Abstract
Rheumatic heart disease (RHD) affects heart-valve tissue and is the most serious consequence of group A Streptococcus infection. Myxomatous degeneration (MXD) is the most frequent valvopathy in the western world. In the present work, key protein expression alterations in the heart-valve tissue of RHD and MXD patients were identified and characterized, with controls from cadaveric organ donors. Proteins were separated by two-dimensional (2D)-electrophoresis and identified by mass spectrometry. We found 17 differentially expressed protein spots, as compared to control samples. We observed an increased expression of ASAP-2 in the RHD patients’ valves, while collagen-VI, haptoglobin-related protein, prolargin, and cartilage oligomeric protein showed reduced expression. Valve tissue of MXD patients, on the other hand, presented lower expression of annexin-A1 and A2, septin-2, SOD (Cu/Zn), and transgelin. Tissue samples from both valvopathies displayed higher expression of apolipoprotein-A1. Biglycan was downexpressed in both diseases. Vimentin and lumican showed higher expression in RHD and lower in MXD. These results suggest that key pathogenetic mechanisms are intrinsically distinct in RHD and MXD.
Keywords: heart-valve proteins, rheumatic heart disease, myxomatous degeneration, 2DE-DIGE, IPA
Introduction
Rheumatic fever (RF) is a post-infectious autoimmune disease that follows sore throat caused by Streptococcus pyogenes in untreated susceptible individuals. Human infection by S. pyogenes, a Gram-positive bacterium, may also have several distinct clinical presentations besides pharyngitis, such as pyoderma, impetigo, toxic shock syndrome, necrotizing fasciitis, and puerperal fever.1 Rheumatic heart disease (RHD) is the most serious complication of RF, leading to chronic valvar lesions. The estimated worldwide prevalence of RHD is more than 15.6 million cases, and it is responsible for approximately 233,000 deaths annually.2 Cases occur almost exclusively in developing countries.
The tissue most commonly suffering from severe damage by the chronic heart autoimmune reactions is the mitral valve, followed by the aortic valve. Although the myocardium can also be affected during acute RF, the lesions are self-limited and disappear in a few days or weeks, while the valve damage is progressive and permanent. The hallmarks of the disease include valve scarring, thickening of the chordae, leaflet verrucae, and angiogenesis. Heart-valves are organized connective tissue structures containing mainly two cell populations, the valve interstitial cells (VIC) that are responsible for extracellular matrix (ECM) turnover and valve endothelial cells (VEC) that surround the valve leaflets.3
Recurrences of acute RF episodes play an important role in valve disease progression because of the inflammatory process, which involves the reactivation of autoreactive T cells. In RHD, CD4+ infiltrating T cells cross-reactively recognize bacterial and human proteins and produce Th1 inflammatory cytokines, such as TNF-α and IFN-γ.4 So, RHD develops as autoimmune reactions in the valvar tissue, in the absence of S. pyogenes, because of a previous throat infection by the bacterium.
Genetic susceptibility is an important feature for the development of RHD. Several gene polymorphisms have been implicated, most of which are involved in the activation of the immune response.5
One recent hypothesis regarding the development of RHD lesions implicates the involvement of the sub-endothelial ECM. Histological findings and immunological studies in various organs, such as heart and vasculature, have suggested that connective tissue may be a common site for many manifestations of the disease.6
Myxomatous degeneration (MXD) is another valvopathy that preferentially affects mitral and aortic valves.7 The disease is characterized by elastic fiber alterations, ultimately inducing valvar dysfunction through valve prolapse, chordae rupture, and mucopolysaccharide accumulation. It also causes collagen disorganization and elastic fiber fragmentation.8 Although it is the most common valve disease in the United States and Europe, its pathophysiology remains largely unknown.9 In contrast to RHD, MXD is not associated with inflammation. Connective tissue dysfunction is likely the most important component of this disease.10 Genetically modified mice with constitutive expression of the highly catabolic metalloproteinase MMP-211 or ablation of the PRKAR1A gene,12 which is involved in protein kinase A signaling, induce myxomatous changes in heart-valves of mice. In spite of the aforementioned information, the valve proteins differentially involved in matrix remodeling between RHD and the non-autoimmune MXD remains elusive.
In the current work, we present a comprehensive set of proteomic analyses that shows a distinctive pattern of molecular signature between RHD and MXD valve lesions, and identify ECM proteins that are differentially expressed in RHD and MXD human mitral valves. To our knowledge, our results present the first high-throughput comparison of protein expression in RHD and MXD human mitral valves.
Methods
Sample collection
Mitral valve fragments were obtained at the time of surgery, from Brazilian adult individuals ethnically heterogeneous. The samples were separated into the two following groups: RHD with mitral valve lesions (n = 6, mean age = 45 years, 33% male, 66% female) and MXD with mitral valve lesions (n = 4, mean age = 60 years, 50% male, 50% female), who underwent surgery for valve replacement. The control group (CTL) consisted of mitral valve biopsies obtained from cadaveric organ donors without history of valvopathies (n = 4, mean age = 43 years, 50% male, 50% female). All valve tissues were dissected and frozen in liquid nitrogen immediately after cerebral death was declared, and the donor organs were removed (heart, liver, and kidney). This study was approved by the Ethics Committee from Heart Institute, School of Medicine, University of São Paulo (number 0053/07). Signed informed consent was obtained from all of the patients or from the relatives of organ donors prior to their inclusion in the study.
Protein extraction and two-dimensional (2D) differential gel electrophoresis (2DE-DIGE)
Unless otherwise stated, the reagents used for 2D-DIGE and mass spectrometry were purchased from GE Healthcare, Upsalla, Sweden. All the biopsied tissues were maintained in liquid nitrogen until processing. The samples were washed three times in ice-cold Tris-buffered saline (TBS) to remove blood contaminants; they were then disrupted in lysis buffer (urea 7 mol/L, thiourea 2 mol/L, CHAPS 4%, Tris–HCl 0.15 mol/L, pH 8.8) at 4 °C. The homogenates were sonicated in an ice bath, centrifuged at 10,000 × g for 45 minutes at 4 °C, and the pellets were discarded. Low molecular weight interferents were removed from the supernatants by dialysis in a 3.5 kDa cut-off membrane for 48 hours at 4 °C. The homogenates were then dried by vacuum centrifugation. The samples were resuspended in the same lysis buffer, and the protein was quantified by a modified Bradford method (Bio Rad Laboratories, Hercules, CA, USA). For accurate detection of differentially expressed protein spots, we used fluorescently labeled cyanines (Cy2™, Cy3™, and Cy5™). This method allows two samples to be run in the same gel, labeled with either Cy3™ or Cy5™, together with a normalizator pool of the samples labeled with Cy2™. SDS-PAGE (10%) analytical gels were loaded with 50 μg of each protein according to the manufacturer’s instructions. Preparative gels were loaded with 650 μg of the pooled samples plus 50 μg of the Cy2™-labeled pooled samples. Prior to electrophoresis, isoelectrofocusing (IEF) separation was performed with 24-cm-long strips (pH range 3–11) loaded with the samples and rehydrated for 12 hours. The gel analyses were conducted with the Decyder™ software to detect differences in spot intensity between the control and sample groups and to calculate the pI (isoelectric point) and molecular mass values of the protein spots.
Differential expression analysis
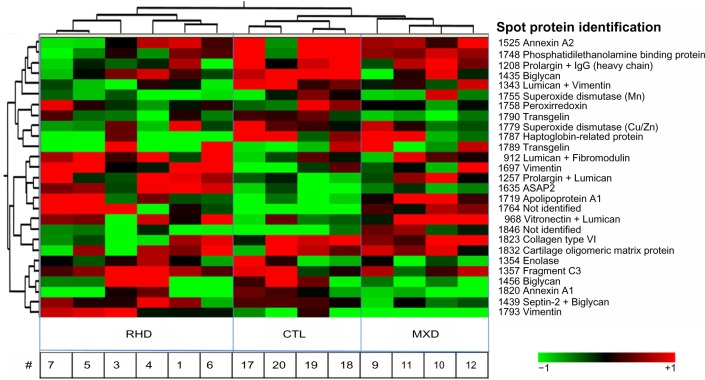
The spots with significant differential expression in the biological variation analysis (BVA) module of Decyder™ (P < 0.05) were chosen for differential in-gel analysis (DIA) in the same software in an initial approach to statistical analysis of differential expression. In the first place, we performed an unsupervised hierarchical clustering algorithm of all the differentially expressed proteins in the samples, using Pearson distance measure, which takes into consideration trends of similarity, and complete linkage of adjacent spots. Each line represents the relative abundance of a differentially expressed protein in each sample. Each column represents the protein extracts from a patient. Then we performed principal component analysis (PCA), with calculation of two principal components for grouping samples (data not shown). Protein spots with differential expression of valve samples from both groups (RHD and MXD) were also calculated in DIA, in which each sample was compared with the control (data not shown). The significance was evaluated using the ANOVA t-test (P < 0.05).
Mass spectrometry
Differentially expressed spots were excised and submitted to controlled proteolysis with trypsin (Promega, Madison, WY, USA). The mass-to-charge ratios of the derived peptides were measured by ESI-MS/MS (Ultima mass spectrometer, Waters™), with fragmentation of the five peaks with greater intensity in each protein spot by electron-transfer dissociation (ETD). The ionic masses were compared to primary sequence databases using the MASCOT search algorithm.13
Ingenuity pathway analysis (IPA)
We used IPA™ (Ingenuity Systems, CA, USA) to evaluate the biological relevance of the quantitatively altered protein dataset in the valve tissue of RHD and for MXD patients. Based on protein–protein interactions, IPA builds networks and generates a P-value by using Fisher’s exact test that fits the network to the user’s set of significant proteins or even genes in a genomic study. This P-value indicates the likelihood that the proteins are linked together only by chance. The IPA™ score is the negative logarithm of the P-value. The resulting network represents the molecules connected directly and indirectly, with experimental validation in the human proteome, according to IPA database.
Results
Protein separation and analysis
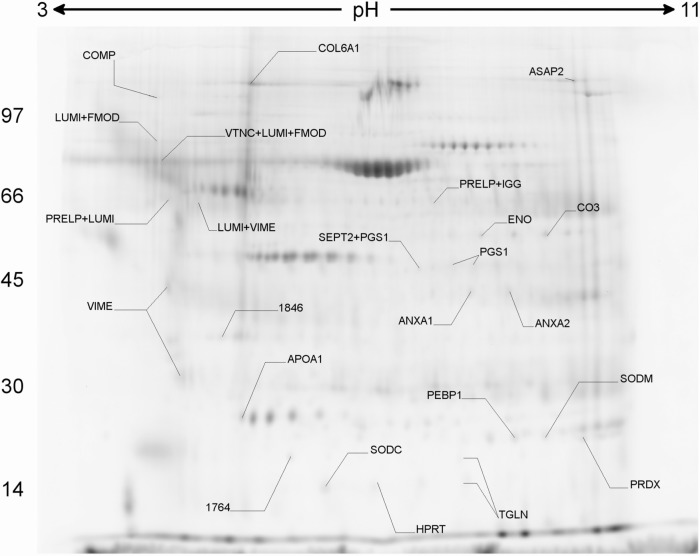
The mitral valve proteins were separated by 2D-DIGE, resulting in 451 detected protein spots. The supplementary table shows all the proteins that could be identified. The BVA analysis allowed the identification of 27 protein spots differentially expressed between gels (P < 0.05). These spots were identified (Fig. 1) and allowed to cluster the valve samples according to clinical type by hierarchical clustering algorithm (Fig. 2) indicating the existence of disease-specific protein expression profiles. In a more accurate analysis, taking the initial set of the 27 highly differentially expressed protein spots between the gels, we could identify 17 proteins with significant difference in expression (P < 0.05) between the groups. Table 1 shows information about these differentially expressed proteins. Exclusively in the RHD group, we found an increased expression of vimentin, lumican, and ASAP-2 (development and differentiation-enhancing factor-2), whereas collagen-VI, haptoglobin-related protein, prolargin, biglycan, and COMP (cartilage oligomeric matrix protein) displayed a lower expression. However, the MXD group showed a reduced expression of vimentin, annexin-A1 and -A2, biglycan, septin-2, superoxide dismutase (Cu/Zn), transgelin, and lumican. Only apolipoprotein-A1 was upregulated in both RHD and MXD patients, compared to control valve tissue.
Figure 1.
Preparative gel and differentially expressed spots. 2D-DIGE analysis of homogenates from a representative gel. The arrows represent the identification of the differentially expressed spots, according to UNIPROT accession codes for human proteins. Differences were considered to be statistically significant for P < 0.05 between groups.
Figure 2.
Differential protein profile. Hierarchical clustering of 27 spots with differential expression between the gels was grouped according to protein expression profile. Each line represents a protein spot, and each column, a sample. Distance measure: Pearson, with complete linkage between neighbor spots and samples. Valve tissue from patients: RHD (n = 6) and MXD (n = 4); CTL (n = 4). The symbol # denotes the sample identifications.
Table 1.
Differentially expressed proteins in the mitral valves with Rheumatic Heart Disease, or Myxomatous Degeneration.
| PROTEIN NAME | FOLD EXPRESSION | CELLULAR LOCATION | GENE ONTOLOGY: MOLECULAR PROCESS | ACCESS CODE | P-VALUE | MASCOT SCORE | COVERAGE (%) | SPOT NUMBER | EXPECTED | OBSERVED | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | pI | MW | pI | |||||||||
| Rheumatic heart disease | ||||||||||||
| Vimentin | 2.45 | Cytoplasm | Structure of cytoskeleton | P08670 | 0.009 | 266 | 14 | 1697 | 54 | 5.1 | 42 | 4.7 |
| Lumican | 1.74 | ECM | Receptor activity | P51884 | 0.034 | 410 | 32 | 1343 | 38 | 6.2 | 75 | 4.9 |
| ASAP-2 | 2.07 | Membrane, cytoplasm, Golgi | Nucleic acid binding | 043150 | 0.010 | 54 | 1 | 1635 | 112 | 6.2 | 37 | 6.2 |
| Collagen-VI | −1.82 | ECM | Receptor activity, extra-cellular matrix structural constituent | P12109 | 0.044 | 837 | 14 | 1823 | 109 | 5.3 | 109 | 5.3 |
| Haptoglobin-related protein | −2.53 | ECM | Oxidoreductase activity | P00739 | 0.032 | 74 | 3 | 1787 | 39 | 6.6 | 39 | 6.6 |
| Prolargin | −1.96 | ECM | Receptor activity | P51888 | 0.026 | 133 | 10 | 1208 | 44 | 9.5 | 61 | 4.3 |
| Biglycan | −1.60 | ECM | Collagen assembly | P21810 | 0.042 | 164 | 17 | 1435 | 42 | 7.2 | 51 | 7.5 |
| COMP | −1,70 | ECM | Collagen and fibronectin binding | P49747 | 0.048 | 114 | 13 | 1832 | 83 | 4.4 | 83 | 4.4 |
| Myxomatous degeneration | ||||||||||||
| Vimentin | −2.81 | Cytoplasm | Structure of cytoskeleton | P08670 | 0.022 | 60 | 4 | 1793 | 54 | 5.1 | 54 | 5.1 |
| Annexin A1 | −2.38 | Membrane | Calcium ion binding | P04083 | 0.007 | 149 | 13 | 1820 | 39 | 6.6 | 39 | 6.6 |
| Biglycan | −2.52 | ECM | Receptor activity | P21810 | 0.035 | 175 | 17 | 1456 | 42 | 7.2 | 51 | 7.5 |
| Septin-2 | −1.57 | Cytoplasm | GTPase activity, structural constituent of cytoskeleton | Q15019 | 0.048 | 70 | 12 | 1439 | 46 | 6.4 | 51 | 7.3 |
| SOD (Cu/Zn) | −2.26 | Mitochondria | Oxidoreductase activity | P04179 | 0.012 | 57 | 16 | 1779 | 16 | 5.7 | 25 | 6.3 |
| Transgelin | −2.09 | Cytoplasm | Structure of cytoskeleton | Q01995 | 0.034 | 138 | 28 | 1790 | 23 | 8.9 | 23 | 8.9 |
| Annexin A2 | −1.87 | ECM | Angiogenesis, fusion of vesicles | P07355 | 0.029 | 245 | 30 | 1525 | 41 | 8.5 | 36 | 8.7 |
| Lumican | −1.98 | ECM | Receptor activity | Q06828 | 0.013 | 330 | 33 | 1343 | 38 | 6.2 | 57 | 5.1 |
| Both valvopathies | ||||||||||||
| Apolipoprotein A1 | 2.36 | ECM | Lipid transport activity | P02647 | 0.040 | 539 | 42 | 1719 | 28 | 5.3 | 31 | 5.1 |
Notes: Mascot score, measures the accuracy of protein identification: values >37 are considered to represent protein identity. a P-value <0.05 is considered significant.
Abbreviations: ASAP-2, development and differentiation-enhancing factor-2; COMP, Cartilage Oligomeric Matrix Protein; SOD, superoxide dismutase; ECM, extracellular matrix; MW, molecular weight; pI, isoelectric point.
Predicted protein–protein interactions in valve damage
The differentially expressed proteins generated distinct lists that were uploaded in IPA (Ingenuity® Systems, www.ingenuity.com) and analyzed based on the content of October 2013. The IPA network maintains a graphical database of networks of interacting genes (Ingenuity Knowledge Base, IKB). The significance of the association between each list and the pathway was measured by Fisher’s exact test. As a result, a score-value was obtained, determining the probability that the association between the genes in our data set and the network generated can be explained by chance alone. Molecules are represented as nodes, and the biological relationship between the two nodes is represented as an edge. All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the IKB. This prediction reinforced the differential altered protein pattern in both RHD and MXD heart-valves. The networks indicate major protein interactions, quantitative alterations, and connecting functional elements that could not be extracted from the experimental data alone in the resulting scale-free topology of the network. These networks for RHD (Fig. 3A) and for MXD (Fig. 3B) presented high likelihood, with scores of 13 and 14, respectively. The protein with the highest centrality (measured by the number of edges linked to it) in the two networks is vimentin, presenting opposite expression changes. An important hub is TGF-beta, in both network predictions. Of note, the proinflammatory cytokine TNF-alpha is part of the network predicted for RHD, but not for MXD. Another crucial difference between the two networks is the involvement of MMP-25 in RHD.
Figure 3.
In silico analysis of the proteins differentially expressed in the valvopathies. IPA® was used to build a network of direct (solid lines) and indirect (dashed lines) interactions between proteins with differential expressions compared to the control group. Symbols in red denote proteins with increased expression and green decreased expression. The molecules in blank were added by the software and are predicted to be altered in each figure. (A) Network of protein–protein interactions for RHD: score of 13 and (B) the same network for MXD: score of 14.
Discussion
The data presented here associated differential expression of structural and ECM proteins with mitral valve damage in patients with end-stage RHD. The significant changes observed in the abundance of key cardiovascular proteins, such as vimentin, lumican, biglycan, and apolipoprotein-A1, strongly suggest that they can be involved in the pathogenesis of both RHD and MXD, but in different ways, and represent a molecular signature in valve lesions.
Among the 451 protein spots detected in the gels, an initial hierarchical clustering of 27 protein spots differentially expressed between the gels clustered the three study groups (RHD, MXD, and CTL). Differential expression mainly in ECM proteins indicates a tissue disordered in RHD valves. However, in the MXD group these alterations were found to be more evident in cytoplasm or plasma membrane proteins, indicating metabolic alterations. Some of these proteins appear in more than one spot, likely reflecting post-translational modifications, isoforms, or protein cleavage.
We observed a 42 kDa form of vimentin differentially expressed in the valves of the RHD patients. Vimentin is a structural protein that forms intermediate filaments throughout a regulated network of serine recognition sites. This protein binds to single-stranded nucleic acids via two repeats of a β-ladder DNA-binding wing located in its head domain.14 Recent studies have demonstrated additional roles for vimentin such as binding and stabilization of collagen mRNAs.15 In previous studies, we demonstrated that vimentin is recognized as an autoantigen by valve-infiltrating T lymphocytes in the valvar tissue.16 In the experiments performed here, most of the sequenced vimentin peptides were derived from the N-terminal portion of its primary sequence. Consistent with these data, 40 kDa vimentin proteolytic fragments have been described as initiators of neoangiogenesis in endothelial cells via a mechanism that depends on the MT1-MMP metalloprotease translocation to endothelial membranes.17 In the context of RHD, vimentin fragments may represent an autoimmune target (as described above) or a neoangiogenesis initiation factor in valvar tissues. However, patients with MXD did not display altered vimentin expression, suggesting that the mechanisms leading to valvar disease are distinct from those observed in MXD.18
Collagen-VI expression, on the other side, was remarkably decreased in the RHD groups. It is a structural component of the ECM whose alpha chain has 108 kDa. Previous studies have demonstrated that PARF, an octapeptide of different collagen regions, can induce antibodies that trigger a response that mimics RHD in rats.19 As collagen-VI and collagen-IV are known to interact,20 one can speculate that collagen type-IV autoantibodies might also be responsible for valve lesion development in humans with RHD. Collagen-IV, whose alpha chain has 161 kDa, could not be evaluated by the method used.
Like collagen, lumican is present in the ECM of human cartilaginous tissues.21 Lumican is a member of the small leucine-rich proteoglycan (SLRP) family and plays a regulatory role by orienting and ordering collagen fibrils, tissue hydration, and tissue repair and regeneration.22,23 Lumican expression has been observed in several tissue types, such as cartilage,24 lung,25 and arteries.26 Although the immature form of lumican is a proteoglycan, it exists predominantly in a glycoprotein form that lacks keratan sulfate. Several studies have demonstrated that lumican and other SLRP members can modulate cell migration and proliferation in tumor growth.27 We observed that a spot in which lumican was differentially expressed migrated with an apparent molecular mass of 72 kDa, while the entire protein has a mass of 38.4 kDa. This mass difference likely reflects glycosylation because lumican contains four glycosylation sites. In addition, molecules such as laminin and N-acetyl β-D-glucosamine have been reported to represent targets of cross-reactive antibodies in RHD-affected valves.28,29 Like collagen-IV, laminin was beyond the molecular mass range of the analysis and could not be evaluated. Laminin antibodies, however, may interfere in the interaction with lumican, decreasing the availability of this protein in the valves to bind to collagen-VI.
We also observed that vitronectin, a glycoprotein with molecular mass of 53 kDa, showed increased expression in the RHD group. These results were consistent with stiffness and fibrosis of valvar inflamed tissues that occurs as a result of vitronectin accumulation,30 as this protein is highly involved in rigidity secondary to remodeling. Although vitronectin regulates adhesion events, it is also involved in a number of proteolytic enzyme cascades, including the complement, coagulation, and fibrinolytic systems.31 Vitronectin represents a unique regulatory link between cell adhesion, humoral defense mechanisms, and cell invasion.32 In addition, increased vitronectin depositions have been observed in areas of tissue injury and necrosis. It is possible that the increased protein levels reflect the process of tissue injury in both diseases.
The in silico analysis predicted the involvement of important molecules in the protein–protein interaction networks generated, such as TNF-alpha and MMP-25 involvement in RHD, but not in MXD. The former is an important proinflammatory cytokine, whose high levels in the valve tissue have already been described in RHD.33 MMP-25 is a metalloprotease highly involved in matrix degradation,34 but its role in RHD has not been described yet. Our current work is the first, to our knowledge, to associate metalloprotease to the development of RHD.
Although we expected similar protein expression profiles in the two valvopathies studied, because fibrosis is a common clinical manifestation of these diseases in end-stage, as a result of remodeling mechanisms in both MXD35 and RHD,36 remarkable molecular differences were observed. The results of our study can show a clear distinct pattern, exclusive in each disease state that may help explain the clinical findings according to molecular alterations, both observed and predicted in silico.
Conclusion
In conclusion, an overall protein imbalance was observed in the valvar constituents, mainly in the ECM. These changes highlight the importance of a differential protein expression profile in the progression to RHD, once the structural integrity of valve tissues is lost, reflecting dysfunction of interstitial cells that are responsible for its maintenance.37 The heart-valve tissues analyzed represent end-stage disease and may not reflect all the pathogenetically relevant steps. Our results promote a deeper understanding of mitral valve end-stage lesions mediated by matrix disorganization and autoimmune responses. Moreover, the specific molecular signatures may represent potential biomarkers for RHD.
Supplementary Data
Supplementary Table 1. All mitral valve proteins that could be identified after separation by 2D-DIGE.
Acknowledgments
We thank Prof. Dr. Francisco Laurindo for helpful discussion and comments on drafts of the manuscript. We thank Dr. Adriana Paes Leme, from LNBio, for providing access to the mass spectrometry facility.
Footnotes
Author Contributions
Conceived and designed the experiments: COM, KSS, PCT, LG, ECN. Analyzed the data: COM, KSS, JK, LG, ECN. Wrote the first draft of the manuscript: COM, KSS, FMF. Contributed to the writing of the manuscript: PMAP, CMAB, ROS, GSS. Agree with manuscript results and conclusions: COM, KSS, FMF, PCT, PMAP, CMAB, ROS, GSS, JK, LG, ECN. Jointly developed the structure and arguments for the paper: COM, FMF, LG, ECN. Made critical revisions and approved final version: JK, LG, ECN. All authors reviewed and approved the final version.
ACADEMIC EDITOR: Thomas Vanhecke, Editor in Chief
FUNDING: The article is supported by CAPES (Coordination for the Improvement of Higher Education Personnel), iii-INCT (Institute of Investigation in Immunology, National Institute for Science and Technology), FAPESP (São Paulo Research Foundation process number 2008/57881–0), and CNPq (National Council for Scientific and Technological Development).
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
REFERENCES
- 1.Ekelund K, Skinhøj P, Madsen J, Konradsen HB. Invasive group A, B, C and G streptococcal infections in Denmark 1999–2002 epidemiological and clinical aspects. Clin Microbiol Infect. 2005;11(7):569–76. doi: 10.1111/j.1469-0691.2005.01169.x. [DOI] [PubMed] [Google Scholar]
- 2.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5(11):685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 3.Hinton RB, Yutzey KE. Heart valve structure and function in development and disease. Annu Rev Physiol. 2011;73:29–46. doi: 10.1146/annurev-physiol-012110-142145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilherme L, Köhler KF, Postol E, Kalil J. Genes, autoimmunity and pathogenesis of rheumatic heart disease. Ann Pediatr Cardiol. 2011;4(1):13–21. doi: 10.4103/0974-2069.79617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guilherme L, Köhler KF, Kalil J. Rheumatic heart disease: mediation by complex immune events. Adv Clin Chem. 2011;53:31–50. [PubMed] [Google Scholar]
- 6.Tandon R, Sharma M, Chandrashekhar Y, Kotb M, Yacoub MH, Narula J. Revisiting the pathogenesis of rheumatic fever and carditis. Nat Rev Cardiol. 2013 doi: 10.1038/nrcardio.2012.197. [DOI] [PubMed] [Google Scholar]
- 7.He Y, Guo Y, Li Z, et al. Echocardiographic determination of the prevalence of primary myxomatous degeneration of the cardiac valves. J Am Soc Echocardiogr. 2011;24(4):399–404. doi: 10.1016/j.echo.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Pellerin D, Brecker S, Veyrat C. Degenerative mitral valve disease with emphasis on mitral valve prolapse. Heart. 2002;88(suppl 4):iv20–8. doi: 10.1136/heart.88.suppl_4.iv20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caira FC, Stock SR, Gleason TG, et al. Human degenerative valve disease is associated with up-regulation of low-density lipoprotein receptor-related protein 5 receptor-mediated bone formation. J Am Coll Cardiol. 2006;47(8):1707–12. doi: 10.1016/j.jacc.2006.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhtar S, Meek KM, James V. Ultrastructure abnormalities in proteoglycans, collagen fibrils, and elastic fibers in normal and myxomatous mitral valve chordae tendineae. Cardiovasc Pathol. 1999;8(4):191–201. doi: 10.1016/s1054-8807(99)00004-6. [DOI] [PubMed] [Google Scholar]
- 11.Mahimkar R, Nguyen A, Mann M, et al. Cardiac transgenic matrix metalloproteinase-2 expression induces myxomatous valve degeneration: a potential model of mitral valve prolapse disease. Cardiovasc Pathol. 2009;18(5):253–61. doi: 10.1016/j.carpath.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Z, Jones GN, Towns WH2nd et al. Heart-specific ablation of Prkar1a causes failure of heart development and myxomagenesis. Circulation. 2008;117(11):1414–22. doi: 10.1161/CIRCULATIONAHA.107.759233. [DOI] [PubMed] [Google Scholar]
- 13.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Shoeman RL, Hartig R, Traub P. Characterization of the nucleic acid binding region of the intermediate filament protein vimentin by fluorescence polarization. Biochemistry. 1999;38(51):16802–09. doi: 10.1021/bi991654r. [DOI] [PubMed] [Google Scholar]
- 15.Challa AA, Stefanovic B. A novel role of vimentin filaments: binding and stabilization of collagen mRNAs. Mol Cell Biol. 2011;31(18):3773–89. doi: 10.1128/MCB.05263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faé KC, Diefenbach daSilva D, Bilate AM, et al. PDIA3, HSPA5 and vimentin, proteins identified by 2-DE in the valvular tissue, are the target antigens of peripheral and heart infiltrating T cells from chronic rheumatic heart disease patients. J Autoimmun. 2008;31(2):136–41. doi: 10.1016/j.jaut.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Kwak HI, Kang H, Dave JM, et al. Calpain-mediated vimentin cleavage occurs upstream of MT1-MMP membrane translocation to facilitate endothelial sprout initiation. Angiogenesis. 2012;15(2):287–303. doi: 10.1007/s10456-012-9262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Demellawy M, El-Ridi R, Guirguis NI, Abdel Alim M, Kotby A, Kotb M. Preferential recognition of human myocardial antigens by T lymphocytes from rheumatic heart disease patients. Infect Immun. 1997;65(6):2197–205. doi: 10.1128/iai.65.6.2197-2205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinkla K, Nitsche-Schmitz DP, Barroso V, et al. Identification of a streptococcal octapeptide motif involved in acute rheumatic fever. J Biol Chem. 2007;282(26):18686–93. doi: 10.1074/jbc.M701047200. [DOI] [PubMed] [Google Scholar]
- 20.Kuo HJ, Maslen CL, Keene DR, Glanville RW. Type VI collagen anchors endothelial basement membranes by interacting with type IV collagen. J Biol Chem. 1997;272(42):26522–9. doi: 10.1074/jbc.272.42.26522. [DOI] [PubMed] [Google Scholar]
- 21.Hocking AM, Shinomura T, McQuillan DJ. Leucine-rich repeat glycoproteins of the extracellular matrix. Matrix Biol. 1998;17(1):1–19. doi: 10.1016/s0945-053x(98)90121-4. [DOI] [PubMed] [Google Scholar]
- 22.Iozzo RV. The family of the small leucine-rich proteoglycans: key regulators of matrix assembly and cellular growth. Crit Rev Biochem Mol Biol. 1997;32(2):141–74. doi: 10.3109/10409239709108551. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarti S, Magnuson T, Lass JH, Jepsen KJ, LaMantia C, Carroll H. Lumican regulates collagen fibril assembly: skin fragility and corneal opacity in the absence of lumican. J Cell Biol. 1998;141(5):1277–86. doi: 10.1083/jcb.141.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grover J, Chen XN, Korenberg JR, Roughley PJ. The human lumican gene. Organization, chromosomal location, and expression in articular cartilage. J Biol Chem. 1995;270(37):21942–9. doi: 10.1074/jbc.270.37.21942. [DOI] [PubMed] [Google Scholar]
- 25.Dolhnikoff M, Morin J, Roughley PJ, Ludwig MS. Expression of lumican in human lungs. Am J Respir Cell Mol Biol. 1998;19(4):582–7. doi: 10.1165/ajrcmb.19.4.2979. [DOI] [PubMed] [Google Scholar]
- 26.Funderburgh JL, Funderburgh ML, Mann MM, Conrad GW. Arterial lumican. Properties of a corneal-type keratan sulfate proteoglycan from bovine aorta. J Biol Chem. 1991;266(36):24773–7. [PubMed] [Google Scholar]
- 27.Vuillermoz B, Khoruzhenko A, D’Onofrio MF, et al. The small leucine-rich proteoglycan lumican inhibits melanoma progression. Exp Cell Res. 2004;296(2):294–306. doi: 10.1016/j.yexcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13(3):470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guilherme L, Faé K, Oshiro SE, Kalil J. Molecular pathogenesis of rheumatic fever and rheumatic heart disease. Expert Rev Mol Med. 2005;7(28):1–15. doi: 10.1017/S146239940501015X. [DOI] [PubMed] [Google Scholar]
- 30.Reilly JT, Nash JR. Vitronectin (serum spreading factor): its localisation in normal and fibrotic tissue. J Clin Pathol. 1988;41(12):1269–72. doi: 10.1136/jcp.41.12.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seiffert D. Constitutive and regulated expression of vitronectin. Histol Histopathol. 1997;12(3):787–97. [PubMed] [Google Scholar]
- 32.Preissner KT. Structure and biological role of vitronectin. Annu Rev Cell Biol. 1991;7:275–310. doi: 10.1146/annurev.cb.07.110191.001423. [DOI] [PubMed] [Google Scholar]
- 33.Guilherme L, Cury P, Demarchi LM, et al. Rheumatic heart disease: proinflammatory cytokines play a role in the progression and maintenance of valvular lesions. Am J Pathol. 2004;165(5):1583–91. doi: 10.1016/S0002-9440(10)63415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kojima S, Itoh Y, Matsumoto S, Masuho Y, Seiki M. Membrane-type 6 matrix metalloproteinase (MT6-MMP, MMP-25) is the second glycosyl-phosphatidyl inositol (GPI)-anchored MMP. FEBS Lett. 2000;480:2–3. 142–6. doi: 10.1016/s0014-5793(00)01919-0. [DOI] [PubMed] [Google Scholar]
- 35.Rabkin E, Aikawa M, Stone JR, Fukumoto Y, Libby P, Schoen FJ. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104(21):2525–32. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- 36.Kim L, Kim doK, Yang WI, et al. Overexpression of transforming growth factor-beta 1 in the valvular fibrosis of chronic rheumatic heart disease. J Korean Med Sci. 2008;23(1):41–8. doi: 10.3346/jkms.2008.23.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu AC, Joag VR, Gotlieb AI. The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology. Am J Pathol. 2007;171(5):1407–18. doi: 10.2353/ajpath.2007.070251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. All mitral valve proteins that could be identified after separation by 2D-DIGE.