Abstract
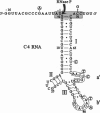
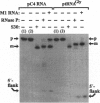
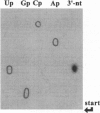
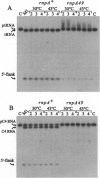
The C4 repressor of the temperate bacteriophages P1 and P7 inhibits antirepressor (Ant) synthesis and is essential for establishment and maintenance of lysogeny. C4 is an antisense RNA acting on a target, Ant mRNA, which is transcribed from the same promoter. The antisense-target RNA interaction requires processing of C4 RNA from a precursor RNA. Here we show that 5' maturation of C4 RNA in vivo depends on RNase P. In vitro, Escherichia coli RNase P and its catalytic RNA subunit (M1 RNA) can generate the mature 5' end of C4 RNA from P1 by a single endonucleolytic cut, whereas RNase P from the E. coli rnpA49 mutant, carrying a missense mutation in the RNase P protein subunit, is defective in the 5' maturation of C4 RNA. Primer extension analysis of RNA transcribed in vivo from a plasmid carrying the P1 c4 gene revealed that 5'-mature C4 RNA was the predominant species in rnpA+ bacteria, whereas virtually no mature C4 RNA was found in the temperature-sensitive rnpA49 strain at the restrictive temperature. Instead, C4 RNA molecules carrying up to five extra nucleotides beyond the 5' end accumulated. The same phenotype was observed in rnpA+ bacteria which harbored a plasmid carrying a P7 c4 mutant gene with a single C-->G base substitution in the structural homologue to the CCA 3' end of tRNAs. Implications of C4 RNA processing for the lysis/lysogeny decision process of bacteriophages P1 and P7 are discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alifano P., Rivellini F., Piscitelli C., Arraiano C. M., Bruni C. B., Carlomagno M. S. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 1994 Dec 15;8(24):3021–3031. doi: 10.1101/gad.8.24.3021. [DOI] [PubMed] [Google Scholar]
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Apirion D., Watson N. A second gene which affects the RNA processing enzyme ribonuclease P of Escherichia coli. FEBS Lett. 1980 Feb 11;110(2):161–163. doi: 10.1016/0014-5793(80)80062-7. [DOI] [PubMed] [Google Scholar]
- Biere A. L., Citron M., Schuster H. Transcriptional control via translational repression by c4 antisense RNA of bacteriophages P1 and P7. Genes Dev. 1992 Dec;6(12A):2409–2416. doi: 10.1101/gad.6.12a.2409. [DOI] [PubMed] [Google Scholar]
- Bothwell A. L., Garber R. L., Altman S. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA. J Biol Chem. 1976 Dec 10;251(23):7709–7716. [PubMed] [Google Scholar]
- Bothwell A. L., Stark B. C., Altman S. Ribonuclease P substrate specificity: cleavage of a bacteriophage phi80-induced RNA. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1912–1916. doi: 10.1073/pnas.73.6.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
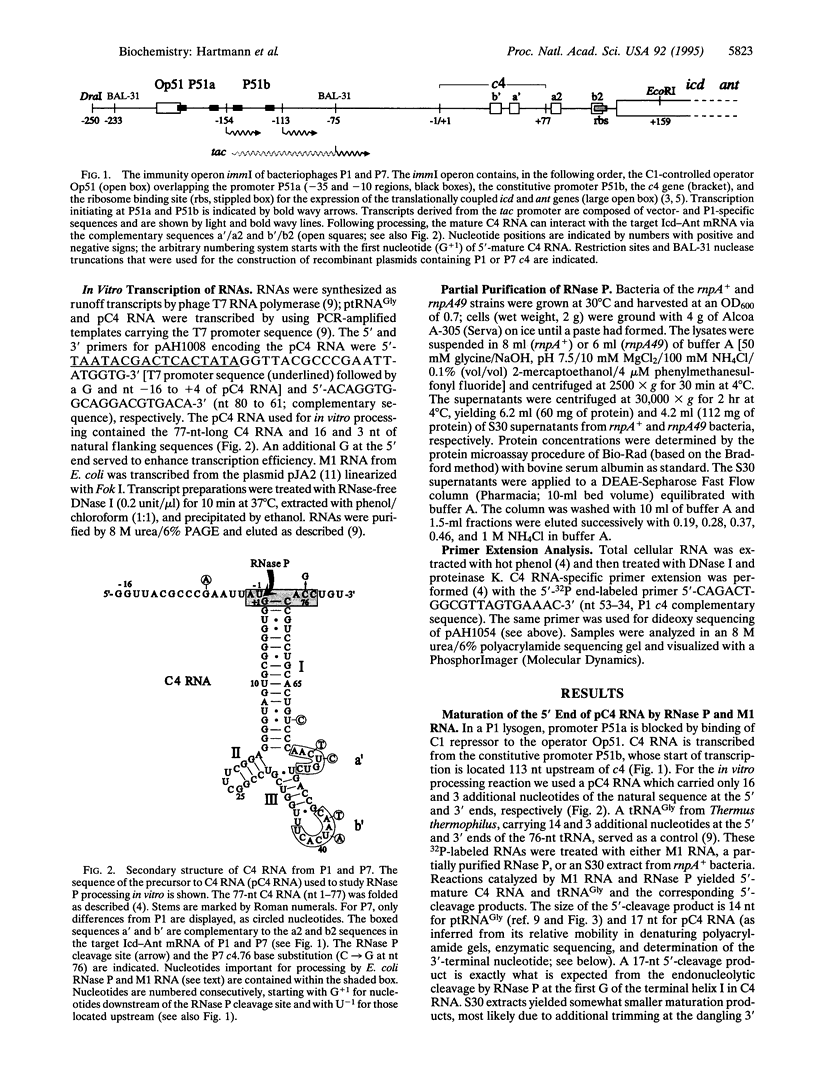
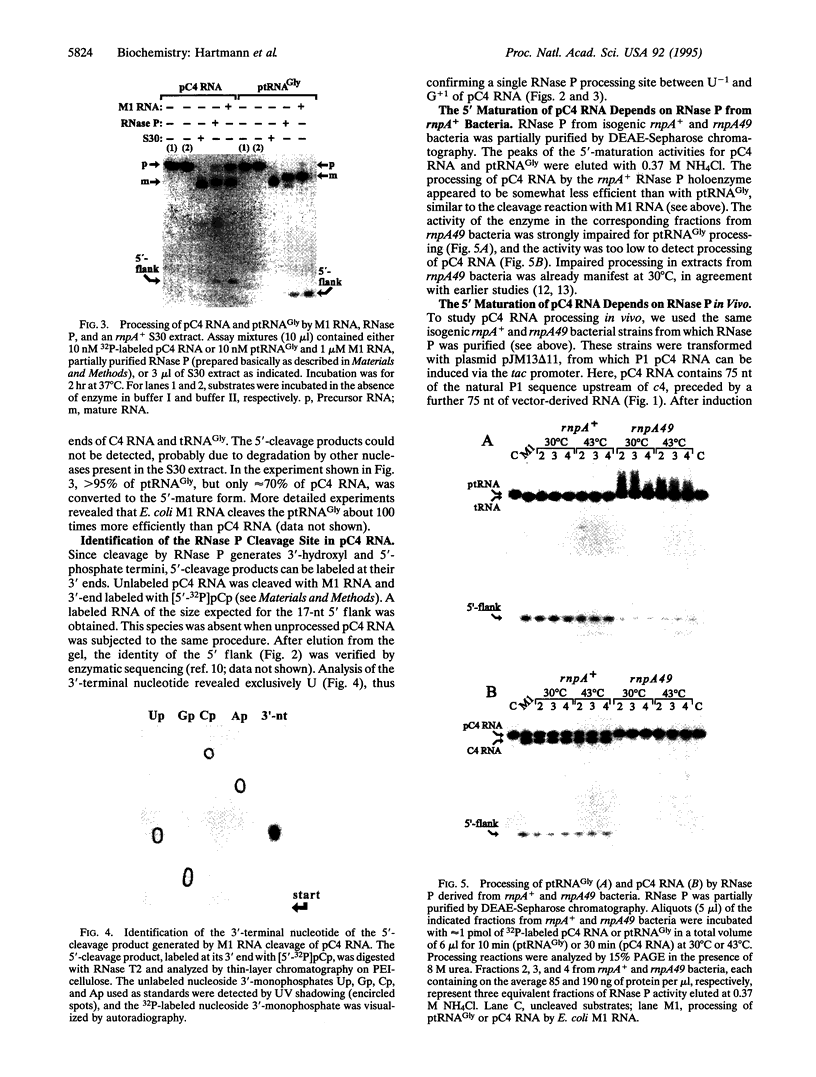
- Citron M., Schuster H. The c4 repressor of bacteriophage P1 is a processed 77 base antisense RNA. Nucleic Acids Res. 1992 Jun 25;20(12):3085–3090. doi: 10.1093/nar/20.12.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron M., Schuster H. The c4 repressors of bacteriophages P1 and P7 are antisense RNAs. Cell. 1990 Aug 10;62(3):591–598. doi: 10.1016/0092-8674(90)90023-8. [DOI] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Reconstitution of enzymatic activity from fragments of M1 RNA. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1266–1270. doi: 10.1073/pnas.89.4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisig A., Riedel H. D., Dobrinski B., Lurz R., Schuster H. Organization of the immunity region immI of bacteriophage P1 and synthesis of the P1 antirepressor. J Mol Biol. 1989 Oct 20;209(4):525–538. doi: 10.1016/0022-2836(89)90591-3. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Baer M. F., Altman S. Differential effects of mutations in the protein and RNA moieties of RNase P on the efficiency of suppression by various tRNA suppressors. J Mol Biol. 1988 Dec 20;204(4):879–888. doi: 10.1016/0022-2836(88)90048-4. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Svärd S. G. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994 Oct 17;13(20):4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y., Kitabatake M., Yokogawa T., Nishikawa K., Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Altman S. Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell. 1994 Jul 1;77(7):1093–1100. doi: 10.1016/0092-8674(94)90448-0. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Peck-Miller K. A., Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991 Sep 5;221(1):1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- Pragai B., Apirion D. Processing of bacteriophage T4 transfer RNAs. Structural analysis and in vitro processing of precursors that accumulate in RNase E-strains. J Mol Biol. 1982 Jan 25;154(3):465–484. doi: 10.1016/s0022-2836(82)80007-7. [DOI] [PubMed] [Google Scholar]
- Schedl P., Primakoff P., Roberts J. Processing of E. coli tRNA precursors. Brookhaven Symp Biol. 1975 Jul;(26):53–76. [PubMed] [Google Scholar]
- Schlegl J., Fürste J. P., Bald R., Erdmann V. A., Hartmann R. K. Cleavage efficiencies of model substrates for ribonuclease P from Escherichia coli and Thermus thermophilus. Nucleic Acids Res. 1992 Nov 25;20(22):5963–5970. doi: 10.1093/nar/20.22.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Svärd S. G., Kirsebom L. A. Determinants of Escherichia coli RNase P cleavage site selection: a detailed in vitro and in vivo analysis. Nucleic Acids Res. 1993 Feb 11;21(3):427–434. doi: 10.1093/nar/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd S. G., Kirsebom L. A. Several regions of a tRNA precursor determine the Escherichia coli RNase P cleavage site. J Mol Biol. 1992 Oct 20;227(4):1019–1031. doi: 10.1016/0022-2836(92)90518-o. [DOI] [PubMed] [Google Scholar]
- Vioque A., Arnez J., Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J Mol Biol. 1988 Aug 20;202(4):835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- Wagner E. G., Simons R. W. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]