Abstract
Rickettsia felis, the agent of flea-borne spotted fever, has a cosmopolitan distribution. Its pathogenic role in humans has been demonstrated through molecular and serologic tests in several cases. The cat flea (Ctenocephalides felis) is considered the main reservoir and the biological vector. The aim of this study was to assess the presence and occurrence of R. felis in fleas collected from dogs and cats in various sites of Palermo (Sicily). Between August and October 2012, 134 fleas were collected from 42 animals: 37 fleas from 13 dogs and 97 fleas from 29 cats. Two species of fleas were identified: 132 Ctenocephalides felis (98.51%) collected on all animals and only two C. canis (1.49%) on one dog. Out of 132 C. felis, 34 (25.76%), 12 from dogs (32.43%) and 22 (22.68%) from cats, were positive for R. felis DNA by a polymerase chain reaction (PCR), confirmed by sequencing. The only two C. canis fleas were negative. About half of examined animals (47.62%, 20/42) were infested with at least one infected flea; in particular 46.15% of dogs (6/13) and 48.28% of cats (14/29). It seems that in the Palermo district there is a peri-domestic cycle, with a relatively high prevalence of R. felis infection in the cat flea, an insect widely diffused in home environments and which can frequently bite humans. The results also suggest that R. felis should be considered in the human differential diagnosis of any spotted-like fever or febrile illness without a clear source of infection in Sicily, especially if the patient is known to have been exposed to flea bites.
Introduction
Rickettsioses are vector-borne zoonotic infections caused by obligate intracellular bacteria of the genus Rickettsia.
Rickettsia felis was probably first detected in Ctenocephalides felis in 1918, and named “Rickettsia ctenocephali” [1]. However, this record was neglected until 1990, when a Rickettsia-like organism was found in C. felis fleas by electron microscopy [2]. At that time, it was referred to as the “ELB agent”, for the original source of the fleas at Elward Laboratory (Soquel, CA, USA). The species R. felis was formally validated by molecular criteria in 2001, and the reference strain was isolated and definitely characterized in 2002 [3]. R. felis has recently been included in the rickettsial transitional group [4].
R. felis is the etiological agent of flea-borne spotted fever (also known as cat flea typhus), described recently as an emerging rickettsiosis of medical importance [5]. Its pathogenic role in humans has been demonstrated through PCR and serology in several cases, prevalently described in hot countries [6]. Typically, the disease presents as a flu-like acute febrile syndrome, very similar to murine typhus [7], associated with headache and rash. Other signs include: asthenia, myalgia, local lymphadenopathy, neurological signs, conjunctivitis, gastrointestinal involvement and cutaneous manifestations, such as generalized maculo-papular exanthema, and in some cases a characteristic inoculation eschar at the site of the flea bite; no fatalities have been reported [5],[8]–[10]. Due to non-specific febrile clinical course, it is thought that many human cases are currently misdiagnosed as other rickettsial and viral infections. In recent years, R. felis has acquired an important role in the etiology of the acute febrile syndrome in many countries [6],[11]. Several cases originally diagnosed as other rickettsial infections, in particular murine typhus, were retrospectively re-evaluated by molecular or serological tools as R. felis infections [6],[12]–[14].
Although only a few confirmed human cases have been described, R. felis is globally distributed and it is primarily associated with cat fleas, Ctenocephalides felis, which appears to be its main vector and reservoir and the only known biological vector. It is also harbored by a variety of hematophagous arthropods [5],[6],[15]–[19].
R. felis transmission is primarily vertical (trans-ovarial and trans-stadial) within a flea population, rather than horizontal between fleas through a bloodmeal [20],[21]. Although not required for the maintenance of the enzootic cycle, mammalian hosts still serve as a mechanical vehicle and bloodmeal source to support flea populations. Within the flea, R. felis infection is disseminated, having been identified by PCR and microscopy in several tissues [22].
The wide distribution of R. felis is related to the worldwide distribution of C. felis. The cat flea is extremely common on cats and dogs in many temperate and tropical regions, but it can also infest other animal species and humans [23]. It represents the great majority of fleas in human homes.
R. felis has now been described in infected arthropods from more than 20 countries all over the world, excepting Antartica [5],[6],[15],[17],[24].
In Italy, only a few studies have been carried out to establish the distribution of R. felis infection in invertebrate hosts [25]–[27], and no clinical case or infection has yet been reported in humans or other mammals.
The aim of our study was to determine the presence and occurrence of this pathogen in fleas collected from dogs and cats in various sites of Palermo (Sicily, Southern Italy).
Materials And Methods
Geographical Area
The study was conducted in the city of Palermo (North coast of Sicily, Southern Italy), between August and October 2012 (temperature: mean value 24,6°C, T min: 18,3°C, T max: 30,8°C; humidity: mean value 66,3%, min 61,8%, max 71,6%).
Flea Collection And Identification
The study was carried out on 42 flea-infested animals, 13 dogs and 29 cats.
Both dogs and cats came from various districts of the urban area of Palermo; they were primarily enrolled at the municipal shelter, but also in private veterinary facilities. Among these, 39 were strays and 3 (2 cats and 1 dog) client-owned animals.
The stray cats and dogs were captured by municipal shelter personnel or by authorized volunteers from animal protection associations, as part of a routine procedure for heath and reproductive control. The cats were captured in special cages and the dogs with painless systems and without the use of leghold traps, poisoned morsels or prods. To recover the fleas, the animals were combed craniocaudally with a plastic fine-toothed flea-comb for at least 15 minutes on the dorsal and ventral trunk. In the stray animals, this was performed after sedation with tiletamine-zolazepam (Zoletil, Virbac) prior to sterilization surgery.
For each animal, the fleas were harvested, kept alive for a few days at room temperature to allow the insects to cleanse themselves of any ingested blood, rinsed with distilled water and stored in 70% ethyl alcohol at room temperature.
The fleas were identified according to previously described identification keys [28],[29].
Dna Extraction From Fleas
After identification, samples of fleas collected from each animal and containing more than one flea were pooled, according to species and sex. Pools ranged from two to six specimens; fleas were grouped in further pools when more than six were collected from the same animal.
The fleas were sectioned longitudinally and one half of each exemplar was immediately subjected to molecular DNA analysis. The other half of each flea was maintained in alcohol, awaiting further analysis. Two specific solutions (180 µL of Genomic Digestion Buffer and 20 µL of Proteinase K) were used overnight, until complete dissolution of flea tissues.
DNA was extracted from each pool using the PureLink Genomic DNA kit (Invitrogen by ©2012 Life Technologies Corporation, Carlsbad, CA, USA) following the manufacturer's instructions. In the case of a positive result in a pool, DNA was extracted from the other half of every single flea present in the positive pool and individually subjected to amplification by polymerase chain reaction (PCR), to evaluate the number of positive fleas in each pool.
Molecular Analysis
The extracted nucleic acids were analyzed to detect the presence of DNA from Rickettsia spp. PCR targeting a 17 KDa gene region [30] was performed using GoTaq Polymerase (Promega, Madison, WI, USA). For each reaction, a positive control, consisting of DNA extracted from Rickettsia conorii Malish 7 cultured in VERO cells, and a negative control, in which DNA was replaced by Nuclease-free water (Promega, Madison, WI, USA), were used.
The fleas which were positive for Rickettsia spp. at the first screening were subjected to PCR amplification of the ompA [31], ompB [32] and gltA genes [33] to identify the Rickettsia species, using a multigene assay.
Electrophoretic migration of PCR products on 1.5% agarose gel containing 10 µL/mL ethidium bromide was performed to detect the presence of the expected amplicons (246 bp for 17 kDa, 425 bp for ompB, 532 bp for ompA and 381 bp for gltA).
PCR products obtained by amplifying the Rickettsia 17 KDa, ompA and ompB genes were purified using the Wizard SV Gel and PCR Clean-up System (Promega, Italy), quantified and sent for sequencing (© 2014 Macrogen Inc., Macrogen Europe, Amsterdam, The Netherlands). The sequences obtained were aligned using Bioedit software (Tom Hall, Ibis Biosciences, Carlsbad, CA, USA) and ClustalW 2.0.10 [34] and analyzed for nucleotide sequence identity by comparing them with the ones present in the GenBank sequence database – provided by the National Center for Biotechnology Information (NCBI) – by means of the Basic Local Alignment Search Tool (BLAST).
Statistical Analysis
An unpaired t-test was applied to compare the differences in average numbers of fleas infesting animals per month and a Pearson Chi square test was applied to assess the correlation between infected and non-infected fleas in host species (cats and dogs). The software application SPSS statistical package (SPSS Inc., USA) for Windows was used.
Ethics Statement
This study and the procedures when employed the animals were approved by the “Ethics Committee” of the Istituto Zooprofilattico Sperimentale (IZS) della Sicilia “A. Mirri”.
The research was conducted on both stray and owned dogs and cats.
All treatments, housing and animal care reported in this study were carried out in accordance with the Companion Animals Protection and Prevention of straying animals Law (15/2000) of the Government of Sicily, based on the EU Directive 2010/63/EU for animal experiments.
Pet owners gave their consent to having their animals involved in this study.
Results
Flea Collection And Identification
A total of 134 fleas were collected from all the animals: 37 fleas from dogs and 97 from cats. The number of fleas varied from 1 to 20 on each animal (3.2±3.6 fleas), with no significant differences (t-test: p>0.05) in average values for month: 3.0±4.1 flea/animal in August, 3.7±3.4 in September and 2.5±1.3 in October.
Two species of flea were identified: 132 (98.51%) belonged to Ctenocephalides felis and only two (1.49%) belonged to Ctenocephalides canis. C. felis fleas were collected on all the animals and were 29 males and 103 females, while C. canis fleas, one male and one female, were collected on one dog.
Molecular Analysis Of Fleas
Out of the 132 C. felis, 34 (25.76%) were positive for Rickettsia spp. at the 17 kDa PCR (Fig. 1). All of these 34 fleas were also positive at the ompB PCR and 20 of them at the ompA PCR. No fleas were positive at the gltA PCR. The two specimens of C. canis were negative and collected from one dog. The same dog also harbored four cat fleas, also negative for Rickettsia spp. DNA. A total of 12 positive fleas (32.43%) were collected from dogs and 22 (22.68%) from cats (Tables 1, 2). Although the positivity was higher in fleas taken from dogs than in those taken from cats, the statistical analysis revealed no significant differences between the two populations (chi-square = 1.29).
Figure 1. Electrophoresis on 1.5% agarose gel of the Rickettsia spp. PCR (246 bp).
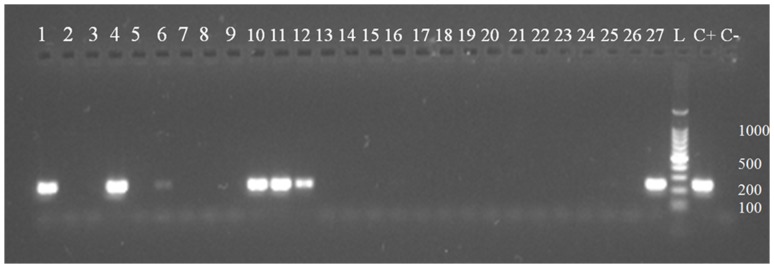
Lines 1–27: DNA from flea samples. Lane L: 100 bp ladder. Lane C+: positive control. Lane C-: negative control.
Table 1. Flea distribution on dogs (§owned dog), with the results of Rickettsia spp. PCR performed on 37 fleas (*two C. canis).
| Dog Id. Nr. | Fleas | ||
| Nr. | Rickettsia spp. PCR (17 kDa) | ||
| + | − | ||
| 1 | 1 | 0 | 1 |
| 2 | 1 | 0 | 1 |
| 3 | 3 | 0 | 3 |
| 4 | 6* | 0 | 6 |
| 5 | 1 | 0 | 1 |
| 6 | 3 | 2 | 1 |
| 7 | 3 | 2 | 1 |
| 8 | 4 | 3 | 1 |
| 9 | 3 | 0 | 3 |
| 10 | 4 | 1 | 3 |
| 11 | 4 | 1 | 3 |
| 12 | 3 | 3 | 0 |
| 13§ | 1 | 0 | 1 |
| Total | 37 | 12 (32.43%) | 25 (67.57) |
Table 2. Flea distribution on cats (§owned cat), with the results of Rickettsia spp. PCR performed on 97 fleas.
| Cat Id. Nr. | Fleas | ||
| Nr. | Rickettsia spp. PCR (17 kDa) | ||
| + | − | ||
| 1 | 1 | 1 | 0 |
| 2 | 2 | 0 | 2 |
| 3 | 1 | 1 | 0 |
| 4 | 1 | 0 | 1 |
| 5 | 1 | 0 | 1 |
| 6 | 1 | 0 | 1 |
| 7 | 3 | 3 | 0 |
| 8 | 1 | 0 | 1 |
| 9 | 6 | 0 | 6 |
| 10 | 4 | 0 | 4 |
| 11 | 1 | 1 | 0 |
| 12 | 1 | 0 | 1 |
| 13 | 2 | 0 | 2 |
| 14 | 4 | 2 | 2 |
| 15 | 1 | 1 | 0 |
| 16 | 3 | 3 | 0 |
| 17 | 1 | 1 | 0 |
| 18 | 1 | 0 | 1 |
| 19 | 13 | 0 | 13 |
| 20 | 11 | 1 | 10 |
| 21 | 20 | 0 | 20 |
| 22 | 3 | 1 | 2 |
| 23 | 2 | 0 | 2 |
| 34 | 2 | 1 | 1 |
| 25 | 2 | 0 | 2 |
| 26 | 3 | 3 | 0 |
| 27 | 2 | 2 | 0 |
| 28 | 2 | 1 | 1 |
| 29 | 2 | 0 | 2 |
| Total | 97 | 22 (22.68%) | 75 (77.32%) |
The positive insects were collected from 20 out of 42 animals (47.62%), 6 dogs (46.15%) and 14 cats (48.28%). All the animals with positive fleas except one (a cat) were strays.
In order to identify the Rickettsia species, 31 out of 34 PCR products were sequenced. For the other three samples the starting material was not of sufficient quantity to proceed with sequencing. Obtained sequences were compared with those deposited in GenBank and all of them showed a very high degree of similarity to R. felis sequences. All the sequences found in this study were submitted to GenBank with the following Accession Numbers: BankIt1735386: KM006831–KM006861 for 17 kDa sequences; BankIt1733537: KM006812–KM006830 for ompB sequences and BankIt1733287: KM006781–KM006811 for ompA sequences (Tables 3, 4).
Table 3. Sequencing results for Rickettsia spp. positive C. felis collected from dogs.
For each gene amplified and sequenced (17 kDa, ompB and ompA), the Accession Number (A.N.) of the corresponding sequence submitted in GenBank and the percentage (%) of identity with respect to reference sequence of R. felis present in GenBank are reported.
Table 4. Sequencing results for Rickettsia positive fleas C. felis collected from cats.
For each gene amplified and sequenced (17 kDa, ompB and ompA), the Accession Number (A.N.) of the corresponding sequence submitted in GenBank and the percentage (%) of identity with respect to reference sequence of R. felis present in GenBank are reported.
Discussion
The results of our research show that almost all fleas collected both on cats and dogs (132/134) were Ctenocephalides felis. This confirms the wider diffusion of the cat flea also in dogs, as previously observed in other studies carried out in Italy and in other countries [25],[35]. Although the two species of Ctenocephalides (C. canis and C. felis) are present throughout the world, the cat flea seems to have a broader distribution than that of the dog [23]. Both species can infest dogs and cats but also other hosts, including humans, especially during the warm months.
In our research we have observed Rickettsia felis infection in 25.37% of the cat fleas, confirming the frequent association between C. felis and the pathogen. The positivity is higher in fleas taken from dogs compared to those taken from cats, although without significant differences.
Although the sample size is small, particularly in the number of dogs and cats enrolled, our data make a contribution to the understanding of the spread of infection not only in Sicily, where information on this matter was not previously available, but also in Italy, where only a few studies have been carried out [25].
Our results differ in part from what has been observed in a previous study carried out in other areas of Italy, where the prevalence was lower on average (11.9%), but similar in the Northeast (23.2%; 26/112 fleas). This study also recorded a higher percentage in fleas from cats (17.6%) than in fleas from dogs (10.2%) [25].
Recent surveys carried out in various European countries showed highly variable prevalence rates of the infection in Ctenocephalides fleas from dogs and cats: 2.76% (10/371 fleas) in Albania [36]; 17.69% (95/537 fleas) in France [37]; 20.66% (25/121 pools of fleas) in the United Kingdom [38]; 19.91% (44/221 pools) in the Netherlands [39]; 43.59% (34/78 fleas) in Catalonia [40] and 54.17% in Andalusia [41]; 8.89% (24/270 fleas) in Germany, where a high positivity in different species of fleas was also reported (100%, of Archeopsylla erinacei) [42].
Our finding of negativity of C. canis cannot lead to any conclusion (only two specimens). In our previous research carried out on fleas collected from foxes in the same area, we observed no positivity in 32 C. canis and in other 76 fleas of various species, while the only two C. felis were both positive [27]. Other authors have reported the absence [25],[41],[42] or a low prevalence [39],[40] in larger sample sizes, in contrast to what has been observed in France, where 27.03% of C. canis (10/37) were infected [37].
In our study, about half the animals examined (47.62%; 20/42) were infested with at least one R. felis infected flea; specifically 46.15% (6/13) of dogs and 48.28% (14/29) of cats. It is noteworthy that the flea specimens were almost all collected from stray dogs and cats and not from kennels or catteries, where the prevalence of R. felis infection is often higher [24]. Hence, it may well be that infection rates in closed animal populations are higher than those presented. Moreover, the presence of an infected flea in one of the three owned animals in our sample, a household cat, is interesting and might suggest an easier transmission to humans. In fact, the seroprevalence of individuals that have reported contact with domestic animals tends to be higher [43].
It will also be interesting to extend the research on pet hosts. The role of mammals as a reservoir of this emerging flea-borne infection needs further confirmatory evidence, because there is to date no consensus on the potential mammalian reservoir(s). The presence of R. felis has been recorded in several peri-domestic species associated with the cat flea, including cats, dogs, opossums and rats [5],[15],[17],[23],[35], but no clinical case has been described in animal hosts, except humans. Recently, it was suggested that the dog [35],[44] and the hedgehog [42] might play a role in the ecology of the pathogen in some areas.
This study shows a potential risk of transmission to humans in the Palermo district, at least in conditions where there is increased contact with stray animals and their ectoparasites (e.g., categories of workers, such as veterinarians, kennel or cattery personnel, volunteers employed in animal capture, etc.).
Although human cases of flea-borne spotted fever are yet to be reported in Italy, it would seem that this infection is more common than is currently recognized. In fact, clinical symptoms are similar to those of other rickettsial diseases (e.g., murine typhus, MSF), making a causal diagnosis difficult. Because antigenic cross-reactivity exists among different species of rickettsiae [14], serological assays are likely to be insufficient to definitively identify R. felis unless other, more sophisticated, serological (Western blot and cross-absorption studies) or molecular assays are performed [6]. Given our findings, we cannot exclude the possibility that R. felis could be implicated in Mediterranean spotted fever, endemic in Southern Italy (Sicily alone accounts more than half of all the national clinical cases) [45]–[47], or in murine typhus due to R. typhi, sporadically reported in Sicily [45]. In fact, diagnosis of MSF in Italy usually depends on clinical evidence supported by serologic confirmation. Current diagnostic tests, however, use R. conorii as the only antigen [47] and hence, due to possible cross-reactions, misdiagnosis may occur.
In light of the above, we conclude that flea-borne spotted fever should be included in the human differential diagnosis of any spotted-like fever or febrile illness without a clear source of infection in Sicily, especially if the patient is known to have been exposed to flea bites. Moreover, careful hygiene and cleaning of pet beddings and a wider use of pesticides in domestic animals, as well as having a direct benefit to the animals themselves, could reduce the risk of transmission of pathogens dangerous to humans.
Acknowledgments
The authors thank Caroline Keir for reviewing the English.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are available from following URL: www.izssicilia.it.
Funding Statement
The authors have no support or funding to report.
References
- 1. Sikora H (1918) Beitrage zur Kenntnis der Rickettsien. Arch Schiffs Tropenhyg Liepzig xxii: 442–446. [Google Scholar]
- 2. Adams JR, Schmidtmann ET, Azad AF (1990) Infection of colonized cat fleas, Ctenocephalides felis (Bouche), with a rickettsia-like microorganism. Am J Trop Med Hyg 43: 400–409. [DOI] [PubMed] [Google Scholar]
- 3. La Scola B, Meconi S, Fenollar F, Rolain JM, Roux V, et al. (2002) Emended description of Rickettsia felis (Bouyer, et al. 2001), a temperature-dependent cultured bacterium. Int J Syst Evol Microbiol 52: 2035–2041. [DOI] [PubMed] [Google Scholar]
- 4.Mansueto P, Vitale G, Cascio A, Seidita A, Pepe I, et al.. (2012) New insight into immunity and immunopathology of rickettsial diseases. Clin Dev Immunol 2012: ID 967852, 26 pages. doi:10.1155/2012/967852. [DOI] [PMC free article] [PubMed]
- 5. Perez-Osorio CE, Zavala-Velazquez JE, Arias Leon JJ, Zavala-Castro JE (2008) Rickettsia felis as emergent global threat for humans. Emerg Infect Dis 14: 1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parola P (2011) Rickettsia felis: from a rare disease in the USA to a common cause of fever in sub-Saharan Africa. Clin Microbiol Infect 17: 996–1000. [DOI] [PubMed] [Google Scholar]
- 7. Eremeeva ME, Karpathy SE, Krueger L, Hayes EK, Williams AM, et al. (2012) Two pathogens and one disease: detection and identification of flea-borne Rickettsiae in areas endemic for murine typhus in California. J Med Entomol 49: 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson K, Wallménius K, Hartwig S, Norlander T, Påhlson C (2013) Bell's palsy and sudden deafness associated with Rickettsia spp. infection in Sweden. A retrospective and prospective serological survey including PCR findings. Eur J Neurol doi:10.1111/ene.12218. [DOI] [PMC free article] [PubMed]
- 9. Renvoisé A, Joliot AY, Raoult D (2009) Rickettsia felis infection in man, France. Emerg Infect Dis 15: 1126–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams M, Izzard L, Graves SR, Stenos J, Kelly JJ (2011) First probable Australian cases of human infection with Rickettsia felis (cat-flea typhus). Med J Aust 194: 41–43. [DOI] [PubMed] [Google Scholar]
- 11. Richards AL, Jiang J, Omulo S, Dare R, Abdirahman K, et al. (2010) Human Infection with Rickettsia felis, Kenya. Emerg Infect Dis 16: 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Civen R, Ngo V (2008) Murine typhus: an unrecognized suburban vectorborne disease. Clin Infect Dis 46: 913–918. [DOI] [PubMed] [Google Scholar]
- 13. Azad AF, Radulovic S, Higgins JA, Noden BH, Troyer JM (1997) Flea-borne rickettsioses: ecologic considerations. Emerg Infect Dis 3: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim MY, Brady H, Hambling T, Sexton K, Tompkin D, et al. (2012) Rickettsia felis infections, New Zealand. Emerg Infect Dis 18: 167–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdad MY, Stenos J, Graves S (2011) Rickettsia felis, an emerging flea-transmitted human pathogen. Emerg Health Threats 4: 7168 DOI: 10.3402/ehtj.v4i0.7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eisen RJ, Gage KL (2012) Transmission of flea-borne zoonotic agents. Ann Rev Entomol 57: 61–82 DOI: 10.1146/annurev-ento-120710-100717 [DOI] [PubMed] [Google Scholar]
- 17. Reif KE, Macaluso KR (2009) Ecology of Rickettsia felis: a review. J Med Entomol 46: 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reif KE, Kearney MT, Foil LD, Macaluso KR (2011) Acquisition of Rickettsia felis by cat fleas during feeding. Vector Borne Zoonotic Dis 11: 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Socolovschi C, Pagés F, Raoult D (2012) Rickettsia felis in Aedes albopictus Mosquitoes, Libreville, Gabon. Emerg Infect Dis 18: 1687–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirunkanokpun S, Thepparit C, Foil LD, Macaluso KR (2011) Horizontal transmission of Rickettsia felis between cat fleas, Ctenocephalides felis . Mol Ecol 20: 4577–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wedincamp J Jr, Foil LD (2002) Vertical transmission of Rickettsia felis in the cat flea (Ctenocephalides felis Bouché). J Vector Ecol 27: 96–101. [PubMed] [Google Scholar]
- 22. Thepparit C, Hirunkanokpun S, Popov VL, Foil LD, Macaluso KR (2013) Dissemination of bloodmeal acquired Rickettsia felis in cat fleas, Ctenocephalides felis . Parasit Vectors 6: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bitam I, Dittmar K, Parola P, Whiting MF, Raoult D (2010) Fleas and flea-borne diseases. Int J Infect Dis 14: e667–e676. [DOI] [PubMed] [Google Scholar]
- 24. Bauer O, Baneth G, Eshkol T, Shaw SE, Harrus S (2006) Polygenic detection of Rickettsia felis in cat fleas (Ctenocephalides felis) from Israel. Am J Trop Med Hyg 74: 444–448. [PubMed] [Google Scholar]
- 25. Capelli G, Montarsi F, Porcellato E, Maioli G, Furnari C, et al. (2009) Occurrence of Rickettsia felis in dog and cat fleas (Ctenocephalides felis) from Italy. Parasit Vectors 2: S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maioli G, Horta MC, Ogrzewalska M, Capelli G, Souza SO, et al. (2009) First detection of Rickettsia felis in Ctenocephalides felis fleas from Italy. Clin Microbiol Infect 15: 222–223. [DOI] [PubMed] [Google Scholar]
- 27. Torina A, Blanda V, Antoci F, Scimeca S, D'Agostino R, et al. (2013) A molecular survey of Anaplasma spp., Rickettsia spp., Ehrlichia canis and Babesia microti in foxes and fleas from Sicily. Transbound Emerg Dis 60: 125–130. [DOI] [PubMed] [Google Scholar]
- 28.Herms WB (1923) Medical and Veterinary Entomology, 2nd edn. The Macmillan Company, New York.
- 29.Chinery M (1998) Guide to Insects of Europe, 3rd edn. Franco Muzzio Editore, Roma.
- 30. Tzianabos T, Anderson BE, McDade JE (1989) Detection of Rickettsia rickettsii DNA in clinical specimens by using polymerase chain reaction technology. J Clin Microbiol 27: 2866–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oteo JA, Portillo A, Santibáñez S, Blanco JR, Pérez-Martínez L, et al. (2006) Cluster of cases of human Rickettsia felis infection from Southern Europe (Spain) diagnosed by PCR. J Clin Microbiol 44: 2669–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi YJ, Jang WJ, Kim JH, Ryu JS, Lee SH, et al. (2005) Spotted fever group and typhus group rickettsioses in humans, South Korea. Emerg Infect Dis 11: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roux V, Rydkina E, Eremeeva M, Raoult D (1997) Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the Rickettsiae. Int J Syst Bacteriol 47: 252–261. [DOI] [PubMed] [Google Scholar]
- 34. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 35. Hii S, Abdad MY, Kopp SR, Stenos J, Rees RL, et al. (2013) Seroprevalence and risk factors for Rickettsia felis exposure in dogs from Southeast Queensland and the Northern Territory, Australia. Parasit Vectors 6: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silaghi C, Knaus M, Rapti D, Shukullari E, Pfister K, et al. (2012) Rickettsia felis and Bartonella spp. in fleas from cats in Albania. Vector Borne Zoonotic Dis 12: 76–77. [DOI] [PubMed] [Google Scholar]
- 37. Gilles J, Just FT, Silaghi C, Pradel I, Lengauer H, et al. (2008) Rickettsia felis in fleas, France. Emerg Infect Dis 14: 684–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shaw SE, Kenny MJ, Tasker S, Birtles RJ (2004) Pathogen carriage by the cat flea Ctenocephalides felis (Bouché) in the United Kingdom. Vet Microbiol 8: 183–188. [DOI] [PubMed] [Google Scholar]
- 39. Tijsse-Klasen E, Fonville M, Gassner F, Nijhof AM, Hovius EK, et al. (2011) Absence of zoonotic Bartonella species in questing ticks: first detection of Bartonella clarridgeiae and Rickettsia felis in cat fleas in the Netherlands. Parasit Vectors 4: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nogueras MM, Pons I, Ortuño A, Lario S, Segura F (2011) Rickettsia felis in fleas from Catalonia (Northeast Spain). Vector Borne Zoonotic Dis 11: 479–483. [DOI] [PubMed] [Google Scholar]
- 41. Márquez FJ, Muniain MA, Rodríguez-Liebana JJ, Del Toro MD, Bernabeu-Wittel M, et al. (2006) Incidence and distribution pattern of Rickettsia felis in peridomestic fleas from Andalusia, Southeast Spain. Ann N Y Acad Sci 1078: 344–346. [DOI] [PubMed] [Google Scholar]
- 42. Gilles J, Just FT, Silaghi C, Pradel I, Passos LM, et al. (2008) Rickettsia felis in fleas, Germany. Emerg Infect Dis 14: 1294–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nogueras MM, Cardeñosa N, Sanfeliu I, Muñoz T, Font B, et al. (2006) Evidence of infection in humans with Rickettsia typhi and Rickettsia felis in Catalonia in the Northeast of Spain Ann N Y Acad Sci. 1078: 159–161. [DOI] [PubMed] [Google Scholar]
- 44. Nogueras MM, Pons I, Ortuno A, Segura F (2009) Seroprevalence of Rickettsia typhi and Rickettsia felis in dogs from North-Eastern Spain. Clin Microbiol Infect 15: 237–238. [DOI] [PubMed] [Google Scholar]
- 45. Torina A, Fernández de Mera IG, Alongi A, Mangold AJ, Blanda V, et al. (2012) Rickettsia conorii Indian tick typhus strain and R. slovaca in humans, Sicily. Emerg Infect Dis 18: 1008–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ciceroni L, Pinto A, Ciarrocchi S, Ciervo A (2006) Current knowledge of rickettsial diseases in Italy. Ann N Y Acad Sci 1078: 143–149. [DOI] [PubMed] [Google Scholar]
- 47. Beninati T, Lo N, Noda H, Esposito F, Rizzoli A, et al. (2002) First detection of Spotted Fever Group rickettsiae in Ixodes ricinus from Italy. Emerg Infect Dis 8: 983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are available from following URL: www.izssicilia.it.


