 1.00 |
| GA/AA | 144(46.5%) | 87 (54.4%) | 1.37 (0.94–2.02) | 1.28 (0.87–1.90) | |
| GG/GAd | 296 (95.5%) | 136 (85.0%) | <0.001a | 1.00 | 1.00 |
| AA | 14 (4.5%) | 24 (15.0%) | (1.0) | 3.73 (1.87–7.46) | 3.60 (1.82–7.52) |
| rs4950928 | |||||
| CCd | 204 (65.8%) | 119 (74.4%) | 0.162 | 1.00 | 1.00 |
| CG | 100 (32.3%) | 39 (24.4%) | 0.67 (0.43–1.03) | 0.75 (0.48–1.16) | |
| GG | 6 (1.9%) | 2 (1.2%) | 0.57 (0.11–2.87) | 0.48 (0.09–2.46) | |
| CCd | 204 (65.8%) | 119 (74.4%) | 0.058 | 1.00 | 1.00 |
| CG/GG | 106 (34.2%) | 41 (25.6%) | 0.66 (0.43–1.02) | 0.73 (0.47–1.12) | |
| CC/CGd | 304 (98.1%) | 158 (98.8%) | 0.586 | 1.00 | 1.00 |
| GG | 6 (1.9%) | 2 (1.2%) | 0.64 (0.13–3.22) | 0.52 (0.10–2.65) | |
| rs880633 | |||||
| TTd | 131 (42.3%) | 75(46.9%) | 0.606 | 1.00 | 1.00 |
| TC | 148 (47.7%) | 69(43.1%) | 0.81 (0.54–1.22) | 0.78 (0.52–1.18) | |
| CC | 31 (10.0%) | 16 (10.0%) | 0.90 (0.46–1.76) | 0.90 (0.46–1.79) | |
| TTd | 131 (42.3%) | 75(46.9%) | 0.339 | 1.00 | 1.00 |
| TC/CC | 179 (57.7%) | 85 (53.1%) | 0.83 (0.57–1.22) | 0.80 (0.54–1.18) | |
| TT/TCd | 279 (90.0%) | 144 (90.0%) | 1.00 | 1.00 | 1.00 |
| CC | 31 (10.0%) | 16 (10.0%) | 1.00 (0.53–1.89) | 1.02 (0.53–1.97) |
Statistical analysis: logistic regression model, Chi-square or Fisher exact tests.
p<0.05; the sample size was estimated with power (1-β), 0.8 and α, 0.05 and the powers were calculated when the comparisons among the subgroups reached statistical significances (p<0.05) with an adequate sample size using WinPepi software, version 10.0.
Cervical neoplasia included pre-cancerous lesions and invasive cancer of the uterine cervix.
The adjusted odds ratios and their 95% confident intervals were estimated by logistic regression model after controlling for age.
Used as a reference for comparisons to evaluate the odds ratio of other genotypes.
When the cervical neoplasia group was further subdivided into subgroups of invasive cancer and pre-cancerous lesions, significant differences existed in the distributions of SNP rs6691378 and rs10399805 of the CHI3L1 gene among the women with cervical invasive cancer and pre-cancerous lesions and the normal women (p = 0.002 and p = 0.003, respectively; Table2). However, no such differences were observed in rs4950928 and rs880633. The mutant homozygous genotypes of both SNPs exhibited different distributions among the patients with cervical invasive cancer and pre-cancerous lesions, and the controls compared to the wild homozygous and heterozygous genotypes (p<0.001 and p<0.001, respectively; Table 2). After adjusting for age, the mutant homozygous AA in both SNPs rs6691378 and rs10399805 not only increased the risk of developing invasive cervical cancer (AOR: 2.37, 95% CI: 1.04–5.38 and AOR: 2.82, 95% CI: 1.20–6.58, respectively) but also pre-cancerous lesions (AOR: 5.26, 95% CI: 2.22–12.50 and AOR: 4.83, 95% CI: 1.90–12.35, respectively) compared to GG/GA (Table 2).
Table 2. Genotype distributions of the single nucleotide polymorphisms of chitinase 3-like 1 gene in the patients with invasive cancer and pre-cancerous lesions of the uterine cervix and normal controls.
| Variables | Normal controls (n = 310) | Pre-cancerous lesions (n = 61) | Invasive cancer(n = 99) | P (power)a | AOR(95% CI)b | AOR (95% CI)c |
| rs6691378 | ||||||
| GGd | 156 (50.3%) | 25 (41.0%) | 42 (42.4%) | 0.002a | 1.00 | 1.00 |
| GA | 138 (44.5%) | 25 (41.0%) | 42 (42.4%) | (0.9) | 1.14 (0.62–.08) | 1.01 (0.60–1.69) |
| AA | 16 (5.2%) | 11 (18.0%) | 15 (15.2%) | 5.62 (2.24–14.08) | 2.38 (1.01–5.62) | |
| GGd | 156 (50.3%) | 25 (41.0%) | 42 (42.4%) | 0.217 | 1.00 | 1.00 |
| GA/AA | 154 (49.7%) | 36 (59.0%) | 57 (57.6%) | 1.51 (0.86–2.64) | 1.15 (0.71–1.88) | |
| GG/GAd | 294 (94.8%) | 50 (82.0%) | 84 (84.8%) | <0.001a | 1.00 | 1.00 |
| AA | 16 (5.2%) | 11 (18.0%) | 15 (15.2%) | (1.0) | 5.26 (2.22–12.50) | 2.37(1.04–5.38) |
| rs10399805 | ||||||
| GGd | 166 (53.5%) | 29 (47.5%) | 44 (44.4%) | 0.003a | 1.00 | 1.00 |
| GA | 130 (42.0%) | 23 (37.7%) | 40 (40.4%) | (0.9) | 1.02 (0.56–1.86) | 1.02 (0.61–1.71) |
| AA | 14 (4.5%) | 9 (14.8%) | 15 (15.2%) | 4.88 (1.85–12.82) | 2.85 (1.18–6.90) | |
| GGd | 166 (53.5%) | 29 (47.5%) | 44 (44.4%) | 0.247 | 1.00 | 1.00 |
| GA/AA | 144 (46.5%) | 32 (52.5%) | 55 (55.6%) | 1.32 (0.76–2.29) | 1.20 (0.74–1.96) | |
| GG/GAd | 296 (95.5%) | 52 (85.2%) | 84(84.8%) | <0.001a | 1.00 | 1.00 |
| AA | 14 (4.5%) | 9 (14.8%) | 15(15.2%) | (1.0) | 4.83 (1.90–12.35) | 2.82 (1.20–6.58) |
| rs4950928 | ||||||
| CCd | 204 (65.8%) | 43 (70.5%) | 76 (76.8%) | 0.361 | 1.00 | 1.00 |
| CG | 100 (32.3%) | 17 (27.9%) | 22 (22.2%) | 0.74 (0.40–1.37) | 0.72 (0.41–1.27) | |
| GG | 6 (1.9%) | 1(1.6%) | 1 (1.0%) | 0.89 (0.10–7.63) | 0.35 (0.04–3.02) | |
| CCd | 204(65.8%) | 43 (70.5%) | 76 (76.8%) | 0.117 | 1.00 | 1.00 |
| CG/GG | 106(34.2%) | 18 (29.5%) | 23 (23.2%) | 0.75 (0.41–1.37) | 0.69 (0.40–1.20) | |
| CC/CGd | 304 (98.1%) | 60(98.4%) | 98 (99.0%) | 0.825 | 1.00 | 1.00 |
| GG | 6 (1.9%) | 1(1.6%) | 1 (1.0%) | 0.96 (0.11–8.20) | 0.38 (0.04–3.27) | |
| rs880633 | ||||||
| TTd | 131 (42.3%) | 34 (55.7%) | 41 (41.4%) | 0.335 | 1.00 | 1.00 |
| TC | 148 (47.7%) | 23 (37.7%) | 46 (46.5%) | 0.60 (0.33–1.06) | 0.96 (0.58–1.61) | |
| CC | 31 (10.0%) | 4 (6.6%) | 12 (12.1%) | 0.50 (0.17–1.52) | 1.31 (0.58–2.96) | |
| TTd | 131 (42.3%) | 34(55.7%) | 41(41.4%) | 0.131 | 1.00 | 1.00 |
| TC/CC | 179 (57.7%) | 27 (44.3%) | 58(58.6%) | 0.58 (0.33–1.01) | 1.02 (0. 63–1.66) | |
| TT/TCd | 279(90.0%) | 57(93.4%) | 87 (87.9%) | 0.523 | 1.00 | 1.00 |
| CC | 31 (10.0%) | 4 (6.6%) | 12 (12.1%) | 0.64 (0.22–1.89) | 1.34 (0.62–2.88) |
Statistical analysis: multiple logistic regression or Chi-square or Fisher exact tests. The AORs with their 95% CIs were estimated by the multiple logistic regression model after controlling for age.
p<0.05; the sample size was estimated with power (1-β), 0.8 and α, 0.05 and the powers were calculated when the comparisons among the subgroups reached astatistical significances (p<0.05) with an adequate sample size using WinPepi software, version 10.0.
Comparison between patients with pre-cancerous cervical lesions and normal controls after adjustments for age.
Comparison between patients with cervical cancer and normal controls after adjustments for age.
Used as a references for comparisons to evaluate the odds ratio of other genotypes.
Abbreviations: AOR, adjusted odds ratio; 95% CI, 95% confidence interval.
Analysis of allele frequencies of chitinase 3-like 1 polymorphisms in the study cohort
Analyzing the allele frequencies of the four CHI3L1 gene polymorphisms in the 470 samples collected, the allele frequencies in CHI3L1 SNPs rs6691378 and rs10399805 polymorphisms were differently distributed among the women with cervical invasive cancer, those with pre-cancerous lesions, and the normal subjects (p = 0.008 and p = 0.012, respectively) (Table 3). There were no such differences in SNPs rs4950928 and rs880633. The mutant alleles A and A in SNPs rs6691378 and rs10399805 increased and tended to increase the risk of developing cervical pre-cancerous lesions (OR: 1.66, 95% CI: 1.11–2.49 and OR: 1.48, 95% CI: 0.98–2.25, respectively; Table 3). They also increased the risk of developing invasive cervical cancer (OR: 1.51, 95% CI: 1.08–2.12 and OR: 1.60, 95% CI: 1.14–2.25, respectively; Table 3).
Table 3. Allele distributions of the single nucleotide polymorphisms of the chitinase 3-like 1 gene in patients with invasive cancer and pre-cancerous lesions of the uterine cervix and normal controls.
| Variables | Normal controls (n = 310) | Pre-cancerous lesions (n = 61) | Invasive cancer(n = 99) | P (power)a | OR (95% CI)b | OR (95% CI)c |
| rs6691378 | ||||||
| Gd | 450 | 75 | 126 | 0.008a | 1.00 | 1.00 |
| A | 170 | 47 | 72 | (0.8) | 1.66 (1.11–2.49) | 1.51 (1.08–2.12) |
| rs10399805 | ||||||
| Gd | 462 | 81 | 128 | 0.012a | 1.00 | 1.00 |
| A | 158 | 41 | 70 | (0.8) | 1.48 (0.98–2.25) | 1.60 (1.14–2.25) |
| rs4950928 | ||||||
| Cd | 508 | 103 | 174 | 0.140 | 1.00 | 1.00 |
| G | 112 | 19 | 24 | 0.84 (0.49–1.42) | 0.63 (0.39–1.00) | |
| rs880633 | ||||||
| Td | 410 | 91 | 128 | 0.144 | 1.00 | 1.00 |
| C | 210 | 31 | 70 | 0.66 (0.43–1.03) | 1.07 (0.76–1.49) |
Statistical analysis: Chi-square test.
p<0.05; the sample size was estimated with power (1-β), 0. 80 and α, 0.05 and the powers were calculated when the comparisons among the subgroups reached statistical significances (p<0.05) with an adequate sample size using WinPepi software, version 10.0.
Comparison between patients with pre-cancerous lesions and normal controls.
Comparison between patients with cervical cancer and normal controls.
Used as a reference to evaluate the odds ratio.
Abbreviations: OR, odds ratio; 95% CI, 95% confidence interval.
Haplotypes of chitinase 3-like 1 SNPs based on Taiwanese women and their involvement in cervical cancer
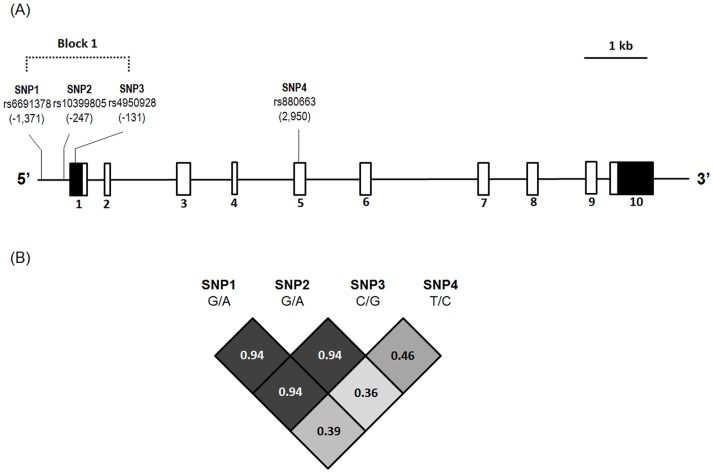
Based on the locations of the analyzed variants (rs6691378, rs10399805, rs4950928 and rs880663), the CHI3L1 gene, locations of the genotyped variants, and their pairwise linkage disequilibrium patterns were plotted (Figure 1). Because the mutant homozygous AA and AA in CHI3L1 SNPs rs6691378 and rs10399805 increased the risk of developing cervical pre-cancerous lesions or invasive cancer, haplotypes containing them (AACC and AACT) were regarded as a risk subgroup, while other haplotypes (i.e., GGCC, GGCT, GGGC, GGGT, GACC, GACT, AGCC, and AGCT) were regarded as a control subgroup. Individuals with haplotypes AACC and AACT had an increased risk of developing cervical neoplasia (p = 0.002). When the cervical neoplasia group was further subdivided into subgroups of invasive cancer and pre-cancerous lesions, significant differences existed in the distributions of CHI3L1 haplotypes among the women with cervical invasive cancer and pre-cancerous lesions and the controls (p = 0.01). The women with the AACC or AACT haplotypes had a higher risk of developing invasive cervical cancer (OR: 1.60, 95% CI: 1.13–2.26; Table 4).
Figure 1. Chitinase 3-like 1 (CHI3L1) gene, locations of the genotyped variants, and their pairwise linkage disequilibrium (LD) patterns.
(A) Schematic presentation of the CHI3L1 (gene ID: 1116) indicates the locations of the analyzed variants (rs6691378, rs10399805, rs4950928, and rs880663). The black and white boxes indicate the un-translated and coding regions, respectively. The exon number labeled below the exons and variant locations begins with the translation start site. (B) The one observed haploblock that the pairwise LD measured is D'labeled and colored in gray scale using the population data from Han Chinese in Beijing, China (CHB) in the HapMap 3.
Table 4. Haplotype distributions of the chitinase 3-like 1 (CHI3L1) gene in the patients with invasive cancer or pre-cancerous lesions of the uterine cervix and normal controls.
| CHI3L3 haplotypesa | Control women | Patients with pre-cancerous lesions | Patients with invasive cancer | P (power)b | OR and 95% CIc | OR and 95% CId |
| AACC and AACT | 153 | 41 | 68 | 0.01 (0.8) | 1.55 (1.02–2.35) | 1.60 (1.13–2.26) |
| Others | 467 | 81 | 130 | 1.00 (reference) | 1.00 (reference) |
Haplotypes containing AACC and AACT were regarded as a risk subgroup; other haplotypes (GGCC, GGCT, GGGC, GGGT, GACC, GACT, AGCC and AGCT) were regarded as a control subgroup.
p<0.05; the sample size was estimated with power (1-β), 0.80 and α, 0.05 and the powers were calculated when the comparisons among the subgroups reached statistical significances (p<0.05) with an adequate sample size using WinPepi software, version 10.0.
Comparison between patients with pre-cancerous lesions and normal controls.
Comparison between patients with cervical cancer and normal controls.
Abbreviations: OR, odds ratio; 95% CI, 95% confidence interval.
Association of chitinase 3-like 1 gene polymorphisms and haplotypes with clinicopathologic variables of cervical cancer, cancer recurrence, and patient survival
There were no associations between CHI3L1 gene polymorphisms and clinicopathologic variables of cervical cancer, cancer recurrence, and patient survival. When the CHI3L1 haplotypes were included in the analysis, they were related to the clinical stage (p = 0.040, OR: 2.31, 95% CI: 0.95–5.72; ≥ stage II vs. stage I; Table 5), tumor diameter (p = 0.054; OR: 2.19, 95% CI: 0.90–5.43; >4 cm vs. ≤4 cm) and vaginal invasion (p = 0.024; OR: 2.66, 95% CI: 0.98–6.83; invasion vs. no invasion). Moreover, significant associations became even more obvious among the CHI3L1 risk haplotypes and clinical stage (p = 0.009; OR: 3.29, 95% CI: 1.18–9.63; ≥ stage II vs. stage I; Table 5), tumor diameter (p = 0.029; OR: 2.75, 95% CI: 0.99–8.04; >4 cm vs. ≤4 cm), and vaginal invasion (p = 0.004; OR: 4.10, 95% CI: 1.40–11.62; invasion vs. no invasion) in SCC specimens, but not in adenocarcinoma tissues.
Table 5. Association of haplotype distribution of chitinase 3-like 1 polymorphisms with clinicopathologic characteristics of the patients with uterine cervical cancer.
| Variablesa | Haplotypes AACC or AACT vs. control haplotypesc | p and odds ratio (95/% confidence interval) | Haplotypes AACC or AACT vs. control haplotypesc in SCC tissues | p and odds ratio (95/% confidence interval) for SCC |
| Clinical stage | 0.04 | 0.009 | ||
| stage Ib | 12 104 | 1.00 | 8 94 | 1.00 |
| ≥stage II | 16 60 | 2.31 (0. 95–5.72) | 14 50 | 3.29 (1.18–9.63) |
| Pathologic type squamous cell | 0.187 | unavailable | unavailable | |
| carcinomab | 22 144 | 1.00 | ||
| adenocarcinoma | 6 20 | 1.96 (0.58–5.81) | ||
| Cell grading | 0.354 | 0.084 | ||
| well (grade 1)b | 8 34 | 1.00 | 8 26 | 1.00 |
| moderate & poor (grades 2/3) | 20 130 | 0.65 (0.25–1.87) | 14 118 | 2.59 (0.84–7.42) |
| Invasion depth of cervical stroma | 0.354 | 0.476 | ||
| ≤10 mmb | 18 90 | 1.00 | 14 80 | 1.00 |
| >10 mm | 10 74 | 0.68 (0.26–1.66) | 8 64 | 0.71 (0.24–1.96) |
| Tumor diameterb | 0.054 | 0.029 | ||
| ≤4 cm | 12 102 | 1.00 | 8 88 | 1.00 |
| >4 cm | 16 62 | 2.19 (0.90–5.43) | 14 56 | 2.75 (0.99–8.04) |
| Parametrium | 0.637 | 0.261 | ||
| no invasionb | 20 124 | 1.00 | 14 108 | 1.00 |
| invasion | 8 40 | 1.24 (0.44–3.22) | 8 36 | 1.71 (0.57–4.80) |
| Vagina | 0.024 | 0.004 | ||
| no invasionb | 18 134 | 1.00 | 16 108 | 1.00 |
| invasion | 10 28 | 2.66 (0.98–6.83) | 6 36 | 4.10 (1.40–11.62) |
| Pelvic lymph node | 0.637 | 0.819 | ||
| no metastasisb | 20 124 | 1.00 | 16 108 | 1.00 |
| metastasis | 8 40 | 1.24 (0.44–3.22) | 6 36 | 1.13 (0.33–3.32) |
Statistical analyses: Chi-square test.
Some clinicopathologic data could not be collected from the patients with cervical cancer due to incomplete medical charts or records.
As a reference.
Haplotypes containing AACC and AACT were regarded as a risk subgroup; other haplotypes (GGCC, GGCT, GGGC, GGGT, GACC, GACT, AGCC and AGCT) were regarded as a control subgroup for comparison.
Abbreviation: SCC, squamous cell carcinoma.
Cervical cancer patients with CHI3L1 risk haplotypes tended to have poor survival event (logistic regression model; p = 0.078; OR:2.99, 95% CI: 0.89–10.08; Table 6). However, cell grading 2/3 (p = 0.049; OR: 5.47, 95% CI: 1.01–29.78) and positive pelvic lymph node metastasis (p = 0.002; OR: 5.58, 95% CI: 1.92–12.63) were independent predictive factors for patient survival event. The five-year survival rate was 66.1% for the patients with the CHI3L1 risk haplotypes, and this increased to 88.1% for those with other haplotypes. However, it could not reach a statistical significance (p = 0.21; OR: 1.68, 95% CI: 0.73–3.85; Table 7). Moreover, CHI3L1 haplotypes also had a tendency to be related to a higher probability of recurrence event (p = 0.081; OR: 3.07, 95% CI: 0.87–10.81; Table 6). Cell grading 2/3 (p = 0.015; OR: 8.73, 95% CI: 1.52–50.29) and positive pelvic lymph node metastasis (p = 0.001; OR: 6.52, 95% CI: 2.25–18.93) were independent predictive factors for cancer recurrence event. In the cervical cancer patients with SCC, CHI3L1 risk haplotypes increased the risk of recurrence event (p = 0.011; OR: 7.50, 95% CI: 1.60–35.16) and tended to increase the risk of poor survival event (p = 0.051; OR: 4.36, 95% CI: 0.99–19.14), in logistic regression model. Deep invasion depth and ≥stage II also increased the risk of recurrence event (p = 0.04; OR: 4.61, 95% CI: 1.08–19.74 and p = 0.047; OR: 6.17, 95% CI: 1.03–37.04, respectively). Positive pelvic lymph node metastasis increased the risk of recurrence event (p = 0.012; OR: 4.49, 95% CI: 1.40–14.46) and tended to increase the risk of poor survival event (p = 0.058; OR: 3.10, 95% CI: 0.96–10.00).
Table 6. Influence of chitinase 3-like 1 (CHI3L1) haplotypes and clinicopathologic parameters on cancer recurrence event and survival event of the patients with uterine cervical cancer.
| Variables | Recurrence event | Survival event | ||
| p value | OR & 95%CIb | p value | OR & 95%CIb | |
| CHI3L1 risk haplotypesa | 0.081 | 3.07 (0.87–10.81) | 0.078 | 2.99 (0.89–10.08) |
| Cell grading 2/3 | 0.015 | 8.73 (1.52–50.29) | 0.049 | 5.47 (1.01–29.78) |
| Positive pelvic lymph node metastasis | 0.001 | 6.52 (2.25–18.93) | 0.002 | 5.58 (1.92–16.23) |
Statistical analysis: logistic regression model.
Haplotypes containing AACC and AACT were regarded as a risk subgroup; other haplotypes (GGCC, GGCT, GGGC, GGGT, GACC, GACT, AGCC and AGCT) were regarded as a control subgroup for comparison.
OR and 95% CI, odds ratio and 95% confidence interval for risk haplotypes AACC and AACT and poor clinicopathologic characteristics, compared to their respective controls.
Table 7. Univariate and multivariate analyses for the correlation of clinicopathologic variables and CHI3L1 risk haplotype expressions with overall survival of the patients with uterine cervical cancer.
| 5-year survival rate (%)b | Hazard ratioc | 95% confidence intervalc | p value | |
| Univariate analysis | ||||
| Stagea | ||||
| I | 91.0 | 1 | Reference | 0.02 |
| others | 77.8 | 2.57 | 1.12–5.91 | |
| Pathologic typea | ||||
| squamous cell | 86.1 | 1 | Reference | |
| carcinoma | 0.18 | |||
| adenocarcinoma | 66.8 | 1.84 | 0.74–4.59 | |
| Depth of stromal | ||||
| invasiona | 0.0024 | |||
| ≤1/2 depth | 90.0 | 1 | Reference | |
| >1/2 depth | 75.1 | 3.28 | 1.44–7.46 | |
| Tumor diametera | ||||
| ≤4 cm | 92.6 | 1 | Reference | 0.0011 |
| >4 cm | 73.4 | 3.55 | 1.56–8.06 | |
| Cell gradinga well | 94.1 | 1 | Reference | 0.0091 |
| moderate and poor | 80.2 | 5.49 | 1.29–23.26 | |
| Parametrialinvasiona | ||||
| no invasion | 88.8 | 1 | Reference | 0.016 |
| invasion | 74.6 | 2.40 | 1.14–5.08 | |
| Vagina invasiona | ||||
| no invasion | 85.6 | 1 | Reference | 0.20 |
| invasion | 80.5 | 1.64 | 0.75–3.57 | |
| Pelvic lymph node metastasisa | ||||
| negative | 90.2 | 1 | Reference | 0.0068 |
| positive | 67.0 | 2.72 | 1.27–5.81 | |
| CHI3L1 risk haplotypes expression | ||||
| negative | 88.1 | 1 | Reference | 0.21 |
| positive | 66.1 | 1.68 | 0.73–3.85 | |
| Multivariate analysis | ||||
| Cell grading | ||||
| well (grade 1)a | 94.1 | 1 | Reference | 0.037 |
| moderate & poor (grades 2/3) | 80.2 | 5.59 | 1.11–28.57 |
Statistical analysis: Kaplan-Meier model and multivariate and univariate Cox regression models.
Some clinicopathologic data could not be collected from the patients with cervical cancer due to incomplete medical charts or records.
Analyzed by the Kaplan-Meier model.
Based on overall survival analyzed by multivariate and univariate Cox regression models.
Abbreviation: CHI3L1, chitinase 3-like 1.
Only cell grading was found to be an independent predictor for overall survival in multivariate analysis (p = 0.037, hazard ratio: 5.59, 95% CI: 1.11–28.57; Table 7). However, CHI3L1 risk haplotypes, depth of stromal invasion, cell grading and pelvic lymph node metastasis were independent predictive factors for the probability of recurrence (Table 8). In multivariate analysis for the probability of recurrence and overall survival of the patients with SCC, CHI3L1 risk haplotypes exhibited a significant association with the probability of recurrence (p = 0.002; hazard ratio: 6.21, 95% CI: 1.90–20.41) and a marginal association with overall survival (p = 0.051; hazard ratio: 3.76, 95% CI: 0.99–14.29) among the clinicopathologic parameters and CHI3L1 haplotypes, using the Cox proportional hazards model (Table 8). However, these findings were not demonstrated in the patients with adenocarcinoma.
Table 8. Influence of chitinase 3-like 1 (CHI3L1) haplotypes and clinicopathologic parameters on cancer recurrence probability and overall survival of the patients with uterine cervical cancer.
| Variables | Recurrence | Overall survival | ||
| p value | OR & 95%CIb | p value | OR & 95%CIb | |
| CHI3L1 risk haplotypesa | 0.009 | 3.57 (1.37–9.35) | 0.174 | 1.99 (0.74–5.35) |
| Depth of stromal invasion | 0.036 | 3.47 (1.09–11.11) | insignificant | |
| Cell grading 2/3 | 0.042 | 5.21 (1.06–25.64) | 0.037 | 5.59 (1.11–28.57) |
| Positive pelvic lymph node metastasis | 0.002 | 5.26 (1.85–15.15) | insignificant | |
Statistical analysis: multivariate Cox regression model.
Haplotypes containing AACC and AACT were regarded as a risk subgroup; other haplotypes (GGCC, GGCT, GGGC, GGGT, GACC, GACT, AGCC and AGCT) were regarded as a control subgroup for comparison.
OR and 95% CI, odds ratio and 95% confidence interval for risk haplotypes AACC and AACT and poor clinicopathologic characteristics, compared to their respective controls.
Discussion
To the best of our knowledge, the present study is the first to show a significant association between CHI3L1 gene polymorphisms and susceptibility to uterine cervical cancer. Both mutant homozygous genotypes AA and AA in the promoter regions −1371 and −247 of CHI3L1 SNPs rs6691378 and rs10399805 not only increased the susceptibility to pre-cancerous lesions but also invasive cancer of the uterine cervix even after controlling for age. No such differences were observed in rs4950928 (−131, promoter region) and rs880633 (+2950, exon 5). Several studies on CHI3L1 SNPs have documented that the genetic variations of CHI3L1 affect circulating YKL-40 levels, both in healthy adults and in patients with diseases [17]–[20]. Kajergarred et al. and Thomsen et al. demonstrated the regulatory effects of some promoter SNPs, including rs6691378, rs10399805, and rs4950928, on plasma YKL-40 levels in the Danish general population [21], [22]. The minor allele of rs10399805 has been reported to enhance the binding of CCAAT enhancer-binding protein (C/EBFα) to the gene promoter and increase CHI3L1 transcription, consequently increasing plasma YKL-40 levels [23], [24]. In contrast, Zheng et al. did not show any significant association between SNP rs10399805 and YKL-40 levels in a Chinese population [17]. Different study populations may affect the results. Verlaan et al. revealed that rs10399805 and rs4950928 modulate CHI3L1 transcription and promoter polymorphisms in CHI3L1 and are associated with asthma [25]. Most of the susceptibility genes for common diseases do not play a vital role in predisposing to the disease, but act as response modifiers to internal or external environmental factors [26], [27].
The YKL-40 protein is encoded by the CHI3L1, and SNPs in the CHI3L1 promoter have been associated with elevated serum YKL-40 levels [20], [28], and differential gene expression [28] and transcript levels [29]. YKL-40 has been reported to initiate the mitogen-activated protein kinase (MAPK) and phosphoinoside-3-kinase (PI3K) signaling pathways, which lead to increased cell proliferation in connective-tissue cells [30]. The murine homologue of YKL-40 is a breast regression protein of 39 kDa (BRP-39) which has been described to be expressed in cancer cells, and YKL-40/BRP-39 has been demonstrated to play a role in cell proliferation, survival, and tissue remodeling [30]–[32]. Serum YKL-40 can be secreted from fully activated macrophages associated with the tumor and can be produced by tumor cells themselves or by non-malignant cells such as activated neutrophils and fibroblasts, chondrocytes, and synovial cells [32]–[34]. Tumor cells have been reported to express and produce YKL-40 in breast and colon cancer [35], while tumor-associated macrophages, but not tumor cells, have been reported to secrete YKL-40 in lung cancer [36]. Mitsuhashi et al. found that pre-treatment serum levels of YKL-40 are elevated in cervical cancer, even in the early stages [15]. Because CHI3L1 SNPs influence YKL-40 expression, they may subsequently influence the development of diseases such as cervical cancer.
The MAFs in rs6691378, rs10399805, and rs4950928 of CHI3L1 promoters in the Taiwanese women in the current study (27.4%, 25.5%, and 18.1%, respectively) are similar to those reported in the Han Chinese in Beijing, China (HCB, 25.6%, 25.6%, and 17.1%, respectively), which are based on the National Center for Biotechnology Information (NCBI) SNP database (dbSNP). The MAF in rs880633 (33.9%) in exon 5 of the CHI3L1 gene is also similar to that in HCB (35.6%), which is also based on the dbSNP. Furthermore, the mutant allele A in SNP rs6691378 increased the risk of developing cervical pre-cancerous lesions and invasive cancer. Although the mutant allele A in SNP rs10399805 also increased susceptibility to cervical cancer, its effect on the development of pre-cancerous lesions was not as strong as that of A in SNP rs6691378. In order to apply the haplotypes, pairwise linkage disequilibrium patterns were established for these SNPs, and the results revealed that rs6691378 has a strong linkage disequilibrium with rs10399805 (D′ = 0.94). The mutant homozygous AA as well as AA in CHI3L1 SNPs rs6691378 and rs10399805 were associated with cervical carcinogenesis. Thus, CHI3L1 haplotypes may be used for further correlation with the development and clinicopathologic variables of cervical cancer and patient prognosis.
Not only the allele distribution of CHI3L1 SNPs but also CHI3L1 risk haplotypes, AACC and AACT, correlated with the susceptibility to cervical pre-cancerous lesions and invasive cancer in the present study. There are few existing studies that have reported the relationships of CHI3L1 haplotypes with diseases. Zhao et al. revealed significant associations of three SNPs in the promoter region of the CHI3L1 gene (rs6691378, −1371; rs10399805, −247 and rs4950928, −131) with schizophrenia [28]. They further constructed these three SNPs as CHI3L1 haplotypes and found that the CHI3L1 haplotypes implicated in schizophrenia susceptibility were associated with altered expression levels of the gene. Genetic variations that change the expression of CHI3L1 may influence some key processes that are CHI3L1-dosage dependent. One is the AKT-mediated signal pathway through PI3K-dependent phosphorylation [30]. This AKT pathway has been associated with cell survival [37], [38] and may regulate cytokine-induced cellular responses [39]. Nonetheless, Zheng et al. could not demonstrate any relationship between CHI3L1 common haplotypes and coronary artery disease or its severity [17].
Mitsuhashi et al. demonstrated that serum YKL-40 level is the best biomarker for uterine cervical cancer compared to the SCC antigens, CA-125, CA19-9, and C-reactive protein [15]. Pre-treatment YKL-40 levels were significantly correlated with FIGO stage and with relapse or persistent disease status, while correlations with nodal status and tumor size were marginal in cervical adenocarcinoma. Cox regression analysis showed that an elevated YKL-40 level is associated with relapse or persistent disease in cervical adenocarcinoma [15]. YKL-40 has been suggested to play a role in cell proliferation, differentiation, and to protect cells from apoptotic signals, and act on extracellular tissue remodeling [30], [40]. Reported as a stimulator of angiogenesis in tumors, YKL-40 is posited to be involved in cancer metastasis [41], [42]. The present study postulates that CHI3L1 SNPs or haplotypes affect the expression of YKL-40. Analyzing the relationship among CHI3L1 SNPs or haplotypes, clinicopathologic variables, cancer recurrence, and patient survival, we could not demonstrate any significant association between CHI3L1 SNPs and these characteristics. However, cervical cancer patients with the CHI3L1 risk haplotypes AACC or AACT had significant associations with clinical stage and vaginal invasion, while the association with tumor size was marginal. In cervical cancer patients with SCC, these relationships were more obvious. Logistic regression model revealed that the patients with the risk haplotypes tend to have higher recurrence event and poorer survival event. The other independent factors were cell grading and pelvic lymph node metastasis. In the patients with SCC, CHI3L1 risk haplotypes more obviously increased the risk of recurrence event and poor survival event. In multivariate Cox proportional hazard model, there was a significant association between CHI3L1 risk haplotypes and the probability of recurrence, and a marginal association between risk haplotypes and overall survival in the patients with cervical SCC. However, these findings were not seen in the patients with adenocarcinoma, implying that CHI3L1 risk haplotypes are particularly applicable for predicting the prognosis in patients with SCC, especially in terms of recurrence.
In conclusion, the CHI3L1 SNPs rs6691378 and rs10399805 and CHI3L1 haplotypes were correlated with the development of pre-cancerous lesions and invasive cancer of the uterine cervix. Cervical cancer patients with the CHI3L1 haplotypes AACC or AACT had poor clinicopathologic characteristics and a poor prognosis. These risk haplotypes were associated with a higher probability of recurrence, especially for those with SCC.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the paper.
Funding Statement
This study was supported by research grants from Chung Shan Medical University Hospital (CSH-2013-D-001, CSH-2014-D-003), the Taiwan National Science Council (NSC 102-2314-B-040-014-MY3) and Chung Shan Medical University Hospital and Chi-Mei Foundation Medical Center (CSMU-CMMC-102-02). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, et al. (2006) DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 355: 983–991. [DOI] [PubMed] [Google Scholar]
- 2. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, et al. (2005) REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer 93: 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sapp H, Goetzl L, Creasman W, Kohler M, Underwood P, et al.. (2008) Combined adverse effect of African American race and deep stromal invasion on survival following radical hysterectomy for cervical cancer. Am J Obstet Gynecol 199: 196 e191–197. [DOI] [PubMed]
- 4. Chabes A, Thelander L (2000) Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem 275: 17747–17753. [DOI] [PubMed] [Google Scholar]
- 5. Schorge JO, Molpus KL, Goodman A, Nikrui N, Fuller AF Jr (1996) The effect of postsurgical therapy on stage III endometrial carcinoma. Gynecol Oncol 63: 34–39. [DOI] [PubMed] [Google Scholar]
- 6. Fuller AF Jr, Elliott N, Kosloff C, Hoskins WJ, Lewis JL Jr (1989) Determinants of increased risk for recurrence in patients undergoing radical hysterectomy for stage IB and IIA carcinoma of the cervix. Gynecol Oncol 33: 34–39. [DOI] [PubMed] [Google Scholar]
- 7. Sevin BU, Nadji M, Lampe B, Lu Y, Hilsenbeck S, et al. (1995) Prognostic factors of early stage cervical cancer treated by radical hysterectomy. Cancer 76: 1978–1986. [DOI] [PubMed] [Google Scholar]
- 8. Wang LM, Lu FF, Zhang SY, Yao RY, Xing XM, et al. (2012) Overexpression of catalytic subunit M2 in patients with ovarian cancer. Chin Med J (Engl) 125: 2151–2156. [PubMed] [Google Scholar]
- 9. Grisaru DA, Covens A, Franssen E, Chapman W, Shaw P, et al. (2003) Histopathologic score predicts recurrence free survival after radical surgery in patients with stage IA2-IB1-2 cervical carcinoma. Cancer 97: 1904–1908. [DOI] [PubMed] [Google Scholar]
- 10. Pecorelli S (2009) Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet 105: 103–104. [DOI] [PubMed] [Google Scholar]
- 11. Morikawa T, Hino R, Uozaki H, Maeda D, Ushiku T, et al. (2010) Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibition in vitro. Hum Pathol 41: 1742–1748. [DOI] [PubMed] [Google Scholar]
- 12. Zhang K, Hu S, Wu J, Chen L, Lu J, et al. (2009) Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: implication of RRM2 in angiogenesis. Mol Cancer 8: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M (2010) Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology 57: 885–892. [DOI] [PubMed] [Google Scholar]
- 14. Duxbury MS, Whang EE (2007) RRM2 induces NF-kappaB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun 354: 190–196. [DOI] [PubMed] [Google Scholar]
- 15. Mitsuhashi A, Matsui H, Usui H, Nagai Y, Tate S, et al. (2009) Serum YKL-40 as a marker for cervical adenocarcinoma. Ann Oncol 20: 71–77. [DOI] [PubMed] [Google Scholar]
- 16. Liu X, Zhou B, Xue L, Yen F, Chu P, et al. (2007) Ribonucleotide reductase subunits M2 and p53R2 are potential biomarkers for metastasis of colon cancer. Clin Colorectal Cancer 6: 374–381. [DOI] [PubMed] [Google Scholar]
- 17. Zheng JL, Lu L, Hu J, Zhang RY, Zhang Q, et al. (2011) Genetic polymorphisms in chitinase 3-like 1 (CHI3L1) are associated with circulating YKL-40 levels, but not with angiographic coronary artery disease in a Chinese population. Cytokine 54: 51–55. [DOI] [PubMed] [Google Scholar]
- 18. Lee H, Kang R, Jee SH, Yoon Y (2010) A promoter polymorphism -2122C>T of CHI3L1 is associated with serum low density lipoprotein cholesterol level in Korean subjects. Clin Biochem 43: 1195–1200. [DOI] [PubMed] [Google Scholar]
- 19. Ober C, Tan Z, Sun Y, Possick JD, Pan L, et al. (2008) Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 358: 1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kruit A, Grutters JC, Ruven HJ, van Moorsel CC, van den Bosch JM (2007) A CHI3L1 gene polymorphism is associated with serum levels of YKL-40, a novel sarcoidosis marker. Respir Med 101: 1563–1571. [DOI] [PubMed] [Google Scholar]
- 21. Kjaergaard AD, Johansen JS, Nordestgaard BG, Bojesen SE (2013) Genetic variants in CHI3L1 influencing YKL-40 levels: resequencing 900 individuals and genotyping 9000 individuals from the general population. J Med Genet 50: 831–837. [DOI] [PubMed] [Google Scholar]
- 22. Thomsen SB, Rathcke CN, Skaaby T, Linneberg A, Vestergaard H (2012) The Association between genetic variations of CHI3L1, levels of the encoded glycoprotein YKL-40 and the lipid profile in a Danish population. PLoS One 7: e47094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sohn MH, Lee JH, Kim KW, Kim SW, Lee SH, et al. (2009) Genetic variation in the promoter region of chitinase 3-like 1 is associated with atopy. Am J Respir Crit Care Med 179: 449–456. [DOI] [PubMed] [Google Scholar]
- 24. Nielsen KR, Steffensen R, Boegsted M, Baech J, Lundbye-Christensen S, et al. (2011) Promoter polymorphisms in the chitinase 3-like 1 gene influence the serum concentration of YKL-40 in Danish patients with rheumatoid arthritis and in healthy subjects. Arthritis Res Ther 13: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verlaan DJ, Ouimet M, Adoue V, Sirois-Gagnon D, Lariviere M, et al. (2012) Promoter polymorphisms in CHI3L1 are associated with asthma. J Allergy Clin Immunol 130: 533–535. [DOI] [PubMed] [Google Scholar]
- 26. Duff GW (2007) Influence of genetics on disease susceptibility and progression. Nutr Rev 65: S177–181. [DOI] [PubMed] [Google Scholar]
- 27. Tiret L (2002) Gene-environment interaction: a central concept in multifactorial diseases. Proc Nutr Soc 61: 457–463. [DOI] [PubMed] [Google Scholar]
- 28. Zhao X, Tang R, Gao B, Shi Y, Zhou J, et al. (2007) Functional variants in the promoter region of Chitinase 3-like 1 (CHI3L1) and susceptibility to schizophrenia. Am J Hum Genet 80: 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, et al. (2007) A genome-wide association study of global gene expression. Nat Genet 39: 1202–1207. [DOI] [PubMed] [Google Scholar]
- 30. Recklies AD, White C, Ling H (2002) The chitinase 3-like protein human cartilage glycoprotein 39 (HC-gp39) stimulates proliferation of human connective-tissue cells and activates both extracellular signal-regulated kinase- and protein kinase B-mediated signalling pathways. Biochem J 365: 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shackelton LM, Mann DM, Millis AJ (1995) Identification of a 38-kDa heparin-binding glycoprotein (gp38k) in differentiating vascular smooth muscle cells as a member of a group of proteins associated with tissue remodeling. J Biol Chem 270: 13076–13083. [DOI] [PubMed] [Google Scholar]
- 32. Hakala BE, White C, Recklies AD (1993) Human cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitinase protein family. J Biol Chem 268: 25803–25810. [PubMed] [Google Scholar]
- 33. Renkema GH, Boot RG, Au FL, Donker-Koopman WE, Strijland A, et al. (1998) Chitotriosidase, a chitinase, and the 39-kDa human cartilage glycoprotein, a chitin-binding lectin, are homologues of family 18 glycosyl hydrolases secreted by human macrophages. Eur J Biochem 251: 504–509. [DOI] [PubMed] [Google Scholar]
- 34. De Ceuninck F, Gaufillier S, Bonnaud A, Sabatini M, Lesur C, et al. (2001) YKL-40 (cartilage gp-39) induces proliferative events in cultured chondrocytes and synoviocytes and increases glycosaminoglycan synthesis in chondrocytes. Biochem Biophys Res Commun 285: 926–931. [DOI] [PubMed] [Google Scholar]
- 35. Johansen JS, Jensen BV, Roslind A, Nielsen D, Price PA (2006) Serum YKL-40, a new prognostic biomarker in cancer patients? Cancer Epidemiol Biomarkers Prev 15: 194–202. [DOI] [PubMed] [Google Scholar]
- 36. Junker N, Johansen JS, Andersen CB, Kristjansen PE (2005) Expression of YKL-40 by peritumoral macrophages in human small cell lung cancer. Lung Cancer 48: 223–231. [DOI] [PubMed] [Google Scholar]
- 37. Brazil DP, Hemmings BA (2001) Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem Sci 26: 657–664. [DOI] [PubMed] [Google Scholar]
- 38. Scheid MP, Woodgett JR (2001) PKB/AKT: functional insights from genetic models. Nat Rev Mol Cell Biol 2: 760–768. [DOI] [PubMed] [Google Scholar]
- 39. Kim AH, Khursigara G, Sun X, Franke TF, Chao MV (2001) Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol Cell Biol 21: 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, et al. (2011) Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol 73: 479–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Faibish M, Francescone R, Bentley B, Yan W, Shao R (2011) A YKL-40-neutralizing antibody blocks tumor angiogenesis and progression: a potential therapeutic agent in cancers. Mol Cancer Ther 10: 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Francescone RA, Scully S, Faibish M, Taylor SL, Oh D, et al. (2011) Role of YKL-40 in the angiogenesis, radioresistance, and progression of glioblastoma. J Biol Chem 286: 15332–15343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the paper.