Abstract
Somatic mutations, often translocations or single nucleotide variations, are pathognomonic for certain types of cancers and are increasingly of clinical importance for diagnosis and prediction of response to therapy. Conventional clinical assays only evaluate 1 mutation at a time, and targeted tests are often constrained to identify only the most common mutations. Genome-wide or transcriptome-wide high-throughput sequencing (HTS) of clinical samples offers an opportunity to evaluate for all clinically significant mutations with a single test. Recently a “desktop version” of HTS has become available, but most of the experience to date is based on data obtained from high-quality DNA from frozen specimens. In this study, we demonstrate, as a proof of principle, that translocations in sarcomas can be diagnosed from formalin-fixed paraffin-embedded (FFPE) tissue with desktop HTS. Using the first generation MiSeq platform, full transcriptome sequencing was performed on FFPE material from archival blocks of 3 synovial sarcomas, 3 myxoid liposarcomas, 2 Ewing sarcomas, and 1 clear cell sarcoma. Mapping the reads to the “sarcomatome” (all known 83 genes involved in translocations and mutations in sarcoma) and using a novel algorithm for ranking fusion candidates, the pathognomonic fusions and the exact breakpoints were identified in all cases of synovial sarcoma, myxoid liposarcoma, and clear cell sarcoma. The Ewing sarcoma fusion gene was detectable in FFPE material only with a sequencing platform that generates greater sequencing depth. The results show that a single transcriptome HTS assay, from FFPE, has the potential to replace conventional molecular diagnostic techniques for the evaluation of clinically relevant mutations in cancer.
Keywords: sequencing, RNA-Seq, cancer, biomarker, archival tissue
High-throughput sequencing (HTS) technology has created tremendous anticipation of clinically relevant discoveries and utilities for oncologic pathology.1 However, to date, the time and financial resources required to use the technology has kept most applications in the research domain. Translation to clinical care is occurring, as the promise of more accessible HTS is being fulfilled by “desktop sequencer” devices such as the Illumina MiSeq and IonTorrent PGM, which offer both rapid and increasingly economical HTS. For instance, desktop sequencing that requires essentially the same amount of time and money as a fluorescence in situ hybridization (FISH) assay is becoming available, and recent reports have used desktop sequencing platforms for HLA genotyping, bacterial genotyping, and targeted SNP sequencing.2-4
To be particularly useful for surgical pathology material, HTS assays need to perform well on formalin-fixed paraffin-embedded (FFPE) tissue, as clinical specimens are not routinely stored as frozen specimens. However, the damage to the nucleic acid by formalin makes sequence analysis difficult in FFPE tissue.5 The vast majority of clinically relevant nucleic acid variations (mutations, fusions, etc.) are evident from the examination of the exonic regions of the genome, which can be assessed either by DNA or RNA sequencing. As formalin fixation is less damaging to DNA than RNA, one approach is to perform whole-genome sequencing. However, sequencing the entire genome requires much more resources than sequencing just the exome, which is estimated to be 1% of the genome, and it can be challenging to obtain equally distributed coverage in coding sequence. Other options for using DNA include targeted polymerase chain reaction (PCR) and exome capture HTS. Targeted PCR requires a pool of specific primers for every possible sequence variation. Exome capture is a technically difficult method of “capturing” a coding sequence of the genome through hybridization and then sequencing these coding sequences. The main challenge with this approach with archival material is obtaining reliable hybridization given that FFPE DNA generally performs poorly in hybridization methods, such as microarrays.6
Sequencing the transcriptome is an attractive alternative as coding sequences are significantly enriched in mRNA. We have shown previously with single-end RNA sequencing that 3′ RNA-Seq (3SEQ) from FFPE tumor material can be used to robustly quantify gene expression6 and identify novel transcribed regions of the genome.7 The challenge in whole-transcriptome sequencing (sequencing of full-length RNA transcripts) is that it is difficult to isolate intact complete RNA transcripts. The vast majority of total RNA is ribosomal RNA, which is of little clinical interest, and this must be removed before sequencing. The characteristic 3′poly-A tail can be used to purify the mRNA; however, if the RNA has been fragmented through formalin damage [low RNA integrity number (RIN)8], the 5′ sequence of the mRNA will be missing for most of the isolated poly-A RNAs. We hypothesize that HTS can overcome poor RNA quality. With enough sequencing, uncommon full-length mRNA from poly-A isolation methods can be sequenced to identify clinically important variations in the exonic regions. In the current study, we show that paired-end, whole-transcriptome HTS of FFPE material using a desktop sequencer with run times of 27 hours can be used to detect pathognomonic fusions in sarcomas.
In this study we focus on gene fusions and sarcomas because of the pathognomonic translocations that characterize many of these cancers, the clinical utility of identifying these translocations, and the resultant battery of molecular tests that have been created to achieve this end. In the past 2 decades, fusions involving over 60 different genes have been described as characteristic molecular events in 19 sarcomas (COSMIC database, November 15, 2011). Detecting these fusions is helpful in making accurate diagnoses and can also have prognostic and therapeutic implications.9-13 Currently in the clinical setting, translocations in sarcomas are diagnosed by FISH or reverse transcriptase PCR (RT-PCR). However, only a very limited number of mutations can be detected in even the most modern laboratories. In addition, keeping even a small number of translocation tests operational for these rare lesions puts a significant burden on molecular laboratories. Here we present a single desktop HTS assay on an FFPE material capable of diagnosing the spectrum of translocations in sarcomas. Advantages of this approach over FISH or RT-PCR include the fact that it is an agnostic approach to mutation detection that can characterize both fusion partners and the exact breakpoint, it can detect unusual rearrangements not captured by the specific FISH probe or PCR primers, the scope of mutation analysis for other mutations can be expanded in the analysis pipeline, and it is a low-cost assay that can be performed in the clinical laboratory by a single technician. This study serves as a proof of the principle that paired-end transcriptome HTS on FFPE material is useful for mutation detection in cancer and can be expected to expand when further improvements in sequencing technology occur.
MATERIALS AND METHODS
Case Selection
The cases chosen for evaluation were from the Stanford pathology archives. Three cases of synovial sarcoma, 3 cases of myxoid liposarcoma, 2 cases of Ewing sarcoma, and 1 case of clear cell sarcoma were selected, all collected within 6 months of RNA isolation. The pathognomonic translocation was confirmed with FISH in all cases. Samples were collected and studied using Health Insurance Portability and Accountability Act–compliant Stanford University Medical Center institutional review board approval.
RNA Isolation
Total RNA was extracted from FFPE material from each FFPE block. The RNA purification was performed using a commercially available kit (Recover All Total Nucleic Acid Isolation Kit; Ambion CAT#AM1975). Total RNA extraction from frozen tissue stored at −80°C was performed by homogenization in Trizol reagent (GibcoBRL/Invitrogen, Carlsbad, CA; CAT#15596-018).
Library Preparation
For TruSeq RNA library construction, total RNA was first analyzed with the BioAnalyzer 2100 (Agilent, Santa Clara, CA). The RIN ranged from 1.0 to 2.4 (Table 1). The sequencing libraries were constructed with the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA) following the manufacturer’s protocol using 10 μg of total RNA from each FFPE specimen and 4 μg of total RNA from each frozen specimen as input RNA. To summarize the work flow, the poly-A-containing mRNA molecules were purified using poly-T oligo-attached magnetic beads and fragmented using divalent cations under elevated temperature, then the first strand cDNA was synthesized using reverse transcriptase and random primers, followed by second-strand cDNA synthesis using DNA polymerase I and RNase H. The cDNA fragments then underwent end repair, addition of a single “A” base, and ligation of the adapters. The ligation products were purified and enriched with PCR to create a final cDNA library. The libraries were quantitated with Qubit 2.0 Fluorometer (Life Technologies, Foster City, CA) and validated with BioAnalyzer 2100 (Agilent).
TABLE 1.
Total Reads, Reads Mapping to Sarcomatome, Read Coverage at Breakpoint (BP), and Breakpoint Spanning Reads (Fusion Reads)
| Sample | Total Reads | Sarcomatome† | BP Coverage | Fusion Reads* |
|---|---|---|---|---|
| SS_F | 8,202,680 | 112,177 | 25 | 18 |
| SS_16 | 7,882,763 | 217,304 | 24 | 16 |
| SS_31 | 6,151,281 | 77,483 | 22 | 8 |
| SS_34 | 5,981,106 | 140,866 | 18 | 10 |
| MLS_09 | 7,388,216 | 37,774 | 24 | 13 |
| MLS_10 | 6,644,348 | 58,769 | 3 | 1 |
| MLS_10_H | 78,267,719 | 652,826 | 35 | 12 |
| MLS_11 | 6,616,442 | 42,326 | 8 | 4 |
| MLS_11_H | 79,086,356 | 478,212 | 40 | 18 |
| EWS_32 | 7,681,341 | 280,025 | 0 | 0 |
| EWS_32_r | 9,307,446 | 101,848 | 12 | 1 |
| EWS_32_H | 214,962,075 | 7,012,482 | 146 | 9 |
| EWS_35 | 6,820,639 | 72,021 | 0 | 0 |
| CCS_33 | 5,450,583 | 37,867 | 3 | 2 |
| CCS_33_H | 202,231,159 | 1,248,442 | 65 | 35 |
Breakpoint spanning reads.
Pairs mapping to sarcomatome (proper and discordant).
_H indicates HiSeq; _r, ribodepleted.
Ribodepletion Library Construction
rRNA was depleted using the Ribo-Zero Magnetic Kit (Epicentre, Madison, WI). Briefly, 1 μg total RNA was incubated with rRNA-removal solution containing rRNA-specific probes according to instructions and incubated at 68°C for 10 minutes. rRNA bound to probes was removed by magnetic bead pull-down. The final ribosomal-depleted RNA was recovered after sodium acetate/glycogen addition and ethanol precipitation overnight. Samples were centrifuged at 10,000g for 30 minutes, washed once per instruction, and resuspended with RNAse-free water.
Sequencing on MiSeq
The validated library for each sample was denatured with NaOH and diluted with the Hyb buffer included in the MiSeq Reagent Kit (Illumina) to 6.5 pM. A phiX 174 control library (Illumina) was also denatured and diluted to the same concentration, then mixed with the sample library at the volume ratio of 1:99. The final library was subsequently loaded to the 300-cycle MiSeq reagent cartridge. The runs were set up as paired-end 150-cycle sequencing on a MiSeq sequencer (Illumina). The sequencing took 27 hours per sample and generated between 5.5 and 9 million reads per sample.
Sequencing on HiSeq
Selected MiSeq libraries were also loaded to the 200-cycle HiSeq cartridge. The runs were set up as paired-end 101-cycle sequencing on a HiSeq 2000 sequencer (Illumina). The sequencing took 11 days per sample and generated between 78 and 214 million reads per sample, dependent on whether a full sequencing lane was used or only half a lane.
Informatic Pipeline for Fusion Detection
The raw data are obtained from the MiSeq machine in fastq format. The fusion detection pipeline starts with the extraction of high-quality 50 mers from the MiSeq data, followed by a screening and then a confirmation stage. The rationale for the 50 mer screening stage is that chimeric 150 mers do not map, and the fusion is missed. The screening stage maps the 50 mer read pairs to the sarcomatome using Tophat-fusion,14 which also flags individual reads that span 2 genes and generates a SAM file.15 Discordantly mapped 50 mer read pairs (read pairs that do not map to the same sarcomatome transcript) are used by fidelity algorithm to identify and rank fusion candidates. The fidelity algorithm applied to the name-sorted SAM alignment file (rather than typical coordinate-sorted) to tally discordant read pair mappings and rank fusion candidates. The algorithm evaluates each gene involved in discordant read pair mapping(s) to determine the number of reads that support each discordant mapping(s), the number of other discordant partners the gene has, and the numbers of discordant reads for each of its partners. Fusion candidates are those in which 1 partner has over 95% of its discordant reads mapped to a single other partner. The metrics of the fidelity algorithm are used to rank gene fusion candidates when >1 are present.
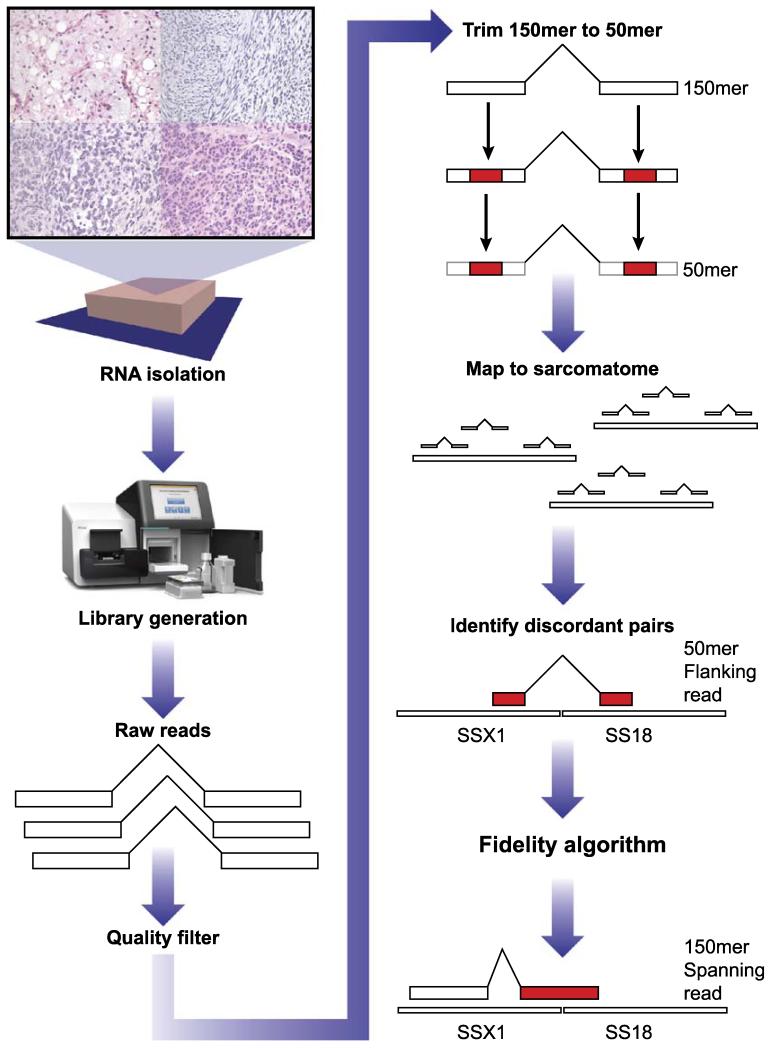
For further evaluation of fusion candidates, we return to the Tophat-fusion alignment SAM file to retrieve all read names for reads supporting the fusion, including both breakpoint flanking pairs and candidate breakpoint spanning reads (Fig. 1) that have been flagged by the Tophat-fusion software. These read names provide the basis for the confirmation stage.
FIGURE 1.
Work flow for analysis. See the Materials and methods section.
The confirmation stage retrieves the full 150 mer reads for the previously identified 50 mer reads that supported the fusion candidates identified in the screening stage and evaluates them against the human genome and transcriptome using BLAST (http://blast.ncbi.nlm.nih.gov). The breakpoint spanning 150 mers are used to characterize the exact sequence of the breakpoint. Furthermore, for highest resolution, the “chimeric percentage” is determined to demonstrate the percentage of a single read mapped to the 2 genes in the fusion (Table 2).
TABLE 2.
MLS_09 Breakpoint Spanning Reads, Columns Show Chimeric Percentage of Each 150 bp Read in the Pair
| Read ID | Read 1 (%FUS)/(%DDIT3) |
Read 2 (%FUS)/(%DDIT3) |
|---|---|---|
| 6490 | 66/26 | 66/26 |
| 12446 | 56/41 | 56/41 |
| 16723 | 50/27 | 50/27 |
| 11235 | 43/39 | 40/42 |
| 14670 | 34/66 | 0/100 |
| 18335 | 47/52 | 68/31 |
| 21806 | 0/100 | 45/55 |
| 13693 | 53/47 | 100/0 |
| 6814 | 59/19 | 59/19 |
| 21289 | 0/100 | 11/89 |
| 12453 | 0/100 | 39/60 |
| 22931 | 100/0 | 16/84 |
| 13972 | 75/25 | 76/23 |
The software used for the bioinformatic analysis, except the fidelity algorithm, has been published and are publicly available.14,15 The fidelity algorithm was written for this project and is available upon request.
RESULTS
Approach
To demonstrate the feasibility of using HTS to replace conventional molecular diagnostics in cancer, we performed transcriptome-wide HTS from RNA isolated from clinical FFPE tissue of sarcoma cases and identified the diagnostic translocations (Fig. 1). We studied 4 types of sarcomas with different translocations: synovial sarcoma, myxoid liposarcoma, Ewing sarcoma, and clear cell sarcoma.
Using archival material within 6 months of collection, the isolated RNA from FFPE material had an RIN range of 2.2 to 2.4, a quality lower than that recommended by the library preparation kit (TruSeq, Illumina). The 1 frozen tissue sample (SS_F) had an RIN of 8.1. The libraries were prepared using a poly-A-directed cDNA and were sequenced to generate between 6 and 9 million 150-nucleotide paired-end reads on the desktop platform (MiSeq) and, for selected cases, between 78 and 214 million 100-nucleotide paired-end reads on the HiSeq platform (Table 1). High-quality reads were aligned to the 83 genes involved in known translocations and mutations in sarcomas (the “sarcomatome”) according to the COSMIC database (date November 15, 2011).16 Fusion candidates were identified using a novel fidelity algorithm created for this project (see the Materials and methods section). Briefly, the highest quality 50 mer paired-end sequences are extracted from each of the 150 mer paired-end reads and used to identify read pairs that map to different mRNAs in the sarcomatome. These “breakpoint flanking” reads are used to identify gene pairs that are candidates for a gene fusion by ranking the fusion partners inversely related to the frequency and magnitude with which each partner forms fusions (see the Materials and methods section). Specifically, fusion candidates are those discordantly mapped gene pairs in which 1 partner has over 95% of its discordant reads mapped to a single other partner. Then, the full 150 mers of the “breakpoint flanking” reads as well as the “candidate breakpoint spanning” reads that were flagged in the alignment are BLASTed against the human genome and transcriptome to confirm specific matches as breakpoint flanking and breakpoint spanning 150 mers. The breakpoint spanning 150 mers are used to characterize the exact sequence of the breakpoint.
Synovial Sarcoma
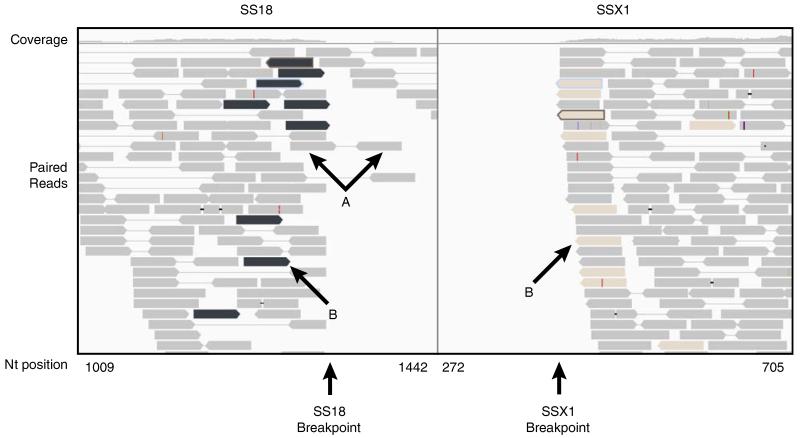
The 3 synovial sarcomas showed the most robust sequencing evidence for the pathognomonic translocation, t(x;18)(p11.2;q11.2), on the desktop platform using FFPE material with the number of breakpoint spanning reads ranging from 8 to 16 in the 3 cases. We determined that 2 of the synovial sarcomas had an SSX2-SS18 fusion, and 1 had an SSX1-SS18 fusion. A fresh frozen synovial sarcoma specimen, with much higher quality RNA (RIN = 8), did not show appreciably more robust evidence for the translocation with 18 reads (Table 1). The case of SS18-SSX1 (SS_16) had particularly robust fusion expression and detection (Fig. 2). The initial 50 mer screening alignment phase identified 18 read pairs flanking the breakpoint. The 150 mer analysis characterized the exact breakpoint and identified 16 read pairs, in which 1 or both reads span the breakpoint by at least 10 nucleotides. Interestingly, there is an absence of reads in the 3′ region of the SSX1 transcript, suggesting that the fusion allele is the only one that is allelicly expressed, and the unaffected SSX1 allele is silenced. There was only 1 other fusion candidate composed of genes involved in sarcoma translocations, EWSR1-COL1A1, represented by 1 breakpoint spanning read. Although this particular fusion has not been described, the fusion involved COL1A1, which is the most highly expressed gene in the sarcomatome. Other than the 3 synovial sarcomas, the SSX1-SS18 and SSX2-SS18 paired-end reads never appeared in any of the other samples.
FIGURE 2.
Split screen for fusion spanning pairs of SS18 and SS1 in sample SS_16. Gray reads represent paired reads that both map to the same transcript, either SS18 (arrows A) or SSX1. The black reads in SS18 window are paired with the tan reads in SSX1 window (arrows B). Notably, there are no reads 5′ to the breakpoint in SSX1, which suggests that the unaffected allele is not expressed. There is some expression of the unaffected SS18 as demonstrated by some reads continuing to map 3′ to the breakpoint. The read coverage is displayed above.
Myxoid Liposarcoma
The FUS-DDIT3 fusion was identified in each of the 3 myxoid liposarcomas sequenced on the desktop platform, with MLS_09 having the highest number of breakpoint spanning reads (13). In MLS_10, the pathognomonic fusion is identified with only 1 supporting read pair from the screening analysis (50 mer), and the same read from the 150 mer confirmation stage is a breakpoint spanning read nearly evenly distributed across the breakpoint. Despite having only 1 supporting read, the FUS-DDIT3 fusion event was top ranked by the fidelity algorithm on the basis of DDIT3 fidelity. The coverage of FUS at the breakpoint is particularly low (3 reads) in this sample, and the coverage of DDIT3 at the breakpoint is 15 reads, which suggests that the expression of the affected allele in this case is particularly low. It is possible that the translocation is different in this tumor: unlike the other myxoid liposarcomas with clear DDIT3-balanced breakaparts on FISH, the FISH for MLS_10 suggested an unbalanced translocation with loss of just the telomeric probe [FUS fuses on the telomeric side (5′) of DDIT3]. One other fusion candidate, ALDH2-LPP had 1 read supporting the fusion event. This was determined to be an artifact as the LPP region involved has spurious, discordant mappings with numerous genes. In MLS_11, FUS-DDIT3 was the top candidate with 2 supporting read pairs in screening. There were 4 pairs of breakpoint spanning 150 mers, with a total read coverage of 8 at the breakpoint. Although the number of reads supporting this fusion event is low, there were no other gene fusion candidates identified. To further evaluate these 2 cases with low support for the gene fusion, we performed HTS on the HiSeq platform for both MLS_10 and MLS_11, generating approximately 10-fold more 101 mer paired-end reads. The coverage at the breakpoint increased with the overall increased sequencing depth, and breakpoint spanning reads were identified with an abundance of 12/35 and 18/40, respectively.
Clear Cell Sarcoma
In the single case of clear cell sarcoma, the pathognomonic fusion, EWSR1-ATF1, was identified in only 3 reads, although 2 of these reads spanned the breakpoint. In this case, a low level of expression of the fusion transcript relative to other transcripts in the sample led to a low coverage at the breakpoint. However, the expression of the fusion gene allele in this tumor approaches 50%, and therefore the fusion can be detected despite the low coverage at the breakpoint. LPP-SS18L1 was the other fusion candidate with 1 read pair. This candidate was determined to be an artifact, as the regions involved for both genes have spurious discordant mappings with numerous genes. Owing to limited read coverage at the breakpoint, HTS on the HiSeq platform was performed on CCS_33 to improve the depth of sequencing at the breakpoint. The coverage at the breakpoint is 65 reads with a full HiSeq lane (202 total million reads) and 35 identified fusion reads.
Ewing Sarcoma
The pathognomonic fusion, EWSR1-FLI1, was not identified in the 2 cases of Ewing sarcoma that we tested (EWS_32 and EWS_35) using the standard poly-A selection method and the desktop platform. The sequencing depth reached 8 to 10× the coverage at the breakpoint, but no chimeric transcripts were identified. To further evaluate the frequency of the fusion transcript and attempt to attain deeper coverage at the breakpoint in EWS_32, we examined 2 alternative methods.
In the first approach, we created a sequencing library using a nonpoly-A selection method. The abundance of ribosomal RNA in a cell typically requires purification of the poly-A RNA to identify features of messenger RNA such as fusion transcripts. An alternate method is to deplete the rRNA such that poly-A isolation, and thus the 3′-end bias in the results, is not required. Sequencing of a library generated from ribosomal-depleted total RNA from EWS_32 detected 1 breakpoint spanning read. However, the depth of coverage at the breakpoint was no greater than in the poly-A selection, because of increased sequencing of intronic sequences characteristic of ribodepleted total RNA libraries,17 and the positive results versus negative results in the 2 approaches likely represents chance. The second approach was to add sequencing depth with the HiSeq platform. The coverage at the breakpoint reached over 100× with HiSeq, and fusion-supporting reads were identified in 6% of the reads across the breakpoint.
For each HiSeq run, the number of reads supporting the fusion event increased proportionally with the sequencing depth (Table 1). Alternate fusion candidates were found in the HiSeq run, but they tended to be different from the original alternate fusion candidates identified in the MiSeq run and were no greater than 4 reads per candidate fusion.
DISCUSSION
Using a desktop HTS platform, the Illumina “MiSeq,” the diagnostic fusion and exact breakpoint could be identified in synovial sarcoma (n = 3), clear cell sarcoma (n = 1), and myxoid liposarcoma (n = 3) without a priori knowledge of the diagnoses associated with the samples. For the samples showing the fewest supporting reads (MLS_10, MLS_11, and CCS_33) and the 2 Ewing sarcoma samples (EWS_32 and EWS_35) in which the fusion was undetectable, we performed HiSeq HTS, and with greater sequencing depth the fusions are all detectable. It is expected that platforms such as the MiSeq will continue to develop, reaching higher numbers of reads in the near future. Thus we expect that the future version of desktop sequencers will be able to cover the translocation where we now only find rare reads.
Our study demonstrates that high specificity and sensitivity can be achieved with a transcriptome-wide HTS approach using clinical archived samples (FFPE material). Although rare false-positive fusions were identified in the initial screening step, none of the 27 described sarcoma fusions in the literature were found in any of the cases other than those of the appropriate diagnosis. Potential sources of false positives include the generation of true chimeric reads as the result of aberrant hybridization during library preparation, the combination of HTS errors that generate new sequences that could map to other genes, and mapping errors that place the read at the wrong location because of sequence homology between 2 genes. In our experience, these events are either rare enough not to impact the results or easily detected by examining the quality of the reads.
The sensitivity of the assay is dependent on the HTS depth and on expression levels of the fusion gene, which appear to vary considerably in different sarcomas in our small sample set. Sequencing depth can be addressed in 2 main ways. First, more reads can be generated for each sample. Improvements in HTS technology have been generating 2- to 5-fold gains in sequencing depth every couple of years. Second, alternative library preparations, like ribosomal-depleted total RNA sequencing, can be used to mitigate the 3′ bias of poly-A-selected mRNA in order to focus sequencing on RNAs that are likely to have clinical relevance. In our hands, these methods do not greatly reduce the depth of sequencing requirement, because much of the reads map to extensive intronic regions of precursor mRNAs, which are typically removed during poly-A selection. Nevertheless, these findings demonstrate that high-quality poly-A RNA with a length that includes the sequence from both fusion partners can be identified in FFPE material that typically have an RIN of <3.
The potential uses of a transcriptome-wide HTS extend beyond fusion transcript identification. In addition to fusion transcripts, genetic variation, somatic mutation, and gene expression levels may be obtained. Extensive work on the gene expression profiling of cancers, such as sarcomas,18,19 can be diagnostically and prognostically useful when combined with variation and mutation data. For example, such data can identify differential usage of the mutant and wild-type alleles. The method can also identify novel and alternate fusion partners. Such advantages are currently not easily available through RT-PCR or FISH. Challenges of data interpretation, data storage, and sequencing costs remain. The desktop sequencing platform costs allow for early adoption in clinical laboratories. Currently, making a library and analyzing the sequencing data is laborious, but we estimate that 10 person hours are involved and that further sophistication and automation of analysis algorithms will bring this down further. At the moment, desktop transcriptome sequencing and analysis costs are approximately 1.5× the cost and time of a single FISH assay. With the ever increasing number of known and clinically important mutations in cancer, it may become prohibitive to maintain validated FISH assays for each translocation and additional molecular assays for other types of mutations. Solutions to the technical and cost parameters of sequencing are rapidly approaching, and advances in molecular diagnostics will likely follow. Translocation detection using FFPE transcriptome HTS is an example of 1 possible advancement in molecular diagnostics. Further studies are warranted to evaluate this assay on FISH-indeterminate cases and other neoplasms with clinically relevant translocations.
The results from this study show that a single desktop transcriptome HTS assay, close in cost and time to a single FISH assay, can be performed on FFPE material to evaluate for translocations involving somatic mutations in sarcomas. These findings reported in this paper serve as proof of the principle that a single HTS assay, particularly as depth of sequencing continues to increase, can be clinically applied to the routine identification of diverse panels of mutations.
Acknowledgments
Source of Funding: Supported by the Department of Pathology, Stanford University Medical Center, Stanford, CA.
Footnotes
Conflicts of Interest: The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
REFERENCES
- 1.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 2.Wang C, Krishnakumar S, Wilhelmy J, et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc Natl Acad Sci USA. 2012;109:8676–8681. doi: 10.1073/pnas.1206614109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harismendy O, Schwab RB, Bao L, et al. Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol. 2011;12:R124. doi: 10.1186/gb-2011-12-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyre DW, Golubchik T, Gordon NC, et al. A pilot study of rapid benchtop sequencing of staphylococcus aureus and clostridium difficile for outbreak detection and surveillance. BMJ Open. 2012;2 doi: 10.1136/bmjopen-2012-001124. pii:e001124. doi: 10.1136/bmjopen-2012-001124. Print 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Ahlfen S, Missel A, Bendrat K, et al. Determinants of RNA quality from FFPE samples. PLoS One. 2007;2:e1261. doi: 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck AH, Weng Z, Witten DM, et al. 3′-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 2010;5:e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunner AL, Beck AH, Edris B, et al. Transcriptional profiling of lncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorensen PH, Lynch JC, Qualman SJ, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 10.de Alava E, Kawai A, Healey JH, et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clin Oncol. 1998;16:1248–1255. doi: 10.1200/JCO.1998.16.4.1248. [DOI] [PubMed] [Google Scholar]
- 11.de Alava E, Panizo A, Antonescu CR, et al. Association of EWS-FLI1 type 1 fusion with lower proliferative rate in Ewing’s sarcoma. Am J Pathol. 2000;156:849–855. doi: 10.1016/S0002-9440(10)64953-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin PP, Brody RI, Hamelin AC, et al. Differential transactivation by alternative EWS-FLI1 fusion proteins correlates with clinical heterogeneity in Ewing’s sarcoma. Cancer Res. 1999;59:1428–1432. [PubMed] [Google Scholar]
- 13.Nielsen TO, West RB. Translating gene expression into clinical care: sarcomas as a paradigm. J Clin Oncol. 2010;28:1796–1805. doi: 10.1200/JCO.2009.26.1917. [DOI] [PubMed] [Google Scholar]
- 14.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunbin Q, Morlan J, Stephans J, et al. Transcriptome profiling from formalin-fixed, paraffin-embedded tumor specimens by RNA-seq. Genome Biol. 2010;11(suppl 1):P31. [Google Scholar]
- 18.Nielsen TO, West RB, Linn SC, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 19.Beck AH, West RB, van de Rijn M. Gene expression profiling for the investigation of soft tissue sarcoma pathogenesis and the identification of diagnostic, prognostic, and predictive biomarkers. Virchows Arch. 2010;456:141–151. doi: 10.1007/s00428-009-0774-2. [DOI] [PMC free article] [PubMed] [Google Scholar]