Abstract
Pulse-chase analysis is a commonly used technique for studying the synthesis, processing and transport of proteins. Cultured cells expressing proteins of interest are allowed to take up radioactively labeled amino acids for a brief interval (“pulse”), during which all newly synthesized proteins incorporate the label. The cells are then returned to non-radioactive culture medium for various times (“chase”), during which proteins may undergo conformational changes, trafficking, or degradation. Proteins of interest are isolated (usually by immunoprecipitation) and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the fate of radiolabeled molecules is examined by autoradiography. This chapter describes a pulse-chase protocol suitable for studies of major histocompatibility complex (MHC) class II biosynthesis and maturation. We discuss how results are affected by the recognition by certain anti-class II antibodies of distinct class II conformations associated with particular biosynthetic states. Our protocol can be adapted to follow the fate of many other endogenously synthesized proteins, including viral or transfected gene products, in cultured cells.
Keywords: MHC class II, biosynthesis, maturation, metabolic labeling, immunoprecipitation
1. Introduction
In the 1960s, Palade et al. devised pulse-chase labeling of secreted proteins in pancreatic tissue slices in order to investigate the role of subcellular compartments in protein trafficking [1–2]. These experiments showed that secretory proteins were sequentially transported from the rough endoplasmic reticulum (ER) to the Golgi complex and then to zymogen granules via small vesicles located in the periphery of the Golgi [1–2]. Since then, pulse-chase labeling has become a widely adopted tool for studying the post-translational fate of proteins.
In a typical experiment, cells of interest are first cultured for a short period (the “pulse”) in the presence of radioactively labeled essential amino acids. This is often a mixture of 35S-labeled cysteine and methionine (Cys/Met). Other essential amino acids may be labeled with 3H or 14C and used in a similar way; this may be useful for proteins with a low Cys/Met content. Nonessential amino acids are not suitable, as their endogenous synthesis would compete with labeling. During the pulse, all proteins synthesized de novo become radiolabeled in proportion to their rate of biosynthesis and their content of amino acids carrying the label. Competition with preexisting pools of unlabeled amino acids in the cells may be reduced, and labeling efficiency increased, by starving the cells of these amino acids prior to pulse-labeling. In order to track the fate of proteins labeled during the pulse, the cells are washed and re-cultured (“chased”) for varying amounts of time in the presence of normal media, containing excess unlabeled amino acids. This ensures that only those proteins that were made during the pulse are radiolabeled; their fate can then be traced during the chase.
The proteomes of mammalian cells are exceedingly complex, so for most purposes, proteins of interest must be enriched by, for example, selective extraction, subcellular fractionation, and/or chromatography. A versatile and specific method uses detergent extraction and small-scale affinity chromatography (i.e., immunoprecipitation; IP) with antibodies (Abs) specific for the protein of interest. The Abs are coupled directly to sepharose beads or are recognized by Ab-binding proteins, such as staphylococcal protein A or G, coupled to sepharose. Monoclonal Abs are often selective for particular protein conformations, and this property must be considered during data interpretation. Such Abs can provide information on the fate of conformationally distinct molecular subsets of the same protein. Ab-bound radiolabeled proteins may be quantified by scintillation counting or visualized after separation on SDS-PAGE gels by autoradiography. The latter method allows quantification by densitometric analysis of gel bands and improves the discrimination between specifically bound proteins of interest and background radioactivity from nonspecific binding. It also reveals intracellular processing steps that alter the molecular weight of proteins of interest (e.g., proteolytic cleavage, modification of carbohydrates, oxidation of disulfide bonds) or their association with other proteins. These capabilities continue to justify the use of radioactivity in the pulse/chase approach, even though non-radioactive techniques for measuring protein synthesis and turnover have become available (for recent a application to MHC proteins, see [3]).
Our laboratories have had a long-standing interest in the analysis of MHC class II (MHC II) glycoproteins, which bind heterogeneous mixtures of peptides in endosomes and present them on the surface of antigen-presenting cells for inspection by CD4+ T helper lymphocytes. Pulse-chase analysis has been invaluable in tracing the complex processes of MHC II protein synthesis, maturation, trafficking, and peptide loading. Newly synthesized MHC II αβ heterodimers are assembled with the invariant chain (Ii) polypeptide in the ER [4–5]. This association aids the correct folding of nascent MHC II molecules and prevents premature binding of misfolded polypeptides [6]. The assembled (αβ)3Ii3 complexes travel through the Golgi apparatus and, targeted by sequence motifs in the cytoplasmic tail of Ii, are directed to late endosomal and pre-lysosomal compartments, which are called MHC class II compartments (MIIC) [7–11]. In these compartments, Ii is progressively degraded by aspartyl and cysteine proteases, including cathepsin S [12–15], via intermediates (p21, p10/12) termed “LIP” (“leupeptin-induced polypeptides”) and “SLIP” (small LIP), respectively, until only small Ii fragments called CLIP (class-II-associated invariant chain peptide) remain bound in the peptide-binding groove of MHC II [16–17]. The subsequent release of CLIP from MHC II and exchange for antigenic peptide are both mediated by HLA-DM [18–20]. The catalytic action of DM also selects for peptides that form kinetically stable complexes with MHC II, and may influence the conformation of MHC II molecules [21]. One unusual biochemical consequence of loading with high-affinity peptides is that the resultant MHC α/β-peptide complexes become resistant to denaturation with SDS in vitro; this feature can be used to track peptide loading in pulse/chase experiments [22]. Some variations in the migration of SDS-stable complexes have been noted, which relate to peptide occupancy and/or conformation (“compact” vs. more slowly migrating “floppy” dimers [23–24]). Peptide-loaded MHC II molecules are transported to the cell surface.
MHC II molecules are encoded at several genetic loci (generating class II isotypes), most of which exhibit extensive allelic polymorphism. The details of the maturational steps described above depend to some extent on the MHC II variant being studied. For example, allelic and isotypic variation in the affinity of MHC II molecules for CLIP results in varying degrees of spontaneous CLIP release in DM-deficient cells, and in varying levels of SDS stability of MHC II complexes with CLIP; this, in turn, affects peptide loading and turnover of MHC II molecules (reviewed in [25]).
Here, we describe a protocol for pulse-chase analysis of MHC class II molecules in human Epstein-Barr virus-transformed B-lymphoblastoid cell lines (B-LCL). We show experimental results for HLA-DR and -DQ molecules (two isotypes of human MHC II), after pulse/chase labeling of B-LCL with defined mutations in DM. These results illustrate what information may be obtained by tracking changes in the molecular associations and abundance of radiolabeled proteins, as well as by use of conformation-sensitive mAbs.
2. Materials
2.1. Cell Culture and Media
Cell culture plasticware and culture media suitable for cells of interest. B-LCL grow well in RPMI1640 medium supplemented with 2 mM L-glutamine and either 10% fetal bovine serum or 10% newborn calf serum.
Complete Cys/Met-free medium: Cell growth medium deficient in Cys/Met (e.g., Cys/Met-free RPMI1640, CellGro; or DMEM, Gibco) supplemented with 2 mM L-glutamine and 10% dialyzed FBS (Invitrogen). Sterile filter (unless prepared from sterile ingredients), store at 4 ° C, warm to 37 °C before use.
Chase medium: Complete growth medium (from 10.2.1.1, above) supplemented with 1 mM each nonradioactive Cys and Met (Sigma). Sterile filter, store at 4 °C, warm to 37 °C before use.
2.2. Radioactive Reagents
Radioactive labeling reagent: [35S] Cys/Met labeling mix (Perkin Elmer) should be stored in a −80°C freezer. Thaw at room temperature before use. Ensure handling with appropriate safety precautions (see Current Protocols in Molecular Biology, Appendix, A.1F for safe use of radioisotopes and specific precautions with 35S-labeled materials).
Radioactive markers: 14C-labeled rainbow protein molecular weight markers (GE Healthcare).
2.3. Preparation and Analysis of Lysates
Lysis Buffer: 50 mM Tris-HCl, 150 mM NaCl, 1% v/v NP-40 (now commercially available as IGEPAL CA-630; see Note 1), pH 8.0. Add 1 mM phenylmethylsulfonyl fluoride (PMSF; from 100 mM stock in dry 100% ethanol, kept at −20 °C; toxic) and 1/50th volume of protease inhibitor cocktail (Roche) into lysis buffer immediately before use. To prepare a 50× stock of the inhibitor cocktail, dissolve a protease inhibitor tablet in 1 ml water. Aliquot and store at −20 °C.
- Beads for immunoprecipitation (IP) and pre-clearing:
- Protein A (Sigma) or protein G (GE Healthcare) sepharose beads: If the beads are supplied as a lyophilized powder, incubate 100 mg of beads in 1 ml PBS at 4 °C for at least one hour to allow swelling. The rehydrated beads may then be washed three times with at least 10 volumes PBS on a sintered glass funnel; alternatively, the washes can be performed by diluting the bead suspension to 50 ml in a conical polypropylene tube, centrifuging for 30 minutes at 920–1430× g in a refrigerated benchtop centrifuge, and carefully aspirating the supernatant. Thereafter, adjust the final volume with PBS to obtain a 50% slurry (i.e., 50 % settled beads by volume) and add sodium azide (toxic) from 10% w/v stock to a final concentration of 0.02%. Store at 4°C. If beads are supplied as a slurry in preservative (e.g., 20% ethanol), the rehydration step may be omitted. However, the washes are recommended to minimize carry-over of preservative into immunoprecipitations.
- (Optional) Pansorbin cells (Calbiochem): protein A-bearing S. aureus cells with IgG-binding capacity. Pansorbin cells may be used for pre-clearing. They are stored as a 10% v/v suspension in PBS with 0.02% sodium azide at 4 °C. Transfer desired amount of Pansorbin cells to a microfuge tube. Centrifuge for 5 min in a benchtop microfuge at 3000 × g or at maximum speed for 1 min at 4°C. Aspirate the supernatant. Resuspend in 1.0 ml of cold lysis buffer and mix gently. Centrifuge again and remove the lysis buffer. Repeat wash.
Pre-immune normal serum with the same animal source as antiserum used for IP, or, for mAbs, a species- and isotype-matched control Ab.
-
Antibodies: The choice of mAbs for IP experiments will be dictated by the experimental goals. Owing to polymorphism and isotypy of the MHC, one important consideration is the selectivity of the mAbs for particular MHC alleles and isotypes. Antibodies raised in mice against human MHC molecules are often, but not invariably, monomorphic (i.e., their binding is relatively unaffected by allelic polymorphism), whereas murine antibodies raised against murine MHC molecules from another strain generally bind to only a subset of alleles. Moreover, mAbs may exhibit conformational preferences, and these preferences often relate to biosynthetic states of class II. See Tables 1 and 2 below for examples of antibodies (anti-human and murine MHC class II proteins, respectively) that have been used successfully and/or frequently in these types of experiments. The specificities of mAbs often become evident in the course of pulse/chase experiments (see Figs. 1 and 2). Antibodies to the cytoplasmic tails of MHC class II α or β chains provide conformation-independent reagents for denatured proteins, and sometimes, but not always, for native proteins (see Ab DA6.147 below).
Monooclonal Abs may be purchased commercially or purified from hybridoma culture supernatants by protein A- or G-sepharose affinity chromatography, using commercial kits. Stock solutions (≈ 1 mg/ml in PBS, quantified by UV absorption spectrophotometry, assuming A280 = 1.4 for a 1 mg/ml solution) can be stored indefinitely at or below −20 °C in aliquots. Thawed aliquots may be kept at 4 °C with addition of sodium azide to 0.05% (from 10% w/v stock) for several weeks.
IP wash buffer: PBS supplemented with 1 mM PMSF (added fresh), 0.05% NP-40, and 0.05% sodium azide.
- SDS Polyacrylamide Gel Components:
- See Current Protocols in Molecular Biology, Chapter 10, Section II, 10.2 for directions on making gels and required buffers, including Laemmli sample loading buffer (SLB), both reducing and non-reducing.
- Equipment and supplies for SDS-PAGE (gel plates, combs, spacers, e.g., BioRad).
Amplify solution (GE Healthcare).
Heated vacuum gel dryer, e.g. BioRad.
Kodak Biomax MR Film (Scientific Imaging film, 20.3 × 25.4 cm, 8 × 10 inches).
Equipment for developing X-ray film.
(Optional) Equipment for film densitometry or phosphorimager system.
Table 1.
Selected antibodies used to study biosynthesis and maturation of human MHC class II molecules.
| Antibody | Isotype (murine) | Specificity in immunoprecipitation | Reference (original and application) |
|---|---|---|---|
| PIN 1.1 | IgG1 | Anti-Ii: Recognizes a cytoplasmic, N-terminal epitope of human Ii and co-immunoprecipitates any Ii-associated HLA class II αβ heterodimer. It also reacts with C-terminally truncated Ii fragments that retain the N terminus (Leupeptin-induced peptides, LIP, but not CLIP). Does not allow detection of Ii on the surface of intact cells by flow cytometry. | [26] |
| BU45 | IgG1 | Anti-Ii: Recognizes the C-terminal lumenal domain of Ii. Co-immunoprecipitates free and class II-associated Ii. Does not react with C-terminally truncated Ii fragments (LIP). Allows detection of Ii on the surface of intact antigen presenting cells (APCs) by flow cytometry. | [27–28] |
| CerCLIP.1 | IgG1 | Anti-CLIP: Recognizes the N-terminus of CLIP (Ii residues 81–105). Immunoprecipitates CLIP-loaded class II αβ heterodimers. Also allows detection of MHC class II-CLIP complexes on intact APC by flow cytometry. Staining intensity, relative to that with anti-class II mAbs, is inversely correlated with DM-catalysed CLIP release. CLIP levels also depend on the affinity of any given MHC allele(s) for CLIP. | [29–30] |
| 6–39 | IgG1 | Recognizes DR3/CLIP complexes, but not free CLIP. | [31] |
| I-5 | IgG1 | Recognizes DR3/CLIP complexes, but not free CLIP. | [32] |
| DA6.147 | IgG1 | Anti-HLA-DR α chain: Recognizes a monomorphic (DR specific) determinant on the C-terminal cytoplasmic tail of DRα. The epitope is accessible for IP in Ii-associated DR but less so for mature DR molecules. It can also be used to immunoprecipitate DRα chain from denatured lysates. Also reacts with dissociated HLA-DR α chains on Western Blots. Not suitable for detection of DR on intact APCs by flow cytometry. | [33–34,30,35] |
| L243 | IgG2a | Anti-HLA-DR: Recognizes a monomorphic, conformational determinant on the α chain of DR αβ heterodimers. DR molecules associated with intact Ii are immunoprecipitated poorly (presumably due to steric masking of the epitope by the Ii C terminus), but DR molecules associated with LIP, CLIP, or endosomal peptides are immunoprecipitated well (see
Fig.10.1). The strength of the residual interaction with DR/Ii complexes may be dependent on the allele and the exact conditions for IP. L243 is poorly suited to the IP of SDS-stable DR/peptide complexes under non-denaturing conditions (without boiling), because these complexes are supershifted by L243, which fails to dissociate in SDS at room temperature. Such complexes (but not dissociated DR chains) are, however, detected by L243 on Western blots. Also detects DR molecules on the surface of intact APCs by flow cytometry and has been used for large-scale affinity chromatography to purify DR. |
[12,34,36–37] |
| ISCR3 | IgG2b | Anti-HLA-DR β chain: immunoprecipitates HLA-DR αβ dimers and is relatively insensitive to conformational maturation. Allows visualization of immunoprecipitated SDS-stable peptide-loaded DR αβ dimers, as it dissociates from antigen in reducing SDS-PAGE sample buffer at room temperature. Also, cross-reacts with MHC II molecules from other species. | [6,38–39] |
| LB3.1 | IgG2a | Anti-HLA-DR α chain: Recognizes both empty and peptide-loaded HLA-DR 1 αβ heterodimer (monomorphic). Has been used for large-scale affinity chromatography of DR. | [40–43,38] |
| MEM-264 | IgG2b | Anti-empty HLA-DR β chain: Recognizes an epitope on the β chain of empty HLA-DR1 αβ heterodimers. Binding of the antibody is abolished by association with CLIP, antigenic peptides, or truncated model peptides. | [40,44,43] |
| 16.23 | IgG3 | Anti-HLA-DR3: Recognizes an allele-specific, DM-dependent epitope present on a subset of DR3 molecules. Empty DR3/DM complexes and DR3/peptide complexes have been reported to bind, but only after DM-mediated CLIP release and not after the alternative self-release exchange mechanism. CLIP-associated DR3 molecules are not recognised. | [45–48,32,49] |
| NFLD.D11 | IgM | Anti-HLA-DR4: Recognizes an allele-specific, DM-dependent determinant on the HLA-DR0401 β chain. | [50] |
| SPVL3 | IgG2a | Anti-HLA-DQ: Recognizes a monomorphic epitope on DQ αβ dimers, but not the denatured individual α or β chains. The epitope is maturation- and DM-dependent (see Fig. 10.2). | [51–53] |
| 2.12.E11 | IgG1 | Anti-HLA-DQ2: Recognizes a conformation-dependent, polymorphic epitope on DQ αβ dimers, but not the denatured individual α or β chains. It is specific for DQB1*0201 or 0202 alleles. | [51,54–55] |
| Ia3 | IgG2a | Anti-HLA-DQ: Recognizes a monomorphic epitope on DQ αβ dimers, but not the denatured individual α or β chains. | [51,56] |
| B7/21.2 | IgG3 | Anti-HLA-DP: Recognizes a monomorphic epitope on HLA-DP α and immunoprecipitates immature and mature DP heterodimers. | [57] |
| XD5.A11 | IgG1 | Anti-MHC class II β chain: Recognizes both intact and denatured DR, DP and DQ β chains and therefore all MHC class II αβ heterodimers. | [58] |
Note that the studies cited here may describe the original derivation or informative uses of these antibodies, or both.
Table 2.
Selected antibodies used to study biosynthesis and maturation of mouse MHC class II molecules
| Antibody | Reactivity | Isotype (murine unless indicated) | Specificity in immunoprecipitation | Reference (Original and application) |
|---|---|---|---|---|
| In-1 | Ii | Rat IgG2b | Recognizes cytoplasmic epitope near N terminus of Ii. Immunoprecipitates free Ii and Ii/class II complexes. Recognizes denatured or non-denatured, full-length and C-terminally truncated Ii fragments, but not CLIP. | [23,59] |
| P4H5 | Ii | Armenian Hamster | Recognizes residues 99–116 of Ii, a lumenal epitope immediately C-terminal of CLIP. | [60–62] |
| M5/114 | I-A/I-E β I-Ab,d,q, I-Ed,k (Not reactive with I-Ak,s,f,g7) | Rat IgG2b | Recognizes β chain epitope (β1 domain = β chain binding groove domain). Competes with MK-D6, but no detectable preference for immature/mature class II. | [60,63,23,61] |
| Y3P | I-Ab,f,p,q,r,s,u,v weakly with I-Ak, also cross-reacts with all rat I-A-like molecules | IgG2a | Recognizes peptide binding domain of mature αβ dimers and αβ/Ii complexes. | [23,61,64] |
| 10-2.16 | I-Ak,r,f,s,g7 β but not reactive with I-Ab,d,p,q | Mouse IgG2b | Recognizes β1 domain. | [65,61,66,24] |
| K24-199 | I-Aα, including I-Ad,k,g7 | IgG2a | Does not recognize newly synthesized class II in presence or absence of Ii, prefers a more mature conformation. | [65,61,67] |
| MKD6 | I-Adβ (not reactive withI-Ak,b,s,f,a) | IgG2a | Recognizes extracellular β chain epitope (β1 domain, competes with M5/114). Prefers more mature (e.g., DM-edited) conformations of I-Ad (IP is poor at early time points, more efficient after ~1 hr chase, and more efficient from DM-expressing than DM-negative cells). | [60,65,68–69] |
| B21-2 | I-Ab,d,g7 β | Rat IgG2b | Recognizes β chain of properly folded αβ dimers. No detectable preference for mature/immature/DM-edited class II. | [68,70] |
| 14-4-4S | Common I-Eα chain (N.B. not all haplotypes express I-E.) | IgG2a | Recognizes α chain of properly folded αβ dimers. | [65,61,71–72] |
Fig.1. Pulse/chase analysis of DR molecules in DM-expressing and non-expressing B-LCL.
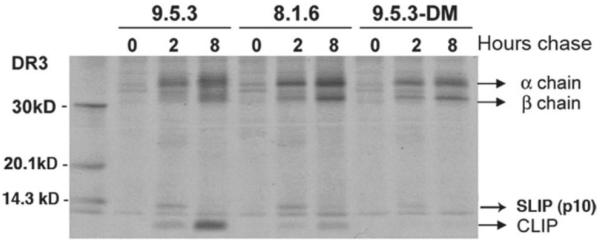
The DM-deficient cell line, 9.5.3, its wild-type progenitor, 8.1.6, and 9.5.3 cells transfected with DM (9.5.3-DM) were pulse-labeled for 1 hr with [35S]-Cys/Met and chased for 0, 2, or 8 hrs. NP-40 cell extracts (3×106 cell equivalents/lane) were immunoprecipitated with the anti-DR mAb L243 and analyzed by 12% SDS-PAGE. The positions of DR α and β chains and of a 10 kDa SLIP fragment of Ii are indicated. Levels of DR-associated CLIP are substantially diminished in DM-expressing 8.1.6 cells at 2 hr and 8 hr of chase, and even further reduced when DM levels are increased by DM transfection of 9.5.3 cells. Also note inefficient IP of immature DR (mostly Ii-associated) in pulse-labeled cells with L243, which accounts for the weak bands at 0 hr chase [53].
Fig.2. Pulse/chase analysis of DQ1 molecules in DM-expressing and non-expressing B-LCL.
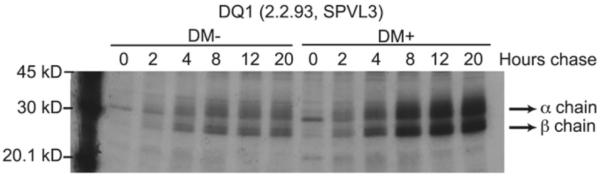
2.2.93 and 2.2.93-DM cells were pulsed for 1 hr with [35S]-Cys/Met and chased for the indicated time periods. Aliquots of cell lysates, normalized for counts, were immunoprecipitated with SPVL-3 for DQ1 and then analysed by SDS-PAGE [44]. This figure illustrates the preferential recognition of a mature confirmation of DQ1 by SPVL3, which is likely peptide-dependent and is significantly increased in the presence of DM.
2.4. Laboratory accommodations for working with radioactivity
Decontamination agent, e.g., Count-Off (PerkinElmer). Obtain appropriate training for dealing with a radioactive spill, in accordance with institutional regulations/standards.
Lab work area equipped with safety equipment for radioactive work (waste containers, disposable absorbent covering, secondary containment, hand-held Geiger counter). Equipment for disposal of radioactive waste sholud be in accordance with institutional regulations/standards.
Refrigerated benchtop centrifuge and microcentrifuge, dedicated for radioactive work.
Tissue culture incubator with dedicated space for radioactive work.
Tissue culture hood that can be reserved for radioactive work.
3. Methods
3.1. Prepare cells for the experiment
Cells should be highly viable and screened for microbial contamination, including mycoplasma. Proliferating cell lines should be in logarithmic growth.
The protocol used here is suitable for suspension cells, such as B-LCL; alternative procedures for adherent cells are in Note 2.
Cells and media are kept as close to 37 °C as possible to minimize interference of the labeling protocol with normal cell metabolism.
Collect sufficient cells for the experiment (typically 106-107 cells per time point; see Note 3) in 50 ml conical polypropylene tubes by centrifuging for 5–10 min at 330–520 × g in a benchtop centrifuge (e.g., 514 × g equivalent to 1500 rpm for 5 min. in a Beckman GS-6R centrifuge) kept at room temperature. Use the same centrifugation conditions in subsequent steps.
3.2. Wash and starve
Wash cells once in at least 5 ml pre-warmed, unsupplemented Cys/Met-free media. The cell density after dilution should be < 1 × 107/ml.
Suspend cells in Cys/Met-free media plus glutamine and dialyzed FBS at a density of 5 × 106 cells/ml (see Note 3) and incubate (starve) at 37 °C in a 50 ml conical tube for 1 hr to deplete endogenous Cys/Met. During this time, place cells in a humidified CO2 incubator and leave the cap partially unscrewed to allow gas exchange. Mix occasionally (see Note 3).
3.3. Pulse labeling
Perform this and subsequent steps with safety precautions for radioactive work (see Current Protocols in Molecular Biology, Appendix, A.1F).
Add 100 to 150 μCi 35S-Cys/Met per ml of starved cell suspension and mix well. Alternatively, cells may be centrifuged after step 3 and suspended in an equal volume of Cys/Met-free media supplemented with glutamine, 10% dialyzed FBS, and 100–150 μCi/ml 35S-Cys/Met. Place the 50 ml conical tube, with the cap partially unscrewed, inside a secondary container inside the CO2 incubator. Incubate at 37°C, mixing occasionally (see Note 3). The duration of pulse-labeling may be varied to suit specific experimental purposes (see Note 4).
3.4. Chase
Collect cells by centrifugation at room temperature. Remove radioactive culture supernatant and dispose using a liquid waste conversion kit or other disposal method, as indicated by institutional regulations. It is preferable to aspirate, rather than decant, radioactive supernatants, to avoid contaminating the outside surfaces or cap thread of the conical tube during decanting.
Wash cells once with 5 ml pre-warmed chase media. Record the time of adding chase media as the start time of the chase.
Resuspend cells in chase media at up to 5× 106 cells/ml. Distribute cells to individual 15 ml or 50 ml conical tubes. Place tubes into a secondary container with caps partially opened. Incubate in a tissue culture incubator at 37°C with occasional mixing (see Note 3). For longer chase times (≥ 8 hours), cells should be diluted to 1× 106 cells/ml in chase media (or lower still, to allow for cell growth, if the chase is continued beyond 24 hours), and appropriately sized tissue culture flasks should be used. Meanwhile, set the temperature of the benchtop centrifuge to 4 °C.
3.5. Harvest
Harvest cells at the end of the pulse and after each time of chase. The timing of sampling should match the time scales of the processes to be investigated; intracellular trafficking, maturation, and peptide loading of MHC class II molecules can be sampled on time scales of hours (see Figs 1 and 2 and Notes 4 and 5).
During collection, keep cells on ice and centrifuge at 4°C. Spin down cells and dispose of media as radioactive waste.
Wash cells once with ice-cold PBS containing 1 mM PMSF (added fresh). Record the time of PBS addition as the time of harvest. Remove the supernatant as completely as possible after spinning down, especially when samples will be lysed in a small volume.
Store the cell pellets from all time points in 15 ml (or 50 ml) tubes in designated boxes (with radioactive hazard warning label) at −20°C (short-term) or −80°C (long-term).
3.6. Detergent extraction
Supplement ice-cold lysis buffer with 1 mM PMSF and Roche Complete protease inhibitors (toxic).
Add lysis buffer (1 ml for up to 108 cells per sample) to frozen cell pellets and mix until cell pellets are dispersed as a turbid but homogenous suspension. Avoid thawing frozen cell pellets before addition of lysis buffer, as this releases active proteases.
Continue lysis by rotating the microcentrifuge tubes at 4°C for 20–60 min or leaving them on ice with occasional vortexing.
Spin down non-solubilized material for 20 min at top speed in a microcentrifuge (13,000 rpm or 10,000 × g) at 4°C.
Transfer the cleared supernatant, which contains the extracted radiolabeled proteins, to a clean tube, or directly to the first pre-clear tube (below).
3.7. Pre-clearing
This step depletes any radiolabeled proteins that bind nonspecifically to the beads and/or antibody used for IP. In the case of B cell lines, pre-clearing also depletes endogenous Ig. Depending on Ab to be used for immunoprecipitation, pre-clearing can be done using Protein A (PAS) or protein G sepharose (PGS) beads (see Note 6). The pre-clearing step may be repeated several times (see Note 7).
For each sample, add to a fresh, labeled tube 30–60 μl 50% slurry of PAS or PGS and 3–5 μl pre-immune normal serum or 10 μg purified, isotype-matched Ig from the same species as the antibody to be used for IP.
Add radiolabeled cell extract (plus any additional lysis buffer) to a final volume of 750-1000 μl and rotate tubes at 4°C for at least one hour.
Spin down beads (full speed in a microfuge, 1–2 min., 4 °C) and transfer supernatant to fresh 1.5 ml Eppendorf tubes.
In some experiments, it may be useful to quantify the amount of radioactivity incorporated into the pre-cleared cell extract and to adjust volumes to match the amount of input radioactivity between samples (for details, see Note 8).
3.8. Immunoprecipitation (IP)
Add PAS or PGS beads (40–100 μl 50% v/v slurry) and desired antibody to each tube (see Note 9). A typical amount of mAb to use is 10 μg = 10 μl of a 1 mg/ml stock in PBS, or a few μl of a polyclonal antiserum; titrating these amounts may improve results.
Rotate tubes for at least 1 hr at 4°C.
Spin down beads. Remove the supernatant (extract depleted of the protein of interest) and transfer to a fresh microfuge tube (see Note 10).
3.9. Washing beads
Wash beads 3–5 times with 1 ml cold IP wash buffer. Each time, remove the supernatant by pipetting or aspiration, without disturbing the pellet (see Note 11). The number of washes may be optimized, and lysis buffer may be used instead of wash buffer. The last wash may be performed in PBS or water to reduce carry-over of nonionic detergent into the elution step.
3.10. Preparation of SDS-PAGE gels
Select the percentage acrylamide of the gel so as to maximize resolution of proteins of interest; gradient gels can be useful if a wide range of molecular weights needs to be analyzed. For MHC class II molecules, 12% gels generally are suitable.
Prepare SDS-PAGE acrylamide gels as described in detail in Current Protocols in Molecular Biology, Chapter 10, Section II, 10.2, or use commercially available pre-cast gels. The best molecular weight resolution is achieved on large gels (14×16×0.15cm), but standard minigels may also be used.
Set up gels in an electrophoresis chamber as per manufacturer's instructions, and add 1× SDS-PAGE running buffer to the buffer compartments.
3.11. SDS-PAGE sample preparation and separation
The conditions for elution of radiolabeled material from beads may be varied for specific purposes. Here, we describe alternative procedures for analysis of dissociated dimers (10.3.11.1) and non-dissociated dimers (for assessment of SDS-stability, 10.3.11.2). Other alternatives not described here include elution in non-reducing conditions to visualize inter-chain disulfide bonding; denaturation followed by re-IP of individual chains, to identify co-precipitated polypeptides within a larger complex [73]; or enzymatic digestion prior to SDS-PAGE separation, for example, using endoglycosaminidase H to remove high-mannose forms of N-linked glycans, which may be used to track intracellular transport through the Golgi apparatus [74].
- Elution of dissociated dimers:
- Resuspend beads in an appropriate amount (limited by the size of gel wells, e.g. 30 μl for a BioRad minigel) of reducing SLB. If buffer volume is large compared to bead volume, 1× SLB may be used, but 1.5× or more concentrated SLB may be used for small elution volumes. This is to compensate for dilution by residual wash buffer in the bead pellet.
- Heat the samples for 10 min to 90–100°C in a heat block to elute protein from beads. Vortex and spin down beads.
- Proceed to step 3.
- Alternative procedure for elution of non-dissociated dimers:
- For visualization of SDS-stable (compact or floppy, see Note 12) dimers, suspend beads in SLB and incubate at room temperature for 10 to 60 min [24]. (Do not boil.) Vortex and spin down beads.
- Elution conditions are required that release immune complexes and dissociate the immunoprecipitating antibody from class II molecules, without also dissociating class II αβ heterodimers associated with peptide. Conditions (SDS concentration, addition of reducing agent, elution time) may need to be varied to optimize this step.
- Note that some mAbs fail to dissociate from SDS-stable class II molecules under typical non-boiled elution conditions and cause super-shifting of bands (see Table 1).
3.12. Sample loading and electrophoresis
Using a Hamilton syringe (dedicated for radioactive work, and rinsed thoroughly between samples) or disposable gel loading tips, load the supernatants from each sample into wells of the SDS-PAGE gel. One well is set aside for radiolabeled molecular weight markers (Amersham/GE Healthcare 14C-labeled rainbow markers), applied as per manufacturer's instructions (see Note 13).
Electrophorese samples until the loading dye front has reached the bottom of the gel (see Note 14).
3.13. Gel fixation and exposure to scintillation cocktail
Although gel fixation is recommended, we have found that it may be skipped. Transfer gel into enough Amplify solution to cover the gel. Incubate gel with gentle rocking for 15–30 min at room temp in the dark (covered with aluminum foil). We have found that the Amplify solution may be reused several times.
3.14. Gel drying
Cut a piece of Whatman 3M filter paper slightly larger than the surface area of the gels. Wet Whatman paper with water. Gently place the gel onto the filter paper and cover it with clear plastic wrap (Saran Wrap). Avoid introducing air bubbles between the layers of this sandwich.
Using a gel dryer according to the manufacturer's instructions, dry the gel for 1.5 hr at 80°C with slow ramp and good vacuum seal. Breaking the seal early or heating the gel too fast will crack the gel.
3.15. Visualize radioactive bands
Remove plastic wrap from the dried gel.
For a high-resolution, publication-quality image, expose autoradiography film to the gel in a film cassette at −80°C, adjusting the exposure time between a few hours and several weeks, depending on the amount of radioactivity being visualized. The exposure time should be varied to achieve detection of radioactive bands of interest whilst minimizing nonspecific background and avoiding overexposure of abundant bands. Allow the frozen film cassettes to return to room temperature before opening them, or the gel and film may stick together and separating them may cause sparks that will leave black spots on the film when developed. Develop the film using an automatic film developer, according to manufacturer's instructions.
Semiquantitative analysis of band intensities may be achieved by film densitometry.
If a phosphorimager is available, a “snapshot” may be obtained by placing the gel on the light-blanked detector screen of a phosphorimager system overnight at room temperature. Phosphorimagers can be more sensitive than film and linear over a wider range of exposures. The data are immediately available for quantitation, but the images obtained are grainier than film and less suitable for publication.
4. Notes
In order to maintain protein-protein interactions that are sensitive to NP-40, or for specific class II alleles thought to have reduced stability (such as I-Ag7), digitonin (1 %) or CHAPS (6 mM) can be used to replace NP-40. Alternative buffer systems may be required, for example, to maintain pH-dependent interactions, such as those between MHC II and DM [75]; note, however, that interactions of antibodies with their antigens and with protein A/G-sepharose may also be pH-dependent.
Steps 1–6 apply to suspension cells. Adherent cells should be split one day before the experiment and allowed to grow in 10-cm or 60-mm tissue-culture dishes overnight. Depending on the abundance of proteins of interest, each chase time point may require one or more dishes of cells. The cells can be washed without centrifugation (by gentle aspiration and addition of media or buffers, as appropriate at each step) and harvested by scraping off cells using disposable cell scrapers, or by detaching with PBS/EDTA, with or without trypsin (unless the protein of interest is sensitive to trypsinization). Detached cells may then be centrifuged and further processed as described in the main protocol (step 6). Alternatively, adherent cells may be extracted without being detached, by washing in PBS followed by addition of ice-cold lysis buffer to culture dishes. After thorough dispersion of cells in lysis buffer, transfer the extract to 1.5 ml microcentrifuge tubes for further processing.
Cell density during starvation and subsequent pulse-labeling and chase may be adjusted according to the metabolism of the cell type under study and the particular experiment. The cell density during starvation and chase should be low enough to avoid nutrient depletion of media, but volumes of radiolabeled media should be minimized to limit disposal cost. Mixing of cells kept in conical tubes is important to avoid depletion of nutrients and oxygen after cells settle under gravity. Moreover, to allow oxygen diffusion and for effective CO2 exchange for appropriate buffering of media, tubes should not be filled with media to a height of more than 2–3 cm. The total cell number per time point may be increased to isolate less abundant or poorly labeled proteins.
The pulse-labeling time varies depending on the experiment and should be adjusted based on the desired time resolution. To track subsequent intracellular trafficking of MHC II molecules on time scales of 2 to 24 hrs, a 30 min to 1 hr pulse is suitable. Very brief pulses (on the order of a minute or even shorter) may be used to track early stages of MHC II assembly in the ER; however, this requires further modifications to this protocol to allow precise timing of the pulse and chase [76–77]. Proteins synthesized at a low rate, or proteins with a low content of Cys/Met, may require longer labeling times to become detectable.
The spin/wash steps between the pulse and chase and the same steps at collection can each take up to 30 min. When using a long (≥ 30 min) pulse with chase time points at intervals of an hour or longer, the time for spinning/washing is not critical, because metabolism slows substantially once cell suspensions are handled in the cold. However, for experiments performed on shorter time scales, the time for manipulation can introduce error to estimated times for particular processing events. In order to ensure accurate timing in these experiments, the pulse may be terminated using protein synthesis inhibitors; labeled and chased cells may be rapidly diluted into a large excess of ice-cold PBS and kept rigorously in the cold thereafter. Centrifugation times may be shortened and g forces increased to reduce processing time.
Protein A and G bind differentially to the different immunoglobulin isotypes, and the choice of protein A- or G-sepharose will be dictated by the mAb(s) used. For binding affinities of protein A and G for various IgG subclasses, see Current Protocols in Molecular Biology, Chapter 10, section VI, Table 10.16.1. A cheaper alternative is to use a 10% suspension of Pansorbin cells (fixed, heat-killed Staphylococcus aureus, bearing protein A) for preclearing. The desired amount of Pansorbin cells (100 ul for each time point) is washed twice before use (see Materials). Add cell lysate and normal pre-clearing serum (plus any additional lysis buffer) directly to the Pansorbin cell pellet to a final volume of 750-1000 μl. Mix thoroughly by pipetting or vortexing. Otherwise proceed as described above.
Additional rounds of pre-clearing may further reduce nonspecific background radioactivity, but each step slightly dilutes the cell extracts and very unstable proteins may gradually denature or degrade, so this step may need to be optimized. When stable proteins are being isolated, one of the pre-clearing steps may be extended overnight. The final preclearing step may be performed with PAS or PGS alone, in order to remove any residual irrelevant antibody before the specific IP.
To quantify cell-associated radioactivity, mix a small aliquot of the extract (e.g., 5 μl) with 1 ml scintillation fluid in a disposable scintillation vial, and count in a scintillation counter. Alternatively, protein-associated radioactivity may be quantified. Spot a small aliquot of the cell extract onto glass filter membranes, soak in 100% w/v aqueous trichloroacetic acid, wash, transfer to scintillation vials, add scintillant, and count. Before proceeding to IP, the volumes of cell extracts may be adjusted to match for equal counts of radioactivity, and lysis buffer added to equalize total volumes. This method can compensate for differences in labeling efficiency between different cell lines for comparison of relative rates of biosynthesis, but is poorly suited for normalizing total radioactivity during an extended chase. This is because radiolabeled proteins will be degraded at very different rates during the chase, so a gradual loss of total radiolabeled proteins over time is expected.
When handling multiple samples, it is simpler to dispense protein A/G sepharose and antibodies into tubes first, before adding the radiolabeled, pre-cleared extract. Further variations of this step may be used. The antibody may be pre-incubated with the pre-cleared extract for an hour and captured by subsequent incubation with protein A/G beads. Alternatively, if hybridoma culture supernatants are to be used as a source of mAbs for IP, the supernatants (0.5–1 ml per sample) may be pre-incubated with protein A or G-sepharose beads for 1 hr in the cold, and washed once in PBS, before addition of radiolabeled cell extracts.
The recovered cell extracts may be disposed of as radioactive waste or recovered into fresh tubes, stored at −80 °C, and used for sequential IPs of other molecules. For sequential IPs, thaw extracts at 4 °C with thorough mixing, spin out any precipitated proteins at ≥ 10,000× g in a microfuge, and preclear at least once with PAS or PGS to minimize any carry-over of antibody or immune complexes into subsequent IPs.
The initial washes contain most of the radiolabeled unbound proteins, and need to be disposed of and accounted for as radioactive waste. Later washes will contain far less radioactivity; as a result, drain disposal may be permissible in some jurisdictions. Processing of multiple samples is streamlined if the later washes are removed by vacuum aspiration, using an aspirator dedicated for radioactive work, fitted with filter traps that prevent escape of radioactivity into house vacuum systems or vacuum pumps.
Floppy dimers (apparent relative molecular mass [Mr] ~63–67K) and compact dimers (Mr, ~56K ) were initially defined in vitro by studies of purified molecules [78]. Size heterogeneity of SDS-stable class II molecules has also been observed in naturally occurring biosynthetic intermediates. Some MHC class II alleles form floppy-sized dimers when loaded with CLIP; others form SDS-unstable complexes. In contrast, DM-edited forms of MHC II loaded with stable peptides generally migrate as compact dimers [24,79,10], but the percentage of SDS-stable complexes varies between alleles and isotypes.
Molecular weight calibration will be most accurate if the markers are diluted in a buffer matched in composition to that of the samples. For immunoprecipitates, a mixture of loading buffer and IP wash buffer, in similar ratios as in samples, works well.
Depending on the type and amount of detergent used in the IP wash buffer, the loading dye front may be broadened. Note that CLIP and other low-molecular weight peptides migrate ahead of the dye front. Their detection therefore requires electrophoresis to be stopped when the dye front is a few mm above the bottom of the gel. Note that such peptides can also be lost during standard gel fixation with methanol/acetic acid.
Acknowledgments
This work was supported by NIH grants F32 AI089090 to TH and AI095813 and AI28809 to E.D.M. R.B. was supported by a Senior Research Fellowship from Arthritis Research UK [ref. 18543].
Abbreviations
- (MHC)
Major histocompatibility complex
- (ER)
endoplasmic reticulum
- (Abs)
antibodies
- (Ii)
invariant chain
- (B-LCL)
EBV-transformed B-lymphoblastoid cell lines
- (SDS-PAGE)
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- (MIIC)
MHC class II compartments
- LIP
(leupeptin-induced polypeptides)
- CLIP
(class-II-associated invariant chain peptides)
- (Cys/Met)
cysteine/methionine
- (PAS)
protein A Sepharose
- (PGS)
protein G Sepharose
- (IP)
immunoprecipitation
- (APCs)
antigen presenting cells
- (SLB)
standard Laemmli sample loading buffer
- (PMSF)
phenylmethylsulfonyl fluoride
References
- 1.Jamieson JD, Palade GE. Intracellular transport of secretory proteins in the pancreatic exocrine cell. I. Role of the peripheral elements of the Golgi complex. J Cell Biol. 1967;34(2):577–596. doi: 10.1083/jcb.34.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamieson JD, Palade GE. Intracellular transport of secretory proteins in the pancreatic exocrine cell. II. Transport to condensing vacuoles and zymogen granules. J Cell Biol. 1967;34(2):597–615. doi: 10.1083/jcb.34.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Riva A, Deery MJ, McDonald S, Lund T, Busch R. Measurement of protein synthesis using heavy water labeling and peptide mass spectrometry: Discrimination between major histocompatibility complex allotypes. Anal Biochem. 2010;403(1–2):1–12. doi: 10.1016/j.ab.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PP, Murphy DB, Hewgill D, McDevitt HO. Detection of a common polypeptide chain in I-A and I-E sub-region immunoprecipitates. Mol Immunol. 1979;16(1):51–60. doi: 10.1016/0161-5890(79)90027-0. [DOI] [PubMed] [Google Scholar]
- 5.Machamer CE, Cresswell P. Biosynthesis and glycosylation of the invariant chain associated with HLA-DR antigens. J Immunol. 1982;129(6):2564–2569. [PubMed] [Google Scholar]
- 6.Busch R, Cloutier I, Sekaly RP, Hämmerling GJ. Invariant chain protects class II histocompatibility antigens from binding intact polypeptides in the endoplasmic reticulum. Embo J. 1996;15(2):418–428. [PMC free article] [PubMed] [Google Scholar]
- 7.Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63(4):707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 8.West MA, Lucocq JM, Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature. 1994;369(6476):147–151. doi: 10.1038/369147a0. [DOI] [PubMed] [Google Scholar]
- 9.Tulp A, Verwoerd D, Dobberstein B, Ploegh HL, Pieters J. Isolation and characterization of the intracellular MHC class II compartment. Nature. 1994;369(6476):120–126. doi: 10.1038/369120a0. [DOI] [PubMed] [Google Scholar]
- 10.Qiu Y, Xu X, Wandinger-Ness A, Dalke DP, Pierce SK. Separation of subcellular compartments containing distinct functional forms of MHC class II. J Cell Biol. 1994;125(3):595–605. doi: 10.1083/jcb.125.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters PJ, Neefjes JJ, Oorschot V, Ploegh HL, Geuze HJ. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature. 1991;349(6311):669–676. doi: 10.1038/349669a0. [DOI] [PubMed] [Google Scholar]
- 12.Blum JS, Cresswell P. Role for intracellular proteases in the processing and transport of class II HLA antigens. Proc Natl Acad Sci U S A. 1988;85(11):3975–3979. doi: 10.1073/pnas.85.11.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maric MA, Taylor MD, Blum JS. Endosomal aspartic proteinases are required for invariant-chain processing. Proc Natl Acad Sci U S A. 1994;91(6):2171–2175. doi: 10.1073/pnas.91.6.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riese RJ, Wolf PR, Bromme D, Natkin LR, Villadangos JA, Ploegh HL, Chapman HA. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity. 1996;4(4):357–366. doi: 10.1016/s1074-7613(00)80249-6. [DOI] [PubMed] [Google Scholar]
- 15.Neefjes JJ, Ploegh HL. Inhibition of endosomal proteolytic activity by leupeptin blocks surface expression of MHC class II molecules and their conversion to SDS resistance alpha beta heterodimers in endosomes. EMBO J. 1992;11(2):411–416. doi: 10.1002/j.1460-2075.1992.tb05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360(6403):474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 17.Sette A, Ceman S, Kubo RT, Sakaguchi K, Appella E, Hunt DF, Davis TA, Michel H, Shabanowitz J, Rudersdorf R, et al. Invariant chain peptides in most HLA-DR molecules of an antigen-processing mutant. Science. 1992;258(5089):1801–1804. doi: 10.1126/science.1465617. [DOI] [PubMed] [Google Scholar]
- 18.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82(1):155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 19.Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain-derived peptide. Immunity. 1995;3(2):197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 20.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375(6534):802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 21.Lovitch SB, Pu Z, Unanue ER. Amino-terminal flanking residues determine the conformation of a peptide-class II MHC complex. J Immunol. 2006;176(5):2958–2968. doi: 10.4049/jimmunol.176.5.2958. [DOI] [PubMed] [Google Scholar]
- 22.Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363(6431):725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 23.Bikoff EK, Huang LY, Episkopou V, van Meerwijk J, Germain RN, Robertson EJ. Defective major histocompatibility complex class II assembly, transport, peptide acquisition, and CD4+ T cell selection in mice lacking invariant chain expression. J Exp Med. 1993;177(6):1699–1712. doi: 10.1084/jem.177.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viville S, Neefjes J, Lotteau V, Dierich A, Lemeur M, Ploegh H, Benoist C, Mathis D. Mice lacking the MHC class II-associated invariant chain. Cell. 1993;72(4):635–648. doi: 10.1016/0092-8674(93)90081-z. [DOI] [PubMed] [Google Scholar]
- 25.Busch R, Rinderknecht CH, Roh S, Lee AW, Harding JJ, Burster T, Hornell TM, Mellins ED. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–260. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 26.Roche PA, Marks MS, Cresswell P. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature. 1991;354(6352):392–394. doi: 10.1038/354392a0. [DOI] [PubMed] [Google Scholar]
- 27.Roche PA, Teletski CL, Stang E, Bakke O, Long EO. Cell surface HLA-DR-invariant chain complexes are targeted to endosomes by rapid internalization. Proc Natl Acad Sci U S A. 1993;90(18):8581–8585. doi: 10.1073/pnas.90.18.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wraight CJ, van Endert P, Moller P, Lipp J, Ling NR, MacLennan IC, Koch N, Moldenhauer G. Human major histocompatibility complex class II invariant chain is expressed on the cell surface. J Biol Chem. 1990;265(10):5787–5792. [PubMed] [Google Scholar]
- 29.Avva RR, Cresswell P. In vivo and in vitro formation and dissociation of HLA-DR complexes with invariant chain-derived peptides. Immunity. 1994;1(9):763–774. doi: 10.1016/s1074-7613(94)80018-9. [DOI] [PubMed] [Google Scholar]
- 30.Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1(7):595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 31.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13(4):517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 32.Stang E, Guerra CB, Amaya M, Paterson Y, Bakke O, Mellins ED. DR/CLIP (class II-associated invariant chain peptides) and DR/peptide complexes colocalize in prelysosomes in human B lymphoblastoid cells. J Immunol. 1998;160(10):4696–4707. [PubMed] [Google Scholar]
- 33.Guy K, Van Heyningen V, Cohen BB, Deane DL, Steel CM. Differential expression and serologically distinct subpopulations of human Ia antigens detected with monoclonal antibodies to Ia alpha and beta chains. Eur J Immunol. 1982;12(11):942–948. doi: 10.1002/eji.1830121109. [DOI] [PubMed] [Google Scholar]
- 34.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184(6):2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AW, Wang N, Hornell TM, Harding JJ, Deshpande C, Hertel L, Lacaille V, Pashine A, Macaubas C, Mocarski ES, Mellins ED. Human cytomegalovirus decreases constitutive transcription of MHC class II genes in mature Langerhans cells by reducing CIITA transcript levels. Mol Immunol. 2011;48(9–10):1160–1167. doi: 10.1016/j.molimm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hitzel C, Gruneberg U, van Ham M, Trowsdale J, Koch N. Sodium dodecyl sulfate-resistant HLA-DR “superdimer” bands are in some cases class II heterodimers bound to antibody. J Immunol. 1999;162(8):4671–4676. [PubMed] [Google Scholar]
- 37.Lampson LA, Levy R. Two populations of Ia-like molecules on a human B cell line. J Immunol. 1980;125(1):293–299. [PubMed] [Google Scholar]
- 38.Guerra CB, Busch R, Doebele RC, Liu W, Sawada T, Kwok WW, Chang MD, Mellins ED. Novel glycosylation of HLA-DRalpha disrupts antigen presentation without altering endosomal localization. J Immunol. 1998;160(9):4289–4297. [PubMed] [Google Scholar]
- 39.Watanabe M, Suzuki T, Taniguchi M, Shinohara N. Monoclonal anti-Ia murine alloantibodies crossreactive with the Ia-homologues of other mammalian species including humans. Transplantation. 1983;36(6):712–718. doi: 10.1097/00007890-198336060-00025. [DOI] [PubMed] [Google Scholar]
- 40.Carven GJ, Chitta S, Hilgert I, Rushe MM, Baggio RF, Palmer M, Arenas JE, Strominger JL, Horejsi V, Santambrogio L, Stern LJ. Monoclonal antibodies specific for the empty conformation of HLA-DR1 reveal aspects of the conformational change associated with peptide binding. J Biol Chem. 2004;279(16):16561–16570. doi: 10.1074/jbc.M314315200. [DOI] [PubMed] [Google Scholar]
- 41.Fu XT, Karr RW. HLA-DR alpha chain residues located on the outer loops are involved in nonpolymorphic and polymorphic antibody-binding epitopes. Hum Immunol. 1994;39(4):253–260. doi: 10.1016/0198-8859(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 42.Knudsen PJ, Strominger JL. A monoclonal antibody that recognizes the alpha chain of HLA-DR antigens. Hum Immunol. 1986;15(2):150–163. doi: 10.1016/0198-8859(86)90023-6. [DOI] [PubMed] [Google Scholar]
- 43.Potolicchio I, Chitta S, Xu X, Fonseca D, Crisi G, Horejsi V, Strominger JL, Stern LJ, Raposo G, Santambrogio L. Conformational variation of surface class II MHC proteins during myeloid dendritic cell differentiation accompanies structural changes in lysosomal MIIC. J Immunol. 2005;175(8):4935–4947. doi: 10.4049/jimmunol.175.8.4935. [DOI] [PubMed] [Google Scholar]
- 44.Rinderknecht CH, Roh S, Pashine A, Belmares MP, Patil NS, Lu N, Truong P, Hou T, Macaubas C, Yoon T, Wang N, Busch R. Mellins ED DM influences the abundance of major histocompatibility complex class II alleles with low affinity for class II-associated invariant chain peptides via multiple mechanisms. Immunology. 131(1):18–32. doi: 10.1111/j.1365-2567.2010.03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson JP, Meo T, Riethmuller G, Schendel DJ, Wank R. Direct demonstration of an HLA-DR allotypic determinant on the low molecular weight (beta) subunit using a mouse monoclonal antibody specific for DR3. J Exp Med. 1982;156(1):104–111. doi: 10.1084/jem.156.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson JP, Wank R. Identification of two cis-encoded HLA-DQ molecules that carry distinct alloantigenic specificities. J Exp Med. 1984;160(5):1350–1359. doi: 10.1084/jem.160.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellins E, Kempin S, Smith L, Monji T, Pious D. A gene required for class II-restricted antigen presentation maps to the major histocompatibility complex. J Exp Med. 1991;174(6):1607–1615. doi: 10.1084/jem.174.6.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanderson F, Thomas C, Neefjes J, Trowsdale J. Association between HLA-DM and HLA-DR in vivo. Immunity. 1996;4(1):87–96. doi: 10.1016/s1074-7613(00)80301-5. [DOI] [PubMed] [Google Scholar]
- 49.Verreck FA, Fargeas CA, Hämmerling GJ. Conformational alterations during biosynthesis of HLA-DR3 molecules controlled by invariant chain and HLA-DM. Eur J Immunol. 2001;31(4):1029–1036. doi: 10.1002/1521-4141(200104)31:4<1029::aid-immu1029>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 50.Patil NS, Pashine A, Belmares MP, Liu W, Kaneshiro B, Rabinowitz J, McConnell H, Mellins ED. Rheumatoid arthritis (RA)-associated HLA-DR alleles form less stable complexes with class II-associated invariant chain peptide than non-RA-associated HLA-DR alleles. J Immunol. 2001;167(12):7157–7168. doi: 10.4049/jimmunol.167.12.7157. [DOI] [PubMed] [Google Scholar]
- 51.Hou T, Macmillan H, Chen Z, Keech CL, Jin X, Sidney J, Strohman M, Yoon T, Mellins ED. An insertion mutant in DQA1*0501 restores susceptibility to HLA-DM: implications for disease associations. J Immunol. 2011;187(5):2442–2452. doi: 10.4049/jimmunol.1100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spits H, Borst J, Giphart M, Coligan J, Terhorst C, De Vries JE. HLADC antigens can serve as recognition elements for human cytotoxic T lymphocytes. Eur J Immunol. 1984;14(4):299–304. doi: 10.1002/eji.1830140404. [DOI] [PubMed] [Google Scholar]
- 53.Fallang LE, Roh S, Holm A, Bergseng E, Yoon T, Fleckenstein B, Bandyopadhyay A, Mellins ED, Sollid LM. Complexes of two cohorts of CLIP peptides and HLA-DQ2 of the autoimmune DR3-DQ2 haplotype are poor substrates for HLA-DM. J Immunol. 2008;181(8):5451–5461. doi: 10.4049/jimmunol.181.8.5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huan J, Meza-Romero R, Mooney JL, Vandenbark AA, Offner H, Burrows GG. Single-chain recombinant HLA-DQ2.5/peptide molecules block alpha2-gliadin-specific pathogenic CD4+ T-cell proliferation and attenuate production of inflammatory cytokines: a potential therapy for celiac disease. Mucosal Immunol. 2011;4(1):112–120. doi: 10.1038/mi.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viken HD, Paulsen G, Sollid LM, Lundin KE, Tjonnfjord GE, Thorsby E, Gaudernack G. Characterization of an HLA-DQ2-specific monoclonal antibody. Influence of amino acid substitutions in DQ beta 1*0202. Hum Immunol. 1995;42(4):319–327. doi: 10.1016/0198-8859(94)00110-c. [DOI] [PubMed] [Google Scholar]
- 56.Shookster L, Matsuyama T, Burmester G, Winchester R. Monoclonal antibody 1a3 recognizes a monomorphic epitope unique to DQ molecules. Hum Immunol. 1987;20(1):59–70. doi: 10.1016/0198-8859(87)90006-1. [DOI] [PubMed] [Google Scholar]
- 57.Robbins PA, Evans EL, Ding AH, Warner NL, Brodsky FM. Monoclonal antibodies that distinguish between class II antigens (HLA-DP, DQ, and DR) in 14 haplotypes. Hum Immunol. 1987;18(4):301–313. doi: 10.1016/0198-8859(87)90077-2. [DOI] [PubMed] [Google Scholar]
- 58.Radka SF, Machamer CE, Cresswell P. Analysis of monoclonal antibodies reactive with human class II beta chains by two-dimensional electrophoresis and Western blotting. Hum Immunol. 1984;10(3):177–186. doi: 10.1016/0198-8859(84)90038-7. [DOI] [PubMed] [Google Scholar]
- 59.Koch N, Koch S, Hämmerling GJ. Ia invariant chain detected on lymphocyte surfaces by monoclonal antibody. Nature. 1982;299(5884):644–645. doi: 10.1038/299644a0. [DOI] [PubMed] [Google Scholar]
- 60.Anderson MS, Miller J. Invariant chain can function as a chaperone protein for class II major histocompatibility complex molecules. Proc Natl Acad Sci U S A. 1992;89(6):2282–2286. doi: 10.1073/pnas.89.6.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Germain RN, Hendrix LR. MHC class II structure, occupancy and surface expression determined by post-endoplasmic reticulum antigen binding. Nature. 1991;353(6340):134–139. doi: 10.1038/353134a0. doi:10.1038/353134a0. [DOI] [PubMed] [Google Scholar]
- 62.Mehringer JH, Harris MR, Kindle CS, McCourt DW, Cullen SE. Characterization of fragments of the murine Ia-associated invariant chain. J Immunol. 1991;146(3):920–927. [PubMed] [Google Scholar]
- 63.Bhattacharya A, Dorf ME, Springer TA. A shared alloantigenic determinant on Ia antigens encoded by the I-A and I-E subregions: evidence for I region gene duplication. J Immunol. 1981;127(6):2488–2495. [PubMed] [Google Scholar]
- 64.Janeway CA, Jr., Conrad PJ, Lerner EA, Babich J, Wettstein P, Murphy DB. Monoclonal antibodies specific for Ia glycoproteins raised by immunization with activated T cells: possible role of T cellbound Ia antigens as targets of immunoregulatory T cells. J Immunol. 1984;132(2):662–667. [PubMed] [Google Scholar]
- 65.Bikoff EK, Germain RN, Robertson EJ. Allelic differences affecting invariant chain dependency of MHC class II subunit assembly. Immunity. 1995;2(3):301–310. doi: 10.1016/1074-7613(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 66.Oi VT, Jones PP, Goding JW, Herzenberg LA. Properties of monoclonal antibodies to mouse Ig allotypes, H-2, and Ia antigens. Curr Top Microbiol Immunol. 1978;81:115–120. doi: 10.1007/978-3-642-67448-8_18. [DOI] [PubMed] [Google Scholar]
- 67.Koch N, Hämmerling GJ, Tada N, Kimura S, Hämmerling U. Cross-blocking studies with monoclonal antibodies against I-A molecules of haplotypes b, d and k. Eur J Immunol. 1982;12(11):909–914. doi: 10.1002/eji.1830121103. [DOI] [PubMed] [Google Scholar]
- 68.Dang LH, Lien LL, Benacerraf B, Rock KL. A mutant antigen-presenting cell defective in antigen presentation expresses class II MHC molecules with an altered conformation. J Immunol. 1993;150(10):4206–4217. [PubMed] [Google Scholar]
- 69.Kappler JW, Skidmore B, White J, Marrack P. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med. 1981;153(5):1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steinman RM, Nogueira N, Witmer MD, Tydings JD, Mellman IS. Lymphokine enhances the expression and synthesis of Ia antigens on cultured mouse peritoneal macrophages. J Exp Med. 1980;152(5):1248–1261. doi: 10.1084/jem.152.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozato K, Mayer NM, Sachs DH. Monoclonal antibodies to mouse major histocompatibility complex antigens. Transplantation. 1982;34(3):113–120. doi: 10.1097/00007890-198209000-00001. [DOI] [PubMed] [Google Scholar]
- 72.Rinderknecht CH, Belmares MP, Catanzarite TL, Bankovich AJ, Holmes TH, Garcia KC, Nanda NK, Busch R, Kovats S, Mellins ED. Posttranslational regulation of I-Ed by affinity for CLIP. J Immunol. 2007;179(9):5907–5915. doi: 10.4049/jimmunol.179.9.5907. [DOI] [PubMed] [Google Scholar]
- 73.Rinderknecht CH, Roh S, Pashine A, Belmares MP, Patil NS, Lu N, Truong P, Hou T, Macaubas C, Yoon T, Wang N, Busch R, Mellins ED. DM influences the abundance of major histocompatibility complex class II alleles with low affinity for class II-associated invariant chain peptides via multiple mechanisms. Immunology. 2010;131(1):18–32. doi: 10.1111/j.1365-2567.2010.03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Busch R, Doebele RC, von Scheven E, Fahrni J, Mellins ED. Aberrant intermolecular disulfide bonding in a mutant HLA-DM molecule: implications for assembly, maturation, and function. J Immunol. 1998;160(2):734–743. [PubMed] [Google Scholar]
- 75.Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19(2):183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 76.Neefjes JJ, Hämmerling GJ, Momburg F. Folding and assembly of major histocompatibility complex class I heterodimers in the endoplasmic reticulum of intact cells precedes the binding of peptide. J Exp Med. 1993;178(6):1971–1980. doi: 10.1084/jem.178.6.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nijenhuis M, Neefjes J. Early events in the assembly of major histocompatibility complex class II heterotrimers from their free subunits. Eur J Immunol. 1994;24(1):247–256. doi: 10.1002/eji.1830240139. [DOI] [PubMed] [Google Scholar]
- 78.Dornmair K, Rothenhäusler B, McConnell HM. Structural intermediates in the reactions of antigenic peptides with MHC molecules. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):409–416. doi: 10.1101/sqb.1989.054.01.050. [DOI] [PubMed] [Google Scholar]
- 79.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84(4):531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]


