Abstract
FAS-associated protein with death domain (FADD) is a major adaptor protein involved in extrinsic apoptosis, embryogenesis, and lymphocyte homeostasis. Although abnormalities of the FADD/death receptor apoptotic pathways have been established in tumorigenesis, fewer studies have analyzed the expression and role of phosphorylated FADD (pFADD). Our identification of FADD as a lymphoma-associated autoantigen in T-cell lymphoma patients raises the possibility that pFADD, with its correlation with cell cycle, may possess role(s) in human T-cell lymphoma development. This immunohistochemical study investigated pFADD protein expression in a range of normal tissues and lymphomas, particularly T-cell lymphomas that require improved therapies. Whereas pFADD was expressed only in scattered normal T cells, it was detected at high levels in T-cell lymphomas (eg, 84% anaplastic large cell lymphoma and 65% peripheral T cell lymphomas, not otherwise specified). The increased expression of pFADD supports further study of its clinical relevance and role in lymphomagenesis, highlighting phosphorylation of FADD as a potential therapeutic target.
Keywords: FADD, pFADD, lymphoma, autoantigen, ALCL, PTCL
Introduction
Autoantibody signatures have provided invaluable information for assisting both in the identification and early detection of disease.1–4 Furthermore autoantigens, such as anaplastic lymphoma kinase (ALK), which are functionally implicated in tumor progression, represent important therapeutic targets.5,6
In a previous study, protein microarrays were screened with sera from lymphoma patients to identify lymphoma-associated antigens. The aim was to identify autoantigens that might represent new tools for identifying high risk patients and/or novel treatment options for patients with systemic ALK-negative anaplastic large cell lymphoma (ALCL), a provisional tumor entity with a poor prognostic outlook. The FAS-associated protein with death domain (FADD), encoded by a gene within 11q13, was identified as an autoantigen recognized by circulating antibodies present not only in the sera of patients with systemic ALCL but also in patients with peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS), as well as B-cell lymphoma.7 PTCL, in common with systemic ALK-negative ALCL, represents a poorly defined lymphoma category with a poor prognostic outlook for which improved therapies are urgently required.8
Multiple roles have been ascribed to the FADD protein. Containing a death domain (DD) and a death effector domain (DED), FADD is a major adaptor protein in the extrinsic apoptotic pathway linking transduction signals from membrane-bound death receptors, eg, CD95 and DR4, with caspase 8.9,10 More recently, interactions of FADD with RIP1 and RIP3 have been identified, with FADD being a negative regulator of a programmed form of necrotic cell death, known as “necrotopsis.”11,12 FADD is also required for embryogenesis and lymphocyte proliferation via pathways that are independent of death receptor-mediated apoptosis,13–16 the DD motif acting as a switch between apoptosis and proliferation.16 The pivotal roles of FADD in cell cycle regulation and proliferation17,18 occur following its phosphorylation (at serine 194 in humans or serine 171 in mice) and nuclear relocalization at the G2/M stage of cell cycle.19 Nuclear pFADD may also be involved in gene surveillance through its regulation with human telomerase reverse transcriptase20 and its association with complexes containing the DNA repair molecules MBD4 and MLH1.21
FADD has been associated with disease development in humans.22–25 The amplification of the FADD gene and its correlation with increased FADD protein expression in head and neck cancers led Gibcus et al. to describe FADD as a possible key gene of functional interest in the 11q13 amplicon.26 Conflicting evidence has, however, been obtained on the relevance of high levels of pFADD to patient outcome in a number of solid tumors such as head and neck cancer,26,27 adenocarcinoma,22,28 gastric cancer,29 and prostate cancer.20 Higher levels of pFADD were, however, found to increase chemosensitization in prostate cancer cells23,30 and HeLa xenografts.31
Of relevance to the current study is that chromosomal abnormalities of the 11q13 region, other than those associated with Cyclin D1, have also been linked to hematological malignancies32,33 thus increasing the possibility of the involvement of FADD in these tumor types. Since pFADD has also been reported to correlate with proliferation in a range of B-cell non-Hodgkin lymphomas (NHLs),34 the current immunohistochemical study on the detection of pFADD focuses on T-cell malignancies to determine its potential relevance as a therapeutic target in these tumors.
Materials and Methods
Tissue samples
Normal tissues
Fresh tonsil was obtained from patients attending the Ear, Nose and Throat Department, John Radcliffe Hospital, Oxford. Paraffin-embedded normal tissues were obtained from the Centro Nacional de Investigaciones Oncologicas (CNIO), Madrid.
Hematological malignancies
Tissue microarrays (TMAs) of a variety of lymphomas were obtained from the CNIO while TMAs of ALK-positive and ALK-negative ALCL and PTCL were obtained from the Department of Pathology, Wüerzburg. The 39 B-cell lymphomas consisted of the following cases: diffuse large B-cell (DLBCL, n = 9), mantle cell lymphoma (MCL, n = 7), follicular lymphoma (FL, n = 7), Burkitt’s lymphoma (BL, n = 6), and Hodgkin’s lymphoma (HL, n = 10). The 227 T-cell tumors consisted of the following cases: PTCL (NOS) (n = 89), ALK-positive ALCL (n = 38), ALK-negative ALCL (n = 54), angioimmunoblastic lymphoma (AITL, n = 15), angiocentric lymphoma (n = 5), T-cell lymphoblastic lymphoma (TCL, n = 7), cutaneous T-cell lymphoma (CTCL, n = 3), T/NK lymphoma (n = 2), mycosis fungoides (MF, n = 6), and T-cell intestinal lymphoma (n = 8).
All normal and neoplastic cells and tissues were obtained only after ethical approval and informed consent had been given.
Cell lines
The Thiel (myeloma), SUDHL-1, Karpas 299 (ALK-positive ALCL), FEPD (ALK-negative ALCL). Jurkat and CEM (T-acute lymphoblastic leukemia), U937 and HL60 (acute myeloid leukemia), germinal center – SUDHL-6, SUDHL-10, and nongerminal center RIVA and HBL-1 (DLBC)-derived cell lines used were obtained and cultured as previously described.5 The cultured cells were used to produce cytocentrifuge preparations, cell pellets, or fixed in 10% formal-saline for paraffin embedding for the preparation of tissue sections of the cell lines.
Antibodies
Monoclonal antibodies
Mouse monoclonal antibodies to CD8 (C8/144), CD45RA (4KB5), and CD68 (KP1) were produced in the authors’ laboratories. Antibodies to CD20 (Clone L26), cytokeratins (Clones Lp34 and Clones MNF116), and β-actin (diluted 1:100) were obtained from DakoCytomation (Glostrup, Denmark).
Polyclonal antibodies
A rabbit polyclonal antibody to FADD phosphorylated at Ser174 was obtained from Cell Signalling Technology (catalog number 2781, Cambridge, UK). Anti-CD3 (DAKO-CD3 – diluted 1:100), anti-mouse Ig conjugated to horseradish-peroxidase (HRP) diluted 1:100, and the Dako EnVision staining kit were obtained from DakoCytomation.
Immunolabeling techniques
Immunoperoxidase
Cytocentrifuge preparations and cryostat sections of tonsil were stained using a two-stage immunoperoxidase technique as previously described.5 Paraffin-embedded tissue sections and microarrays were de-waxed, and antigen retrieval was carried out in 50 mM Tris:2 mM EDTA pH 9.0 by microwave pressure cooking for 2 minutes. The tissue sections were then incubated with a primary antibody for 60 minutes at room temperature (or in the case of anti-pFADD overnight at 4 °C) before being washed in phosphate-buffered saline (PBS) for 5 minutes. Antibody binding was detected using the ChemMate™ DAKO EnVision™ Detection Kit, Peroxidase/DAB using the manufacturer’s instructions. Cells were counterstained using hematoxylin and mounted in Aquamount (VWR International, Lutterworth, UK).
Double immunoperoxidase labeling
pFADD staining was performed as described above but with omission of the counterstaining step. The sections were then incubated with one of the following: anti-CD3, anti-CD20, anti-cytokeratin, anti-CD8, or anti-CD68 for 30 minutes at room temperature before using the Envision™ anti-mouse/rabbit Dual Link reagent. After washing in PBS for 5 minutes, the slides were incubated with the vector SG Peroxidase (HRP) substrate kit (Vector Laboratories, Peterborough, UK). After washing, the slides were air-dried overnight and mounted in VectorMount (Vector Laboratories).
Results
pFADD distribution in normal tissues
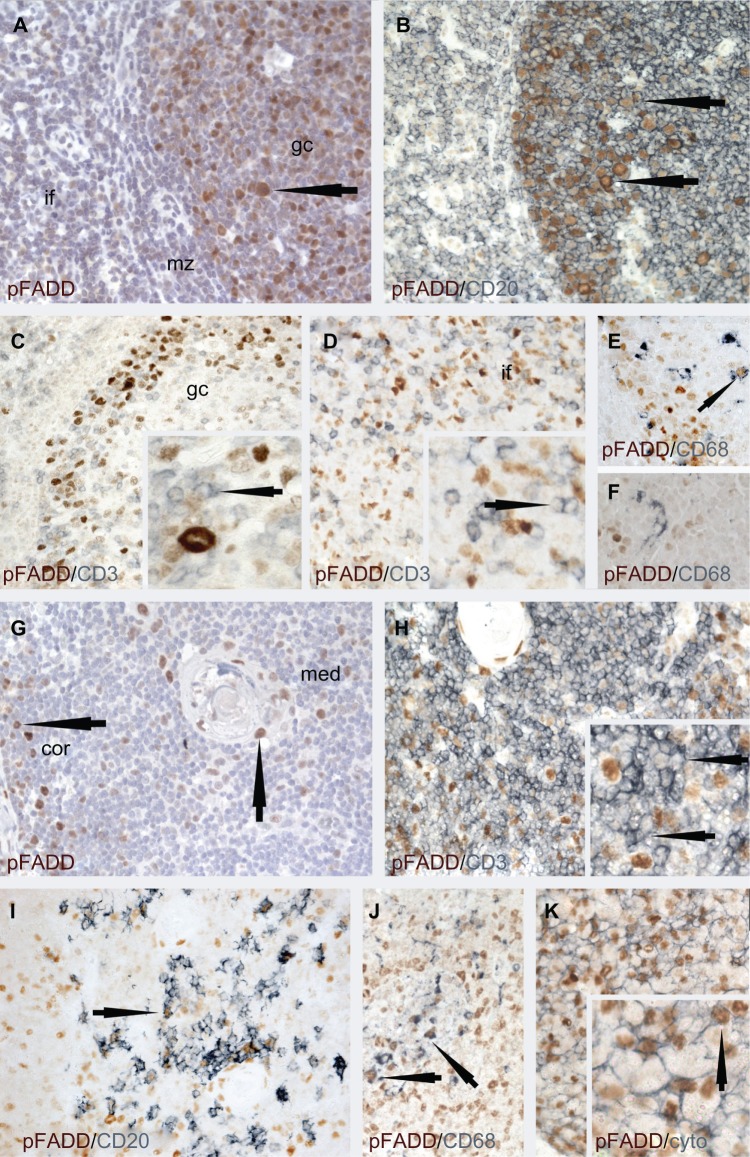
The antibody recognizing pFADD has been used in a number of previous studies.22,26,34,35 Its use in the current study showed pFADD to have a limited nuclear distribution in normal tissues, as summarized in Table 1. Of particular interest was the differential expression of pFADD in populations of hematopoietic cells. High levels of pFADD were detected only in subpopulations of tonsillar germinal center and interfollicular cells (Fig. 1A). Double labeling studies confirmed the majority of these cells to be CD20-positive B cells (Fig. 1B). In contrast, pFADD was undetectable in the majority of the CD3-positive T cells (Figs. 1C and D), with only low levels of pFADD observed in a minority of CD3-positive T cells. Although a small subpopulation of CD68-positive tingible body macrophages expressed pFADD (Fig. 1E), no interfollicular cells were observed that were stained for both pFADD and CD68 (Fig. 1F).
Table 1.
Expression of pFADD in normal cells and tissues.
| TISSUE | *pFADD DISTRIBUTION |
|---|---|
| Hematopoietic | |
| Bone marrow | Megakaryocytes and occasional large blast cells |
| Tonsil | Epithelium, subpopulations of germinal center B-cells, tingible body macrophages and subpopulations of interfollicular T and B cells |
| Thymus | Subpopulations of B cells, macrophages, epithelium, and scattered T cells |
| Non-Hematopoietic | |
| Brain | No labeling detected |
| Thyroid | Occasional nucleus of epithelial cells |
| Lung | No labeling detected |
| Breast | No labeling detected |
| Colon | Subset of crypt epithelial cells |
| Liver | Occasional hepatocyte |
| Kidney | Occasional cell in tubule |
| Pancreas | Acinar and islet cells |
| Fallopian tubes | Subset of epithelial cells |
| Uterus | Scattered epithelial cells |
| Testis | Majority of spermatogonia and weak labeling of Leydig cells. |
| Placenta | Scattered syncitiotrophoblast |
| Prostate | Subset of epithelial cells |
| Bladder | Epithelial cells |
Note:
Nuclear staining.
Figure 1.
Immunolabeling studies to show pFADD protein expression in tonsil (A–F) and thymus (G–J).
Notes: Tonsil: (A) Single immunoperoxidase labeling shows heterogeneity of pFADD expression in the nuclei of subpopulations of tonsil cells. Strong labeling is present in a subpopulation of germinal center (gc) cells (arrowed). Scattered cells with weaker labeling are present in the mantle zone (mz) and interfollicular (if) areas. (B) It can be seen from the double immunoperoxidase labeling studies that pFADD (brown) is heterogeneously expressed in the majority of CD20-positive (blue) germinal center B cells (arrowed) but (C) absent from the majority of CD3-positive (blue) T cells in the germinal center (inset, arrowed) and (D) interfollicular areas (inset, arrowed). (E) Nuclear pFADD expression (brown) is also present in the occasional CD68-positive tingible body macrophage (blue, arrowed) while (F) pFADD was not detected in the interfollicular CD68-positive macrophages (arrowed). Thymus: From (G) it can be seen that the expression of pFADD is heterogenous and limited to subpopulations of cells (arrowed) in the thymic medulla (med) and cortex (cor). (H) pFADD was undetectable in the majority of the CD3-positive 9blue) thymocytes (inset, arrowed), pFADD was undetectable in the majority of the CD3-positive (blue) thymocytes (inset, arrowed). (I) Strong pFADD expression (brown) was, however, detected in (I) some CD20-positive B cells (blue, arrowed), (J) CD68-positive macrophages (blue, arrowed), and (K) the cytokeratin-positive (blue) thymic epithelial cells (inset, arrowed).
In the thymus, strong pFADD staining was limited to subsets of cells in the medulla and cortex (Fig. 1G). Double labeling studies showed only an occasional CD3-positive cell to be pFADD-positive, with the majority of CD3-positive T cells lacking detectable pFADD (Fig. 1H, inset, arrowed). The remaining pFADD-positive cells included subpopulations of CD20-positive B cells (Fig. 1I), CD68-positive macrophages (Fig. 1J) as well as thymic epithelial cells (Fig. 1K).
pFADD expression in hematological malignancies
pFADD expression was also detected in T-cell derived as well as B-cell and myeloid-derived cell lines. The staining was nuclear and generally strong. Cytoplasmic staining was also seen in the myeloid (K562, HL60, and U937), megakaryocytic (HEL), and the T-acute lymphoblastic leukemia (Jurkat and CEM) cell lines tested. The results are summarized in Table 2.
Table 2.
pFADD protein expression in cell lines derived from hematological malignancies.
| CELL LINE | *pFADD |
|---|---|
| T-cell | |
| FEPD | Nuclei ++ >95% |
| SUDHL-1 | Nuclei + >95% |
| Karpas 299 | Nuclei heterogenous +/++ >50% |
| Jurkat | Nuclei ++ >90% |
| CEM | Nuclei/cytoplasm ++ <50% cells |
| B-cell | |
| SUDHL-6 | Nuclei + >75% |
| SUDHL-10 | Nuclei ++ 90% |
| RIVA | Nuclei + 20% |
| HLY-1 | Nuclei +/− 80% |
| HBL-1 | Nuclei + 40% |
| Thiel | Nuclei +/− 1% |
| Myeloid/megakaryotic | |
| K562 | Nuclei/cytoplasm ++ <95% |
| HL60 | Nuclei/cytoplasm ++ <95% |
| U937 | Nuclei/cytoplasm ++ <95% |
| HEL | Nuclei/cytoplasm ++ <95% |
Notes:
Cytoplasmic and nuclear intensity of labeling is as follows: ++ = strong, + = moderate and +/− = weak, % refers to proportion of cells labeled. FEPD – ALK-negative anaplastic large cell lymphoma; SUDHL-1 and Karpas 299 – ALK-positive anaplastic large cell lymphoma; Jurkat and CEM – T-acute lymphoblastic leukemia; SUDHL-6 and SUDHL-10 – germinal center-derived diffuse large B-cell lymphoma; RIVA and HBL-1 – non-germinal center-derived diffuse large B-cell lymphoma; Thiel – myeloma; K562 – chronic myeloid leukemia; U937 and HL60 – acute myeloid leukemia; HEL – megakaryocytic-derived cell lines.
A preliminary staining study was performed on mixed lymphoma TMAs, and the results are summarized in Table 3. pFADD proteins were detected across a range of B-cell lymphomas. Heterogeneity in the intensity of staining of pFADD was also seen both within and between cases of DLBCL and MCL (not shown).
Table 3.
pFADD expression in hematological malignancies.
| IMMUNOLABELING RESULTS | ||||
|---|---|---|---|---|
| TOTAL CASES | NEGATIVE | <30% POSITIVE CELLS N (% TOTAL CASES) |
>30% POSITIVE CELLS N (% TOTAL CASES) |
|
| B-cell lymphomas | ||||
| Diffuse large B-cell lymphoma | 9 | 2 (22%) | 4 (43%) | 3 (33%) |
| Mantle cell lymphoma | 7 | 1 (14%) | 4 (57%) | 2 (29%) |
| Follicular lymphoma (n = 7) | 7 | 4 (57%) | 3 (43%) | 0 (0%) |
| Burkitt lymphoma | 6 | 3 (50%) | 2 (33%) | 1 (17%) |
| Hodgkin’s lymphoma Lymphocyte predominant | 3 | 0 (0%) | 1 (33%) | 2 (66%) |
| Hodgkin’s lymphoma Mixed cellularity | 2 | 0 (0%) | 0 (0%) | 2 (100%) |
| Hodgkin’s lymphoma Nodular sclerosis | 2 | 0 (0%) | 0 (0%) | 2 (100%) |
| Hodgkin’s lymphoma Lymphocyte rich | 3 | 0 (0%) | 0 (0%) | 3 (100%) |
| T-cell lymphomas | ||||
| Peripheral T-cell lymphoma (not otherwise specified) | 89 | 31 (35%) | 49 (55%) | 9 (10%) |
| ALK+ Anaplastic large cell lymphoma | 38 | 6 (16%) | 8 (21%) | 24 (63%) |
| ALK− Anaplastic large cell lymphoma | 54 | 9 (17%) | 5 (9%) | 40 (74%) |
| Angioimmunoblastic lymphoma | 15 | 5 (33.3%) | 5 (33.3%) | 5 (33.3%) |
| Angiocentric lymphoma | 5 | 0 (0%) | 3 (60%) | 2 (40%) |
| T lymphoblastic | 7 | 3 (43%) | 3 (43%) | 1 (14%) |
| Cutaneous T-cell lymphoma | 3 | 1 (33.5%) | 2 (66.5%) | 0 (0%) |
| Mycosis Fungoides | 6 | 3 (50%) | 2 (33%) | 1 (17%) |
| T/NK-cell lymphoma | 2 | 0 (0%) | 2 (100%) | 0 (0%) |
| T-cell intestinal lymphoma | 8 | 0 (0%) | 3 (37%) | 5 (63%) |
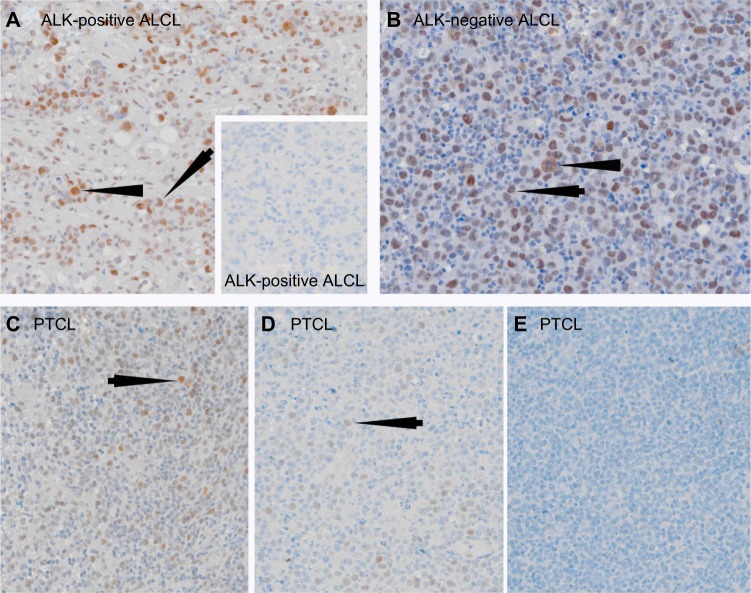
In view of the importance of FADD in normal T-cell survival and proliferation,17 the previously reported expression of pFADD in a number of B-cell malignancies,34 and the presence of an anti-FADD immune response in patients with T-cell malignancies,7 we analyzed pFADD expression in 227 T-cell tumors, with particular emphasis on ALCL and PTCL (NOS). The results are shown in Table 3. Strong labeling of the tumor cells was observed in 32/38 (84%) ALK-positive and 45/54 (83%) ALK-negative ALCL. Examples of results obtained are shown in Figures 2A and 2B. The numbers of tumor cells expressing pFADD varied from tumor biopsy to tumor biopsy (range 10%–99%). Moderate to weak pFADD labeling was also observed in other T-cell tumors. A total of 58/89 (65%) cases of PTCL (NOS) were positive for pFADD. Labeling of more than 30% of cells was detected in 9 (10%) of these cases while 49 (55%) cases showed less than 30% of the tumor cells to be pFADD-positive. pFADD expression was not detected in 31 (35%) cases of PTCL (NOS). Examples of staining are shown in Figures 2C–2E). Heterogeneity in the levels of staining of pFADD was observed within tumors in both ALCL and PTCL (NOS). pFADD expression was detected in a range of other T-cell lymphomas, including 67% (10/15) AITL, 57% (4/7) T-ALL, 50% (3/6) MF, and 66% (2/3) CTCL cases.
Figure 2.
Single immunoperoxidase labeling demonstrating pFADD expression in ALCL and PTCL (NOS).
Notes: (A) An example of a case of ALK-positive ALCL where the tumor cells express pFADD (arrowed). Note the presence of pFADD in the cytoplasm of mitotic cells (arrowhead). The inset shows an example of a pFADD-negative case of ALK-positive ALCL. (B) Strong nuclear pFADD expression is also present in a case of ALK-negative ALCL (arrowed). Cytoplasmic pFADD is also present in a mitotic cell (arrowhead). Different examples of pFADD in three cases of PTCL are shown in (C–E). While the majority of tumor cells are pFADD-positive (arrowed) in (C), the percentage drops to less than 30% in (D) while the PTCL in (E) lacks detectable pFADD.
Additional evidence to support the dysregulation of FADD in T-cell tumors was obtained by analyzing levels of FADD transcripts from the publicly available Oncomine database (www.oncomine.org). Analysis of two independent gene expression profiling (GEP) datasets indicated that the FADD transcripts were significantly more highly expressed in T-cell lymphomas compared to nonmalignant T-cell populations (Fig. 3). Thus T-cell malignancies exhibit both elevated FADD transcription and FADD phosphorylation.
Figure 3.
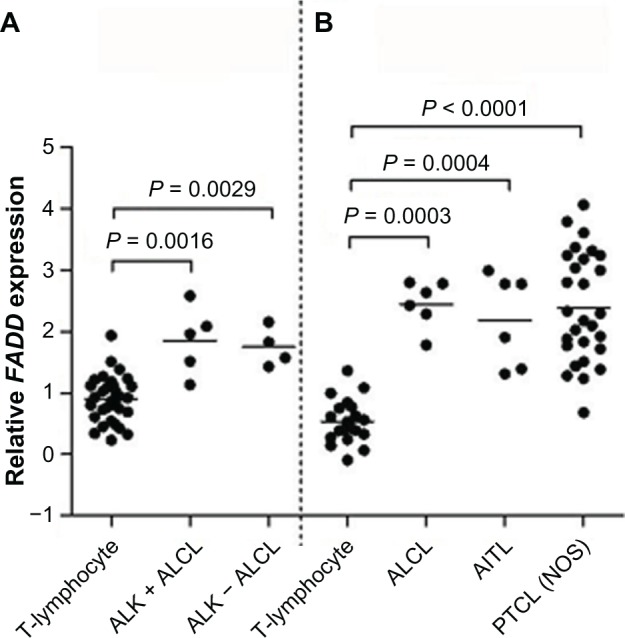
FADD transcript expression in T-cell lymphomas and ALCL.
Notes: FADD transcript was expressed at significantly higher levels in T-cell lymphomas than in nonmalignant T-cell populations in two independent gene expression profiling datasets: (A) normal T-lymphocytes, n = 31; ALCL (anaplastic large cell lymphoma), n = 9 (five cases of ALK-positive ALCL and four cases of ALK-negative ALCL);38 and (B) normal T-lymphocytes, n = 20 (including five samples each of CD4-positive and CD8-positive T-lymphocytes); ALCL, n = 6; AITL (angioimmunoblastic T-cell lymphoma), n = 6; PTCL (NOS) (peripheral T-cell lymphoma, not otherwise specified), n = 28.8
Discussion
Although pFADD has been studied in a number of solid tumors, investigations of its expression in human hematological malignancies are limited. We previously used sera from ALCL patients to screen a protein microarray containing more than 12,000 different proteins with the aim of identifying lymphoma-associated antigens that could identify high-risk patients or that might represent novel therapeutic targets. Four antigens, FADD, transcription elongation factor A protein-like 2 (TCEAL2), lipid phosphate phosphate-related protein type 3 (LPPR3), and ribonuclease T2 (RNASET2), were identified as ALK-negative ALCL-associated autoantigens. Further validation studies also confirmed FADD, TCEAL2, and RNASET2 to be PTCL-associated autoantigens recognized by circulating antibodies in patients.7 This result, combined with the interest in the role of pFADD in oncogenesis, indicated the importance of performing an immunohistochemical study of pFADD protein expression, in a range of T-cell lymphomas.
High levels of pFADD protein had a limited distribution in normal tissues and were restricted to the nucleus. These results are in agreement with previous studies reporting nuclear pFADD protein in subpopulations of human gastric epithelial cells, hepatocytes, kidney, and tonsillar cells.28,29,34,36 The current study using double labeling techniques is, however, the first not only to confirm high levels of pFADD expression in subpopulations of normal CD20-positive B cells but also to identify the absence or restricted low-level expression of pFADD in normal tonsillar T cells and thymocytes. Since the presence of pFADD has been linked to T-cell activation and proliferation in human cell lines and in mice,17,18,37 the lack of pFADD in T cells in the current study suggests that there are only low numbers of activated T cells in the G2/M stage of cell cycle in normal tonsil and thymus.
pFADD was observed in B-cell-derived cell lines and tumors, a result in keeping with a previous study where pFADD expression was found to correlate with the proliferation of B-lymphoma cells.34 pFADD expression was, however, also detected in a wide variety of T-cell leukemia-derived cell lines and in primary T-cell lymphoma biopsies. 65% PTCL (NOS) and 83% ALCL expressed levels of pFADD. No significant difference was found between ALK-positive and ALK-negative ALCL, with high levels of pFADD being detected in 84% and 83% of ALK-positive and ALK-negative ALCL, respectively. Although pFADD protein expression was previously reported in ALCL and CTCL, Clarke et al investigated only nine cases of ALCL and three cases of CTCL.36 The increased expression of pFADD protein in T-cell malignancies compared to normal T-cells studied here suggests that pFADD is accumulated possibly as a result of the constitutive activation of the tumor cells. The increase in pFADD could also result from increased expression of FADD or its increased phosphorylation as part of the disease process. The evidence from two GEP datasets8,38 (shown in Fig. 3), where increased levels of FADD transcripts were observed in PTCL, AITL, ALK-positive and ALK-negative ALCL, compared to normal T cells, suggests that the increased pFADD protein expression in some patients is likely to reflect the upregulation of FADD transcription and thus, most likely, increased FADD protein.
A major factor in lymphoma growth is resistance to apoptosis. Examples include abnormalities in the Fas/FasL and CD30-mediated apoptotic pathways as well as c-FLIP overexpression in T-cell lymphomas such as ALK-positive ALCL,39 and ALK-negative tumors such as CTCL and mycosis fungoides.40,41 It is possible that re-localization of FADD to the nucleus following its phosphorylation19,42 may play a major role in the dysregulation of apoptosis in human lymphomas as reported for murine T-ALL.43 pFADD overexpression has also been linked to increased activity of the anti-apoptotic NF-κB protein.22,44 and activation of the c-jun NH2-terminal kinase (JNK) signaling pathway,24,30 both of which are implicated in T-cell lymphomagenesis.45,46 Aberrant pFADD expression has also been linked with proliferation in human B-cell lymphomas34 and to cell cycle defects in murine T cells,37 with hematopoietic as well as solid tumor cells arresting at the G2/M stage.18,19,23,24
Further investigations are required to determine the prognostic relevance of pFADD in T-cell malignancies, which has been linked with varying outcomes in malignancies in other studies. Increased levels of pFADD and NF-κB have been linked to poorer survival in head and neck26 and lung cancers,22,28 while others have described low pFADD expression to be associated with poorer prognosis in myeloid leukemia.47 There is also a reported link between higher levels of pFADD and improved local tumor control in early stage larynx cancer27 and prostate cancer.20 Of interest is that the increased transition from FADD to pFADD has been associated with an improved sensitivity to treatment with chemotherapy in prostate cancer23,30 and radiotherapy in head and neck cancer.27
Although ALK-positive ALCL is associated with a good prognosis,48 the same cannot be said for ALK-negative ALCL and PTCL-NOS, and improved therapies are urgently needed for these lymphomas.49–52 FADD and its interacting proteins, such as RIP1, are expressed in both ALK-negative and ALK-positive ALCL53 and may thus represent exciting therapeutic targets. Although there is evidence for a tumor suppressor role of pFADD in nonhematological tumors,31 it will be important to study further the biological activity of pFADD in T-cell malignancies. Of interest was a report that a novel inhibitor of FADD phosphorylation resulted in decreased levels of the anti-apoptotic NF-κB,44 a factor implicated in lymphogenesis,54 suggesting the potential of pFADD as a therapeutic target in T-cell malignancies.
In conclusion, the results from the current study provide the first detailed immunohistochemical study of the expression of pFADD in normal tissues and hematological malignancies, with particular emphasis on T-cell tumors. The increased expression of pFADD observed in T-cell lymphomas not only suggests a possible role in lymphoma development but also further highlights phosphorylation of the lymphoma-associated autoantigen FADD as a potential therapeutic target.
Acknowledgments
We would like to thank Maite Cabes, Dr Graham Collins, and Dr Andrew Campbell for their help in obtaining clinical data and samples.
Footnotes
Author Contributions
SP, DM, HC, ZAA, EG, EH, KKW, APA and KP performed the research and analyzed the results. GR and CSRH contributed essential reagents. KP, AHB, and DM designed the research and analyzed the data. All authors made an intellectual input and contributed to writing the paper. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: Steve Myers, Editorial Board
FUNDING: This research was funded by Julian Starmer-Smith Lymphoma Fund, Leukaemia & Lymphoma Research, Sam Foye Fund, National Institute for Health Research (NIHR) Oxford Biomedical Research Center Programme, and the Science Foundation Ireland and Enterprise Ireland (PC/2007/193). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
REFERENCES
- 1.Horn S, Lueking A, Murphy D, et al. Profiling humoral autoimmune repertoire of dilated cardiomyopathy (DCM) patients and development of a disease-associated protein chip. Proteomics. 2006;6(2):605–13. doi: 10.1002/pmic.200401293. [DOI] [PubMed] [Google Scholar]
- 2.Kijanka G, Murphy D. Protein arrays as tools for serum autoantibody marker discovery in cancer. J Proteomics. 2009;72(6):936–44. doi: 10.1016/j.jprot.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Kijanka G, Hector S, Kay EW, et al. Human IgG antibody profiles differentiate between symptomatic patients with and without colorectal cancer. Gut. 2010;59(1):69–78. doi: 10.1136/gut.2009.178574. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Yu J, Sreekumar A, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–35. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 5.Cooper CD, Liggins AP, Ait-Tahar K, Roncador G, Banham AH, Pulford K. PASD1, a DLBCL-associated cancer testis antigen and candidate for lymphoma immunotherapy. Leukemia. 2006;20(12):2172–4. doi: 10.1038/sj.leu.2404424. [DOI] [PubMed] [Google Scholar]
- 6.Ait-Tahar K, Damm-Welk C, Burkhardt B, et al. Correlation of the autoantibody response to the ALK oncoantigen in pediatric anaplastic lymphoma kinase-positive anaplastic large cell lymphoma with tumor dissemination and relapse risk. Blood. 2010;115(16):3314–9. doi: 10.1182/blood-2009-11-251892. [DOI] [PubMed] [Google Scholar]
- 7.Patel S, Chen H, Monti L, et al. RNASET2 – an autoantigen in anaplastic large cell lymphoma identified by protein array analysis. J Proteomics. 2012;75(17):5279–92. doi: 10.1016/j.jprot.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Piccaluga PP, Agostinelli C, Califano A, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. 2007;117(3):823–34. doi: 10.1172/JCI26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldin MP, Varfolomeev EE, Pancer Z, Mett IL, Camonis JH, Wallach D. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J Biol Chem. 1995;270(14):7795–8. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 10.Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81(4):505–12. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 11.Green DR, Oberst A, Dillon CP, Weinlich R, Salvesen GS. RIPK-dependent necrosis and its regulation by caspases: a mystery in five acts. Mol Cell. 2011;44(1):9–16. doi: 10.1016/j.molcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471(7338):373–6. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh WC, de la Pompa JL, McCurrach ME, et al. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science. 1998;279(5358):1954–8. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Cado D, Chen A, Kabra NH, Winoto A. Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature. 1998;392(6673):296–300. doi: 10.1038/32681. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Rosenberg S, Wang H, Imtiyaz HZ, Hou YJ, Zhang J. Conditional Fas-associated death domain protein (FADD): GFP knockout mice reveal FADD is dispensable in thymic development but essential in peripheral T cell homeostasis. J Immunol. 2005;175(5):3033–44. doi: 10.4049/jimmunol.175.5.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imtiyaz HZ, Zhou X, Zhang H, Chen D, Hu T, Zhang J. The death domain of FADD is essential for embryogenesis, lymphocyte development, and proliferation. J Biol Chem. 2009;284(15):9917–26. doi: 10.1074/jbc.M900249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua ZC, Sohn SJ, Kang C, Cado D, Winoto A. A function of Fas-associated death domain protein in cell cycle progression localized to a single amino acid at its C-terminal region. Immunity. 2003;18(4):513–21. doi: 10.1016/s1074-7613(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 18.Alappat EC, Volkland J, Peter ME. Cell cycle effects by C-FADD depend on its C-terminal phosphorylation site. J Biol Chem. 2003;278(43):41585–8. doi: 10.1074/jbc.C300385200. [DOI] [PubMed] [Google Scholar]
- 19.Scaffidi C, Volkland J, Blomberg I, Hoffmann I, Krammer PH, Peter ME. Phosphorylation of FADD/MORT1 at serine 194 and association with a 70-kDa cell cycle-regulated protein kinase. J Immunol. 2000;164(3):1236–42. doi: 10.4049/jimmunol.164.3.1236. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura Y, Shimada K, Tanaka N, Fujimoto K, Hirao Y, Konishi N. Phosphorylation status of Fas-associated death domain-containing protein regulates telomerase activity and strongly correlates with prostate cancer outcomes. Pathobiology. 2009;76(6):293–302. doi: 10.1159/000245895. [DOI] [PubMed] [Google Scholar]
- 21.Screaton RA, Kiessling S, Sansom OJ, et al. Fas-associated death domain protein interacts with methyl-CpG binding domain protein 4: a potential link between genome surveillance and apoptosis. Proc Natl Acad Sci U S A. 2003;100(9):5211–6. doi: 10.1073/pnas.0431215100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Bhojani MS, Heaford AC, et al. Phosphorylated FADD induces NF-kappaB, perturbs cell cycle, and is associated with poor outcome in lung adenocarcinomas. Proc Natl Acad Sci U S A. 2005;102(35):12507–12. doi: 10.1073/pnas.0500397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimada K, Matsuyoshi S, Nakamura M, Ishida E, Konishi N. Phosphorylation status of Fas-associated death domain-containing protein (FADD) is associated with prostate cancer progression. J Pathol. 2005;206(4):423–32. doi: 10.1002/path.1791. [DOI] [PubMed] [Google Scholar]
- 24.Matsuyoshi S, Shimada K, Nakamura M, Ishida E, Konishi N. FADD phosphorylation is critical for cell cycle regulation in breast cancer cells. Br J Cancer. 2006;94(4):532–9. doi: 10.1038/sj.bjc.6602955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cimino Y, Costes A, Damotte D, et al. FADD protein release mirrors the development and aggressiveness of human non-small cell lung cancer. Br J Cancer. 2012;106(12):1989–96. doi: 10.1038/bjc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibcus JH, Menkema L, Mastik MF, et al. Amplicon mapping and expression profiling identify the Fas-associated death domain gene as a new driver in the 11q13.3 amplicon in laryngeal/pharyngeal cancer. Clin Cancer Res. 2007;13(21):6257–66. doi: 10.1158/1078-0432.CCR-07-1247. [DOI] [PubMed] [Google Scholar]
- 27.Schrijvers ML, Pattje WJ, Slagter-Menkema L, et al. FADD expression as a prognosticator in early-stage glottic squamous cell carcinoma of the larynx treated primarily with radiotherapy. Int J Radiat Oncol Biol Phys. 2011;83(4):1220–26. doi: 10.1016/j.ijrobp.2011.09.060. [DOI] [PubMed] [Google Scholar]
- 28.Bhojani MS, Chen G, Ross BD, Beer DG, Rehemtulla A. Nuclear localized phosphorylated FADD induces cell proliferation and is associated with aggressive lung cancer. Cell Cycle. 2005;4(11):1478–81. doi: 10.4161/cc.4.11.2188. [DOI] [PubMed] [Google Scholar]
- 29.Yoo NJ, Lee SH, Jeong EG, et al. Expression of nuclear and cytoplasmic phosphorylated FADD in gastric cancers. Pathol Res Pract. 2007;203(2):73–8. doi: 10.1016/j.prp.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Shimada K, Matsuyoshi S, Nakamura M, Ishida E, Kishi M, Konishi N. Phosphorylation of FADD is critical for sensitivity to anticancer drug-induced apoptosis. Carcinogenesis. 2004;25(7):1089–97. doi: 10.1093/carcin/bgh130. [DOI] [PubMed] [Google Scholar]
- 31.Jang MS, Lee SJ, Kim CJ, Lee CW, Kim E. Phosphorylation by polo-like kinase 1 induces the tumor-suppressing activity of FADD. Oncogene. 2011;30(4):471–81. doi: 10.1038/onc.2010.423. [DOI] [PubMed] [Google Scholar]
- 32.Wong KF. 11q13 is a cytogenetically promiscuous site in hematologic malignancies. Cancer Genet Cytogenet. 1999;113(1):93–5. doi: 10.1016/s0165-4608(98)00285-4. [DOI] [PubMed] [Google Scholar]
- 33.Thorns C, Bastian B, Pinkel D, et al. Chromosomal aberrations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma unspecified: a matrix-based CGH approach. Genes Chromosomes Cancer. 2007;46(1):37–44. doi: 10.1002/gcc.20386. [DOI] [PubMed] [Google Scholar]
- 34.Drakos E, Leventaki V, Atsaves V, et al. Expression of serine 194-phosphorylated Fas-associated death domain protein correlates with proliferation in B-cell non-Hodgkin lymphomas. Hum Pathol. 2011;42(8):1117–24. doi: 10.1016/j.humpath.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee EW, Kim JH, Ahn YH, et al. Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat Commun. 2012;3:978. doi: 10.1038/ncomms1981. [DOI] [PubMed] [Google Scholar]
- 36.Clarke LE, Bayerl MG, Bruggeman RD, et al. Death receptor apoptosis signaling mediated by FADD in CD30-positive lymphoproliferative disorders involving the skin. Am J Surg Pathol. 2005;29(4):452–9. doi: 10.1097/01.pas.0000155154.46434.93. [DOI] [PubMed] [Google Scholar]
- 37.Osborn SL, Sohn SJ, Winoto A. Constitutive phosphorylation mutation in Fas-associated death domain (FADD) results in early cell cycle defects. J Biol Chem. 2007;282(31):22786–92. doi: 10.1074/jbc.M703163200. [DOI] [PubMed] [Google Scholar]
- 38.Eckerle S, Brune V, Döring C, et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23(11):2129–38. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]
- 39.Oyarzo MP, Medeiros LJ, Atwell C, et al. c-FLIP confers resistance to FAS-mediated apoptosis in anaplastic large-cell lymphoma. Blood. 2006;107(6):2544–7. doi: 10.1182/blood-2005-06-2601. [DOI] [PubMed] [Google Scholar]
- 40.Contassot E, Kerl K, Roques S, et al. Resistance to FasL and tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in Sezary syndrome T-cells associated with impaired death receptor and FLICE-inhibitory protein expression. Blood. 2008;111(9):4780–7. doi: 10.1182/blood-2007-08-109074. [DOI] [PubMed] [Google Scholar]
- 41.Braun FK, Hirsch B, Al-Yacoub N, et al. Resistance of cutaneous anaplastic large-cell lymphoma cells to apoptosis by death ligands is enhanced by CD30-mediated overexpression of c-FLIP. J Invest Dermatol. 2010;130(3):826–40. doi: 10.1038/jid.2009.299. [DOI] [PubMed] [Google Scholar]
- 42.Alappat EC, Feig C, Boyerinas B, et al. Phosphorylation of FADD at serine 194 by CKIalpha regulates its nonapoptotic activities. Mol Cell. 2005;19(3):321–32. doi: 10.1016/j.molcel.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Villa-Morales M, Gonzalez-Gugel E, Shahbazi MN, Santos J, Fernandez-Piqueras J. Modulation of the Fas-apoptosis-signalling pathway by functional polymorphisms at Fas, FasL and Fadd and their implication in T-cell lymphoblastic lymphoma susceptibility. Carcinogenesis. 2010;31(12):2165–71. doi: 10.1093/carcin/bgq201. [DOI] [PubMed] [Google Scholar]
- 44.Schinske KA, Nyati S, Khan AP, et al. A novel kinase inhibitor of FADD phosphorylation chemosensitizes through the inhibition of NF-kappaB. Mol Cancer Ther. 2011;10(10):1807–17. doi: 10.1158/1535-7163.MCT-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui J, Wang Q, Wang J, et al. Basal c-Jun NH2-terminal protein kinase activity is essential for survival and proliferation of T-cell acute lymphoblastic leukemia cells. Mol Cancer Ther. 2009;8(12):3214–22. doi: 10.1158/1535-7163.MCT-09-0408. [DOI] [PubMed] [Google Scholar]
- 46.Odqvist L, Sánchez-Beato M, Montes-Moreno S, et al. NIK controls classical and alternative NF-kappaB activation and is necessary for the survival of human T-cell lymphoma cells. Clin Cancer Res. 2013;19(9):2319–30. doi: 10.1158/1078-0432.CCR-12-3151. [DOI] [PubMed] [Google Scholar]
- 47.Tourneur L, Delluc S, Lévy V, et al. Absence or low expression of Fas-associated protein with death domain in acute myeloid leukemia cells predicts resistance to chemotherapy and poor outcome. Cancer Res. 2004;64(21):8101–8. doi: 10.1158/0008-5472.CAN-04-2361. [DOI] [PubMed] [Google Scholar]
- 48.Delsol G, Jaffe ES, Falini B, et al. Anaplastic large cell lymphoma (ALCL), ALK-positive. In: Swerdlow SH, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer; 2008. pp. 312–6. [Google Scholar]
- 49.Savage KJ. Therapies for peripheral T-cell lymphomas. Hematology Am Soc Hematol Educ Program. 2011;2011:515–24. doi: 10.1182/asheducation-2011.1.515. [DOI] [PubMed] [Google Scholar]
- 50.Ferreri AJ, Govi S, Pileri SA, Savage KJ. Anaplastic large cell lymphoma, ALK-negative. Crit Rev Oncol Hematol. 2013;85(2):206–15. doi: 10.1016/j.critrevonc.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Bisig B, Gaulard P, de Leval L. New biomarkers in T-cell lymphomas. Best Pract Res Clin Haematol. 2012;25(1):13–28. doi: 10.1016/j.beha.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Armitage JO. The aggressive peripheral T-cell lymphomas: 2013. Am J Hematol. 2013;88(10):910–8. doi: 10.1002/ajh.23536. [DOI] [PubMed] [Google Scholar]
- 53.Hirsch B, von der Wall E, Hummel M, Durkop H. RIP1 expression is necessary for CD30-mediated cell death induction in anaplastic large-cell lymphoma cells. Lab Invest. 2013;93(6):677–89. doi: 10.1038/labinvest.2013.50. [DOI] [PubMed] [Google Scholar]
- 54.Zhao WL. Targeted therapy in T-cell malignancies: dysregulation of the cellular signaling pathways. Leukemia. 2010;24(1):13–21. doi: 10.1038/leu.2009.223. [DOI] [PubMed] [Google Scholar]