Abstract
Immunoglobulin G (IgG) is a central mediator of host defense due to its ability to recognize and eliminate pathogens. The recognition and effector responses are encoded on distinct regions of IgGs. The diversity of the antigen recognition Fab domains accounts for IgG's ability to bind with high specificity to essentially any antigen. Recent studies have indicated that the Fc effector domain also displays considerable heterogeneity, accounting for its complex effector functions of inflammation, modulation and immune suppression. Therapeutic anti-tumor antibodies, for example, require the pro-inflammatory properties of the IgG Fc to eliminate tumor cells, while the anti-inflammatory activity of Intravenous Immunoglobulin G (IVIG) requires specific Fc glycans for activity. In particular, the anti-inflammatory activity of IVIG is ascribed to a small population of IgGs in which the Asn297-linked complex N-glycans attached to each Fc CH2 domain include terminal α2,6-linked sialic acids. We used chemoenzymatic glycoengineering to prepare fully di-sialylated IgG Fc and solved its crystal structure. Comparison of the structures of asialylated Fc, sialylated Fc, and F241A Fc, a mutant that displays increased glycan sialylation, suggests that increased conformational flexibility of the CH2 domain is associated with the switch from pro- to anti-inflammatory activity of the Fc.
Keywords: sialylated IgG Fc, inflammation, IVIG, N-linked glycan, X-ray crystallography
Introduction
IVIG is used worldwide as an antibody replacement therapy and as a treatment for a variety of inflammatory disorders [1]. IVIG is pooled IgG from the serum of healthy donors. It contains the four different subtypes of human IgG (IgG1, IgG2, IgG3, and IgG4) and over 30 glycovariants [2]. Although the beneficial effects of IVIG as an antiinflammatory preparation are apparent, the mechanism of action has only recently begun to be understood as being correlated with the relatively constant portion, or Fc region, of IgGs [3]. The Fc region of IgG interacts with a variety of receptors, determined by the amino acid and glycan composition of the Fc. Among these receptors are the canonical FcγRs: four are activating (FcγRI, FcγRIIA, FcγRIIIA, FcγRIIIB) and one is an inhibitory receptor (FcγRIIB) [3]. The simultaneous expression of these receptors on effector cells sets a threshold for inflammatory responses [4]. IVIG causes an up-regulation of the inhibitory receptor FcγRIIB, initiating anti-inflammatory responses [5].
A downside of IVIG therapy is that it is only effective when administered at high doses (1-3 g/kg) when treating inflammatory diseases [6]. Rationalizing the high dose requirements, the anti-inflammatory activity of IVIG has been attributed to a small population of IgGs with α2,6-sialyated N-linked glycans attached to a conserved asparagine (Asn297) in the Fc CH2 domain [7]. The Asn297-linked carbohydrate is a complex, usually core-fucosylated, biantennary glycan containing a pentasaccharide Man3GlcNAc2 core (Man, mannose; GlcNAc, N-acetylglucosamine), which can be modified by addition of terminal galactose or sialic acid [8] (Fig. 1). Approximately 10% of the Fc regions in IVIG terminate in sialic acid, of which 1-3% are disialylated [2; 9; 10]. In vivo studies have shown that Fc sialylation is the main determinant in IVIG antiinflammatory activity [9; 11]. For example, IVIG that was enriched in terminally-sialylated Fc glycans showed a 10-fold increase in anti-inflammatory activity [7; 12], and if IVIG was treated with neuraminidase to remove terminal sialic acids or with PNGase F to remove the entire N-linked glycan, the anti-inflammatory activity of IVIG was abrogated [9]. Similar results were found for recombinant, 2,6 sialylated IgG1 [7]. In mouse models, including a serum-induced arthritis model in mice, sialylated Fc (sFc; defined here as referring to a mixture of sialylated glycoforms) decreased inflammation of ankle joints [9], and IVIG activity was shown to be dependent upon terminally-sialylated Fc regions in four independent in vivo model systems of autoimmune disease mediated by auto-antibodies [11].
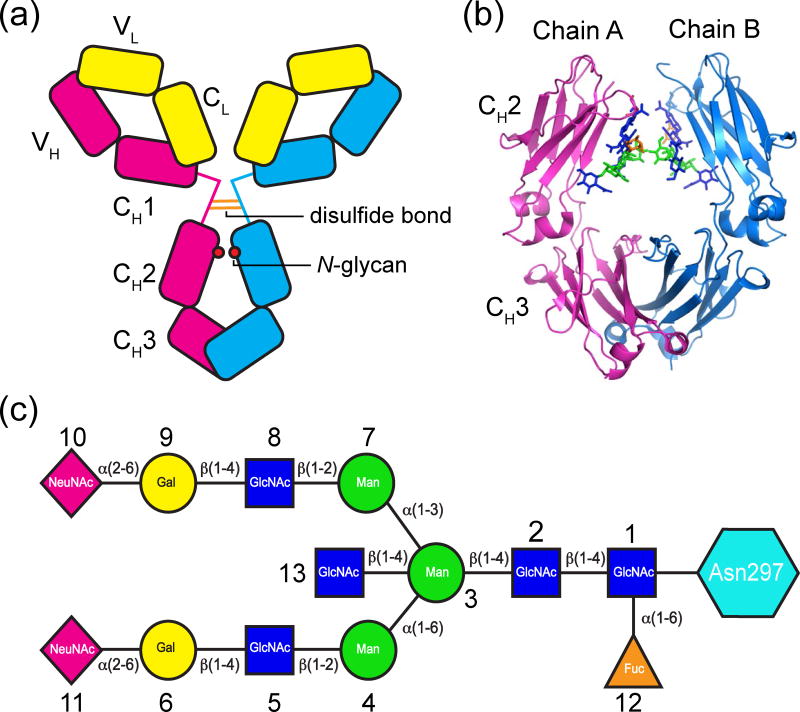
Fig. 1.
IgG, Fc and glycan structures. (a) Schematic structure of IgG1. (b) Ribbon diagram of wtFc (PDB: 3AVE) with Asn297-linked N-glycans shown as sticks. Glycan residues are colored as in panel c. (c) Schematic of an Asn297-linked N-glycan. The biantennary glycan consists of a mannose (Man) and N-acetylglucosamine (GlcNAc) core that can be modified by addition of core fucosylation (Fuc), a bisecting GlcNAc, terminal galactose (Gal) and sialic acid (NeuNAc).
The anti-inflammatory properties of sFc have been attributed to the effect of sialylation switching the specificity of Fc binding to its receptors. In vivo experiments showed that the anti-inflammatory effects of sFc required expression of the C-type lectin-like receptor specific intracellular adhesion molecule-grabbing non-integrin R1 (SIGN-RI) (the mouse homolog of human dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin; DC-SIGN), leading to a model in which conformational changes in Fc resulting from sialylation of the Asn297-attached glycan allowed interactions with members of the SIGN receptor family [13]. Indeed, previous structural studies demonstrated that modification of the Asp297-linked glycan can affect Fc structure. For example, it was shown that the glycan contributed to an “open” conformation of IgG Fcs, in which the CH2 domains were separated by a larger distance than in deglycosylated Fc structures [14]. If the IgG Fc glycan was enzymatically removed, the CH2 region adopted a “closed” state [15]. Conformational changes have also been observed when individual sugar residues on the Fc-linked glycan were modified. When fucose was removed, a subtle change involving Tyr296 was observed in X-ray crystallographic and NMR structures [16; 17]. This modification resulted in an increased affinity for the activating receptor FcγRIIIa, leading to enhanced antibody-dependent cellular cytotoxicity (ADCC) activity [18; 19]. Solution NMR studies have reported increased mobility of the glycan arms upon sialylation, further supporting the contention that alterations in the glycan composition can influence the structure of the Fc [20].
Here we solved the crystal structure of a chemically-homogeneous disialylated Fc (di-sFc) and compared it to new structures of a partially sialylated Fc (F241A Fc) and wtFc, as well as to wtFc and glycomutant Fc structures available in the protein data bank (PDB). We found that di-sFc and F241A Fc show increased conformational heterogeneity in crystals compared to wtFc, a characteristic that may relate to sialylation and anti-inflammatory properties.
Results
Glycan analysis of purified Fc proteins
Proteins were produced by transient transfection in HEK 293-6E cells as IgG1 Fc fragments (wtFc, F241A Fc and F243A Fc) or in a stably-transfected Chinese hamster ovary (CHO) cell line (wtFc) [21]. Disialylated sFc (di-sFc) was prepared by chemoenzymatic glycoengineering [22] of an Fc fragment isolated after papain cleavage of Rituximab, a human IgG1.
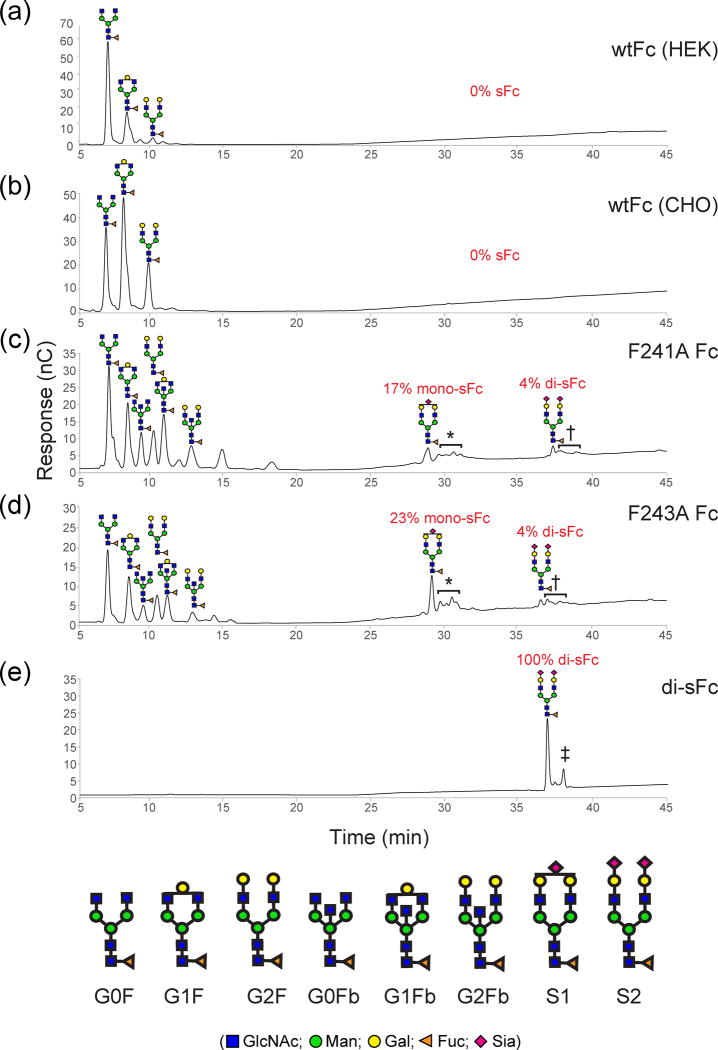
Carbohydrate analyses of N-glycans released from the Fc proteins revealed that the N-glycans attached to Asn297 of wtFc expressed in either HEK293 or CHO cells consisted of only three glycoforms: G0F, G1F, and G2F (Fig. 2a,b,). This result was consistent with previous studies investigating glycoforms of IgGs expressed in HEK cells [23]. Neither mono- nor disialylated N-glycans on either form of wtFc were detected. By contrast, the glycans attached to the F241A and F243A Fc mutants contained more diverse glycoforms: in addition to the G0F, G1F, and G2F glycoforms, significant portions of the N-glycans carried a bisecting GlcNAc moiety and contained terminal sialic acids (Fig. 2c,d,). The sialylated forms included about 20% of terminally monosialylated N-glycans (S1G2F and other monosialylated glycoforms) and about 4% of disialylated N-glycans (S2G2F and other disialylated glycoforms). These results were consistent with a recent study of the F241A and F243A Fc mutants [23], suggesting that these substitutions promote further processing of Fc glycans resulting in generation of sialylated and bisecting GlcNAc-containing glycoforms. The chemoenzymatically remodeled Fc (di-sFc) was almost homogeneous: >95% of the N-glycans were the disialylated glycoform (Fig. 2e, Fig. S1).
Fig. 2.
HPAEC-PAD analysis of PNGase F-released N-glycans of IgG Fc proteins. (a) wtFc expressed in HEK cells. (b) wtFc expressed in CHO cells. (c) F241A Fc expressed in HEK cells. (d) F243A expressed in HEK cells. (e) di-sFc prepared by chemoenzymatic glycoengineering of an Fc fragment isolated after papain cleavage of Rituximab. The amounts of mono- and disialylated N-glycans are shown. * indicates a mixture of heterogeneous monosialylated glycoforms. † indicates a mixture of heterogeneous disialylated glycoforms. ‡ indicates a satellite peak due to epimerization of reducing end GlcNAc to ManNAc.
We also compared the Fc proteins in a Western blot using Sambucus Nigra (SNA), a lectin that binds preferentially to α2,6-linked sialic acid attached to a terminal galactose [24]. As expected, SNA blots of wtFc, F241A, F243A and di-sFc proteins demonstrated sialylation of di-sFc, F241A Fc and F243A Fc, but not wtFc (Fig. 3). These results were consistent with previous reports of partial sialylation of the N-glycans of F241A and F243A Fc [25].
Fig. 3.
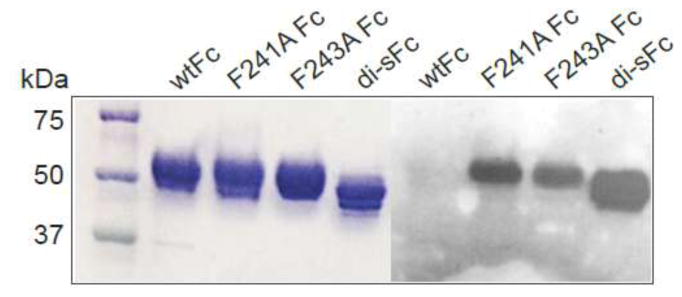
SDS-PAGE (left) and SNA blot (right) of Fc proteins expressed in HEK cells. wtFc, F241A Fc and F243A Fc were expressed as Fc fragments; di-sFc was prepared by papain cleavage from Rituximab IgG.
Liquid chromatography/electrospray ionization mass spectrometry (LC-ESI-MS) was used to analyze wtFc that was expressed as an Fc fragment in HEK cells before and after complete removal of the N-glycans by PNGase F treatment. The analysis revealed two major species that were 950 and 1315 Da larger than the calculated molecular mass of the polypeptide backbone following PNGase F treatment, suggesting the existence of additional posttranslational modifications (Fig. S2a). A similar analysis of wtFc expressed in CHO cells revealed three major species that were 269, 922, and 1218 Da larger than the calculated molecular mass of the polypeptide backbone (Fig. S2b). The results for the F241A Fc and F243A Fc proteins that were expressed in HEK cells were the same as the HEK-expressed wtFc (Fig. S2c,d). In contrast, an Fc fragment prepared from papain cleavage of an intact IgG1 (Rituximab) did not contain posttranslational modifications other than the N-glycans (Fig. S2e). A monosaccharide composition analysis on the recombinant Fcs after removal of the N-glycans with PNGase F revealed the presence of GalNAc, GlcNAc, Gal, and Neu5Ac (Fig. S3b,c), suggesting the existence of O-linked glycans. In contrast, the Fc fragment prepared from papain cleavage of Rituximab did not contain any monosaccharides after PNGase F cleavage (Fig. S3d). On the basis of the additional molecular mass (950 Da) in comparison with the polypeptide backbone, the major O-glycan on the HEK-expressed Fcs appeared to be a core 1 structure carrying two sialic acid residues [calculated mass for (Neu5Ac)2-Gal-GalNAc-H2O = 947.86 Da]. In the CHO-expressed wtFc, an additional mass of 1218 Da on the polypeptide backbone was observed. However, this additional mass does not correspond to typical O-glycan structures, implicating potential additional modifications on the core structures; e.g., acetylation on the sialic acid moieties. The detailed structures of the O-glycans remain to be determined. However, the lack of reactivity of HEK-expressed wtFc in the SNA blot (Fig. 3) demonstrated that there were no α2,6-linked sialic acids on the putative O-glycans. Since the hinge region of IgG where the O-glycans would likely be located is usually disordered in Fc crystal structures, these glycans (if present) would not be visible in published structures of recombinantly-expressed Fc fragments [23; 26; 27; 28]. Nor are there apparent differences in the structural properties of Fcs that correlate with whether they were expressed as Fc fragments versus prepared by cleavage of intact IgG (Table 1).
Table 1. Distance measurements between CH2 domains in IgG1 Fc structures.
| PDB ID |
Protein | Category | How expressed | Space Group | Cell Dimensions a,b,c (Å) [β angle; deg] |
Resolution (Å) |
Pro238 Cα (Å) |
Phe241 Cα (Å) |
Arg301 Cα (Å) |
Pro 329 Cα (Å) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1H3X | Fc (G0F)2 | wtFc | IgG | P212121 | 50, 80, 139 | 2.4 | 19.7 | 21.9 | 32.9 | 22.6 |
| 3AVE | Fc Fucosylated | wtFc | IgG | P212121 | 50, 79, 144 | 2.0 | 19.3 | 21.8 | 32.3 | 25.1 |
| 2DTS | Fc Defucosylated | wtFc | IgG | P212121 | 50, 78, 144 | 2.2 | 18.6 | 21.5 | 32.2 | 24.2 |
| 3DO3 | Fc | wtFc | IgG | P212121 | 50, 80, 139 | 2.5 | 20.1 | 22.4 | 32.8 | 23.5 |
| 1FC1 | Fc | wtFc | IgG | P212121 | 50, 80, 146 | 2.9 | 19.6 | 23.5 | 34.2 | 26.8 |
| N/A | Fc | wtFc | IgG | P212121 | 50, 80, 138 | 2.4 | 18.6 | 21.6 | 32.6 | 22.3 |
| 1HZH | Intact IgG1 | wtFc | IgG | H32 | 271, 271, 175 | 2.7 | 21.1 | 22.2 | 33.6 | 23.9 |
| 1H3Y | Fc high salt | Fc high salt | IgG | P6122 | 153, 153, 117 | 4.1 | 16.7 | 22.3 | 25.9 | 29.6 |
| 4KU1 | Fc (G2F)2 | glycovariant | IgG | P212121 | 49, 80, 128 | 1.9 | 21.3 | 23.3 | 33.2 | 29.1 |
| 1H3V | Fc (G2F)2 | glycovariant | IgG | P212121 | 50, 81, 142 | 3.1 | 19.3 | 23.2 | 32.9 | 26.9 |
| 1H3W | Fc (G2F)2 | glycovariant | IgG | C2221 | 50, 149, 76 | 2.9 | 22.9 | 26.1 | 35.9 | 33.8 |
| 1H3U | Fc (M3N2F)2 | glycovariant | IgG | P212121 | 50, 80, 144 | 2.4 | 19.2 | 22.0 | 32.7 | 24.2 |
| 2WAH | Fc (Man9NAG2) | glycovariant | Fc | P212121 | 49, 75, 149 | 2.5 | 30.2 | 29.8 | 42.8 | 36.4 |
| 4B7I | Fc Hybrid glycan | glycovariant | Fc | P212121 | 49, 73, 141 | 2.4 | 36.1 | 33.2 | 47.0 | 41.1 |
| 4ACP | Fc (GlcNAc) | glycovariant | Fc | C2 | 69, 111, 78, 108° | 2.5 | 11.4 | 17.8 | 26.2 | N/A |
| 3DNK | Fc deglycosylated | glycovariant | IgG | P212121 | 49, 78, 147 | 2.8 | 24.8 | 25.0 | 37.4 | 27.6 |
| 3S7G | Fc deglycosylated | glycovariant | Fc | P1 | 66, 66, 78 | 3.1 | 10.7 | 16.8 | 25.8 | 18.9 |
| 3S7G | Fc deglycosylated | glycovariant | Fc | P1 | 66, 66, 78 | 3.1 | 12.1 | 17.8 | 27.1 | 19.6 |
| 2QL1 | Fc DLE mutant | mutant Fc | Fc | C2221 | 50, 148, 74 | 2.5 | 27.6 | 28.5 | 37.2 | 39.3 |
| 3FJT | Fc YTE mutant | mutant Fc | Fc | P212121 | 50, 80, 146 | 2.5 | 20.0 | 22.1 | 33.4 | 25.3 |
| 3C2S | Fc FES mutant | mutant Fc | IgG | C2221 | 50, 147, 75 | 2.3 | 28.1 | 28.8 | 38.1 | 35.9 |
| 3D6G | Fc: protein A | complex | IgG | P212121 | 50, 79, 139 | 2.3 | 19.2 | 22.2 | 32.9 | 24.3 |
| 1L6X | Fc: Z34C | complex | IgG | C2221 | 54, 125, 90 | 1.7 | 22.2 | 22.8 | 33.9 | 26.6 |
| 1OQO | Fc (G0): mini Z | complex | ? | P21212 | 86, 126, 55 | 2.3 | 20.0 | 22.1 | 33.1 | 24.3 |
| 1OQX | Fc (G2): Z34C | complex | ? | P21212 | 87, 126, 54 | 2.6 | 20.1 | 21.8 | 33.0 | 24.7 |
| 1DN2 | Fc: peptide | complex | IgG | P21 | 68, 61, 68, 104° | 2.7 | 26.1 | 27.0 | 37.4 | 31.9 |
| 1FCC | Fc: Protein G | complex | IgG | P43212 | 111, 111, 160 | 3.2 | 22.1 | 26.0 | 34.5 | 34.7 |
| 1FC2 | Fc: protein A | complex | IgG | P3121 | 71, 72, 147 | 2.8 | 18.3 | 22.0 | 31.4 | 27.7 |
| 1E4K | Fc :FcgRIIIB | complex | IgG | P6522 | 115, 115, 299 | 3.2 | 25.0 | 25.7 | 36.7 | 30.3 |
| 1T83 | Fc :FcgRIIIB | complex | IgG | P212121 | 73, 102, 123 | 3.0 | 24.8 | 26.1 | 36.9 | 31.3 |
| 1T89 | Fc :FcgRIIIB | complex | IgG | P6522 | 115, 115, 301 | 3.5 | 24.5 | 25.9 | 37.3 | 30.6 |
| 3AY4 | Fc afucosylated: FcγRIIIA | complex | IgG | P41212 | 77, 77, 350 | 2.2 | 25.2 | 25.7 | 36.3 | 30.8 |
| 3RY6 | Fc: FcγRIIa | complex | IgG | C2221 | 153, 256, 58 | 3.8 | 20.5 | 23.1 | 34.4 | 31.4 |
| 3SGK | afucosylated Fc: FcγRIIIA | complex | IgG | P212121 | 67, 88, 141 | 2.4 | 25.6 | 26.6 | 37.4 | 32.6 |
| 3SGJ | fucosylated Fc: FcγRIIIA | complex | IgG | P212121 | 67, 88, 140 | 2.2 | 25.6 | 26.3 | 36.9 | 32.3 |
| N/A | di-sFc closed | N/A | IgG | P21 | 53, 154, 66, 111° | 3.0 | 13.0 | 18.5 | 26.7 | 21.6 |
| N/A | di-sFc open | N/A | IgG | P21 | 53, 154, 66, 111° | 3.0 | 20.2 | 23.9 | 31.9 | N/A |
| 4BYH | sFc | N/A | Fc | P6122 | 153, 153, 112 | 2.3 | 17.6 | 21.8 | 26.2 | 30.5 |
| N/A | F241A Fc | N/A | Fc | P212121 | 58, 75, 114 | 2.2 | 22.9 | 24.9 | 33.8 | 31.1 |
| 4BM7 | F241A Fc | N/A | Fc | P212121 | 49, 73, 136 | 2.0 | 34.0 | 31.7 | 45.1 | N/A |
The finding of putative O-glycans on proteins expressed as Fc fragments, but not on an Fc fragment derived by cleavage of intact IgG, could be explained if expression of Fc as a fragment allows access during biosynthesis by O-glycan-adding enzymes in the Golgi apparatus to a region of the IgG hinge that is inaccessible when intact IgG is trafficking through the Golgi. Although there were no apparent structural differences in Fcs as a function of whether or not they were produced in a form predicted to contain the putative O-glycan, these results suggest that functional studies employing Fcs expressed directly as fragments should compare results with Fcs derived by enzymatic cleavage of intact IgGs to rule out potential effects of unnatural O-glycosylation.
wtFc structures are relatively similar
In order to assess potential conformational differences in Fc structures, we first compared wtFc structures available in the Protein Data Bank (PDB) and a 2.4 Å resolution wtFc structure solved in this study (space group P212121; one molecule per asymmetric unit) (Supplementary Table 1). We defined “wild type” as those Fc structures that were derived from IgGs or Fc fragments containing the naturally-occurring mixture of N-glycans attached to Asn297 when the protein was produced in mammalian cells, which included our new wtFc structure and PDB entries 1H3X, 3AVE, 2DTS, 3DO3, 1FC1 and 1HZH. Fc fragments are homodimers in which the N-terminal CH2 domains contact each other through their Asn297-linked N-glycans, and the C-terminal CH3 domains form a stable interface that is relatively constant between structures (Fig. 1b and 4a). Although the CH2 domain N-glycans are generally heterogeneous in wtFc proteins (Fig. 2), ordered electron density is usually observed for eight or nine carbohydrate residues of the biantennary complex glycan attached to Asn297 of each CH2 domain (PDB codes: 1H3X, 3AVE, 2DTS, 3DO3, 1FC1 and 1HZH).
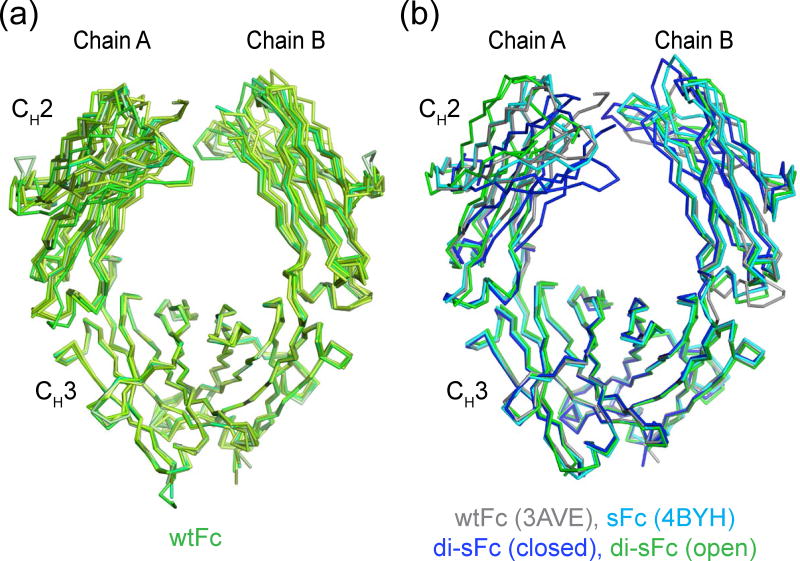
Fig. 4.
Comparison of wtFc and sFc structures. Fc structures were aligned by superimposing the CH3 domains. (a) Alignment of wtFc structures (the wtFc structure solved in this study plus PDB entries 1HZH, 1FC1, 2DTS, 3AVE, 1H3X, and 3DO3). (b) Alignment of closed and open structures of di-sFc (this study), sFc (PDB 4BYH), and wtFc (PDB 3AVE).
In order to evaluate potential differences in wtFc and sialylated Fc structures (Fig. 4), the CH3 domains of the wtFc structures were aligned, and we monitored the separation of the CH2 domains as previously described [29] by measuring distances between the Cot atoms at CH2 positions 238, 241, 301, and 329. The differences in CH2 domain separation (Table 1) relative to a published wtFc structure (3AVE) were plotted for each set of paired Cot positions (Fig. 5). Despite differences in crystal packing, the wtFc structures exhibited only small deviations in CH2 domain separation distances, demonstrating relatively little structural variability. By contrast, a low-resolution wtFc structure solved from crystals grown under high salt conditions [14] (PDB: 1H3Y) showed deviations from the other wtFc structures that were crystallized in lower salt concentrations (Fig. 5, Table 1). Structures of Fc proteins with different defined glycoforms showed a larger degree of variability in CH2 domain separation than wtFc structures, as was also true for Fc structures with mutations introduced to enhance effector functions, and Fc structures solved as complexes with other proteins (Fig. 5). There were no systematic differences in CH2 domain separation or structure based on whether or not the Fc fragment was predicted to contain an O-glycan in its hinge region (Table 1; Fig. 5).
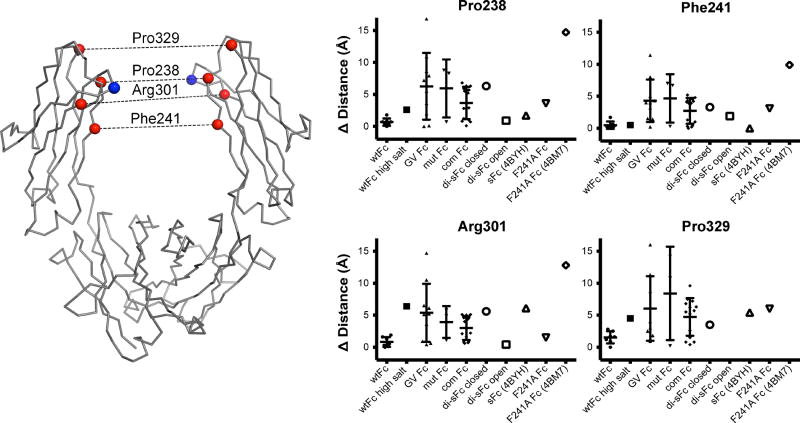
Fig. 5.
Comparison of CH2 domain separation in Fc structures. Left: location of Cα atoms for Pro238, Phe241, Arg301, and Pro329 (red spheres) and Asn297 (blue spheres) indicated on the CH2 domains of wtFc (PDB ID: 3AVE). CH2 domain separation in individual Fc structures was evaluated by measuring distances (dotted lines) between the corresponding red spheres on each chain. Right: differences in CH2 domain separation relative to a reference wtFc structure (3AVE) plotted for the each set of paired Cα positions in the indicated category of Fc structure. When a structure category had more than one member, the mean and standard deviation are superimposed upon the values. Fc structure categories are as follows: wtFc (structure solved in this study plus PDB entries 1H3X, 3AVE, 2DTS, 3DO3, 1FC1, and 1HZH), wtFc high salt (1H3Y), glycovariant (GV) Fcs (1H3W, 1H3U, 1H3V, 2WAH, 4B7I, 4ACP, 3DNK, 4KU1, and 3S7G), mutant (mut) Fcs (2QL1, 3FJT, and 3C2S), Fcs complexed with other proteins (com Fc) (3D6G, 1L6X, 1OQO, 1OQX, 1DN2, 1FCC, 1FC2, 1E4K, 1 T83, 1 T89, 3AY4, 3RY6, 3SGK, and 3SGJ), di-sFc closed, di-sFc open, sFc (4BYH), F241A Fc (this study), and F241A Fc (4BM7). Pro329 on one chain of the di-sFc open and 4BM7 structures was disordered; therefore these distance values were not measured or plotted.
Structures of two forms of di-sFc
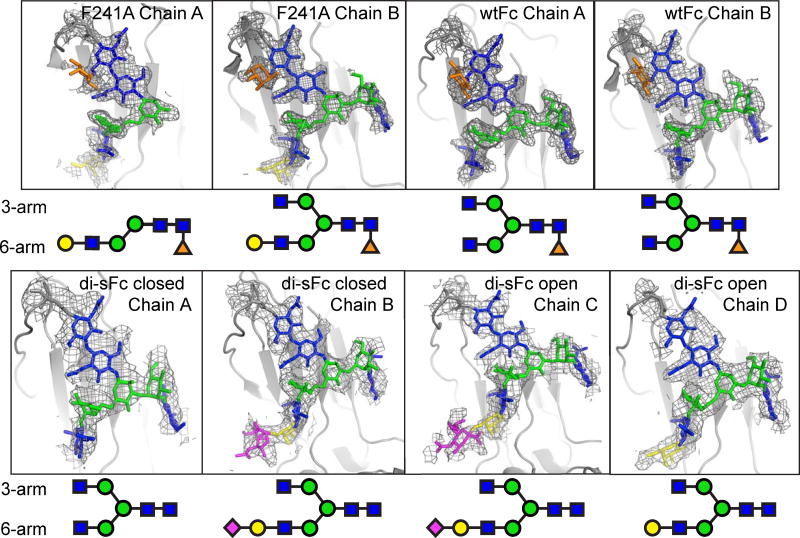
A 3.0 Å resolution of di-sFc was solved in space group P21 with two di-sFc molecules per asymmetric unit. The structure was compared with wtFc structures and a previously described structure of sFc that included a mixture of di- and monosialylated Fc (PDB: 4BYH) [30]. In the di-sFc structure, electron density for biantennary N-linked glycans was visible for all four di-sFc chains in the two di-sFc dimers, but although the di-sFc protein was fully disialylated (Fig. 2e), electron density for a terminal sialic acid was found only on the 6-arm of chains B and C, with the 6-arm of chains A and D showing ordered glycan residues up to a GlcNAc or galactose, respectively, and the 3-arm showing electron density for glycan moieties up to the GlcNAc (chains A, B, C and D) (Fig. 6). Similarly, sialic acid was visible only on the 6-arm of chain A in the 4BYH sFc structure [30]. In all three sFc structures (in 4BYH as well as in the two copies of di-sFc in the new sFc structure), the 6- and 3-arms of the biantennary glycans projected away from the protein surface and were solvent exposed; thus the ordered sialic acid on the glycan 6-arm was not positioned into a pocket formed by Glu258 and Lys290, as previously proposed [13].
Fig. 6.
Comparison of ordered glycans on Fc structures. 2Fo-Fc electron density (contoured at 0.7σ) from an annealed omit map in the region of the Asn297-linked N-glycan is shown for both chains of di-sFc closed, di-sFc open, F241A Fc, and wtFc (this study). Ordered glycan residues are shown schematically below each map using the colors and shapes defined in Fig. 1c.
Interestingly, the two di-sFc dimers in the asymmetric unit of the P21 crystals exhibited differences in the arrangement of their CH2 domains. One di-sFc (chains A and B) exhibited a relatively “closed” conformation, as defined by the separation between the two CH2 domains, whereas the second di-sFc (chains C and D) exhibited a relatively “open” conformation (Table 1 and Fig. 4b). As compared with a representative wtFc structure (3AVE), the open di-sFc exhibited a 6.8 Å root mean square deviation (RMSD) (calculated for the 103 Cα atoms of one CH2 domain after alignment of the partner CH2 domain in each structure), as compared a 15.0 Å RMSD between the wtFc and closed di-sFc CH2 domains, a 7.9 Å RMSD between the open di-sFc and the 4BYH sFc, and a 8.8 Å RMSD between the closed di-sFc and the 4BYH sFc. To quantitatively evaluate CH2-CH2 domain separation distances, which cannot be easily defined by a single number reflecting an average separation, we used a previously-defined metric in which the distance between four partner Cot atoms on each CH2 domain are measured (Pro238, Pro329, Phe241 and Arg301) and compared between structures [29]. In addition to determining CH2-CH2 separation distances for these Cα residues (Table 1), we calculated the differences relative to a reference wtFc structure (3AVE) to determine how greatly the various structures deviated from wtFc and other Fc structures. As defined by the distances between these Cα atoms, the closed di-sFc exhibited a greater difference in CH2 domain separation when compared to wtFc structures than 4BHY sFc, whereas the open di-sFc Cα exhibited a difference within the range observed for other wtFc structures (Fig. 5).
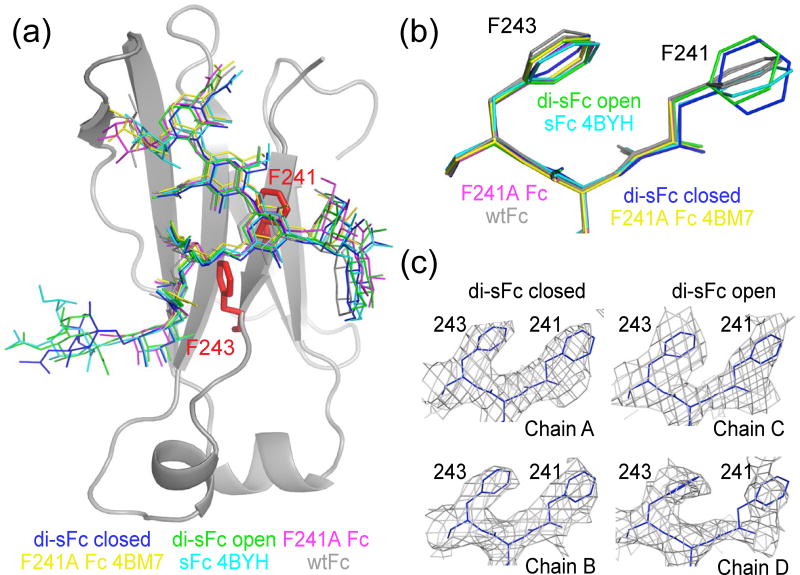
We next aligned individual CH2 domains of the closed di-sFc, open di-sFc, wtFc (3AVE) and the 4BYH sFc structures to compare the orientations of hydrophobic side chains (Phe241 and Phe243) adjacent to the N-linked glycan attached to Asn297 (Fig. 7a). No major changes were observed in the overall fold of the CH2 domains, and only minor changes were found in the Phe243 side chain orientation. However, larger changes were observed in the Phe241 side chain, which was rotated ∼90° compared to Phe241 in other Fc structures (Fig. 7b). Typically the aromatic rings of the Phe241 and Phe243 side chains were stacked with respect to each other; however in both CH2 domains of the closed and open di-sFc structures, Phe241 adopted a different conformation such that it was no longer stacked with Phe243 (Fig. 7b,c and Fig. S4). The Phe241 side chain was not involved in crystal contacts in any of the structures (data not shown); thus these differences were not related to crystal packing. Although the Asn297-linked N-glycan is adjacent to Phe241 and Phe243, no major differences were observed in the glycan conformation (Fig. 7a), thus the Phe241 side chain orientation did not affect the glycan conformation.
Fig. 7.
Comparison of glycan and protein residue conformations in sFc and wtFc structures. (a) Alignment of Asn297-linked N-glycans of di-sFc closed (blue), di-sFc open (green), F241A Fc (magenta), F241A Fc 4BM7 (yellow), sFc 4BYH (cyan), wtFc from this study and wtFc 3AVE (both gray). The side chains of Phe241 and Phe243 are shown in red on the CH2 domain of wtFc. (b) Alignment of Phe(Ala)241–Leu242–Phe243 region in the structures of di-sFc closed (blue), di-sFc open (green), F241A Fc (magenta), F241A Fc 4BM7 (yellow), sFc 4BYH (cyan), wtFc from this study and wtFc 3AVE (both gray).(c) 2Fo-Fc electron density (contoured at 0.7σ) from an annealed omit map in the region Phe243 and Phe241 side chains for the di-sFc structures. See also Fig. S5.
Structure of F241A Fc
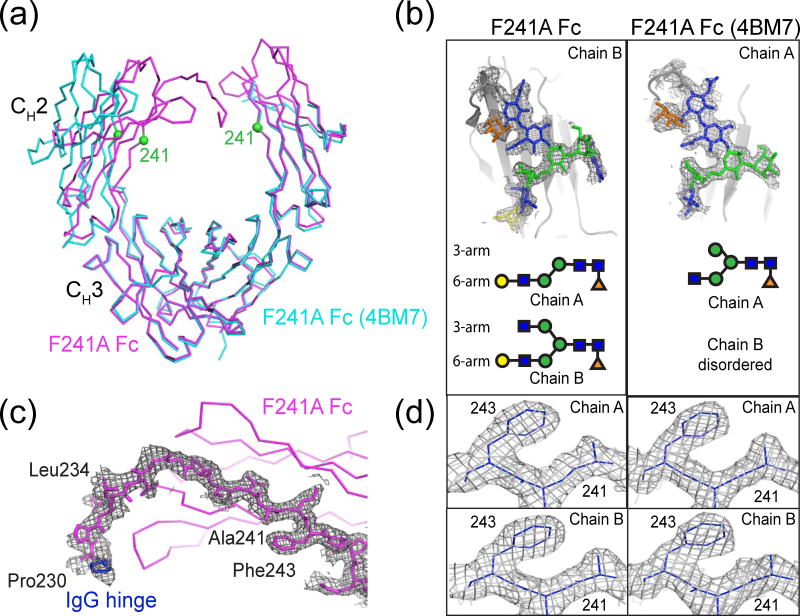
We also solved a 2.2 Å resolution crystal structure of F241A Fc (space group P212121; one molecule per asymmetric unit) and compared it to wtFc and sFc structures and to PDB code 4BM7, a previous F241A Fc structure [23] (Fig. 8a, b). On chain A of the new F241A Fc structure, there was ordered electron density for residues Pro230 to Leu443, which included part of the hinge region (Glu216 to Pro230 for human IgG1 proteins) that normally links the Fab to the N-terminus of the Fc region (Fig. 8c). By contrast, the first ordered residue on both chains of wtFc structures was usually Leu234, Leu235, Gly236, Gly237, or Pro238 (PDB codes: 1H3X, 3AVE, 2DTS, 3DO3, 1FC1 and 1HZH), and the first ordered residue of the F241A B chain was Gly237. The hinge region was also disordered in the 4BM7 F241A Fc structure (the first ordered residues were Gly237 on chain A and Pro238 on chain B). At Asn297, seven ordered glycan residues were modeled on chain A and nine residues were modeled on chain B of the new F241A structure (Fig. 6), but sialic acids were not visible in the new F241A or the 4BM7 F241A structures although the F241A glycans included both mono-sialylated and disialylated species [23] (Fig. 2 and Fig. 8b). Because N-glycans were not completely resolved even in the di-sFc structure, which was solved from crystals containing Fc protein with completely homogeneous N-glycans, we cannot use the F241A Fc structures to determine the compositions of N-glycans in the F241A Fc crystals. Therefore it is not possible to determine whether a defined glycan species crystallized from the heterogeneous protein preparations used to produce both of the F241A Fc structures and all of the wtFc structures.
Fig. 8.
Comparison of F241A Fc structures. (a) Superimposition of Cα traces of F241A Fc (this study) (magenta) and F241A Fc (PDB 4BM7) (cyan). Locations of Cα atoms at position 241 of each chain are shown as green spheres. (b) 2Fo-Fc electron density (contoured at 0.7 σ) from annealed omit maps for F241A Fc (this study) and F241A Fc (PDB: 4BM7). Ordered glycan residues are shown schematically below each map using the colors and shapes defined in Fig. 1c. (c) 2Fo-Fc electron density (annealed omit map) of an ordered portion of the hinge region (blue) on chain A contoured at 0.6σ. (d) 2Fo-Fc electron density (contoured at 0.7σ) from annealed omit maps in the region of the Phe243 and Ala241 side chains for the structures of F241A Fc (this study) (left) and F241A Fc (PDB: 4BM7) (right).
Comparison of the CH2 domain separations of the F241A Fc structure with other Fc structures showed that the new F241A Fc structure was similar to wtFc structures and the open di-sFc structure, but substantially more closed than the 4BM7 F241A Fc (Fig. 5, Table 1). (Because portions of the chain B CH2 domain of the 4BM7 F241A Fc structure were disordered, including the Asn297-linked glycan and residues 264-274 (BC loop), 296-300 (C′E loop), and 324-332 (FG loop), we could not measure all of the interchain distances in the 4BM7 F241A Fc structure.) This large degree of difference in F241A Fc conformations is similar to that observed for di-sFc conformations but distinct from wtFc conformations. Since both of the F241A Fc structures (the structure reported here and the 4BM7 structure) were derived from proteins that were partially sialylated, this observation is consistent with the suggestion that a greater degree of conformational heterogeneity is available to sialylated Fc proteins than to wtFc with conventional glycosylation species. Alternatively, or perhaps in addition, the F241A substitution itself could result in greater conformational heterogeneity.
Discussion
The Fc regions of IgGs can function to either promote or suppress the inflammatory response. Previous studies have shown that the Asn297-linked N-glycan composition is a determining factor in function [12; 31]. Both biochemical and in vivo studies have shown that sialylation of the Fc glycan is essential for the anti-inflammatory activity of IVIG [2; 9; 12]. Here we compared structures of wtFc, which carries mostly asialylated N-glycans, to sialylated forms of Fc, both a fully disialylated form produced by chemoenzymatic glycoengineering, and an F241A mutant form of Fc. Compared with wtFc, the Asn297-linked N-glycan of F241A and F243A Fc showed increased mono- and disialylated products when the protein was expressed in mammalian cells. This suggests that IgGs with an F241A mutation may be effective anti-inflammatory molecules due to increased sialylation.
Our analyses of the wtFc proteins produced in two different mammalian cell lines demonstrated that the Asn297-linked N-glycans were mainly G0F, G1F and G2F glycoforms, consistent with previous studies [23], thus we assume a similar composition for the N-glycans on the relatively structurally-invariant wtFc structures in the pdb. Not only are these glycans mostly asialylated, they also contain only a small percentage of digalactosylated forms. In a systematic comparison of wtFc structures reported here and previously, we found only small deviations in the “openness” of the Fc, evaluated by the distance between the partner CH2 domains in the Fc dimer, despite different crystal packing environments, however, we noted that certain glycan species cause increased variation in crystal structures. For example, the F241A Fc structure reported here and one described previously [23] showed a larger degree of variation in CH2 domain orientations than seen in a survey of seven wtFc structures. Other glycovariants of Fc, including a recent structure of a digalactosylated form of wtFc [32], also showed a larger degree of conformational variability as assessed by differential CH2 domain separations.
Similar to the partially sialylated F241A Fc structures, crystal structures of sialylated wtFc (sFc) have been found in several conformations. In our crystal structure of fully disialylated Fc, we found both open and closed CH2 domain conformations in the same crystal, and a previously published sFc structure [30] revealed a conformation similar to the canonical wtFc structures, again suggesting a large degree of inter-domain flexibility of sialylated forms of Fc compared to the relatively invariant structures of asialylated wtFc.
Recent molecular dynamics simulations showed a wide range of conformations that Fc can adopt [32]. Here we have documented a greater flexibility of glycovariant forms of Fc that may be key to the ability of the Fc region of IgG to modulate inflammatory and anti-inflammatory activities mediated by glycan differences. Further structural investigations of how glycan composition and conformational flexibility affect binding to Fc receptors that mediate differential activities will be critical to understanding the significance of these results.
Methods
Protein Expression and Purification
Fc proteins were expressed in transiently-transfected HEK 293-6E suspension cells (F241A Fc and F243A Fc) or stably-transfected Chinese hamster ovary cells (wtFc) as described [21; 33]. Fc proteins were isolated from harvested supernatants using protein A chromatography (GE Healthcare) followed by size exclusion chromatography in 25 mM Tris-Cl pH 7.5, 100 mM NaCl on a Superdex 16/60 gel-filtration column (GE Healthcare). Fractions corresponding to the Fc were pooled and used for crystallization and glycan analysis.
Fc protein for di-sFc production was prepared by papain cleavage of Rituximab (Rituxan®) (Genentech, Inc., California), a human IgG1 Fc. Rituximab (40 mg) was buffer exchanged into 10 mM Tris-HCl pH 7.5 overnight at 4°C. Dialyzed material was concentrated to 20 mg/ mL (∼2 mL). Immobilized papain (Thermo Scientific) was prepared following the manufacturer's protocol. 1 mL of immobilized papain slurry (50% immobilized papain) was added to the Rituximab solution. The reaction was incubated at 37°C for 5 hours while continuously inverting the sample tube. The resin was removed using a spin column and the filtrate was loaded over a protein A column (2 × 5 mL) that was pre-equilibrated with 10 mM Tris-HCl, pH 7.5. The column was washed with 10 column volumes of 10 mM Tris-HCl, pH 7.5 until no protein was detected in the flow through. Bound Fc was eluted in 4 mL of a 100 mM citric acid solution pH 3.5. Eluted fractions were quickly neutralized using a 1.5 M solution of Tris-HCl pH 8.8. Purified Fc was combined, concentrated and injected over a 40 mL (4 × 10 mL) HiTrap desalting column (GE Healthcare) pre-equilibrated in a phosphate buffered saline solution (pH 7.4) to give 10 mg of Fc. wtFc for crystallization was prepared similarly by papain digestion of the anti-HIV-1 IgG1 antibody 21c (expressed in HEK 293-6E cells as described [33]).
Preparation of disialylated Fc (di-sFc)
Purified Fc (10 mg) prepared by papain cleavage of Rituximab IgG was treated with 30 μg of recombinant S. pyogenes EndoS [22] for 1 hr at 37°C. Analysis by LC-MS showed complete cleavage of the glycan. The deglycosylated Fc was isolated using a Sephacryl S-200 HR size exclusion column (GE Healthcare) while monitoring UV absorbance and collecting peaks. The fractions containing deglycosylated Fc were pooled and concentrated to give 9 mg of the intermediate (Fucα1,6)GlcNAc-Fc. A solution of (Fucα1,6)GlcNAc-Fc (5 mg) and sialoglycan-oxazoline (5 mg) buffered with Tris-HCl (100 mM, pH 7.0, 0.5 mL) was incubated with EndoS-D233A (200 μg) [22] at 30°C. Aliquots were taken at time intervals for LC-MS analysis of reaction progression. Quantitative conversion was achieved in 2 hours. The product was purified using size exclusion chromatography as described above. Product fractions were pooled and concentrated to give di-sFc (4.5 mg). LC-MS data: calculated for (Fucα1,6)GlcNAc-Fc monomer, MW = 25287.3 Da; found MW = 25289 Da (deconvolution data obtained from the original spectrum). Calculated for fully sialylated N-glycan-Fc monomer, MW = 27288.9 Da; found MW = 27289 Da (deconvolution data).
N-glycan analysis
25 μg of each Fc fragment was treated with PNGase F (NEB) in a G7 buffer (0.5 M sodium phosphate, pH 7.5) following the manufacturer's instructions. Samples were purified over a CarboPac PGC column and analyzed using HPAEC-PAD on a Dionex ICS 5000 with a CarboPac PA200 column. The N-glycans were eluted in 100 mM NaOH with the following gradient conditions: 0-10 mM NaOAC in 20 min and then 10-100 mM NaOAc in 20 min. Peaks were assigned using N-glycan standards purified from PNGase F digestions of Rituximab, sheep IgG (Sigma), and sialylglycopeptide (SGP) from chicken egg yolk.
Monosaccharide compositional analysis of putative O-glycans
PNGase F-treated Fc proteins lacking N-glycans were dissolved in 3M TFA in screw cap vials. The tubes were sealed and incubated at 100 °C for 4 hours. Tubes were then cooled to 25 °C, and the reaction mixture was evaporated to dryness in a SpeedVac. The dried samples were dissolved in water and subjected to HPAEC-PAD analysis on a CarboPac PA10 column. Monosaccharides were eluted with 18 mM NaOH and detected with pulsed amperic detection with a gold electrode using a standard monosaccharide waveform. For analysis of sialic acids from putative O-glycans, the de-N-glycosylated Fc proteins were treated with neuraminidase (NEB) for 16 hours in the desialylation buffer provided by the manufacturer (NEB). The samples were then subjected to HPAEC-PAD analysis on a CarboPac PA10 column using a sialic acid standard (Sigma) for detection and quantification.
LC-ESI-MS Analysis of Fc fragments
Fc fragments and PNGase F-treated Fc fragments were reduced with 50 mM TCEP at 37 °C for 15 minutes and subjected to LC-ESI-MS analysis on an Agilent C8 column eluted with a gradient of 0-60% acetonitrile in 10 min.
Crystallization and data collection
Crystals of di-sFc (space group P21; a = 52.73 Å, b = 154.22 Å, c = 66.12 Å, β = 110.78°; two Fc dimers per asymmetric unit) were grown in sitting drop vapor diffusion by mixing equal volumes of di-sFc (6.15 mg/ml) with a solution containing 0.2 M magnesium chloride, 0.1 M sodium acetate pH 5, and 20% (w/v) PEG 6000 at 20°C. Crystals were cryopreserved in well solution supplemented with 30% glycerol. Data were collected to 3.0 Å resolution at beamline 8.2.1 of the Advanced Light Source (ALS) at Lawrence Berkeley National Laboratory (LBNL).
Crystals of wtFc (space group P212121; a = 49.56 Å, b = 79.69 Å, c = 138.06 Å; one Fc dimer per asymmetric unit) were grown in sitting drop vapor diffusion by mixing equal volumes of protein with a solution containing 0.1 M sodium cacodylate pH 6.5 and 18% (w/v) PEG 3350 at 20°C. The protein solution contained mainly 21c Fab produced by papain digestion of 21c IgG, but contaminating amounts of the Fc crystallized. The Fc crystals were cryopreserved in well solution containing 30% glycerol. F241A Fc crystals (space group P212121; a = 57.91 Å, b = 74.70 Å, c = 113.71 Å; one Fc dimer per asymmetric unit) were grown in hanging drops by mixing F241A Fc (4.4 mg/ml) with 30% PEG 1500 at 20°C and cryopreserved in well solution containing 25% glycerol. Data were collected for both wtFc (2.4 Å resolution) and F241A Fc (2.2 Å resolution) at beamline 12-2 of the Stanford Synchrotron Radiation Lightsource (SSRL).
Data Processing and Structure Determination
Diffraction data were processed and indexed using iMosflm [34] and integrated and scaled using POINTLESS and SCALA [35; 36]. In addition to considering I/σI and completeness of the highest resolution shell, we used the CC1/2 statistic [37] (correlation coefficient between two random halves of the data set where CC1/2> 10%) to determine the high-resolution cutoff for our data. We used Phenix [38] to compute CC1/2 values. Structures were solved by molecular replacement using PHASER [39] and published Fc structures as search models (PDB 1H3X for F241A Fc and wtFc; 3D03 for di-sFc). Modeling was done using COOT [40] using composite omit maps for building glycans and nearby protein residues in the CH2 domains. Crystallographic refinement was done using the Phenix crystallography package [38] by refining individual B factors for the higher resolution structures (F241A Fc and wtFc; 2.2 Å and 2.4 Å resolution, respectively). For the lower resolution di-sFc structure (3.0 Å resolution), we refined group B factors and used non-crystallographic symmetry (NCS) restraints during refinement. Because the CH2 domain orientations differed between the two di-sFc molecules in the asymmetric unit, the NCS restraints were applied to individual domains (CH2 domain residues Phe240 – Ile336, and residues CH3 domain Pro343 – Leu443 on each Fc chain). We used PyMol [41] for superposition calculations and molecular representations.
The F241A Fc model (Rfree = 23.2%; Rwork = 20.6%) included 421 protein residues in the F241A Fc dimer (Pro230 - Leu443 on chain A and Gly237- Leu443 on chain B), 16 glycan residues (GlcNAc1-Gal6, Fuc12 on chain A and GlcNAc1-GlcNAc8, Fuc12 on Chain B), and 351 water molecules. No electron density was observed for residues Thr225 and Cys226 of the Fc hinge or for Man7-NeuNAc11 or Gal9-NeuNAc11 in the N-linked glycan of chain A or B, respectively. The wtFc model (Rfree = 26.4%; Rwork = 21.1%) included 415 protein residues in the wtFc dimer (Pro238-Ser444 on chain A and Gly236-Pro443 on chain B), 16 glycan residues (GlcNAc1-GlcNAc5, Man7, GlcNAc8, Fuc12 on both chains A and B), and 165 water molecules. No electron density was observed for residues Cys226 to -Pro230 of the Fc hinge or for Gal6, Gal9, or NeuNAc11 on the N-glycan. Side chains were disordered for residues 269 and 311 on chain A and 246, 269, 311, and 326 on chain B. The di-sFc structure (Rfree = 28.0%; Rwork = 26.0%) included two conformationally distinct Fc dimers (chains A and B in the closed dimer and chains C and D in the open dimer). The closed di-sFc dimer model included 414 protein residues (Gly236 - Ser444 and Pro238 - Ser444 on chains A and B, respectively) and 16 glycan residues (GlcNAc1- GlcNAc5, Man7, GlcNAc8 on chain A and GlcNAc1 - GlcNAc8, NeuNAc11 on chain B). No electron density was observed for residues Glu225 to Pro230 of the Fc hinge or for Gal6, Gal9 - NeuNAc10 (chain A) or Gal9, NeuNAc11 on (chain B) in the N-linked glycan. Side chains were disordered for residues 246, 251, 258, 268, 269, 272, 274, 280, 288, 292, 294, 295, 296, 301, 307, 317, 326, 340, 355, 360, 365, 386, 410, 414, 416, 418, and 433 on chain A and 248, 269, 272, 274, 283, 298, 314, 326, 340, 355, 384, and 386 on chain B. The open di-sFc dimer model included 410 protein residues (residues Pro238 - Leu443 on each Fc chain) and 17 glycan residues (GlcNAc1-GlcNAc8, NeuNAc11 on chain C and,GlcNAc1-GlcNAc8 on chain D). No electron density was observed for residues Cys226 to Pro230 of the Fc hinge, Ala327-Ile332 (chain C), Val264-Asp270 (Chain D) or for Gal9, NeuNAc10 (chain C) or Gal9-NeuNAc11 (chain D) in the N-linked glycan. Side chains were disordered for residues 242, 246, 268, 269, 270, 272, 274, 285, 288, 290, 293, 295, 300, 301, 309, 311, 312, 320, 340, 342, 360, 380, 384, 386, 416, 418, 419, and 433 on chain C and 248, 263, 272, 276, 286, 288, 290, 295, 301, 302, 323, 332, 334, 360, 377, 384, 386, and 439 on chain D.
Supplementary Material
Highlights.
What structural changes occur with sialylation cause IgG anti-inflammatory state?
Sialylated Fc structures exist in open and closed conformations.
The sidechain of Phe241 can adopt an altered conformation in sialylated Fc.
F241A mutation causes increased inter-domain flexibility in Fc crystal structures.
Acknowledgments
We thank Jost Vielmetter and the Caltech Protein Expression Center for protein production, Beth Stadtmueller and members of the Bjorkman, Ravetch, and Wang laboratories for critical reading of the manuscript, Marta Murphy for help making figures, and Jens Kaiser and members of the staff at the Stanford Synchrotron Radiation Lightsource (SSRL) and the Advanced Light Source (ALS) for help with data collection and processing. Operations at SSRL are supported by the US Department of Energy and the National Institutes of Health. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This research was supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health Grant HIVRAD P01 AI100148 (P.J.B. and J.V.R.); (the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health), the National Institutes of Health (R01 AI041239 to P.J.B.), Award DP1OD006961 from the Office of The Director, National Institutes of Health (to P.J.B.), R01 GM096973 (to L.X.W.), R56 AI034662 (to J.V.R.), and the Molecular Observatory at Caltech supported by the Gordon and Betty Moore Foundation. P.J.B. is a Howard Hughes Medical Institute investigator.
Footnotes
Accession Numbers: Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4Q7D (wtFc), 4Q6Y (di-sFc), and 4Q74 (F241A Fc).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nimmerjahn F, Ravetch JV. Anti-inflammatory actions of intravenous immunoglobulin. Annu Rev Immunol. 2008;26:513–33. doi: 10.1146/annurev.immunol.26.021607.090232. [DOI] [PubMed] [Google Scholar]
- 2.Anthony RM, Ravetch JV. A novel role for the IgG Fc glycan: the anti-inflammatory activity of sialylated IgG Fcs. J Clin Immunol. 2010;30(Suppl 1):S9–14. doi: 10.1007/s10875-010-9405-6. [DOI] [PubMed] [Google Scholar]
- 3.Nimmerjahn F, Ravetch JV. FcgammaRs in health and disease. Curr Top Microbiol Immunol. 2011;350:105–25. doi: 10.1007/82_2010_86. [DOI] [PubMed] [Google Scholar]
- 4.Samuelsson A, Towers TL, Ravetch JV. Anti-inflammatory activity of IVIG mediated through the inhibitory Fc receptor. Science. 2001;291:484–6. doi: 10.1126/science.291.5503.484. [DOI] [PubMed] [Google Scholar]
- 5.Tackenberg B, Jelcic I, Baerenwaldt A, Oertel WH, Sommer N, Nimmerjahn F, Lunemann JD. Impaired inhibitory Fcgamma receptor IIB expression on B cells in chronic inflammatory demyelinating polyneuropathy. Proc Natl Acad Sci U S A. 2009;106:4788–92. doi: 10.1073/pnas.0807319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelfand EW. Differences between IGIV products: impact on clinical outcome. Int Immunopharmacol. 2006;6:592–9. doi: 10.1016/j.intimp.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–6. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nimmerjahn F, Ravetch JV. The antiinflammatory activity of IgG: the intravenous IgG paradox. J Exp Med. 2007;204:11–5. doi: 10.1084/jem.20061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–3. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 10.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 11.Schwab I, Mihai S, Seeling M, Kasperkiewicz M, Ludwig RJ, Nimmerjahn F. Broad requirement for terminal sialic acid residues and FcgammaRIIB for the preventive and therapeutic activity of intravenous immunoglobulins in vivo. Eur J Immunol. 2014 doi: 10.1002/eji.201344230. [DOI] [PubMed] [Google Scholar]
- 12.Anthony RM, Kobayashi T, Wermeling F, Ravetch JV. Intravenous gammaglobulin suppresses inflammation through a novel T(H)2 pathway. Nature. 2011;475:110–3. doi: 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sondermann P, Pincetic A, Maamary J, Lammens K, Ravetch JV. General mechanism for modulating immunoglobulin effector function. Proc Natl Acad Sci U S A. 2013;110:9868–72. doi: 10.1073/pnas.1307864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol. 2003;325:979–89. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 15.Feige MJ, Nath S, Catharino SR, Weinfurtner D, Steinbacher S, Buchner J. Structure of the murine unglycosylated IgG1 Fc fragment. J Mol Biol. 2009;391:599–608. doi: 10.1016/j.jmb.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 16.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Umana P, Benz J. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci U S A. 2011;108:12669–74. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, Kato K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol. 2007;368:767–79. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 18.Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 2005;310:1510–2. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 19.Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277:26733–40. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 20.Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, Prestegard JH. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry. 2012;51:4618–26. doi: 10.1021/bi300319q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sprague ER, Martin WL, Bjorkman PJ. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J Biol Chem. 2004;279:14184–93. doi: 10.1074/jbc.M313281200. [DOI] [PubMed] [Google Scholar]
- 22.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc. 2012;134:12308–18. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, Baruah K, Harvey DJ, Vasiljevic S, Alonzi DS, Song BD, Higgins MK, Bowden TA, Scanlan CN, Crispin M. Engineering hydrophobic protein-carbohydrate interactions to fine-tune monoclonal antibodies. J Am Chem Soc. 2013;135:9723–32. doi: 10.1021/ja4014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2-6)Gal/GalNAc sequence. J Biol Chem. 1987;262:1596–601. [PubMed] [Google Scholar]
- 25.Lund J, Takahashi N, Pound JD, Goodall M, Jefferis R. Multiple interactions of IgG with its core oligosaccharide can modulate recognition by complement and human Fc gamma receptor I and influence the synthesis of its oligosaccharide chains. J Immunol. 1996;157:4963–9. [PubMed] [Google Scholar]
- 26.Crispin M, Bowden TA, Coles CH, Harlos K, Aricescu AR, Harvey DJ, Stuart DI, Jones EY. Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. J Mol Biol. 2009;387:1061–6. doi: 10.1016/j.jmb.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 27.Oganesyan V, Damschroder MM, Leach W, Wu H, Dall'Acqua WF. Structural characterization of a mutated, ADCC-enhanced human Fc fragment. Mol Immunol. 2008;45:1872–82. doi: 10.1016/j.molimm.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 28.Oganesyan V, Damschroder MM, Woods RM, Cook KE, Wu H, Dall'acqua WF. Structural characterization of a human Fc fragment engineered for extended serum half-life. Mol Immunol. 2009;46:1750–5. doi: 10.1016/j.molimm.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 29.Teplyakov A, Zhao Y, Malia TJ, Obmolova G, Gilliland GL. IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface. Mol Immunol. 2013;56:131–9. doi: 10.1016/j.molimm.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Crispin M, Yu X, Bowden TA. Crystal structure of sialylated IgG Fc: implications for the mechanism of intravenous immunoglobulin therapy. Proc Natl Acad Sci U S A. 2013;110:E3544–6. doi: 10.1073/pnas.1310657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony RM, Wermeling F, Karlsson MC, Ravetch JV. Identification of a receptor required for the anti-inflammatory activity of IVIG. Proc Natl Acad Sci U S A. 2008;105:19571–8. doi: 10.1073/pnas.0810163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank M, Walker RC, Lanzilotta WN, Prestegard JH, Barb AW. Immunoglobulin G1 Fc Domain Motions: Implications for Fc Engineering. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diskin R, Marcovecchio PM, Bjorkman PJ. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol. 2010;17:608–13. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leslie AGW, Powell HR. Processing DIffraction Data with Mosfilm. In Evolving Methods for Macromolecular Crystallography. 2007;245:41–51. [Google Scholar]
- 35.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–92. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 37.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–3. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrödinger L. The PyMOL Molecular Graphics System 1.2r3pre edit. The PyMOL Molecular Graphics System 2011 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.