Abstract
The ALK tyrosine kinase inhibitor (TKI), crizotinib, shows significant activity in patients whose lung cancers harbor ALK fusions but its efficacy is limited by variable primary responses and acquired resistance. In work arising from the intriguing clinical observation of a patient with ALK fusion+ lung cancer who had an ‘exceptional response’ to an IGF-1R antibody, we define a therapeutic synergism between ALK and IGF-1R inhibitors. Similar to IGF-1R, ALK fusion proteins bind to the adaptor, IRS-1, and IRS-1 knockdown enhances the anti-tumor effects of ALK inhibitors. In models of ALK TKI resistance, the IGF-1R pathway is activated, and combined ALK/IGF-1R inhibition improves therapeutic efficacy. Consistent with this finding, IGF-1R/IRS-1 levels are increased in biopsy samples from patients progressing on crizotinib therapy. Collectively, these data support a role for the IGF-1R/IRS-1 pathway in both ALK TKI-sensitive and TKI-resistant states and provide biological rationale for further clinical development of dual ALK/IGF-1R inhibitors.
Keywords: ALK, ALK fusions, IGF-1R, IRS-1, tyrosine kinase inhibitor, crizotinib, ceritinib, cancer, lung cancer, targeted therapeutics, drug resistance, exceptional responder
Introduction
Genomic alterations in the anaplastic lymphoma kinase (ALK) gene are found in numerous malignancies1. In a phase I trial, the ALK tyrosine kinase inhibitor (TKI), crizotinib, induced an objective radiographic response (ORR) of 60.8% and a median progression-free survival of 9.7 months2. Unfortunately, patients invariably develop therapeutic resistance. Mechanisms of resistance to ALK blockade are only beginning to be elucidated3-6.
Analysis of ‘exceptional responders’ has revealed important insights into drug sensitivity7. We describe a patient with ALK+ lung cancer who had an ‘exceptional response’ to an IGF-1R inhibitor prior to ALK TKI therapy. Using this patient's case as a paradigm, we uncover an association between IGF-1R/IRS-1 and ALK signaling. Collectively, our studies suggest that this rationally selected combination of inhibitors may be an effective strategy to delay or overcome resistance to therapeutic ALK inhibition.
Results
Exceptional response to an IGF-1R inhibitor
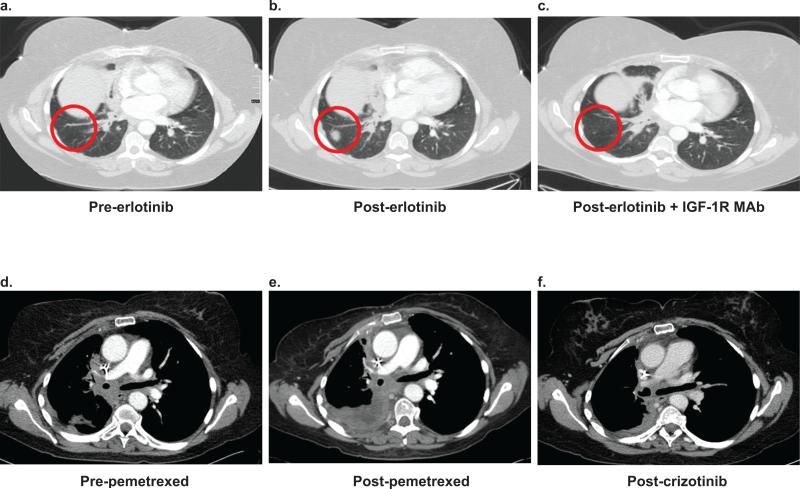
A 50 year-old female with stage IV lung adenocarcinoma received standard first line platinum-based chemotherapy. She then enrolled in a clinical trial of erlotinib followed at progression by erlotinib plus an IGF-1R monoclonal antibody (MAb). At the time of enrollment, her tumor mutation status was unknown. She developed progressive disease after 1 month of erlotinib (Fig. 1a,b). Per protocol, the IGF-1R MAb was added, and she then experienced a partial response lasting 17 months (Fig. 1c). She remained on this treatment longer than any other patient enrolled in the trial.
Figure 1. Exceptional response to an IGF-1R inhibitor prior to ALK TKI therapy in a patient with ALK+ lung cancer.
Representative images from serial CT scans of the chest in a 50 year-old female with ALK+ lung cancer documenting responses to the indicated therapies. Images are labeled a–f in temporal sequence. The red circles in a–c represent a new lesion in the right lung that developed after 1 month of erlotinib and then responded to erlotinib plus an IGF-1R antibody. The scale bar in Figure 1a indicates 4 cm and is representative for all images.
At the time of progression on erlotinib plus the IGF-1R MAb, her tumor was sent for molecular profiling. As expected, based on the lack of response to erlotinib, the tumor did not contain an EGFR mutation (Supplementary Table 1); surprisingly, it was found to harbor an ALK rearrangement. Subsequently, she enrolled in the phase III trial of crizotinib versus chemotherapy and was randomized to pemetrexed. After four cycles, she had disease progression (Fig. 1e), was started on crizotinib per protocol, and had a partial response (Fig. 1f).
Previous studies have reported a 0% response rate for ALK+ lung cancer patients treated with erlotinib alone8. Thus, we hypothesized that in this patient, either the combination of erlotinib plus the IGF-1R inhibitor was synergistic against ALK+ lung cancer, or the IGF-1R inhibitor alone was somehow responsible for the tumor response. To address this hypothesis, we treated H3122 cells, which harbor an EML4-ALK E13;A20 fusion, with erlotinib, an IGF-1R inhibitor, or the combination. We observed no therapeutic synergism between erlotinib and the IGF-1R inhibitors (Supplementary Fig. 1a,b), suggesting that this patient's tumor response was more likely due to the IGF-1R antibody. Based on this clinical observation, we hypothesized that there is cross-talk between IGF-1R and ALK which may be exploited therapeutically to improve anti-tumor responses.
Therapeutic synergism between ALK and IGF-1R inhibitors
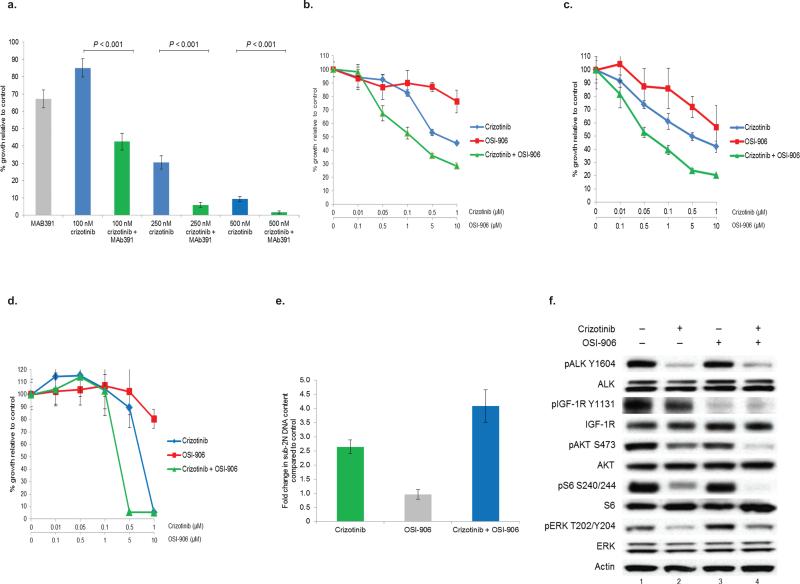
We tested the ability of IGF-1R inhibitors alone or in combination with ALK inhibitors to impede the growth of ALK+ lung cancer cells. The IGF-1R specific MAb, MAb391, had modest, but reproducible, single agent activity in H3122 cells. However, MAb391 sensitized H3122 cells to the anti-proliferative effects of crizotinib (Fig. 2a). When IGF-1R was inhibited with MAb391, sensitivity to crizotinib was also enhanced in STE-1 (EML4-ALK E13;A20) cells, a novel lung adenocarcinoma cell line we developed from a patient with ALK+ lung cancer (Supplementary Fig. 1c). Similar results were also seen when H3122 cells were treated with the dual IGF-1R/insulin receptor TKI, OSI-906, plus crizotinib (Fig. 2b). We extended this finding to other ALK+ lung cancer cell lines, including H2228 (EML4-ALK E6a/b;A20) (Fig. 2c) and STE-1 (Fig. 2d). Co-treatment with an ALK TKI plus an IGF-1R TKI also induced better anti-tumor responses in SUDHL-1 lymphoma cells, which harbor an NPM-ALK fusion, suggesting that this effect is not specific to ALK-mutant lung cancer (Supplementary Fig. 1e). The combination of crizotinib plus OSI-906 was confirmed to be synergistic using the Mix-Low method9 (Supplementary Fig. 1d). OSI-906 has no off-target activity against ALK at the doses used in these experiments10.
Figure 2. Combination therapy with an IGF-1R inhibitor plus an ALK inhibitor promotes cooperative inhibition of cell growth in TKI sensitive ALK+ lung cancer cells.
(a) H3122 (EML4-ALK E13;A20) lung cancer cells were treated with crizotinib or crizotinib + MAb391. Soft agar assays were performed to assess growth inhibition. Each point represents hextuplicate biological replicates. Data are presented as the percentage of viable cells compared to control (vehicle only) cells and are representative of three independent experiments. P values were determined with the Student's T-test. (b–d) H3122 (EML4-ALK E13;A20) (b), H2228 (EML4-ALK 6a/b;A20) (c), and STE-1 (EML4-ALK E13;A20) (d) lung cancer cells were treated with increasing amounts of crizotinib, OSI-906, or the combination for 72h. Cell titer blue assays were performed to assess growth inhibition. Each point represents hextuplicate biological replicates. Data are presented as the percentage of viable cells compared to control (vehicle only) cells and are representative of three or more independent experiments. (e) STE-1 cells were treated with one dose of 1 μM crizotinib, 2 μM OSI-906, or the combination for a total of 72h prior to harvest. Cells were stained with propidium iodide (PI) and counted on a FACSCalibur machine. (f) H3122 cells were treated with crizotinib, OSI-906, or the combination for 2h prior to harvest. Lysates were subjected to immunoblotting with antibodies specific for the indicated proteins. Select images were quantified using a Bio-Rad Gel Doc XR and Image Lab software (Supplementary Fig. 1i and Supplementary Fig. 2b).
Compared to crizotinib alone, the combination of crizotinib plus OSI-906 resulted in increased levels of apoptosis (Fig. 2e) and decreased phosphorylation of downstream targets (Fig. 2f). Furthermore, the combination of crizotinib plus MAb391 was more effective at delaying the growth of ALK+ xenografts (Supplementary Fig. 1f). Collectively, these data show that the combination of ALK plus IGF-1R inhibitors results in an enhanced anti-tumor response in ALK+ lung cancer cells.
To ascertain the specificity of this effect, we examined whether inhibitors of other tyrosine kinases could produce analogous results. Neither the EGFR inhibitor, erlotinib (Supplementary Fig. 1g), nor the dual HER2/EGFR inhibitor, lapatinib (Supplementary Fig. 1h), was able to sensitize H3122 cells to the effects of crizotinib. These data suggest that the synergistic anti-proliferative effect described above is specific to IGF-1R blockade.
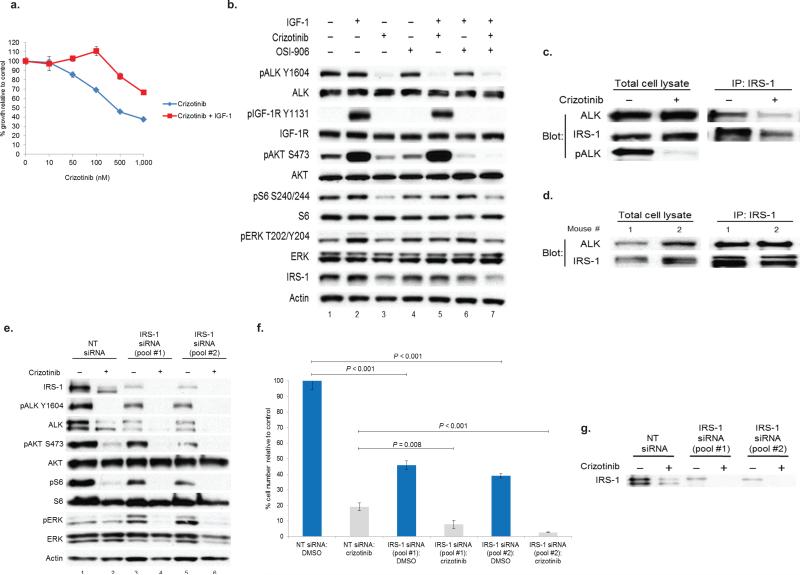
To assess if ligand induced activation of IGF-1R could influence the anti-proliferative effects of ALK blockade, we treated H3122 cells with crizotinib alone or in combination with IGF-1. Addition of IGF-1 induced resistance to the growth inhibitory effects of crizotinib (Fig. 3a). IGF-1 ligand stimulated phosphorylation of IGF-1R but not ALK (Fig. 3b and Supplementary Fig. 2a), suggesting no direct cross-talk between the two kinases. When cells pre-treated with crizotinib were stimulated with IGF-1, ALK phosphorylation was inhibited; however, downstream signaling was sustained as evidenced by continued phosphorylation of AKT (Fig. 3b). OSI-906 was able to inhibit this response. Taken together, these data suggest that signaling through IGF-1R may be a compensatory mechanism for the growth inhibitory effects of ALK inhibitor therapy.
Figure 3. IRS-1 knock-down impairs downstream signaling and blocks proliferation of ALK+ lung cancer cells.
(a) H3122 cells were treated with crizotinib or crizotinib + IGF-1 for 72h. Cell titer blue assays were performed to assess growth inhibition. Each point represents hextuplicate biological replicates. Data are presented as the percentage of viable cells compared to control. (b) H3122 cells were serum starved overnight and then treated with the indicated TKIs for 6h. As indicated, cells were then stimulated with IGF-1 for 10min. Lysates were subjected to immunoblotting with antibodies specific for the indicated proteins. (c) H3122 cells were treated with vehicle or crizotinib. Lysates were subjected to immunoprecipitation (IP) for IRS-1 and western blotting for the indicated antibodies. (d) Tumor containing lung tissue from two different EML4-ALK E13;A20 transgenic mice were pulverized, lysed, and subjected to immunoprecipitation (IP) for IRS-1 and western blotting for the indicated antibodies. (e) STE-1 cells were transfected with the non-targeting siRNA (“NT”) or with two distinct pools of IRS-1 siRNA and treated with 500nM crizotinib for 72h . Lysates were subjected to immunoblotting with antibodies specific for the indicated proteins. (f) STE-1 cells were transfected with the indicated siRNAs and treated with 500 nM crizotinib for 72h. Triplicate biological replicates for each sample were counted on Coulter Counter. P values were determined with the Student's T-test. Data are representative of three independent experiments. (g) Western blot showing IRS-1 knockdown in the experiment shown in Fig. 3f.
IRS-1 knock-down impedes growth of ALK+ lung cancer cells
We investigated molecular mechanisms underlying the cooperative anti-tumor response between ALK and IGF-1R inhibitors. IRS-1 is a well-known substrate and adaptor protein for IGF-1R11, and IRS-1 has been demonstrated to be a primary adaptor for PI3K activation in H3122 cells12. However, the precise mechanism whereby ALK fusion proteins link to effector pathways remains undefined. We observed that IRS-1 levels decreased with crizotinib treatment (Fig. 3b). Using lysates from H3122 cells, we found that ALK and IRS-1 co-immunoprecipitated and that the interaction decreased after the addition of crizotinib (Fig. 3c). We also validated that this interaction occurs in vivo using tissue from two different EML4-ALK E13;A20 transgenic mice13 (Fig. 3d).
Next, we hypothesized that if IRS-1 is an adaptor protein for ALK, then knock-down of IRS-1 would sensitize cells to the effects of ALK inhibition. Consistent with our hypothesis, IRS-1 knock-down sensitized STE-1 cells to the effects of crizotinib (Fig. 3e). Levels of phosphorylated AKT, S6, and ERK were lower in IRS-1 siRNA transfected, crizotinib treated cells compared to crizotinib treated controls. Finally, IRS-1 knockdown impaired the proliferation of STE-1 cells in the absence of crizotinib and also sensitized these cells to the anti-proliferative effects of ALK inhibition (Fig. 3f,g). Analogous results were seen in H2228 cells (Supplementary Fig. 3a,b). Taken together, these data suggest that IRS-1 is an adaptor protein which links both IGF-1R and ALK to downstream signaling pathways.
IGF-1R pathway up-regulation in ALK TKI resistant cells
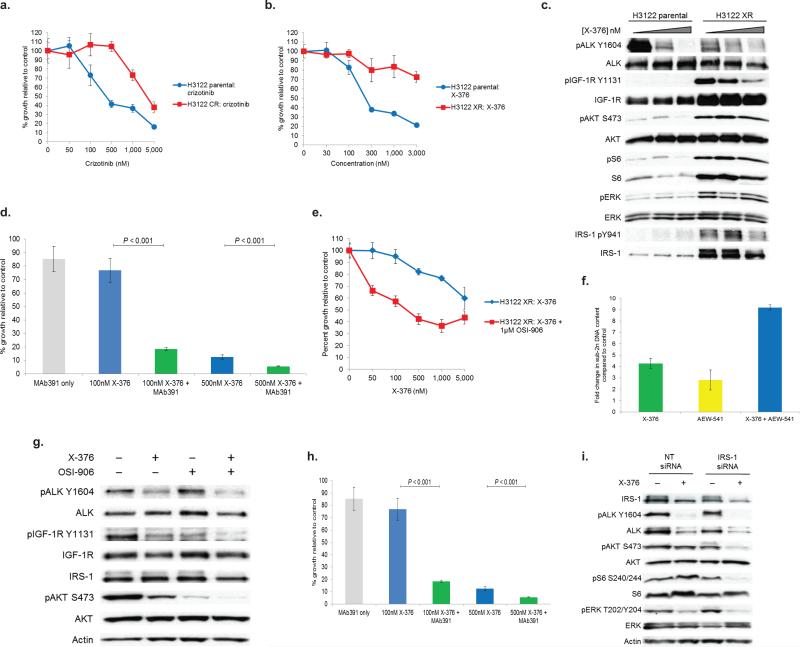
Starting with drug sensitive (‘parental’) cells, we derived H3122 cells that were resistant to crizotinib (Fig. 4a) or to X-376 (Fig. 4b), a more potent and more specific ALK inhibitor14. Notably, a derivative of X-376 (X-396) is currently in phase I clinical trials (NCT01625234). We analyzed these resistant cell lines by a variety of methods. H3122 crizotinib-resistant cells (‘H3122 CR’) displayed amplification of the EML4-ALK E13;A20 fusion by ALK FISH (Supplementary Fig. 4a,b), as previously described15. These cells did not have any ‘second-site’ ALK kinase domain mutations (data not shown).
Figure 4. The IGF-1R pathway is activated in models of ALK TKI resistance.
Isogenic pairs of H3122 parental (i.e. TKI sensitive), crizotinib-resistant (“CR”), or X-376-resistant (“XR”) cells were treated with crizotinib (a) or X-376 (b). Cell titer blue assays were performed with hextuplicate biological replicates. Data shown are representative of ≥ 3 independent experiments. (c) H3122 XR cells were treated with X-376 for 4h. Lysates were subjected to immunoblotting with antibodies specific for the indicated proteins. (d) H3122 XR cells were treated with X-376 or X-376 + MAb391. Soft agar assays were performed using hextuplicate biological replicates. Data are representative of two independent experiments. (e) H3122 XR cells were treated with X-376 or X-376 + OSI-906 for 72h. Cell titer blue assays were performed with hextuplicate biological replicates. Data are representative of three independent experiments. (f) H3122 XR cells were treated with X-376, AEW-541, or the combination daily for 72h. Cells were stained with propidium iodide (PI) and counted on a FACSCantoII machine. (g) H3122 XR cells were treated with the indicated inhibitors for 4h. Lysates were subjected to immunoblotting with antibodies specific for the indicated proteins. (h) H3122 XR cells were transfected with the indicated siRNAs and treated with 500 nM X-376 for 72h. Quadruplicate biological replicates for each sample were counted on Coulter Counter. Data are representative of three independent experiments. (i) Western blots confirming IRS-1 knockdown in the experiment shown in Fig. 4h. All P values shown were determined with the Student's T-test.
H3122 X376-resistant cells (‘H3122 XR’) harbored neither ALK amplification (Supplementary Fig. 4c) nor second-site mutations. However, H3122 XR cells maintained phosphorylation of AKT, S6, and ERK, even in the continued presence of X-376 (Fig. 4c). We hypothesized that an alternative upstream kinase(s) must be activated in these cells in order to maintain signaling. Phospho-RTK arrays revealed an increase in IGF-1R phosphorylation (Supplementary Fig. 4d). Indeed, IGF-1R phosphorylation and total protein levels were elevated in the ALK-TKI resistant compared to the ALK-TKI sensitive (i.e. ‘parental’) cells (Fig. 4c). H3122 XR cells also exhibited increased phosphorylation and total protein levels of IRS-1. Overall, these results suggest that the IGF-1R/IRS-1 pathway plays a role in maintaining downstream signaling in the presence of continuous ALK inhibition and therefore may represent a mechanism whereby cells evade ALK blockade.
Finally, we sought to determine how IGF-1 signaling is up-regulated in ALK TKI resistant cells. IGF-1 ligand levels were increased in the conditioned media from H3122 XR cells (Supplementary Fig. 4d). Gene expression profiling revealed that IGF binding protein 3 (IGFBP3) was down regulated in resistant versus parental cells (Supplementary Tables 2 and 3). IGFBP3 is known to block IGF-1 induced activation of IGF-1R16,17. Overall, these studies show that the IGF-1R signaling pathway is activated by multiple mechanisms in H3122 XR cells.
We evaluated the effects of IGF-1R inhibition in ALK TKI resistant cells. The combination of X-376 with either MAb391 or OSI-906 (Fig. 4d,e) partially restored X-376 sensitivity in H3122 XR cells. Apoptosis was also enhanced in H3122 XR cells treated with X-376 and the IGF-1R TKI, AEW-54118 (Fig. 4f). In accord with these data, combination treatment with ALK and IGF-1R inhibitors in H3122 XR cells inhibited AKT phosphorylation to a greater extent than either inhibitor alone (Fig. 4g). The addition of OSI-906 also partially restored the sensitivity of H3122 CR cells to the growth inhibitory effects of crizotinib (Supplementary Fig. 4c).
Since IRS-1 levels were increased in H3122 XR cells (Fig. 4c), we examined whether IRS-1 knock-down would also affect signaling and proliferation in the resistant cells (Fig. 4h,i). We transfected H3122 XR cells with IRS-1 or control siRNAs and then treated cells with X-376. IRS-1 knockdown sensitized these cells to the anti-proliferative effects of ALK inhibition (Fig. 4h) and resulted in a further decline in phosphorylation of downstream targets compared to X-376 alone or IRS-1 knockdown alone (Fig. 4i).
Previous studies have suggested that the IGF-1R pathway can drive EGFR inhibitor resistance19,20. We tested the efficacy of combined EGFR/IGF-1R inhibition in 4 different isogenic pairs of EGFR TKI- sensitive and -resistant cell lines21,22 (Supplementary Table 4). The addition of the IGF-1R TKI, OSI-906, was not synergistic with the EGFR TKI, erlotinib, in any of these cells (Supplementary Fig. 5a–d). Furthermore, in contrast to the ALK TKI-resistant cells, there was no increase in IGF-1R or IRS-1 in the EGFR TKI-resistant cell lines (Supplementary Fig. 5e). These data suggest that the effects seen with the ALK/IGF-1R inhibitor combinations in the ALK cell lines are true differences.
Increased IGF-1R in tumor samples at the time of resistance
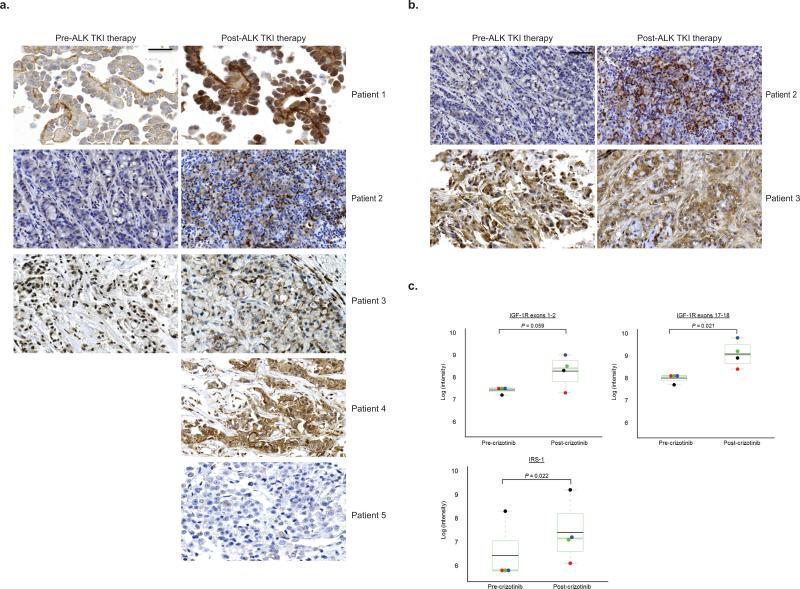
To validate the clinical implications of our in vitro findings, we evaluated phospho-IGF-1R (pIGF-1R) and IRS-1 levels in patient tumor biopsy samples. Three sets of paired pre-/post-crizotinib tumor samples as well as two post-crizotinib tumor samples from five different patients were examined by immunohistochemistry (IHC) for pIGF-1R and blindly evaluated by pathologists. As a control, we also performed pIGF-1R IHC on lung cancer tissue microarrays (TMAs); representative examples are shown in Supplementary Fig. 6a–c. Four of five tumor biopsies taken at the time of acquired resistance displayed increased levels of pIGF-1R (Fig. 5a, patients 1–4). For two of the paired tumor samples, we had sufficient tissue to examine IRS-1 levels by IHC (Fig. 5b); one post-treatment sample (patient 2) had increased IRS-1 expression. These five samples were also assessed for ALK kinase domain mutations associated with crizotinib resistance (Supplementary Table 5). Patient 4's post-crizotinib tumor harbored an ALK G1202R mutation.
Figure 5. Increased IGF-1R and IRS-1 in patient biopsy samples at the time of acquired resistance to crizotinib.
(a–b) Tumor samples taken before and at the time of resistance to ALK TKI therapy were analyzed for IGF-1R pY1161 expression (a) and for IRS-1 expression (b) by immuno-histochemistry. All images viewed correspond to a magnification of 40x. The scale bar indicates 200 micometers. (c–d) RNA was extracted from formalin-fixed, paraffin embedded tumor biopsy samples prior to and at the time of progressive disease on crizotinib and run on the NanoString assay. Expression levels of IGF-1R (c) and IRS-1 (d) are compared pre- and post- crizotinib. NanoString target sequences for IGF-1R have been previously reported23. The colored dots within each box plot represent distinct pairs of matched pre- and post- crizotinib samples. The black dots indicate the patient sample. The red dots and the green dots represent H3122 parental (TKI sensitive cells) compared with H3122 CR cells at 1× crizotinib resistance (1 μM final concentration of crizotinib, red dot) or H3122 CR cells at 2× crizotinib resistance (2 μM final concentration of crizotinib, green dot). The blue dots represent H3122 parental compared with H3122 XR cells. P values were determined with a modified paired T-test using the limma package.
As an orthogonal approach, we performed mRNA expression analysis for IGF-1R and IRS-1 using Nanostring23 on matched patient samples and on isogenic pairs of ALK TKI-sensitive and resistant cell lines. In the one case with enough pre- and post-treatment tissue for available for analysis, IGF-1R and IRS-1 (Fig. 5c,d) mRNA levels were increased in the post-crizotinib relative to the pre-crizotinib tumor sample. Similar results were obtained with the cell lines. In contrast, Nanostring analysis of 11 matched pairs of EGFR mutant lung tumor biopsies revealed no significant change in IRS-1 levels post EGFR TKI therapy (Supplementary Fig. 6d–f), suggesting that the changes observed in IRS-1 were specific to ALK+ lung cancer. Overall, these data validate our pre-clinical findings, showing that levels of IGF-1R and IRS-1 can be increased post ALK TKI therapy in patient-derived samples.
LDK-378 inhibits phosphorylation of ALK and IGF-1R in vitro
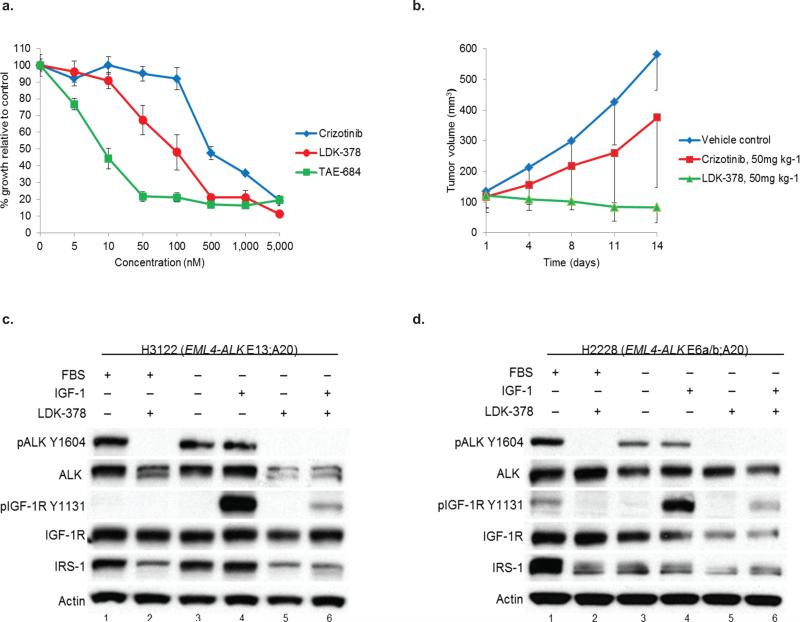
The second-generation ALK TKI, LDK-378 (ceritinib), has demonstrated a 56% ORR in patients with ALK+ lung cancer who have progressed on crizotinib24. Yet, only a minority of patients had ALK alterations, suggesting the possibility of alternative ‘bypass’ mechanisms which are sensitive to LDK-378. Interestingly, LDK-378 and the structurally related ALK inhibitor, TAE-684, can inhibit both ALK and IGF-1R in vitro25. We hypothesized that the efficacy of LDK-378 may be due to this drug's ability to simultaneously block both ALK and IGF-1R. LDK-378 was more potent than crizotinib in H3122 (Fig. 6a), STE-1 (Supplementary Fig. 7a), and H3122 XR cells (Supplementary Fig. 7b). LDK-378 was also more potent at inducing apoptosis in H3122 cells (Supplementary Fig. 7c). Furthermore, LDK-378 was significantly more effective at delaying the growth of H3122 xenografts compared to the equivalent dose crizotinib (Fig. 6b).
Figure 6. The second generation ALK inhibitor, LDK-378, blocks phosphorylation of both ALK and IGF-1R.
(a) H3122 lung cancer cells containing the EML4-ALK E13;A20 fusion were treated with increasing amounts of crizotinib, LDK-378, or TAE-684 for 72h. Cell titer blue assays were performed to assess growth inhibition. Each point represents hextuplicate biological replicates. Data are presented as the percentage of viable cells compared to control (vehicle only treated) cells and are representative of three or more independent experiments. (b) Athymic nu/nu female mice were injected subcutaneously with H3122 lung cancer cells harboring the EML4-ALK E13;A20 fusion. When tumors reached an average volume of 100mm3, mice were randomized to receive crizotinib alone (50 mg kg−1 p.o. daily × 5 days), LDK-378 alone (50 mg kg−1 p.o. daily × 5 days), or vehicle control (n = 5 for crizotinib and LDK-378, n = 4 for vehicle control). Tumor volumes were assessed every 3-4 days. *P = 0.0159 based on the Wilcoxon rank sum test. (c–d) H3122 (c) and H2228 cells (d) were grown overnight in the presence or absence of serum and then treated with LDK-378 for 1 hour. As indicated, cells were then stimulated with IGF-1 for 10min and harvested. Lysates were subjected to immunoblotting with antibodies specific for the indicated proteins.
Next, we tested LDK-378's ability to inhibit IGF-1R phosphorylation. H3122 (Fig. 6c) and H2228 (Fig. 6d) cells were treated with LDK-378 alone or in combination with IGF-1. Importantly, LDK-378 treatment inhibited ALK phosphorylation and was also able to overcome the IGF-1 ligand induced increase in IGF-1R phosphorylation in both ALK+ cell lines (Figures 6c,d compare lanes 4 and 6). These data suggest that LDK-378's potency in vivo may be due to this agent's combined ability to block both ALK and IGF-1R.
Discussion
We report that ALK and IGF-1R inhibitors have cooperative anti-proliferative effects. IGF-1R inhibitors sensitized tumor cells to the effects of ALK inhibition. The therapeutic synergism between ALK and IGF-1R inhibitors was observed in both the ALK TKI-sensitive and ALK TKI-resistant settings. Chronic ALK inhibition was associated with enhanced IGF-1R signaling. The ALK TKI resistant cells utilized numerous mechanisms to activate IGF-1R signaling. Importantly, the addition of an IGF-1R inhibitor sensitized the resistant cells to the effects of ALK blockade.
We propose drug combinations co-targeting ALK and IGF-1R as a novel therapeutic approach in patients with ALK+ lung cancer. This rationally selected combination of targeted therapies should be effective in both the ALK TKI-naïve and TKI-resistant setting. Our data may also in part explain the surprising 56% ORR for the ‘second generation’ ALK TKI, LDK-378, in patients with ALK+ lung cancer who had progressed on crizotinib24. Since responses to LDK-378 were observed in both patients with and without ‘second-site’ ALK mutations, the increased ‘on-target’ potency of LDK-378 towards ALK is alone not enough to explain all of the responses seen to this agent. We hypothesize that the potency of this agent is due to its ability to simultaneously inhibit both ALK and IGF-1R, and our in vitro experiments confirm that LDK-378 does inhibit phosphorylation of both ALK and IGF-1R. Further studies will be necessary to validate this hypothesis in clinical samples.
Mechanisms of acquired resistance to ALK inhibitors are just beginning to be understood. Target alterations have only been found in a minority of resistant tumors examined to date. ‘Bypass’ signaling has also been reported3-6. Interestingly, target alterations and bypass signaling do not appear to be mutually exclusive. One of the patients in our study had both an ALK G1202R mutation and increased pIGF-1R at the time of crizotinib resistance. Previous reports have also documented the co-occurrence of ALK kinase domain mutations with increased EGFR phosphorylation or focal KIT amplification in two separate patients with crizotinib resistance3. The frequency of the various ALK TKI resistance mechanisms and the degree to which the ‘off-target’ effects of the ALK TKI may dictate both clinical efficacy and also mechanism of resistance remains to be more precisely defined.
In conclusion, provoked by observations from an ‘exceptional responder’, we have identified the IGF-1R/IRS-1 signaling axis as a potential therapeutic target in ALK+ lung cancer. Since ALK is as an oncogenic ‘driver’ in a growing number of hematologic and solid organ tumors, an improved understanding of ALK signaling as well as mechanisms of ‘escape’ to ALK inhibition may have direct therapeutic implications for other ALK-driven malignancies.
ONLINE METHODS
Cell culture
All cell lines were maintained in a humidified incubator 5% CO2 at 37 °C. The human lung adenocarcinoma cell lines H3122 and H2228 have been described previously and were verified to harbor their reported genetic alterations by direct cDNA sequencing14. Derivation of the human lung adenocarcinoma cell line, STE-1, is described below. The human anaplastic lymphoma cell line, SUDHL-1, has been previously described26. The isogenic pairs of EGFR TKI sensitive and EGFR TKI resistant cell lines used in these studies, including PC-9/PC-9 ERc1, HCC4006/HCC4006 ER, HCC827/HCC827 ER, and HCC2279/HCC2279 ER, have been previously described21,22. The EGFR TKI resistant cells (denoted ‘ER’ for Erlotinib Resistant) were continuously grown in 1 μM erlotinib. All cell lines were maintained in RPMI 1640 medium (Mediatech, Inc., Manassas, VA, USA) supplemented with 10% heat inactivated fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA) and penicillin-streptomycin (Mediatech, Inc., Manassas, VA, USA) to final concentrations 100 U ml−1 and 100 μg ml−1, respectively. All cell lines were routinely evaluated for mycoplasma contamination.
Derivation of STE-1 cell line
Pleural fluid was obtained with informed consent from a patient with crizotinib-naïve ALK+ metastatic lung adenocarcinoma. After pelleting the cells and washing 3x in sterile PBS, red blood cells were lysed in ACK buffer (Lonza INC, Allendale, NJ, USA). After lysis, the remaining cell pellet was washed 3x in sterile PBS. The remaining mixture of cells was then distributed into 10cm dishes. Cells were cultured in RPMI supplemented with 10% heat inactivated fetal bovine serum and penicillin-streptomycin as described above. The medium was changed every 1-3 days for approximately 3 months. To verify that the established cell line (named STE-1) harbored an ALK fusion, ALK FISH (Supplementary Fig. 8a) and cDNA sequencing (Supplementary Fig. 8b) of the ALK fusion was performed as described below.
Whole Genome Sequencing of the STE-1 cell line
Paired-end sequencing of tumor and matched blood genomic DNA was conducted on an Illumina Genome Analyzer IIx platform. The reads were aligned to the Human Genome (UCSC hg19) using BWA27. The default arguments of BWA were applied to the alignment. After the alignment, we ran the software SAMtools 28 to convert the alignment files to a sorted, indexed binary alignment map (BAM) format. Then, we used the Picard Web Site [http://picard.sourceforge.net/index.shtml] to mark duplicate reads. To obtain the best call set possible, we also used the software GATK29 to do realignment and recalibration. The recalibrated alignment files were then used for SNV detection. The sequencing data from this study can be accessed at the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRP044308 (tumor: SRR1514863 and blood: SRR1514948 blood).
Generation of TKI-resistant cell lines
To create ALK TKI resistant lines, parental (TKI sensitive) cells were cultured with increasing concentrations of TKIs starting with the IC30. Doses were increased in a stepwise pattern when normal cell proliferation patterns resumed. Fresh drug was added every 72-96 hours. Resistant cells that grew in 1μM crizotinib and 4μM X-376 were derived after approximately 6 months of culturing in the continuous presence of drug. DNA identity testing on both the parental and resistant cells confirmed that the cells were derived from the same origin. Resistant cells were maintained initially as polyclonal populations under constant TKI selection.
Compounds
X-376 was prepared as described previously14. Crizotinib (ChemieTek, Indianapolis, IN, USA), OSI-906 (Selleck Chemicals, Houston, TX, USA), AEW-541 (Selleck Chemicals, Houston, TX, USA), LDK-378 (Selleck Chemicals, Houston, TX, USA) and Lapatinib (Selleck Chemicals, Houston, TX, USA) were dissolved in DMSO. Erlotinib was synthesized by the Memorial Sloan-Kettering Cancer Center (MSKCC) Organic Synthesis Core. MAb391 (R&D systems, Minneapolis, MN, USA) was dissolved in PBS.
Cell viability, soft agar, and apoptosis assays
For viability experiments, cells were seeded in 96-well plates at 25%-33% confluency and exposed to drugs alone or in combination the following day. At 72 hours post drug addition, Cell Titer Blue reagent (Promega, Madison, WI, USA) was added and fluorescence was measured on a Spectramax spectrophotometer (Molecular Devices, Sunnyvale, CA, USA) according to the manufacturer's instructions. All experimental points were set up in hextuplicate replicates (except for the data presented in Figure 4b, which was set up in triplicate) and were performed at least three independent times. Data are presented as the percentage of viable cells compared to control (vehicle only treated) cells. Drug synergism was assessed using the Mix-Low method9. For soft agar assays, cells were seeded in 96 well plates and treated with drug according to the manufacturer's instructions (Cell Biolabs, Inc., San Diego, CA, USA). The absorbance at 570nm was measured 7 to 9 days post cell seeding and drug treatment on a Synergy MX microplate reader (Biotek, Winooksi, VT, USA). All experimental points were set up in hextuplicate replicates and were performed at least two independent times. Data are presented as the percentage of viable cells compared to control (vehicle only treated) cells. P values were determined with the Student's T-test. For STE-1 apoptosis experiments, cells were seeded in 6 well plates at approximately 50% confluency and treated the indicated inhibitors (one dose of inhibitor only). At 72 hours post drug addition, cells were collected, washed in PBS, and fixed in 100% ethanol. Fixed cells were stained with propidium iodide (working solution: 40 μg mL−1 propidium iodide plus 3.8 mM sodium citrate in PBS). For H3122 XR apoptosis experiments, cells were seeded in 6 well plates at approximately 75% confluency and treated the indicated inhibitors once every 24 hours for a total of 3 doses. At 72 hours post drug addition, cells were collected, washed in PBS, and fixed in 100% ethanol. All fixed cells were stained with propidium iodide (working solution: 40 μg mL−1 propidium iodide plus 3.8 mM sodium citrate in PBS). Data were collected on a FACSCantoII™ (BD Biosciences, San Jose, CA, USA) with FACSDiVa™ software to collect the data and Winlist™ software for the analysis (Verity Software House).
Antibodies and immunoblotting
The following antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA): ALK D5F3 (cat # 3333, 1:1000 dilution), ALK 31F12 (cat # 3791, 1:1000 dilution), phospho-ALK tyrosine 1604 (cat # 3341, 1:500–1:750 dilution), ribosomal protein S6 (cat # 2317, 1:1000–1:2000 dilution), phospho-S6 serine 240/244 (cat # 5364, 1:5000–1:6000 dilution), ERK (cat # 9102, 1:2000–1:3000 dilution), phospho-ERK threonine 202/tyrosine 204 (cat # 9101, 1:2000–1:3000 dilution), AKT (cat # 9272, 1:1000–1:2000 dilution), phospho-AKT serine 473 (cat # 9271, 1:500 dilution), IGF-1R β (cat # 3027, 1:2000 dilution), phospho-IGF-1R tyrosine 1131 (cat # 3021, 1:500–1:1000 dilution), IRS-1 D23G12 (cat # 3407, 1:1000 dilution), HRP-conjugated anti-mouse (cat # 7076, 1:1000–1:2000 dilution), and HRP-conjugated anti-rabbit (cat # 7074, 1:1000–1:2000 dilution). The actin antibody (cat # A2066, 1:5000 dilution) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
For immunoblotting and immunoprecipitation, cells were harvested, washed in PBS, and lysed in 50 mM Tris·HCl, pH 8.0/150 mM sodium chloride/5 mM magnesium chloride/1% Triton X-100/0.5% sodium deoxycholate/ 0.1% SDS/40 mM sodium fluoride/1 mM sodium orthovanadate and complete protease inhibitors (Roche Diagnostics, Indianapolis, IN, USA). Lysates were subjected to SDS-PAGE followed by blotting with the indicated antibodies and detection by Western Lightning ECL reagent (PerkinElmer, Waltham, MA, USA). Select images were quantified using a Bio-Rad Gel Doc XR and Image Lab software (Bio-Rad, Hercules, CA, USA) and normalized to the actin signal. Data are represented as band signal intensity compared to vehicle only control. For immunoprecipitation experiments, lysates were incubated with the primary antibody overnight at 4 °C. Protein A dynabeads (Invitrogen, Carlsbad, CA, USA) were then added and incubated in the lysate for 1 hour at 4 °C. Immobilized beads were washed 4× with lysis buffer.
cDNA sequencing of ALK
Total RNA was isolated from cell pellets using the RNeasy mini kit (Qiagen, Germantown, MD, USA). SuperScript III one-step RT-PCR system with platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) was used to perform both cDNA synthesis and PCR amplification with gene specific primers: EML4 E18F (aligned on EML4 exon 13, 5’ – TTAGCATTCTTGGGGAATGG- 3’) and ALK_kinase domain_R (5’-GCCTGTTGAGAGACCAGGAG-3’). The 1223bp PCR product, which includes the EML4-ALK fusion point and the entire ALK kinase domain, was sequenced in both directions by Sanger dideoxynucleotide sequencing. Sequencing data confirmed that H3122 parental, H3122 CR, H3122 XR, and STE-1 all harbored the EML4-ALK E13:A20 (variant 1) fusion and that H2228 harbored the EML4-ALK E6a/b:A20 (variant 3) fusion. No kinase domain mutations were found in any of these cell lines compared with Homo sapiens mRNA for EML4-ALK E13;A20 variant 1 (GeneBank: AB274722.1).
RTK proteome array and IGF-1 ELISAs
Proteome Profiler Human Phospho-RTK Array Kits (R&D Systems, ARY001B) and human IGF-1 Quantikine ELISA kits (R&D Systems, DG100) were processed according to manufacturer protocols.
siRNA experiments
Cells were reverse transfected with siRNAs using the Lipofectamine RNAimax reagent (Life Technologies, Grand Island, NY, USA). Non-targeting (“NT”) and IRS-1 siRNAs pools were purchased from Dharmacon (Lafayette, CO, USA) and Santa-Cruz (Dallas, TX, USA).
NanoString nCounter analysis
NanoString nCounter analysis of RNA isolated from formalin-fixed and paraffin embedded (FFPE) samples was performed as previously described23. The raw data were normalized to the nCounter system spike-in positive and negative controls in each sample. The normalized results are expressed as the relative mRNA level. The comparison between pre-/post-treatment groups was performed by modified paired T-test using the limma package.
Fluorescence in situ hybridization (FISH)
Samples were tested for ALK rearrangements by FISH (Vysis ALK Break Apart FISH Probe Kit) as previously described30.
Gene expression analysis
RNA from cell lines and controls was prepared, labeled, and hybridized to Illumina HT-12 array. For analysis, the sample probe profile data in the GeneSpring export format was transformed to log-2 scale and normalized using quantile method by applying beadarray package31 in BioConductor 2.11. Four duplicated samples were averaged to reduce the total sample size to 9 across three groups. The comparison between drug sensitive and drug resistant groups was performed using limma package. The significantly changed probes were identified by moderated t statistics. The p-values from moderated t-tests were adjusted by Benjamini and Hochberg's method to control false discovery rate. The microarray data reported in this article have been deposited in NCBI's Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (Accession no. GSE49508, URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49508).
Xenograft studies
Nude mice (nu/nu; Harlan Laboratories) were used for in vivo studies and were cared for in accordance with guidelines approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee and Research Animal Resource Center. 8 week old female mice were injected subcutaneously with 5 million H3122 cells together with matrigel. Once tumors reached an average volume of 100 mm3, mice were randomized to the different treatment cohorts. For the experiment in Supplementary Fig. 1f, mice were randomized to receive either crizotinib alone, MAb391 alone, crizotinib + MAb391, or vehicle control (n = 6 for vehicle alone, crizotinib, and crizotinib + MAb391. n = 5 for MAb391 only). Crizotinib was administered at 50 mg kg−1 p.o. daily × 5 days. MAb391 was administered at 1 mg i.p. every 3 days. For the experiment in Fig. 6b, mice were randomized to receive crizotinib alone (50 mg kg−1 p.o. daily × 5 days), LDK-378 alone (50 mg kg−1 p.o. daily × 5 days), or vehicle control (n = 5 for Crizotinib and LDK-378, n = 4 for vehicle control). Mice were observed daily throughout the treatment period for signs of morbidity/mortality. Tumors were measured twice weekly using calipers, and volume was calculated using the formula: length × width2 × 0.52. Body weight was also assessed twice weekly. P values were determined with the Wilcoxon Rank Sum test.
EML4-ALK (E13;A20) transgenic mice
Genetically engineered mice harboring the EML4-ALK E13;A20 fusion variant has been previously reported13. Three month old mice from both genders were used in the described studies. All animal treatment studies were reviewed and approved by the IACUC at the Dana-Farber Cancer Institute.
Tumor Biopsy Samples
All patient tumor biopsy samples were obtained under Institutional Review Board (IRB) approved protocols (Vanderbilt University IRB# 050644, Memorial Sloan-Kettering Cancer Center IRB #10-136, University Hospital of Cologne IRB #06037, Peter MacCallum Cancer Center IRB#08/71). Written informed consent was obtained from all patients. All samples were deidentified and protected health information was reviewed according to the Health Insurance Portability and Accountability Act (HIPAA) guidelines. For the index patient's tumor, clinical genotyping was performed using the SNaPshot platform as previously described32.
Immunohistochemistry
Tumors were pathologically identified and classified according to WHO guidelines. Briefly, 3 μm thick sections of FFPE tumors were deparaffinized. For pIGF-1R staining, antigen retrieval was performed by boiling the section in citrate buffer at pH 6 for 25 minutes. For IRS-1 staining, no pretreatment was necessary. Primary antibodies were used as follows: pIGF-1R (pY1161) (Ab39398, 1:100, pH 6, Abcam Inc.), IRS-1 (Ab40777, 1:100, Abcam Inc.). Corresponding secondary antibodies and detection kits were used (Enhancer: post antibody blocking for Bright Vision plus; Immuno Logic c-DPVB blocking and Polymer: Poly-HRP-GAM/R/R IgG; Immuno Logic c-DPVB999HRP, BrightVision+ cat #DPVB999HRP, ImmunoLogic, Duiven, The Netherlands, www.immunologic.nl) and stained on an automated stainer (LabVision Autostainer 480S, Thermo Scientific). Staining intensities were individually evaluated by 3 independent observers.
Supplementary Material
Acknowledgements
This work was supported by the VICC Cancer Center Core grant (P30-CA68485), a career development award from the Vanderbilt Specialized Program of Research Excellence in Lung Cancer grant (CA90949), NCI grants R01CA121210 and P01CA129243, and by the Joyce Family Foundation. CML was additionally supported by an NIH K12 training grant (K12 CA9060625), an ASCO Young Investigator Award, a Uniting Against Lung Cancer grant, and a Damon Runyon Clinical Investigator Award. CML was the Carol and Jim O'Hare chief fellow from 7/1/2011 through 6/30/2012. LCH and RB were supported by the Deutsche Forschungsgemeinschaft (SFB 832, Tumormicromilieu) and the German Cancer Aid (CIO Köln Bonn). MB was supported by the European Regional Development Fund, grant number FKZ:005-111-0027. GVM was supported by the Victorian Cancer Agency grant TS10_01. KKW is supported by the NIH CA122794, CA140594, CA163896, CA166480 and CA154303 grants. PKP was supported by a Uniting Against Lung Cancer grant. RKT is supported by the EU-Framework Programme CURELUNG (HEALTH-F2-2010-258677), by the Deutsche Forschungsgemeinschaft through TH1386/3-1 and through SFB832 (TP6), by the German Ministry of Science and Education (BMBF) as part of the NGFNplus program (grant 01GS08100), and by the Deutsche Krebshilfe as part of the Oncology Centers of Excellence funding program. SP was supported by a grant from the Rudolph Becker Foundation. JW was supported by the German Cancer Aid (CIO Köln Bonn), the Federal Ministry of Education and Research (NGFNplus), and the Ministry of Economy, Energy, Industry and Craft of North Rhine-Westfalia in the PerMed. NRW framework program. ZZ was supported by the NIH R01LM011177. We thank J. Sosman and C. Arteaga for their critical review of this manuscript, C. Liang for providing X-376, and A. Nashabi for administrative assistance. Australian specimens were processed by the Victorian Cancer Biobank. The human anaplastic lymphoma cell line, SUDHL-1, was a generous gift from Dr. S. Morris of St. Jude Children's Research Hospital.
Footnotes
Author Contributions:
C.M.L. and W.P. conceived the project and wrote the manuscript. C.M.L., N.T.M., Y.Y., H.J., and M.R.B performed the molecular biology experiments. H.C., P.L., X.C., and R.S. performed the statistical analysis. S.C.O., L.C.H., A.F., and R.K.T. performed all the IGF-1R and IRS-1 immunohistochemistry experiments. S.C.O., L.O., P.K.P., R.B., S.A., S.P., M.B., M.B., J.W., M.G., G.V.M., B.S., P.A.R., T.M.R., and R.K.T. provided clinical samples. D.H.J. and L.H. provided clinical care for the index patient. Z.C. and K.K.W. provided the EML4-ALK E13;A20 transgenic mice. D.L., L.W., Y.S., and M.L. performed all the FISH and nanostring experiments. R.T. and E.D.S. performed the xenograft studies. Q.W. and Z.Z. analyzed the whole-genome sequencing data.
Conflicts of Interest:
CML and WP have filed a provisional patent describing the improved efficacy of combined treatment of ALK inhibitors plus IGF-1R inhibitors compared to ALK inhibitors alone (serial number 61/768,072). CML has served on an Advisory Board for Pfizer and has served as a speaker for Abbott and Qiagen. WP has done consulting for MolecularMD, AstraZeneca, Bristol-Myers Squibb, Symphony Evolution, Clovis Oncology, Exelixis, Clarient, Champions, Clarient (MDOutlook), and WebMD. WP has received research funding from Enzon, Xcovery, AstraZeneca, Symphogen, Clovis Oncology, and Bristol-Myers Squibb. Rights to EGFR T790M testing were licensed on behalf of WP and colleagues to MolecularMD. RKT is a founder and shareholder of Blackfield AG, a company focused on cancer genome diagnostics and cancer genomics-based drug discovery. LCH received lecture fees (Roche, Novartis, Pfizer, Qiagen). RKT received consulting and lecture fees (Sanofi-Aventis, Merck, Roche, Lilly, Boehringer Ingelheim, Astra-Zeneca, Atlas-Biolabs, Daiichi-Sankyo, Blackfield AG) as well as research support (Merck, EOS and AstraZeneca). RB is a cofounder and owner of Targos Molecular Inc. and served on advisory boards for Pfizer, Roche, Boehringer Ingelheim, Merck Serono, Novartis and Lilly. JW has served as a Consultant or Advisory Role for Roche, Novartis, Pfizer, Boehringer-Ingelheim, AstraZeneca, Bayer Pharmaceuticals, Eli Lilly, and Merck. GVM has served on an Advisory board for Pfizer. BS has served on Advisory Boards for Pfizer and Novartis. KKW received sponsored research grants from Takeda, AstraZeneca, Roche and Infinity Pharmaceuticals. He is also a consultant for G1 Therapeutics. PKP has served on an Advisory Board for Bristol-Myers-Squibb. SP received consulting and lecture fees (Novartis, Curagita, Roche, Abbot, Definiens) as well as research support (Ventana-Roche). JW has served as a Consultant or Advisory Role for Roche, Novartis, Pfizer, Boehringer-Ingelheim, AstraZeneca, Bayer Pharmaceuticals, Eli Lilly, Merck. GVM has served on an Advisory board for Pfizer. BS has served on Advisory Boards for Pfizer and Novartis. LH has served on Advisory Boards for Bristol-Myers-Squibb (compensated), PUMA (uncompensated), and HelixBio (uncompensated). LH has also received research grants from Astellas and has served as a speaker for Boehringer Ingelheim.
References
- 1.Grande E, Bolos MV, Arriola E. Targeting oncogenic ALK: a promising strategy for cancer treatment. Mol Cancer Ther. 2011;10:569–579. doi: 10.1158/1535-7163.MCT-10-0615. [DOI] [PubMed] [Google Scholar]
- 2.Camidge DR, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012 doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4:120ra117. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doebele RC, et al. Mechanisms of Resistance to Crizotinib in Patients with ALK Gene Rearranged Non-Small Cell Lung Cancer. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovly CM, Pao W. Escaping ALK inhibition: mechanisms of and strategies to overcome resistance. Sci Transl Med. 2012;4:120ps122. doi: 10.1126/scitranslmed.3003728. [DOI] [PubMed] [Google Scholar]
- 6.Tanizaki J, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0392. [DOI] [PubMed] [Google Scholar]
- 7.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AT, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boik JC, Newman RA, Boik RJ. Quantifying synergism/antagonism using nonlinear mixed-effects modeling: a simulation study. Stat Med. 2008;27:1040–1061. doi: 10.1002/sim.3005. [DOI] [PubMed] [Google Scholar]
- 10.Mulvihill MJ, et al. Discovery of OSI-906: a selective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor. Future Med Chem. 2009;1:1153–1171. doi: 10.4155/fmc.09.89. [DOI] [PubMed] [Google Scholar]
- 11.Metz HE, Houghton AM. Insulin receptor substrate regulation of phosphoinositide 3-kinase. Clin Cancer Res. 2011;17:206–211. doi: 10.1158/1078-0432.CCR-10-0434. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, et al. Using tandem mass spectrometry in targeted mode to identify activators of class IA PI3K in cancer. Cancer Res. 2011;71:5965–5975. doi: 10.1158/0008-5472.CAN-11-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 2010;70:9827–9836. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovly CM, et al. Insights into ALK-driven cancers revealed through development of novel ALK tyrosine kinase inhibitors. Cancer Res. 2011;71:4920–4931. doi: 10.1158/0008-5472.CAN-10-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama R, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A. 2011;108:7535–7540. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guix M, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortot AB, et al. Resistance to irreversible EGF receptor tyrosine kinase inhibitors through a multistep mechanism involving the IGF1R pathway. Cancer Res. 2013;73:834–843. doi: 10.1158/0008-5472.CAN-12-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Echeverria C, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5:231–239. doi: 10.1016/s1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- 19.Morgillo F, et al. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–2803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 20.Vazquez-Martin A, et al. IGF-1R/epithelial-to-mesenchymal transition (EMT) crosstalk suppresses the erlotinib-sensitizing effect of EGFR exon 19 deletion mutations. Scientific reports. 2013;3:2560. doi: 10.1038/srep02560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chmielecki J, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohashi K, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suehara Y, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–6608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw AT, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galkin AV, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci U S A. 2007;104:270–275. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris SW, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gainor JF, et al. ALK Rearrangements Are Mutually Exclusive with Mutations in EGFR or KRAS: An Analysis of 1,683 Patients with Non-Small Cell Lung Cancer. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 32.Su Z, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.