Abstract
Asymmetric cell divisions allow stem cells to balance proliferation and differentiation. During embryogenesis, murine epidermis expands rapidly from a single layer of unspecified basal layer progenitors to a stratified, differentiated epithelium. Morphogenesis involves perpendicular (asymmetric) divisions and the spindle orientation protein LGN, but little is known about how the apical localization of LGN is regulated. Here, we combine conventional genetics and lentiviral-mediated in vivo RNAi to explore the functions of the LGN-interacting proteins Par3, mInsc and Gαi3. Whereas loss of each gene alone leads to randomized division angles, combined loss of Gnai3 and mInsc causes a phenotype of mostly planar divisions, akin to loss of LGN. These findings lend experimental support for the hitherto untested model that Par3–mInsc and Gαi3 act cooperatively to polarize LGN and promote perpendicular divisions. Finally, we uncover a developmental switch between delamination-driven early stratification and spindle-orientation-dependent differentiation that occurs around E15, revealing a two-step mechanism underlying epidermal maturation.
Between embryonic day 13.5 (E13.5) and E17.5 of murine epidermal development, a single layer of basal progenitors develops into a stratified, differentiated epithelium. Increasing evidence suggests that cell division orientation is essential for establishing this proper tissue architecture1-3. However, the molecular mechanisms governing spindle positioning in skin and other tissues are still unfolding4. In Caenorhabditis elegans zygotes and Drosophila melanogaster neuroblasts, a group of cortical proteins polarize in such a way that the mitotic spindle aligns along a precise axis5-7. This frequently results in an asymmetric cell division (ACD), where cell fate determinants are unequally distributed between daughter cells. Mutations that perturb this balance can affect not only normal development and growth, but also result in overgrowth associated with cancers8-10.
In many epithelia, cell polarity and spindle orientation are inextricably linked. The PDZ scaffold protein Par3 (Baz in Drosophila) is enriched at the apical cell cortex, and associates with another scaffolding protein, Par6, and atypical protein kinase C (aPKC), whose substrates have been linked to segregation of cell fate determinants11. Baz can also interact with the adapter protein Inscuteable (mInsc in mammals), which binds directly to Pins (partner of Inscuteable), homologue of vertebrate LGN/Gpsm2 and AGS3/Gpsm1. In turn, Pins can bind membrane-anchored GDP-bound heterotrimeric G proteins (Gαi and/or Gαo) through carboxy-terminal GoLoco motifs, and the microtubule-binding protein Mud (NuMA in vertebrates) through amino-terminal TPR repeats. NuMA/Mud, in connection with the motor protein dynactin, generates pulling forces on astral microtubules that reorient the mitotic spindle2,6,9,12. Recent evidence, however, suggests that LGN cannot simultaneously bind both mInsc and NuMA13-16. Thus, how apical localization of LGN is controlled in vivo by upstream regulators such as mInsc–Par3 and G proteins remains poorly understood, particularly for mammalian systems.
Using a combination of traditional genetics and in vivo RNA-mediated interference (RNAi), we examine the consequences of removing mInsc, Pard3 (Par3) and Gnai3 (Gαi3) function in developing epidermis. Rather than causing a shift to planar (symmetric) divisions as when LGN or Numa1 are knocked down, division orientation is randomized following mInsc, Pard3 or Gnai3 loss. We identify one of three mammalian Gαi homologues, Gαi3, as pivotal for promoting apical localization of LGN, non-planar divisions and epidermal differentiation. Moreover, combined loss of mInsc and Gnai3 leads to a phenotype resembling LGN loss unveiling their cooperativity in promoting perpendicular divisions. Finally, we show that early stratification does not require the spindle orientation machinery, instead relying more extensively on differentiation through delamination of basal cells. These studies thus reveal how delamination and oriented cell divisions play distinct roles in promoting epithelial differentiation at different developmental stages.
RESULTS
LGN expression correlates with division orientation but is developmentally restricted
LGN and its downstream effector NuMA couple cortical polarity cues to changes in the microtubule cytoskeleton that reorient the mitotic spindle and promote perpendicular divisions. When either of these genes are knocked down in developing epidermis, most divisions occur with a planar orientation, rather than the normal ‘bimodal’ distribution of ~60% perpendicular and ~40% planar17. Although LGN localizes to the apical cortex of mitotic epidermal progenitors undergoing a perpendicular division, in neural progenitors, LGN localizes laterally and promotes planar divisions18-20. This suggests that LGN might be differentially localized in perpendicular versus planar divisions.
We used the cleavage furrow marker survivin to identify late-stage mitotic cells and unambiguously characterize epidermal division angles (Fig. 1a). In perpendicular divisions with a division angle >45° relative to the basement membrane, LGN was nearly always enriched over the more apical daughter (Fig. 1a,b). Apical LGN was observed in 78% of cells at telophase (n = 51), similar to what has been reported at earlier stages of mitosis17,21. These are likely to be asymmetric divisions, as supported by genetic lineage tracing4,22. Conversely, in planar divisions (<45°), LGN was not detected in most cells (64%, n = 77). These data reveal that LGN is generally apical in perpendicular divisions, and unpolarized (absent or evenly distributed) in planar divisions.
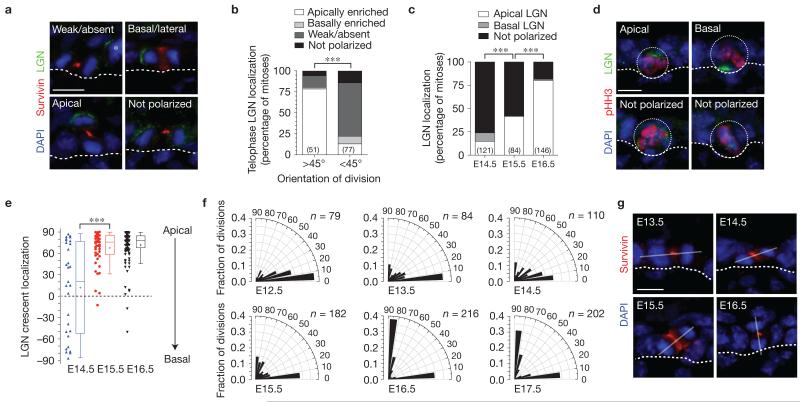
Figure 1.
LGN promotes perpendicular divisions in a developmentally restricted manner. (a) In telophase cells at E16.5, LGN can localize in one of four different patterns: absent (undetectable), not polarized (distributed evenly between daughter cells), basal/lateral (distributed preferentially over the more basal daughter nucleus), or apical. Survivin (red) labels the cleavage furrow and facilitates identification of late-stage mitotic cells. Asterisk shows a neighbouring prometaphase cell with normal apical LGN. (b) Polarized apical LGN is a hallmark of perpendicular divisions (>45° relative to the basement membrane), occurring in 78% of mitoses. In planar divisions (<45°), LGN is generally absent. (c) Apical polarization of LGN during mitosis is inefficient until ~E16.5. (d) Examples of LGN localization in E14.5–E15.5 prometaphase/metaphase cells (dotted circles depict cell boundary). (e) Quantification of LGN crescent orientation (degrees relative to basement membrane) in mitotic basal cells from E14.5–E16.5, shown as dot plots of individual cells (left, n > 30 per condition) and Tukey box-and-whisker population plots (right). Box boundaries indicate the 25% and 75% quartiles, the middle bar the median and the plus symbol the mean. Though LGN can infrequently polarize in E14.5 basal cells, its orientation is random. By E15.5–E16.5 its distribution trends towards apical. (f) Radial histograms depicting the orientation of cell divisions in late-stage (anaphase/telophase) mitotic cells in wild-type embryos at different ages. Orientation of cell divisions is almost exclusively planar in the early single-layered epithelium. Oblique and vertical divisions begin to appear from E13.5 to E15.5 as the first suprabasal cells appear. By E16.5, oblique divisions are rare. Nearly all (86%) divisions occur within 20° of perpendicular or planar at E16.5, compared with 62% at E15.5 and 56% at E14.5. n indicates cells from 5 to 12 independent animals. (g) Examples of typical telophase cells at different ages. Solid lines indicate the axis of division. Location of basement membrane in a,d,g is indicated by a dashed line. Scale bars, 10μm. P values (*** P <0.0001) determined by chi-square in b,c and two-tailed Student’s t-tests in e. Chi-square P values related to f can be found in Supplementary Table 1. n values in b,c represent the number of cells analysed from between 3 and 5 independent animals.
We next investigated whether apical LGN correlates with stratification onset at ~E13.5. Surprisingly, LGN was rarely detected in phospho-histone-H3 (pHH3+) mitotic basal cells before E14.5, when <25% of cells examined (n = 121) exhibited polarized LGN (Fig. 1c,d). Although early LGN-positive cells exhibited variable LGN orientation, apical bias became more pronounced by E16.5 (Fig. 1e). Thus, stratification precedes the ability of basal cells to polarize LGN.
In agreement with previous reports2,17,21-23, most divisions at E12.5 occurred parallel to the basement membrane, whereas at E16.5 and later, they were largely bimodal (Fig. 1f,g). However, careful inspection of divisions between E13.5 and E15.5 revealed a high incidence of oblique angles, which had previously been unrecognized. Statistical analyses revealed that the most significant change in division angle distribution occurred between E15.5 and E16.5 (Mann–Whitney test, P = 0.0002; Supplementary Table 1 for chi-square P values related to this figure and all other division orientation data). Importantly, this shift from ‘randomized’ to ‘bimodal’ division orientations coincides with the timing when LGN becomes efficiently apically polarized, highlighting E15.5 as a critical transition period in epidermal maturation.
Early stratification relies on delamination rather than oriented cell divisions
In vitro studies suggested that differentiation can occur by ‘delamination’, whereby basal keratinocytes co-expressing suprabasal markers detach from their underlying substratum24. However, evidence that delamination occurs in vivo is lacking. As a first step, we investigated potential co-expression of basal (keratins 5 and 14; K5/K14) and suprabasal (keratins 1 and 10; K1/K10) markers, and found that at E13.5, sporadic basal cells indeed expressed both K10 and K5 (Supplementary Fig. 1a). The epidermis stratifies in a posterior → anterior gradient, and at E14.5 head skin remains a single layer, now almost completely K5+/K10+ (Supplementary Fig. 1b). In addition, the hemidesmosome marker β4-integrin, required for efficient adherence to underlying basement membrane25, was diffusely distributed up until ~E15.5, becoming basally polarized concomitant with keratin marker segregation (Supplementary Fig. 1b,c). These findings suggest that before epidermal maturation, basal cells may be less adhesive and prone to delamination.
Even at E15.5, occasional K10+ cells, also positive for K5 and β4-integrin, were observed with downward V-shaped protrusions into the basal layer (Supplementary Fig. 1c and Fig. 1a). Suggestive of active basement membrane detachment, such cells were observed at all ages of stratification examined, but were most abundant at E15.5 (Fig. 2b). Intriguingly, fewer mitoses were observed at E15.5 than at E16.5, when asymmetric cell divisions become prevalent (Fig. 2c)2,17.
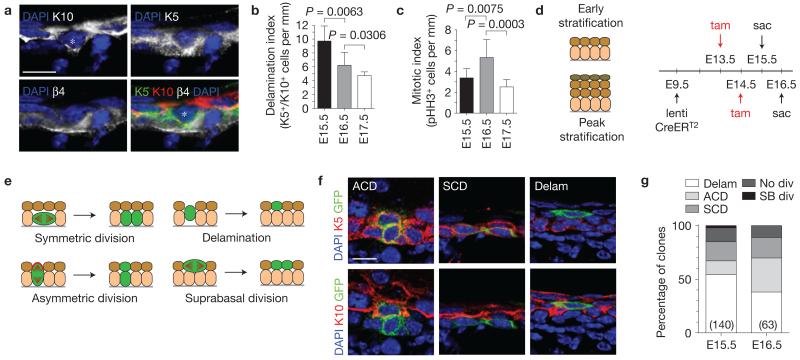
Figure 2.
Early stratification is driven by delamination. (a) Co-expression of spinous (K10) and basal (K5) keratins characterizes some basally positioned (K5+/β4-integrin+) cells at E15.5 (marked by an asterisk). (b) Quantification of K5+/K10+ double-positive basal cells during epidermal stratification, which peaks at E15.5 and decreases thereafter (n = 8 animals per age). (c) Quantification of mitotic (pHH3+) basal cell density during the same period reveals a peak at E16.5 (n = 8 animals per age). (d) Lentiviral lineage tracing strategy used to label small clones of cells during early (13.5–E15.5) and peak (E14.5–E16.5) stratification. CreERT2 is injected into Rosa26-confetti28 embryos at E9.5, and 2 mg tamoxifen (tam) is administered by gavage to induce sporadic clones. (e) Clonal analysis of lineage tracing clones following a 48 h induction period. (f) Examples of clones obtained at E15.5. Note that two-cell clones with one suprabasal K10+ cell and a basal K5+ cell represent the product of an ACD, whereas two-cell clones with 2 K5+ basal cells represent an SCD. Single K10+ cells in the first spinous layer represent the likely product of a basal cell delamination. (g) Quantification of clone types observed at E15.5 and E16.5. Delamination events predominate at E15.5 and diminish by E16.5. Among mitotic clones, SCDs outnumber ACDs at E15.5 although the converse is true at E16.5, consistent with analyses of division orientation at these ages. See Supplementary Table 2 for statistical analyses. n indicates number of clones scored from >5 animals from 3 separate litters per age. Scale bars, 10 μm. P values in b,c determined by two-tailed Student’s t-tests. Error bars in b,c represent the s.d.
Next, we employed short-term lineage tracing using ultrasound-guided in utero delivery26,27 of a lentiviral tamoxifen-inducible CreERT2 to the amniotic fluid surrounding E9.5 Rosa26-confetti28 reporter embryos. This allowed us to examine earlier time points than possible with transgenic K14::CreERT2, and to more carefully discriminate neighbouring clones owing to the four unique fluorophores expressed by marked epidermal cells and their clonal progeny. Small (1–3 cell), sporadic clones (<1% of epidermis) were induced by administering tamoxifen to pregnant females at either E13.5 or E14.5, and embryos were collected 48 h later (Fig. 2d). Reasoning that clone size and position reflected past behaviour (Fig. 2e), K5+ basal 2-cell clones were interpreted to reflect a symmetric cell division (SCD), whereas clones with one K5+ basal and one K10+ suprabasal cell suggested an ACD (Fig. 2f). At E15.5, SCDs outnumbered ACDs by a ratio of ~4:3, shifting to ACDs over SCDs (~5:3) at E16.5 (Fig. 2g and Supplementary Table 2). In agreement with a previous report22, a number of 3-cell clones consisted of two basal and one suprabasal cell, confirming that individual basal cells can divide both symmetrically and asymmetrically (Supplementary Table 2). More importantly, at both ages, single-labelled K10+ suprabasal cells were seen directly above unmarked basal cells, suggesting that they had delaminated and differentiated without division. As discussed in the Methods, it is unlikely that these cells were already suprabasal at the time they were marked. Moreover, although these single cell clones comprised the majority at E15.5 (76/140, ~54%), they were significantly less abundant at E16.5 (24/63, ~38%, P = 0.0137 by chi-square test). Together, these data support the notion that early stages of stratification (up to ~E15.5) are driven by delamination, whereas later stages increasingly rely on ACDs.
Spindle orientation genes are dispensable during early stages of stratification
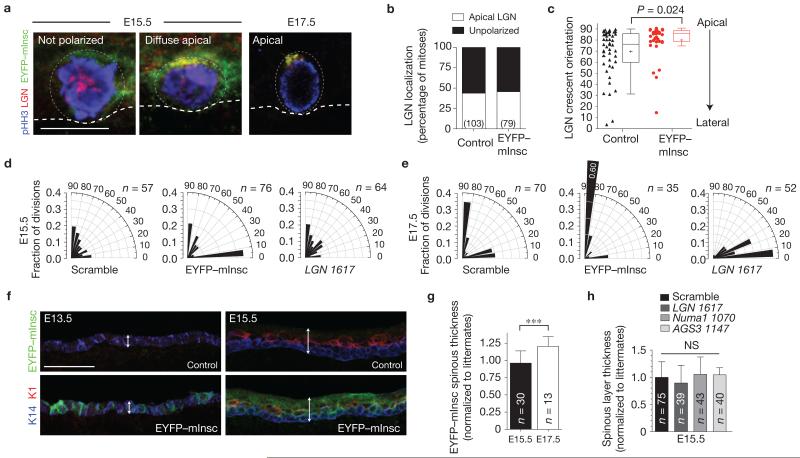
As expression of the direct binding partner of LGN, mInsc, is developmentally regulated and increases as stratification progresses22, we reasoned that the inability of LGN to polarize efficiently before E15.5 might be due to insufficient mInsc. Therefore, we used lentivirus to induce precocious mInsc expression. Validating its functionality, apical EYFP–mInsc co-localized with LGN at E16.5–E17.5. However, at E15.5, neither EYFP–mInsc nor LGN polarized efficiently, exhibiting diffuse crescents (Fig. 3a,b), although when LGN did polarize, EYFP–mInsc significantly enhanced its apical localization (Fig. 3c). Moreover, although it did not affect division angle orientation at E15.5 cells, EYFP–mInsc accentuated perpendicular divisions at E17.5 (Fig. 3d,e). Finally, although EYFP–mInsc enhanced differentiation at E17.5, it could not at E13.5 or E15.5 (Fig. 3f,g). Thus, at E15.5, when delamination provides the driving force for stratification, epidermis seems refractory to the spindle-reorienting activity of mInsc.
Figure 3.
Loss- or gain-of-function in spindle orientation genes does not alter early stratification behaviour. (a) Lentiviral overexpression of EYFP–mInsc in E15.5 (left, centre) and E17.5 (right) mitotic basal cells. Although EYFP–mInsc relocalizes LGN to the apical domain in ~90% of cells at E17.5, it is markedly less efficient at E15.5. (b) Quantification of LGN localization in control (YFP−) and EYFP–mInsc+ mitotic basal cells at E15.5, showing no significant difference between groups. (c) Quantification of LGN crescent orientation from lateral (0°) to apical (90°), similar to Fig. 1e. Although EYFP–mInsc polarizes infrequently at E15.5, it can promote apical LGN localization when present. n = 36 cells for control, n = 45 for EYFP–mInsc from 4 to 5 independent animals. Box boundaries indicate the 25% and 75% quartiles, the middle bar the median, and the plus symbol the mean. (d,e) Radial histograms of division orientation at E15.5 (d) and E17.5 (e). At E15.5, division angles are essentially randomized in controls (Scramble shRNA), as in wild-type embryos (Fig. 1f). Neither knockdown of LGN nor overexpression of EYFP–mInsc has any effect at E15.5. However, strong division reorientation phenotypes are observed at E17.5, with EYFP–mInsc promoting perpendicular divisions and loss of LGN inducing planar divisions. Chi-square P values can be found in Supplementary Table 1. n indicates cells from 4 to 6 independent animals. (f) Ectopic expression of EYFP–mInsc (green) does not accelerate differentiation at early ages. The differentiation marker Keratin 1 (K1, red) is expressed at low levels at E13.5 (left), and by E15.5 (right), K1 becomes restricted to spinous layer cells, while K14 (blue) marks basal cells. Epidermal thickness is indicated by the double arrow. (g) Quantification of spinous (K10+) layer thickness in skin transduced with EYFP–mInsc, normalized to age-matched littermates. There is no statistically significant difference between EYFP–mInsc and controls at E15.5, whereas at E17.5, EYFP–mInsc promotes spinous thickening. n indicates sections analysed from >3 animals per age. (h) Quantification of spinous layer thickness at E15.5, normalized to littermates. Knockdown of LGN, Numa1 or the LGN homologue AGS3/Gpsm1 does not affect early stratification. n indicates sections analysed from n≥4 embryos for knockdown and littermate controls. Scale bars, 10 μm (a), 50 μm (f). Error bars (g,h): s.d. P values determined by Mann–Whitney non-parametric test in c and two-tailed Student’s t-tests in g,h; *** P < 0.0001; NS, not significant.
To further explore how spindle orientation machinery contributes to early stratification, we investigated the roles of LGN, NuMA and Notch signalling, which are critical mediators of differentiation at E16.5 (ref. 17). Interestingly, although LGN loss compromised non-planar spindle orientations at E17.5, it had no effect at E15.5 (Fig. 3d,e). LGN perdurance is unlikely, because immunoblots from sorted cells confirmed efficient knockdown by E15.5 (ref. 17). Early spinous differentiation was also unaffected by LGN or Numa1 loss (Fig. 3h and Supplementary Fig. 2c). Moreover, the LGN homologue AGS3 did not seem to function as a surrogate to promote early perpendicular divisions, because spinous differentiation was unaltered following AGS3 knockdown (Fig. 3h).
Notch signalling functions downstream of spindle orientation in many systems, including skin17,29-31. Using a Notch reporter lentivirus17, we noted that Notch activity was barely detectable in differentiating K10+ suprabasal cells at E15.5, whereas at E17.5 suprabasal Notch activity was high (Supplementary Fig. 2a,b). At E17.5, ~30% of suprabasal cells were Notch+, compared with only ~5% of basal cells, mostly concentrated in nascent hair follicles. This contrasts with E15.5, where only 5–6% of cells were GFP+, and there was no suprabasal bias.
The RBPJ transcription factor functions with Notch intracellular cleaved domain (NICD) in canonical Notch target expression in epidermis32. We created skin-conditional RBPJ mutants using lentiviral Cre–mRFP1 (ref. 17) to directly examine whether initial spinous layers form independently of Notch. Spinous layers developed normally by E15.5 in LGN-1617 short hairpin RNA (shRNA) knockdowns and Rbpj knockouts (Supplementary Fig. 2c). However, although control (Scramble shRNA) epidermis continued to thicken thereafter, further differentiation was impaired following LGN or Rbpj targeting (Supplementary Fig. 2d). Thus, the same loss- and gain-of-function mutations that affect division orientation and stratification at E16.5 and beyond show little impact on earlier stages, pointing to E15.5 as a critical period when basal cells acquire a competency to use the spindle orientation machinery to promote differentiation.
LGN and mInsc play distinct roles in spindle orientation
In many systems, including Drosophila neuroblasts and distal lung bud epithelium, Insc, Pins and their homologues play similar roles in spindle orientation30,33-35. In others, such as murine neural progenitors, mInsc loss has opposite and more severe effects than LGN loss on spindle orientation and cell fate decisions36. Thus, we were surprised to find that conditional deletion of mInsc in epidermis using transgenic K14::Cre caused no overt abnormalities in either epidermal morphology or barrier function.
LGN could still form apical crescents in mInsc conditional knockout (cKO) animals (Fig. 4a, top left), albeit with reduced efficiency, which may explain why loss of mInsc does not phenocopy LGN loss. However, aberrant LGN localization was evident in about half of the mitotic basal cells. In some instances, cortical recruitment of LGN failed; in others, LGN was cortical but either evenly distributed or polarized at abnormal locations (Fig. 4a,b). Similar results were observed with a mInsc hairpin (Fig. 4b and Supplementary Fig. 3). Following mInsc loss, a high proportion of crescents formed at oblique rather than apical orientations (Fig. 4c,d). Abnormal LGN localization was even observed at telophase, resulting in a failure of LGN to segregate into the appropriate apical daughter (Fig. 4e). These findings reveal some interesting differences between murine epidermis and Drosophila neuroblasts. Though Pins/LGN localization defects are more severe in insc mutant neuroblasts33,34, paradoxically, apical complex localization defects are rescued in telophase fly insc mutants6,9, but not in epidermis.
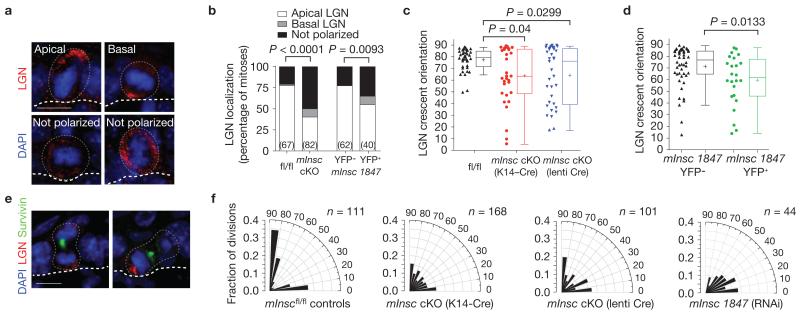
Figure 4.
mInsc cKOs show impaired LGN localization and randomized spindle orientation. (a) In E17.5 mInsc cKOs, LGN localizes apically as in wild-type cells in a minority of cases (top left), but more frequently is mislocalized in one of three ways: mispositioned basal crescents (top right), weakly expressed (bottom left), or cortical but unpolarized (bottom right). (b) Quantification of LGN localization patterns in E17.5 mitotic basal cells. LGN is nearly always apical in control cells (76%), whereas this is less frequently observed (40%) in mInsc cKOs. Similar results are observed in mInsc knockdowns, comparing transduced (H2B–YFP+) with non-transduced internal (H2B–YFP−) controls. n (cells from >4 independent animals) indicated in parentheses. (c) Dot and Tukey box-and-whisker plots of LGN crescent orientation in E17.5 controls and mInsc cKOs. LGN crescents show a strong apical bias in controls, but are more broadly distributed in mInsc cKOs, regardless of whether transgenic K14–Cre (middle, red) or lentiviral-delivered Cre–mRFP1 (right, blue) is used. (d) LGN crescent orientation in mInsc-1847 shRNA H2B–YFP+ (right, green) and H2B–YFP− internal controls (left, black), as in c. (e) Abnormal LGN orientation persists into telophase in mInsc cKOs. Examples show LGN segregating equally into both daughter cells even in a perpendicular division (left), or erroneously segregating into the basal daughter (right). (f) Radial histograms of division angle orientations at E16.5–E17.5. Littermate mInscfl/fl controls (left) show a typical, largely ‘bimodal’ distribution with most divisions occurring close to perpendicular (~55–60%) or planar (30–35%) and few at oblique angles. When mInsc is deleted using either transgenic or lentiviral Cre, or knocked down by mInsc-1847 shRNA, division orientation becomes more randomized, with a higher proportion of oblique division angles. n indicates cells from 4 to 6 independent animals. Scale bars in a,e are 10 μm. P values were calculated by chi-square tests (b), or Mann–Whitney tests (c,d). Chi-square P-values related to (f) can be found in Supplementary Table 1. In c,d each data point represents one cell; data collected from n > 4 independent animals. Box boundaries indicate the 25% and 75% quartiles, the middle bar the median, and the plus symbol the mean.
On examining division orientation, we discovered that in contrast to mInscfl/fl controls, mInsc-null or -knockdown basal cells exhibited many oblique division angles (Fig. 4f). This increase in oblique divisions is highly significant (P < 0.0001 by chi-square test), and was observed throughout stratification, irrespective of whether transgenic K14::Cre or lentiviral Cre–mRFP1 was used. Thus, basal cells lacking mInsc behaved in many ways like immature (E14.5–E15.5) wild-type basal cells, underscoring the important role of mInsc in refining random divisions towards the mature bimodal pattern. However, mInsc is not necessary for perpendicular divisions to occur, and is sufficient only during peak stratification.
Loss of Pard3 phenocopies mInsc
In both Drosophila and mammalian cells including keratinocytes, Insc binds to Par3. Moreover, in fly neuroblasts, baz (Par3), pins (LGN) and insc (mInsc) mutants exhibit similar phenotypes of abnormal spindle orientation and mislocalized apical and basal polarity components2,16,33,34,37,38. In mammalian simple epithelia, Par3 is an important regulator of apicobasal polarity, as demonstrated in three-dimensional Madin-Darby canine kidney (MDCK) cell cultures, where Par3 knockdown causes defective lumen formation and spindle orientation39. That said, a germline knockout of Pard3, although embryonic lethal owing to epicardial defects, shows normal epithelial architecture in most tissues examined, including stratified epithelia40. Given these findings on Par3 and our own with mInsc, we wondered whether loss of Pard3 might cause alterations in spindle orientations and epidermal differentiation in vivo that could have been overlooked previously.
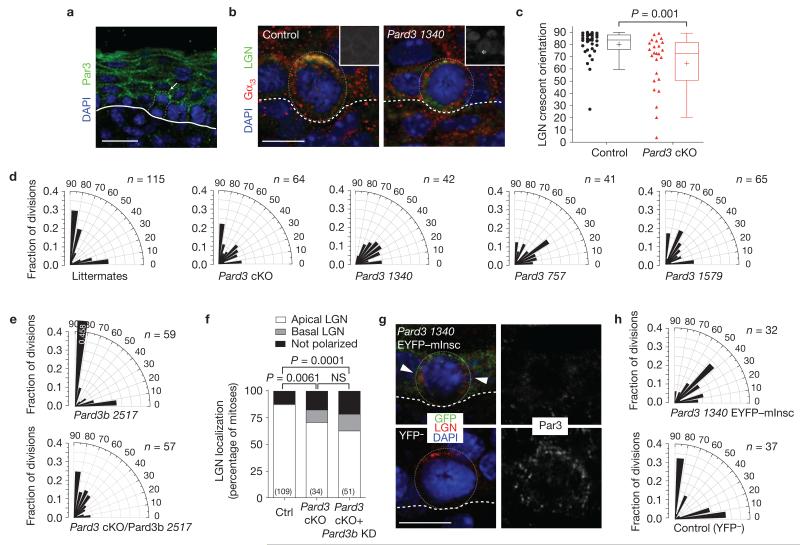
Par3 is enriched apically in basal epidermal cells2, whether in interphase or mitosis (Fig. 5a,g). To assess the consequences of Pard3-deficiency on epidermal development, we generated lentiviral Cre–mRFP1 Pard3 cKOs as well as three independent knockdown lines. Immunofluorescence microscopy of skin sections from both cKO and knockdown embryos revealed that Par3 was selectively lost in transduced regions (Supplementary Fig. 3c,d). As with mInsc cKOs, Pard3-depleted epidermis exhibited LGN localization defects and increased frequency of oblique division angles (Fig. 5b–d).
Figure 5.
Pard3 knockout or knockdown phenocopies loss of mInsc. (a) Par3 expression in E17.5 epidermis. Dotted line depicts outline of telophase cell undergoing a perpendicular division. Arrow denotes enrichment of Par3 in apical daughter. (b) Knockdown of Pard3 frequently results in mislocalization of LGN and Gαi3 away from the apical cortex. Insets show RFP expression, with the asterisk marking the outlined mitotic cell. (c) Dot and Tukey box-and-whisker plots of LGN crescent orientation in E17.5 control (red) and Pard3 cKO mitotic basal cells (black), showing reduced apical bias (compare with Fig. 4c,d). n > 25 cells per genotype from n > 5 independent animals. Box boundaries indicate the 25% and 75% quartiles, the middle bar the median, and the plus symbol the mean. (d) Radial histograms depicting division angle orientation in E17.5 controls (left), Pard3 cKOs (second panel) and Pard3 knockdowns (third–fifth panels). Loss of Pard3 results in an increase in oblique division angles and reduction in perpendicular divisions. (e,f) Knockdown of Pard3b on a Pard3 cKO background. (e) Radial histograms depicting division angle orientation following loss of Pard3b alone (top) or following combined loss of Pard3/Pard3b (bottom). Pard3b loss has no apparent phenotype on its own and does not enhance the Pard3 phenotype. (f) LGN localization errors are observed in both single (Pard3 knockout) and double (Pard3 knockout/Pard3b knockdown) mutants, but at similar frequency. (g) When EYFP–mInsc is expressed on a Pard3 knockdown background (top), it fails to be apically polarized as efficiently as in Scramble shRNA controls (Supplementary Table 2). LGN (arrowheads) also frequently fails to co-localize with ectopic mInsc. Both Par3 and LGN are generally apically enriched in YFP− controls (bottom). (h) The ability of EYFP–mInsc to promote perpendicular divisions (Fig. 2e) requires Pard3. Radial histograms of division orientation at E17.5 reveal that Pard3 1340 EYFP–mInsc basal cells (top) show a similar propensity towards oblique divisions as Pard3 mutants (d). Scale bars, 20 μm in a, 10 μm in b,g. P values in were determined by Mann–Whitney test (c) or by chi-square (f). NS, not significant. Chi-square P values related to d,e,h can be found in Supplementary Table 1. n values in d–f,h represent cells from 4 or more animals.
Humans and mice have a second Par3 gene, termed PAR3L or Pard3b (refs 41,42). To test for possible compensation, we generated a double mutant (Pard3 cKO/Pard3b knockdown (KD)), by transducing Par3fl/fl embryos with a lentivirus harbouring both Cre–mRFP1 and Pard3b-2517 shRNA, which reduced Pard3b messenger RNA levels by >99%. As in Pard3 cKO epidermis, LGN localization was frequently abnormal in the double mutant; however, Pard3b-deficiency did not significantly exacerbate the Pard3 phenotype (Fig. 5e,f). Moreover, Pard3b shRNA knockdown alone showed no division angle errors (Fig. 5e). These data argue against functions for Pard3b in controlling oriented epidermal divisions or acting redundantly with Pard3.
To test whether similarities in mInsc and Pard3 phenotypes reflected a critical role for Par3 in promoting apical localization of mInsc, we overexpressed EYFP–mInsc while knocking down Pard3. This strategy circumvented the hurdle we faced in identifying a suitable antibody for endogenous mInsc. Although EYFP–mInsc was effective in recruiting LGN to the apical cortex of wild-type (Scramble shRNA) E17.5 epidermis (Fig. 3a), without Par3, neither EYFP–mInsc nor LGN showed efficient apical cortical recruitment (Fig. 5g and Supplementary Table 3). Moreover, LGN and mInsc co-localized in only 28% of pHH3+ mitotic cells lacking Par3, compared with 89% of controls. Finally, division orientation in Pard3 EYFP–mInsc basal cells mirrored that of Pard3 mutants (Fig. 5h), demonstrating that mInsc requires Par3 to reorient spindles efficiently.
Loss of Gnai3 but not Gnai2 results in cell division defects and impaired differentiation
LGN interacts with mInsc and NuMA through its N-terminal TPR motifs, whereas its C-terminal GoLoco motifs bind heterotrimeric G proteins of the Gαi and Gαo family15,16,43,44. Mutations in C. elegans Gαi homologues goa-1/gpa-16, or the single Drosophila homologue Gαi (G-iα65A) show spindle orientation defects resembling those of LGN homologue loss (gpr-1/2 or pins; refs 45–49). Although indirect evidence from mouse neural progenitors supports a role for Gβγ subunits50, functional studies on mammalian Gαi proteins are lacking to link these proteins to oriented cell divisions.
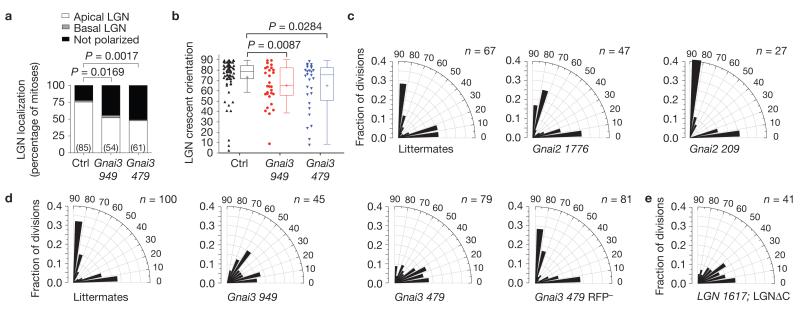
Although all three murine Gαi homologues are expressed in keratinocytes, only Gαi3 localized to the cell cortex in mitotic cells (Fig. 5b and Supplementary Fig. 3f). In vitro, lentiviruses targeting Gnai3 (test) or Gnai2 (control) achieved >97% knockdown of their messenger RNA targets (Supplementary Fig. 3c). In vivo, Gnai3-479 shRNA achieved more complete knockdown than Gnai3-949 shRNA, which occasionally left some residual cortical Gαi3 (Supplementary Fig. 3e,f). In both Gnai3 knockdowns, LGN was frequently mislocalized, either failing to be recruited to the cortex, or forming weaker apical and/or ectopic crescents (Figs 6a,b and 7c,d).
Figure 6.
Gαi3 promotes apical LGN localization and perpendicular divisions. (a) Following Gnai3 knockdown, a significantly higher proportion of basal cells mislocalize LGN. n values represent cells from 3 to 5 animals. (b) Dot and Tukey box-and-whisker plots of LGN crescent orientation in Gnai3-949 shRNA (red, n = 28 cells) and Gnai3-479 shRNA (blue, n = 28) mitotic basal show a weaker apical bias in knockdowns compared with controls (black, n = 64). Data were pooled from 4 independent experiments. Box boundaries indicate the 25% and 75% quartiles, the middle bar the median, and the plus symbol the mean. (c,d) Radial histograms of division angle orientation in E17.5 Gnai2 (c) and Gnai3 (d) knockdowns. Both Gnai3 hairpins result in fewer perpendicular and more oblique division angles, with the more severe phenotype being observed with the stronger Gnai3-479 shRNA. Note the specificity and cell autonomy of the division orientation defect, as a normal distribution of division angles is observed in RFP− basal cells in mosaic Gnai3-479-shRNA-transduced embryos (d, far right). n indicates cells from >4 animals per condition in 2–3 independent experiments. (e) Replacement of endogenous LGN with a C-terminal deletion mutant lacking the Gαi -binding GoLoco motifs results in a division angle profile very similar to loss of Gαi3 , demonstrating that this interaction is critical for the apical cortical localization of LGN. n indicates cells analysed from 5 animals in 2 independent experiments. P values were calculated by chi-square (a), or Mann–Whitney tests (b). Chi-square P values related to c–e can be found in Supplementary Table 1.
Figure 7.
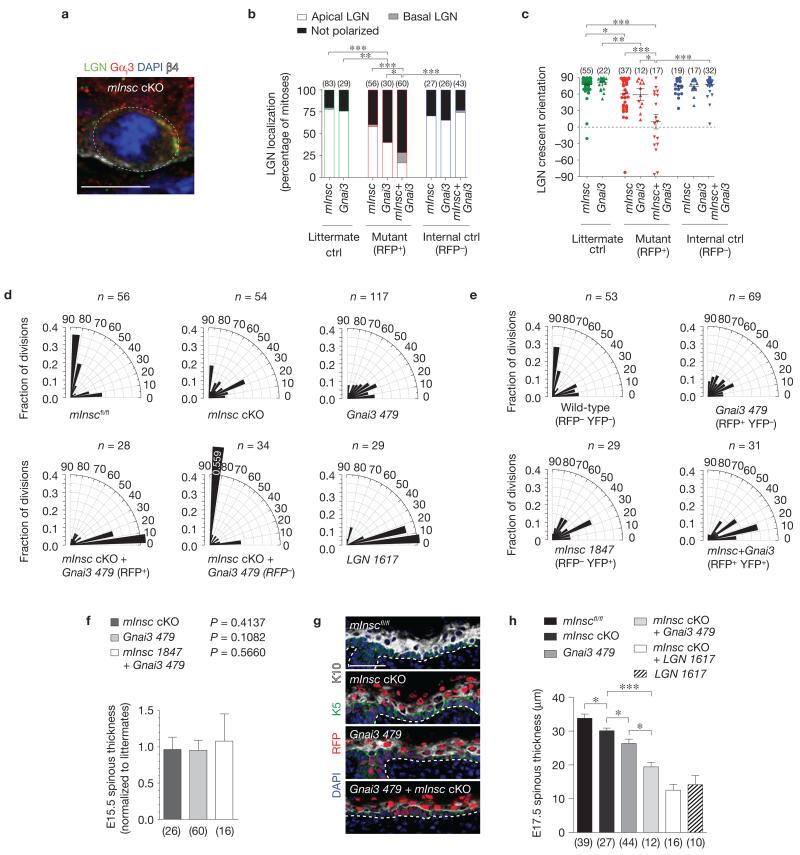
Gαi3 and mInsc act cooperatively to localize LGN to the apical cortex. (a) Co-localization of LGN and Gαi3 in E16.5 mInsc cKO mitotic basal cells. Where LGN is polarized (n = 59 of 79 cells), Gαi3 co-localizes in the same cortical domain in 88% of cases, even when in ectopic locations as in b. (b,c) LGN localization at E17.5 becomes progressively impaired following loss of mInsc, Gnai3, and combined loss of mInsc+Gnai3. Littermates are shown in green, knockdown (RFP+) in red, and internal controls (RFP−) in blue. There is near complete loss of apical LGN in mInsc+Gnai3 mutants, and the few cells that can polarize LGN do so randomly. n indicates cells per condition from 3 to 6 independent animals. Mean ± s.e.m. shown in black. (d) Division angle orientation in E17.5 back skin (n indicates cells from 3 to 6 animals per group). Loss of mInsc or Gnai3 alone induces oblique divisions, whereas mInsc + Gnai3 loss shows a more severe phenotype of mostly planar divisions, as seen in LGN knockdowns. RFP− internal controls in the double mutant show a compensatory increase in perpendicular divisions. (e) E17.5 division orientation in embryos transduced with a mixture of Gnai3-479 shRNA H2B–RFP and mInsc-1847 shRNA H2B–YFP (n = cells from 4 animals). Double-knockdown (RFP+ YFP+) basal cells show an enhanced bias towards planar divisions as in mInsc cKO; Gnai3-479 shRNA basal cells. (f) As with LGN, Numa1 and AGS3 (Fig. 3h), loss of mInsc and Gnai3 alone or in combination does not alter early stratification, as measured by E15.5 spinous thickness. n indicates sections analysed from >4 animals per group. (g,h) Spinous differentiation (K10, white) in E17.5 head skin. Small but significant decreases in differentiation are observed in single mutants, whereas defects in mInsc; Gnai3 double mutants are comparable to LGN knockdowns. Note that loss of mInsc + LGN does not further enhance the LGN phenotype, consistent with LGN being epistatic to mInsc. n indicates sections analysed from >4 animals per genotype. Scale bars, 10 μm in a, 50 μm in g. Error bars: s.d. (f,h). P values: * P < 0.05, ** P < 0.01, *** P < 0.0001, as determined by chi-square (b), Mann–Whitney test (c) or two-tailed Student’s t-tests (f,h). Chi-square P values related to d,e are in Supplementary Table 1.
Division angles were unaffected by Gnai2 loss, but were altered in Gnai3 knockdowns (Fig. 6c,d), with a median division angle of 30° for Gnai3-479 shRNA (stronger hairpin) and 38° for Gnai3-949 shRNA (weaker hairpin). The bias towards oblique and planar divisions was stronger for Gnai3 knockdowns than for Pard3 (median division angle = 48°) or mInsc cKOs (46°), but weaker than LGN knockdowns (19°). Although this might reflect a more important role for Gαi3 over mInsc in apical cortical recruitment of LGN, it may also in part be attributable to differences in the interaction stoichiometry (mInsc::LGN is 1:1, whereas Gαi–GDP::LGN is 4:1; ref. 13).
To probe the importance of the Gαi–LGN interaction further, we used an LGN mutant that lacks its C-terminal GoLoco motifs (LGNΔC), predicted to be incapable of binding Gα subunits. A genetic knock-in mouse engineered in this way showed spindle orientation defects in developing forebrain, similar to what was observed using LGN RNAi (ref. 19). We showed previously that this mutant was probably hypomorphic, as it can partially rescue differentiation defects caused by LGN knockdown17. Expression of LGNΔC in place of endogenous LGN resulted in a phenotype remarkably similar to Gnai3 knockdown, with many oblique and planar divisions, and a median division angle of 31° (Fig. 6e). These data underscore the importance of the interaction of Gαi3 with LGN for its proper apical localization.
Combined loss of Gnai3 and mInsc phenocopies LGN knockdown
Genetic data from Drosophila neuroblasts support a model whereby Gαi and Pins operate in one arm of the spindle orientation pathway and Insc and baz act in another48,49,51. Given these similar functions in Drosophila, it was surprising that in mammalian epidermis, Gnai3 loss produced a weaker phenotype than LGN loss. As our results with other Gαi proteins did not support redundancy, we turned to the possibility that mInsc, which can also bind LGN at its N terminus, might act cooperatively with Gαi3 to polarize LGN. Examining Gαi3 expression in mInsc mutants, we found that in mitotic basal cells where LGN remained polarized, Gαi3 co-localized with LGN in 88% of cases (52/59 cells), even when LGN localized to ectopic sites (Fig. 7a).
As Gαi3 was capable of polarizing LGN independently of mInsc, we next created a double mutant of Gnai3 with mInsc by injecting mInscfl/fl embryos with a lentivirus dually harbouring Gnai3-479 shRNA and Cre–mRFP1. As with K14::Cre mInsc cKOs (Fig. 4b), the percentage of mitotic basal cells with unpolarized LGN increased in lenti-Cre mInsc cKOs (40%) compared with mInscfl/fl littermates (20%, Fig. 7b). The stronger Gnai3-479 shRNA hairpin increased this fraction further (60%), and in the double mutant, 86% of basal cells showed LGN localization errors. Examination of residual LGN crescent orientation (Fig. 7c) revealed weakening of apical bias in both mInsc and Gnai3 single mutants (mean orientation angle of 60° and 59°, respectively), whereas LGN was essentially randomized in the double mutant (mean orientation of 6°). In addition, nearly all divisions were planar in double mutants (median angle = 11°), akin to loss of LGN (median angle = 13°, Fig. 7d). In embryos co-infected with a mixture of mInsc-1847 shRNA H2B–YFP and Gnai3-479 shRNA H2B–RFP, doubly transduced cells (RFP+YFP+) exhibited a phenotype similar to mInsc cKO; Gnai3 knockdown (Fig. 7e).
Finally, we examined the impact of altering division orientation on spinous differentiation. As with LGN, Numa1 and AGS3 knockdowns, loss of mInsc, Gnai3 or both had no effect on K10 expression at E15.5, further arguing that the spindle orientation pathway is dispensable at this time (Fig. 7f). At E17.5, mInsc cKOs showed a significant but mild decrease in spinous differentiation compared with mInscfl/fl controls. Gnai3 knockdowns had a more severe defect, but there was the most profound deficit in double mutants (Fig. 7g,h). These genetic data are consistent with mInsc and Gnai3 acting in parallel pathways that converge on LGN. In further support of LGN being epistatic to mInsc, the double mutant phenocopies loss of LGN alone. Although LGN knockdown embryos showed further thinning of spinous layers compared with mInsc; Gnai3 mutants, this could be explained by residual cortical LGN expression (Fig. 7b) and/or a compensatory increase in perpendicular divisions in RFP− cells within these mosaic embryos (Fig. 7d). It is also formally possible that an additional pathway could converge on LGN involving another protein other than mInsc or Par3.
DISCUSSION
A two-step mechanism for epidermal stratification and differentiation
Our studies provide several insights into how oriented cell divisions function in mammalian stratified epithelial development. First, we show that vertical divisions are quite rare at early stages of epidermal stratification (E14.5–E15.5). Surprisingly, none of the classical spindle orientation genes is required for establishing the first spinous layers, which instead rely on delamination to promote differentiation. We unearth the existence of a subsequent discrete step in epidermal maturation that involves competency of basal cells to polarize mInsc and LGN, undergo perpendicular divisions and execute Notch signalling.
Further, we provide insights into mechanisms by which LGN is recruited to the apical cortex through the combined activity of Par3–mInsc and Gαi3 (Fig. 8). Whereas knockdown of LGN leads primarily to planar divisions, loss of either Pard3, mInsc or Gnai3 alone results in a weaker phenotype with a high incidence of oblique divisions, mediated by errors in LGN polarization. Moreover, combined loss of both Gnai3 and mInsc recapitulates the more severe LGN phenotype, demonstrating that these pathways converge on LGN in epidermal oriented divisions, rather than on NuMA/Mud, as in Drosophila neuroblasts.
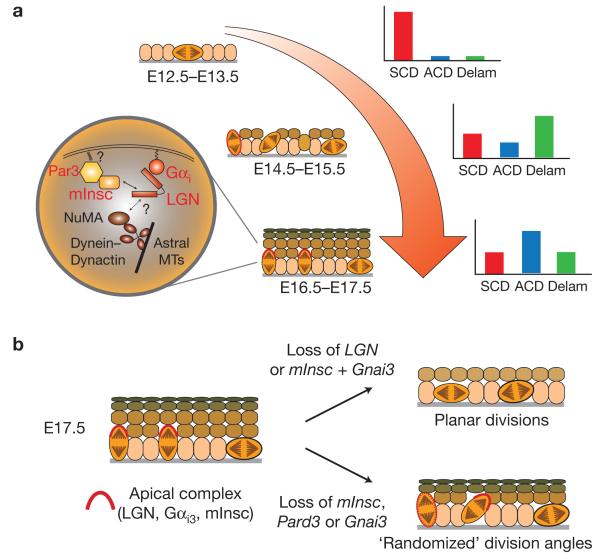
Figure 8.
Model of epidermal stratification mechanisms. (a) Schematic diagram depicting stages of epidermal stratification from the early single-layered epithelium (E12.5–E13.5) through formation of the first suprabasal spinous layers (E14.5–E15.5) and into the peak stage of stratification (E16.5–E17.5). The relative contributions of planar symmetric divisions (SCDs, red), perpendicular asymmetric divisions (ACDs, blue) and basal cell delamination events (Delam, green) are shown in the schematic graphs at the right. The orientation of mitotic figures are shown by the orange mitotic cells, and the localization of the apical complex consisting of LGN, Gαi3 and mInsc is shown as a red crescent. When delocalized, as at E14.5, it is depicted as a dashed oval. In the circular inset, a schematic of the spindle orientation pathway is shown. The uncertainty around how Gαi and mInsc impact the interaction of LGN with NuMA is indicated by a question mark, as is the source of the cue that localized Par3 to the apical cortex. (b) Summary of phenotypes observed on compromising LGN, or the protein complexes that regulate its apical localization. (Left) the normal E17.5 fully stratified epidermis, where perpendicular divisions outnumber planar ones by roughly 2:1, and the LGN complex is localized to the apical cortex of most perpendicularly oriented dividing cells. (Right, top) Loss of LGN or mInsc+Gnai3 leads to severe thinning of the epidermis, with largely planar divisions. (Right, bottom) Loss of Par3, mInsc or Gnai3 alone leads to LGN localization defects, an increase in oblique divisions, and mild differentiation defects.
Unique and shared functions of mInsc, LGN and Par3 function in spindle orientation
Our findings on E12.5→E15.5 embryonic mammalian epidermis resemble those of E11 mammalian neurogenesis. Radial glia progenitor divisions transition from planar (symmetric) to oblique (asymmetric; refs 52,53). However, loss or gain of mInsc function has no apparent effect on radial glia division orientation at E11.5 (ref. 36), as with epidermal basal cells before E15.5. That said, in radial glia, loss of mInsc at E14.5 reduces oblique divisions, whereas transgenic overexpression of mInsc–GFP promotes them. Thus, in both neural and epidermal progenitors, the function of mInsc is developmentally restricted, but in the former, it promotes oblique divisions but in the latter it suppresses them.
LGN, on the other hand, promotes planar neurogenic divisions in developing cortex and neural tube, and its loss increases oblique divisions19,20,54. These apparent discrepancies could be explained by differences in the subcellular localization of these proteins in different cell types. In neural progenitors, mInsc and LGN are thought to localize to exclusive domains—LGN laterally and mInsc apically—which could explain their opposing effects on division orientation 4,55. In epidermis, both are enriched apically, such that loss of either gene reduces perpendicular divisions. We propose that the ability of Gαi3 to partially recruit LGN apically probably accounts for why mInsc loss results in a less severe epidermal phenotype than LGN loss.
Recent structural studies of Insc–Pins(LGN) and Pins–Mud(NuMA)–Gαi–GDP complexes have revealed that LGN cannot bind Insc and NuMA simultaneously, and that Insc has a higher affinity for LGN than NuMA (refs 13–16). This favours a model whereby early in mitosis, Par3–mInsc and Gαi3 promote apical recruitment of LGN—facilitating its transfer to NuMA—and LGN–NuMA then acts to reorient the mitotic spindle, analogous to passing a baton. As LGN binds mInsc and NuMA through its N-terminal TPR repeats and Gαi–GDP through its C-terminal GoLoco motifs, it seems likely that these proteins can form a tripartite complex, although structural data are lacking because this complex has been refractory to crystallization13. Future work will be required to determine the mechanism by which NuMA displaces mInsc from the LGN complex, and whether Gαi facilitates this displacement by altering the interaction of LGN with NuMA.
Like mInsc, Par3 functions in promoting asymmetric cell fates and Notch signalling in cortical neuronal progenitors31. Whether Par3 does so by altering the plane of division of radial glia remains unknown. Thus, to our knowledge, ours is the first study to directly show that mammalian Par3 influences spindle orientation. A recent study in fly intestine reveals several interesting parallels to murine epidermis56. Like basal cells, intestinal stem cells polarize Baz/Par3, Par6 and aPKC to their apical cortex at mitosis and Par3 promotes non-planar (differentiative) divisions. Moreover, as with skin, integrins are required for proper division orientation2, and overexpression of active Notch can rescue differentiation defects caused by altered spindle orientation17. It is interesting to note, however, that epidermal knockout of Prkci, one of two mammalian atypical protein kinase C genes, seems to result in increased perpendicular, but not oblique divisions57. Although the protein encoded by Prkci, aPKCι/γ, localizes to the apical cortex, Par3 localization is unaffected by its loss, suggesting that the Par3–mInsc–LGN spindle orientation pathway remains intact in Prkci mutants. It will be interesting to see whether redundancy with the other mammalian aPKC homologue, Prkcz, or alternative spindle orientation pathways are at play here.
Spindle orientation and cell fate
Spindle orientation can influence binary cell fate choices4,58. Indeed, we previously demonstrated a functional link between spindle orientation and epidermal cell fate acquisition by showing that LGN and NuMA promote perpendicular divisions, and when depleted, fewer suprabasal (differentiating) and more basal (self-renewing) cells are generated17. As we did not observe pronounced effects on differentiation in mInsc, Par3 or Gnai3 mutants, this suggests that many oblique divisions, like perpendicular ones, may be operationally asymmetric. Although Notch signalling lies downstream of ACDs in developing epidermis, as it does in many other systems31,56,59,60, fate determinants that segregate asymmetrically between suprabasal and basal daughters have yet to be identified. It is possible that they may not exist, as loss of adhesion to basement membrane or to other cells may be sufficient to trigger differentiation, as demonstrated by our findings on delamination in early stratification. Clarification of these important questions must await elucidation of fate determinants and/or live imaging approaches in embryonic skin similar to those that have been recently reported for adult hair follicles61. □
METHODS
Animals
Mice were housed in an AAALAC-certified animal facility and all procedures were performed using IACUC-approved protocols. Wild-type CD1 mice (Charles River) were used for most experiments except in the case of conditional (floxed) allele lines, which were maintained on a mixed 129/C57Bl6 background. Pard3 (refs 40,62), mInsc (ref. 36) and Rbpj (ref. 63) mice have been reported previously. Females were maintained as homozygous fl/fl, bred with homozygous or heterozygous males, and were injected with lentiviral Cre–mRFP1 (ref. 17) to conditionally delete the gene of interest. The procedure for producing, concentrating, and micro-injecting lentivirus into the amniotic fluid surrounding E9.5 embryos has been described previously17,27.
Constructs and RNAi
For each gene targeted, between five and ten shRNAs were screened for knockdown efficiency in primary mouse keratinocytes. Clones were obtained from The RNAi Consortium (TRC) Mission shRNA library (Sigma) versions 1.0, 1.5 and 2.0, present in either the pLKO.1–puro or TRC2–pLKO–puro vector. Alternately, shRNAs were cloned from complementary annealed oligonucleotides from sequences obtained from the TRC or RNAi consortium/Codex databases, with AgeI and EcoRI linkers. We use the following nomenclature for identifying shRNA clones: the gene name, hyphen, the number of the nucleotide base (NCBI Accession number) where the 21-nucleotide target sequence begins. Lentivirus was packaged in 293FT cells (Invitrogen) using the second-generation helper plasmids pMD2.G and psPAX2 from D. Trono (EPFL, Lausanne, Switzerland; Addgene plasmids numbers 12259 and 12260). Keratinocytes were transduced at an MOI ≈1 using ~25 μl of lentiviral supernatant collected in UltraCulture (Lonza) + 10% chelated FBS + 5 μg ml−1 Polybrene. Infections were typically performed in 6-well plates seeded with 150,000 keratinocytes, which were centrifuged at 1,100g for 30 min at 37 ° C, then transferred to E low calcium medium64. Two days following infection, 1 μg ml−1 puromycin was added to generate stable cells lines. At 4 days following the addition of puromycin, cells were lysed and RNA isolated using the Absolutely RNA Microprep (Agilent). cDNA was generated from ~10–100 ng of total RNA using Superscript VILO (Invitrogen). Levels of mRNA knockdown were quantified by RT–qPCR (Applied Biosystems 7500 Fast or 7900HT) using two independent primer sets for each target gene, and Hprt1 and cyclophilin B (Ppib2) as reference genes. cDNA from stable cell lines transduced with the non-targeting Scramble shRNA hairpin was used as a reference control. Primer efficiencies were calculated by dose–response curves and were required to be >1.8, and relative abundance was calculated using the ΔΔCT method. RT–qPCR runs were performed in triplicate; mean knockdown values were determined by calculating the geometric mean of ΔΔCT values for at least 2 runs (>6 samples). The following primer sequences were used: Pard3 (5′-GA CAAAGCCGGAAAGGATAAGAAG-3′, 5′-CATGCCCCCATACAGCAACTC-3′; 5′-CGGCAGAACATTGGGGTTGT-3′, 5′-TTCTCGCGTTGGGACAGTTGT-3′), Pard3b (5′-CACCTCCAGGCGAAATGATAGTAGT-3′, 5′-GATCCGCCAGCGA CTGAACT-3′; 5′-CAGGCAATGAAATCTCCCAGTGT-3′, 5′-TCAGGGCCTTTC TTTAGGTCAATC-3′), Gnai2 (5′-CTTTGGCCGCTCACGAGAATA-3′, 5′-TAA GCGCTCAAGGCGACACA-3′; 5′-AGGCCACAGTCCTTCAGGTTGT-3′, 5′-GT CGCCTTGAGCGCTTATGAC-3′) Gnai3 (5′-ATGCCCGACAGTTATTTGTTTTA GC-3′, 5′-TCGGATCTTTGGCCACCTACA-3′; 5′-TGCTCGGCGCTGGAGA AT-3′, 5′-ATAACTGTCGGGCATCATCTGCT-3′); mInsc (5′-AGCCGTGGTA GCCCAGGTTAC-3′, 5′-AGGCCTCTTGGCACAGTTTGAT-3′; 5′-GGCCCTCA CACGGGAAGTG-3′, 5′-CTGCCGGCAGACCTTGAAGA-3′). Sequences for Scramble, LGN/Gpsm2, Hprt1 and Ppib2 primers have been described previously17.
Lentiviral lineage tracing
CreERT2 from R26R–CreER (Addgene 12168, courtesy of T. Jacks, MIT, Cambridge, Massachusetts, USA) was cloned by PCR into pLKO PGK MCS (ref. 17). In our hands, this construct was found to have no detectable leakiness in the absence of tamoxifen both in vitro and in vivo. As a reporter for Cre activity, the R26R-confetti mouse28 (Jax strain 017492, B6.129P2-Gt(ROSA)26Sortm1(CAG−Brainbow2.1)Cle/J) was chosen, as Cre-mediated recombination at the locus results in constitutive expression of one of 4 different fluorophores. This allowed us to unambiguously discriminate neighbouring clones in rare instances where multiple labelled cells were found nearby. The fluorescent signal was detected using a polyclonal antibody against GFP, which recognizes 3 of the 4 fluorophore variants present in the confetti cassette: membrane-tethered Cerulean (mCFP), nuclear GFP (nGFP), and cytoplasmic YFP, which could then be discriminated on the basis of their subcellular localization. High-titre viruses containing the CreERT2 cassette were transduced into E9.5 embryos derived from a R26R-confetti homozygote male crossed with CD1 females. Clonal recombination was induced by administering a single dose of tamoxifen (50 μg g−1 of dam weight) by oral gavage, as intraperitoneal injection of tamoxifen at doses sufficient to induce recombination frequently led to aborted litters in our hands. Forty-eight hours following tamoxifen administration was empirically determined to be sufficient to allow most clones labelled to consist of 1–3 cells. Tamoxifen was administered at E13.5 to monitor early stratification, and at E14.5 for peak stratification. Although CreERT2 was delivered at the time when the epidermis was a single layer of basal cells, it will be expressed in all of their descendants owing to its ubiquitous PGK promoter. Thus, we did observe periderm clones at both ages, as well as granular layer cells. These were excluded from our counts because it is likely that these cells were already suprabasal at the time the labelling was initiated. Only cells present in the first spinous layer were counted as it is unlikely that they were already at a suprabasal position at the time of labelling given the rapid rate of differentiation at this age. Further, the epidermis in regions where clones were counted (head and upper back skin) is still largely a single layer of basal cells (plus periderm) at the ages (E13.5–E14.5) when tamoxifen was administered. Clones were imaged as z-stacks, with basal and suprabasal cells identified as K5+ and K14+ , respectively, and were scored independently by two observers, with ambiguous or discordant clones discarded. Clones were scored from 3 to 6 embryos, arising from 2 to 3 independent experiments at each age.
Antibodies, immunohistochemistry and imaging
Embryos were either embedded whole (<E16.5) or skinned and flat-mounted on Whatmann paper. For the latter, both head and back skin were collected and both control and knockdown skins were mounted together in single OCT (Tissue Tek) blocks to reduce potential variability due to differences in immunostaining conditions between slides. When whole embryos were used, control and knockdown embryos were collected on the same slide and stained together. Skin/embryos were embedded unfixed, sectioned at 8 μm on a Leica CM3050S or CM1950 cryostat and mounted on SuperFrost Plus slides (Fisher Scientific). Sections were fixed for 10 min at room temperature in 4% EM grade paraformaldehyde (Electron Microscopy Sciences), rinsed several times with PBS, and blocked for 1 h in PBS + 5% normal donkey serum (NDS) + 1–3% BSA + 0.2% Triton-X 100 (with or without 8% gelatin). Primary antibodies were diluted in block and incubated at 4 ° C overnight. After rinsing with PBS, secondary antibodies were added for 2 h at room temperature, rinsed with PBS, stained 5 min with DAPI, and then mounted in ProLong Gold antifade mounting medium (Invitrogen). Primary antibodies used: monoclonal rabbit anti-survivin clone 71G4B7 (Cell Signaling 2808S, 1:1,000), chicken anti-GFP (Abcam ab13970, 1:5,000), affinity-purified GP anti-LGN (Fuchs laboratory17, 1:500), rabbit anti-LGN (Fuchs laboratory, licensed to EMD Millipore ABT174, 1:2,000), Rb anti-Gαi3 (EMD Millipore 371726), GP anti-K5 (Fuchs laboratory, 1:200), rabbit anti-K10 (Covance PRB-159P, 1:1,000), rabbit anti-K1 (Fuchs laboratory, 1:1,000), GP anti-K14 (Acris BP5009, 1:500), rabbit anti-phospho histone H3 (Cell Signaling 3377P, or Upstate/Millipore 06–570, 1:1,000), rabbit anti-Par3 (Upstate/EMD Millipore 07–330, 1:500). The Par3 antibody used detected all three major isoforms (relative molecular masses of 100,000, 150,000 and 180,000) by western blot analysis.
Images were acquired using either a Zeiss AxioPlan 2 wide-field epifluorescent microscope with an EXFO/Lumen Dynamics X-Cite metal halide lamp and ×20/0.8 air and ×100/1.4−0.7 oil Plan-Apochromat objectives, or a Leica TCS SPE-II 4 laser confocal system on a DM5500 upright scope with ACS Apochromat ×20/0.60 multi-immersion, ACS Apochromat ×40/1.15 oil ACS Apochromat ×63/1.30 oil, and HCX PlanApo ×100/1.40−0.70 oil objectives.
Measurements, quantification, graphing and statistics
Measurements of LGN orientation angle and division angle were performed using MetaMorph or Fiji (ImageJ), as previously described17. Early stage mitotic cells were identified using pHH3 and/or condensed chromatin as markers, whereas late stage mitotic cells were identified using the spindle midbody/cleavage furrow marker survivin. In cases where LGN localization was quantified, mitotic cells were first identified by DAPI and pHH3, then scored for LGN (whether LGN+ or LGN−). In this way, images were collected blind to the LGN expression state. To reduce variability in all immunostaining protocols, particularly where discriminations had to be made between expression levels or states (for example, LGN present or absent), head or back skins from the entire litter/cohort were embedded in single OCT blocks so that they could be sectioned and stained together. For younger embryos (for example, E15.5) where it is difficult to remove the skin, whole embryos were embedded. In this case, 2–3 embryos of different genotypes were mounted and stained on the same slide, again to reduce variability. Where representative images were selected from quantified data sets, they were chosen to most closely match the mean for the data set (for example, Figs 3f,g, 7g,h and Supplementary Fig. 2a,b). In addition, as mentioned above, immunostaining protocols were controlled within cohorts, were repeated at least twice, and exposure and contrast settings were kept constant for comparable images in a figure panel.
The method for measurement of division angles has been described previously17. Briefly, late-stage mitotic cells were identified by the presence of survivin immunoreactivity at the midbody/cleavage furrow. Cells were scored only if both daughter nuclei surrounding the survivin staining could be unambiguously identified. Angles were measured by drawing a line through the centres of the two nuclei, and parallel to the basement membrane. In mInsc; K14::Cre cKOs and littermates, images were captured and measurements were quantified blind to genotype. As it was not possible to be blinded to genotype in cases when RFP expression marked knockdown/knockout cells, in these instances mitotic cells were identified first by survivin staining, and imaged blind to whether they were RFP+ or RFP− . In this way, RFP− internal controls served as a blinded control in lentiviral-transduced tissue. To reduce any bias in data collection, all data from each group were not analysed until all images were collected. n values are indicated in each radial histogram and indicate the number of cells analysed; each experiment was repeated with at least two independent injections/litters, and data from at least 3 embryos (typically 5 or more) were collected. No statistical method was used to predetermine sample size, but data were collected from all available embryos of the indicated genotypes (no samples were excluded unless, for example, lentiviral transduction efficiency was very low). Similar procedures were implemented for quantification of LGN/apical marker expression.
Radial histograms were generated using Origin 9.0 software from data binned into 10° increments from 0 to 90° using GraphPad Prism 5. All other graphs and statistical analyses (Mann–Whitney, Student’s t-tests and chi-square tests) were produced using Prism. The appropriate statistical test was determined with consideration for whether the data fit a normal distribution or not. For example, a bimodal distribution of division angles does not, so in this case we categorized division angles into perpendicular (70–90°), oblique (20–70°) and planar (0–20°), and compared data sets by chi-square tests. All categorical data were also analysed by chi-square tests (for example, Figs 1c, 4b, 5f, 6a and 7c). When comparing distributions of division angles for different ages throughout development (Fig. 1f), a Mann–Whitney test was used because the data sets compared consisted of both normal and non-normal distributions, and the Mann–Whitney test is more robust than Student’s t -test in this setting. Similarly, for comparison of distributions of LGN crescent orientation, a Mann–Whitney test was also used because the data do not fit a normal distribution. For Tukey box-and-whisker plots, the dimensions of the box represent the 25th–75th percentile, the horizontal bar represents the median, the ‘+’ represents the mean, and the whiskers represent 1.5× the interquartile difference. Spinous thickness using the marker K10 was measured using Metamorph or Fiji as previously described17.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Stokes, D. Oristian and A. Aldeguer (Fuchs laboratory) and T. Anthony Curtis (Williams laboratory), for their expert technical assistance. We thank K. Byrd, K. Lough, and members of the Williams and Fuchs laboratories for critical reading of the manuscript and K. Lough for valuable input into the model presented in Fig. 8. We are grateful to S. Ohno and T. Hirose (both at Yokohama City University of Medicine, Japan) for sharing the Pard3 floxed mouse line. S.E.W. was supported by an American Cancer Society postdoctoral fellowship and E.F. is an investigator in the Howard Hughes Medical Institute. Work in the Fuchs laboratory was supported by a grant from the National Institutes of Health (E.F. R37-27883).
Footnotes
Note: Supplementary Information is available in the online version of the paper
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at www.nature.com/reprints
References
- 1.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 2.Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smart IH. Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. Br. J. Dermatol. 1970;82:276–282. doi: 10.1111/j.1365-2133.1970.tb12437.x. [DOI] [PubMed] [Google Scholar]
- 4.Williams SE, Fuchs E. Oriented divisions, fate decisions. Curr. Opin. Cell Biol. 2013;25:749–758. doi: 10.1016/j.ceb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 7.Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- 8.Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer. 2012;12:23–38. doi: 10.1038/nrc3169. [DOI] [PubMed] [Google Scholar]
- 9.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 2007;8:462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- 11.Homem CC, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- 12.Prehoda KE. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb. Perspect. Biol. 2009;1:a001388. doi: 10.1101/cshperspect.a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culurgioni S, Alfieri A, Pendolino V, Laddomada F, Mapelli M. Inscuteable and NuMA proteins bind competitively to Leu-Gly-Asn repeat-enriched protein (LGN) during asymmetric cell divisions. Proc. Natl Acad. Sci. USA. 2011;108:20998–21003. doi: 10.1073/pnas.1113077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mauser JF, Prehoda KE. Inscuteable regulates the Pins-Mud spindle orientation pathway. PLoS ONE. 2012;7:e29611. doi: 10.1371/journal.pone.0029611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuzawa S, Kamakura S, Iwakiri Y, Hayase J, Sumimoto H. Structural basis for interaction between the conserved cell polarity proteins Inscuteable and Leu-Gly-Asn repeat-enriched protein (LGN) Proc. Natl Acad. Sci. USA. 2011;108:19210–19215. doi: 10.1073/pnas.1110951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, et al. LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Gαi/LGN/NuMA pathways. Mol. Cell. 2011;43:418–431. doi: 10.1016/j.molcel.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Hashash AH, Warburton D. Cell polarity and spindle orientation in the distal epithelium of embryonic lung. Dev. Dyn. 2011;240:441–445. doi: 10.1002/dvdy.22551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konno D, et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 20.Peyre E, et al. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J. Cell Biol. 2011;193:141–154. doi: 10.1083/jcb.201101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luxenburg C, Amalia Pasolli H, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat. Cell Biol. 2011;13:203–214. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulson ND, Lechler T. Robust control of mitotic spindle orientation in the developing epidermis. J. Cell Biol. 2010;191:915–922. doi: 10.1083/jcb.201008001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladden AB, Hebert AM, Schneeberger EE, McClatchey AI. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev. Cell. 2010;19:727–739. doi: 10.1016/j.devcel.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watt FM, Green H. Stratification and terminal differentiation of cultured epidermal cells. Nature. 1982;295:434–436. doi: 10.1038/295434a0. [DOI] [PubMed] [Google Scholar]
- 25.Dowling J, Yu QC, Fuchs E. Beta4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beronja S, Fuchs E. RNAi-mediated gene function analysis in skin. Methods Mol. Biol. 2013;961:351–361. doi: 10.1007/978-1-62703-227-8_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Kawaguchi D, Furutachi S, Kawai H, Hozumi K, Gotoh Y. Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat. Commun. 2013;4:1880. doi: 10.1038/ncomms2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Hashash AH, et al. Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development. 2011;138:1395–1407. doi: 10.1242/dev.058479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Bultje RS, et al. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demehri S, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer M, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Gα-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- 34.Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- 35.Parmentier ML, et al. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 2000;20 doi: 10.1523/JNEUROSCI.20-14-j0003.2000. RC84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postiglione MP, et al. Mouse inscuteable induces apical-basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron. 2011;72:269–284. doi: 10.1016/j.neuron.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- 38.Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- 39.Hao Y, et al. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirose T, et al. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development. 2006;133:1389–1398. doi: 10.1242/dev.02294. [DOI] [PubMed] [Google Scholar]
- 41.Gao L, Macara IG, Joberty G. Multiple splice variants of Par3 and of a novel related gene, Par3L, produce proteins with different binding properties. Gene. 2002;294:99–107. doi: 10.1016/s0378-1119(02)00681-9. [DOI] [PubMed] [Google Scholar]
- 42.Kohjima M, et al. PAR3beta, a novel homologue of the cell polarity protein PAR3, localizes to tight junctions. Biochem. Biophys. Res. Commun. 2002;299:641–646. doi: 10.1016/s0006-291x(02)02698-0. [DOI] [PubMed] [Google Scholar]
- 43.Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 44.Pan Z, et al. An autoinhibited conformation of LGN reveals a distinct interaction mode between goloco motifs and TPR motifs. Structure. 2013;21:1007–1017. doi: 10.1016/j.str.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Gotta M, Ahringer J. Distinct roles for Gα and Gβγ in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat. Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- 46.Colombo K, et al. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- 47.Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai Y, Yu F, Lin S, Chia W, Yang X. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell. 2003;112:51–62. doi: 10.1016/s0092-8674(02)01170-4. [DOI] [PubMed] [Google Scholar]
- 49.Yu F, Cai Y, Kaushik R, Yang X, Chia W. Distinct roles of Gαi and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 2003;162:623–633. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- 52.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shitamukai A, Matsuzaki F. Control of asymmetric cell division of mammalian neural progenitors. Dev. Growth Differ. 2012;54:277–286. doi: 10.1111/j.1440-169X.2012.01345.x. [DOI] [PubMed] [Google Scholar]
- 54.Morin X, Jaouen F, Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat. Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- 55.Lancaster MA, Knoblich JA. Spindle orientation in mammalian cerebral cortical development. Curr. Opin. Neurobiol. 2012;22:737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goulas S, Conder R, Knoblich JA. The par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell. 2012;11:529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niessen MT, et al. aPKClambda controls epidermal homeostasis and stem cell fate through regulation of division orientation. J. Cell Biol. 2013;202:887–900. doi: 10.1083/jcb.201307001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 59.Coumailleau F, Furthauer M, Knoblich JA, Gonzalez-Gaitan M. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 2009;458:1051–1055. doi: 10.1038/nature07854. [DOI] [PubMed] [Google Scholar]
- 60.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rompolas P, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iden S, et al. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell. 2012;22:389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 63.Tanigaki K, et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 64.Nowak JA, Fuchs E. Isolation and culture of epithelial stem cells. Methods Mol. Biol. 2009;482:215–232. doi: 10.1007/978-1-59745-060-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.