Abstract
Influenza is an acute respiratory disease in mammals and domestic poultry that emerges from zoonotic reservoirs in aquatic birds and bats. Although influenza viruses are among the most intensively studied pathogens, existing control options require further improvement. Influenza vaccines must be regularly updated because of continuous antigenic drift and sporadic antigenic shifts in the viral surface glycoproteins. Currently, influenza therapeutics are limited to neuraminidase inhibitors; novel drugs and vaccine approaches are therefore urgently needed. Advances in vaccinology and structural analysis have revealed common antigenic epitopes on hemagglutinins across all influenza viruses and suggest that a universal influenza vaccine is possible. In addition, various immunomodulatory agents and signaling pathway inhibitors are undergoing preclinical development. Continuing challenges in influenza include the emergence of pandemic H1N1 influenza in 2009, human infections with avian H7N9 influenza in 2013, and sporadic human cases of highly pathogenic avian H5N1 influenza. Here, we review the challenges facing influenza scientists and veterinary and human public health officials; we also discuss the exciting possibility of achieving the ultimate goal of controlling influenza’s ability to change its antigenicity.
Keywords: influenza virus, antigenic changes, vaccines, antiviral drugs, H1N1, H7N9, H5N1, pandemic, zoonosis
Introduction
Influenza viruses are notoriously variable. They have a segmented negative-sense RNA genome that lacks proofreading mechanisms and thus mutations are continuously accumulated. Antigenic drift and antigenic shift are continuous processes that result in influenza viruses existing as a quasispecies.1 In their natural reservoirs in aquatic birds and bats, influenza A viruses show minimal evolution and cause inapparent disease; but once they transfer to other species, influenza A viruses can evolve rapidly and cause pandemics and epidemics of acute respiratory disease in domestic poultry, lower mammals, and humans.2
The realization that all influenza A viruses emerge from zoonotic reservoirs has led to a the concept of one world–one health, which means that the problem of controlling emerging influenza viruses is a challenge to both veterinary and public health officials worldwide.3 The emergence of a pandemic H1N1 influenza in 2009, at the same time that the influenza community was preparing for the spread of highly pathogenic H5N1 influenza, emphasizes the unpredictability of this pathogen. The control strategies then depended on the use of a single class of drugs, the neuraminidase (NA) inhibitors (NAIs), because a homologous H1N1 vaccine was not available during the first pandemic wave. Further challenges arose with the emergence of an H7N9 influenza virus in China in 2013, which caused inapparent disease in wild life and poultry, but severe respiratory disease in humans. The current challenge is to be prepared if either the H2N2 influenza virus—absent from humans for 50 years—re-emerges from the aquatic bird reservoir, or the H5N1 or H7N9 viruses acquire efficient human-to-human transmissibility.
This review considers the challenges that veterinary and public health officials encounter when attempting to control not only novel emerging influenza subtypes with pandemic potential, but also continuous antigenic variation in circulating epidemic strains. Antiviral, vaccine, prevention, and eradication strategies are considered from the perspective of current challenges. We focus mainly on influenza A viruses, with limited discussion of influenza B viruses.
Biology of influenza
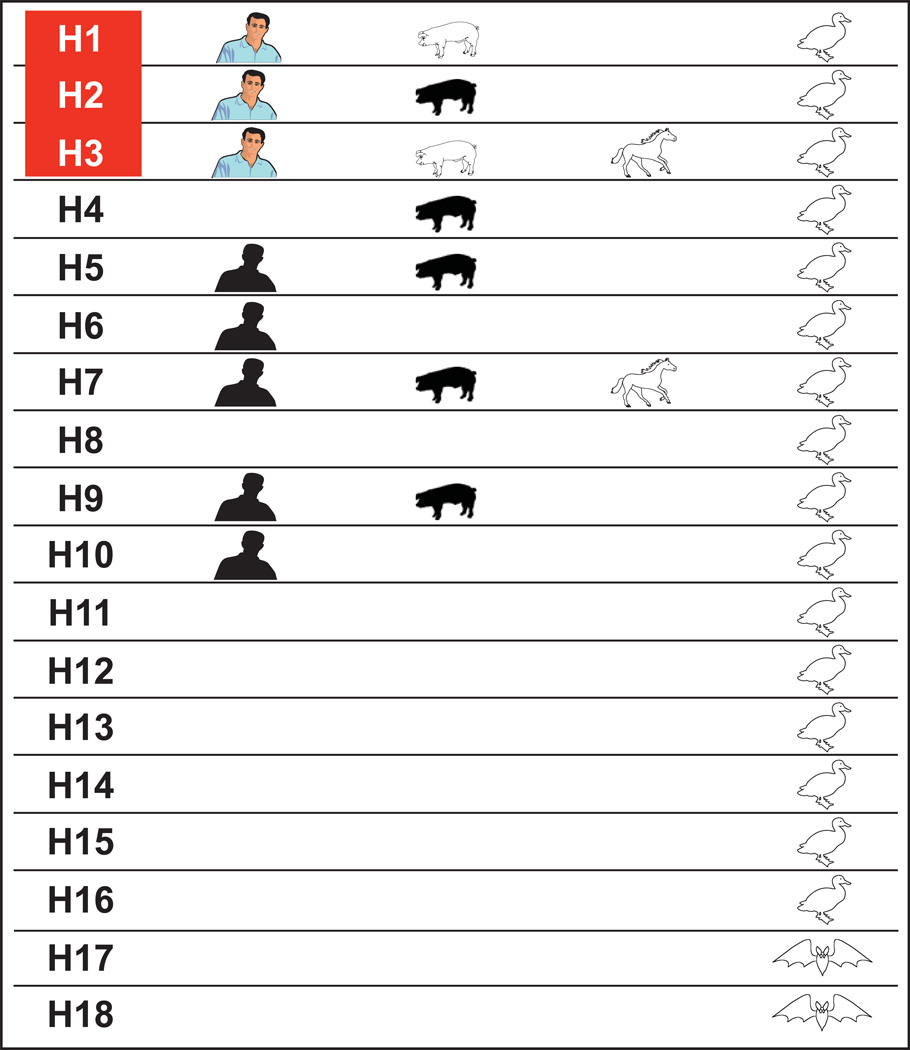
Eighteen hemagglutinin (HA) and 11 NA subtypes of influenza A viruses are found in nature (Fig. 1). HA subtypes are further subdivided into two groups: group 1 includes H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18; group 2 includes H3, H4, H7, H10, H14, and H15.4 Sixteen HA and nine NA subtypes circulate in the aquatic birds of the world, where they replicate primarily in the intestinal tract at the higher body temperature of birds (40 °C), have a target-cell receptor preference for α2,3 sialic acid, and are spread primarily by fecal–oral transmission through water. Two HA and NA subtypes of influenza A virus are found in bats; they have not been cultured but can be detected by polymerase chain reaction (PCR), cause inapparent disease, and do not attach to sialic acid residues.5 Influenza B viruses have a single subtype with two lineages, Victoria and Yamagata. Although influenza B viruses have been detected in seals, they are primarily confined to humans.
Figure 1.
A diagram of the host ranges of the 18 hemagglutinin (HA) subtypes of influenza A viruses. Open figures indicate hosts in which stable lineages are established. Filled figures indicate hosts in which sporadic but not consistent interhost transmission of influenza has occurred. Colored figures indicate influenza subtypes that have caused pandemic or epidemic influenza. The 16 HA subtypes of influenza A viruses in aquatic birds and the two subtypes in bats cause inapparent disease and are shed in feces. In mammalian hosts, influenza causes upper respiratory disease. In horses, two subtypes—Equine 1 (H7N7) and Equine-2 (H3N8)—have circulated; however, Equine-1 has not been detected in horses for the past 2 decades. Sporadic transmission of H2 in pigs in China was reported in 1957, when the Asian H2N2 pandemic occurred but did not establish a stable lineage. Sporadic transmissions of H4, H5, H6, H7, H9, and H10 to humans and pigs have increased since the mid-1990s, but no consistent interhost transmission has occurred.
Only three HA subtypes of influenza A viruses (H1N1, H2N2, and H3N2) have caused pandemics in humans in the past century. Thus, these may be the only subtypes that can establish lineages in humans. However, the occasional transmission of avian influenza viruses to humans (i.e., H5, H6, H7, H9, and H10) results in mild-to-severe disease but without consistent human-to-human transmission, which raises doubt about the hypothesis that only H1N1, H2N2, and H3N2 can establish infection in humans. The influenza subtype H7N7, which circulated in horses for many years (Equine-1), is an example of an influenza A subtype that established a stable lineage in mammals.
The two HA subtypes H5 and H7, nonpathogenic in their aquatic bird reservoir species, are unique in their ability to become highly pathogenic in domestic poultry.6 One diagnostic characteristic of high pathogenicity is the acquisition of multiple basic amino acids in the connecting peptide of the HA molecule. The resulting HA is cleaved by the furin family of enzymes, resulting in virus that can spread systematically in infected hosts, causing hemorrhaging and neurological symptoms. This high pathogenicity of avian H5 and H7 influenza viruses is a polygenic property that requires a specific grouping of viral gene segments. Since the 1990s, H5 and H7 viruses have transmitted to mammals, including humans, multiple times; the current challenge is to control the outbreaks of H5N1 and low-pathogenic H7N9 viruses before they acquire the ability to consistently transmit between humans.
The transmission of avian influenza viruses from the aquatic bird reservoir to mammals requires multiple changes that favor zoonotic spread. These include the optimal temperature of virus replication decreasing from 40 °C (avian) to 37 °C (mammalian); the site of replication changing from intestinal to respiratory; and the receptor specificity switching from α2,3 sialic acid to α2,6 sialic acid.7 Intermediate hosts that possess dual-specificity sialic acid receptors on their cells, (e.g., swine and domestic poultry (quails and turkeys)) may be involved in the interspecies transfer of influenza viruses. Changes in animal husbandry practices from small family farms to large poultry- and swine-raising farms to meet increasing human demands, and the practice of live poultry and animal marketing, contribute substantially to the increased threat of RNA viruses emerging from their zoonotic reservoirs. The possible relevance of domestic cats and dogs in the interspecies transmission of influenza viruses to humans requires additional study.
Challenges of continuous antigenic drift and antigenic shift
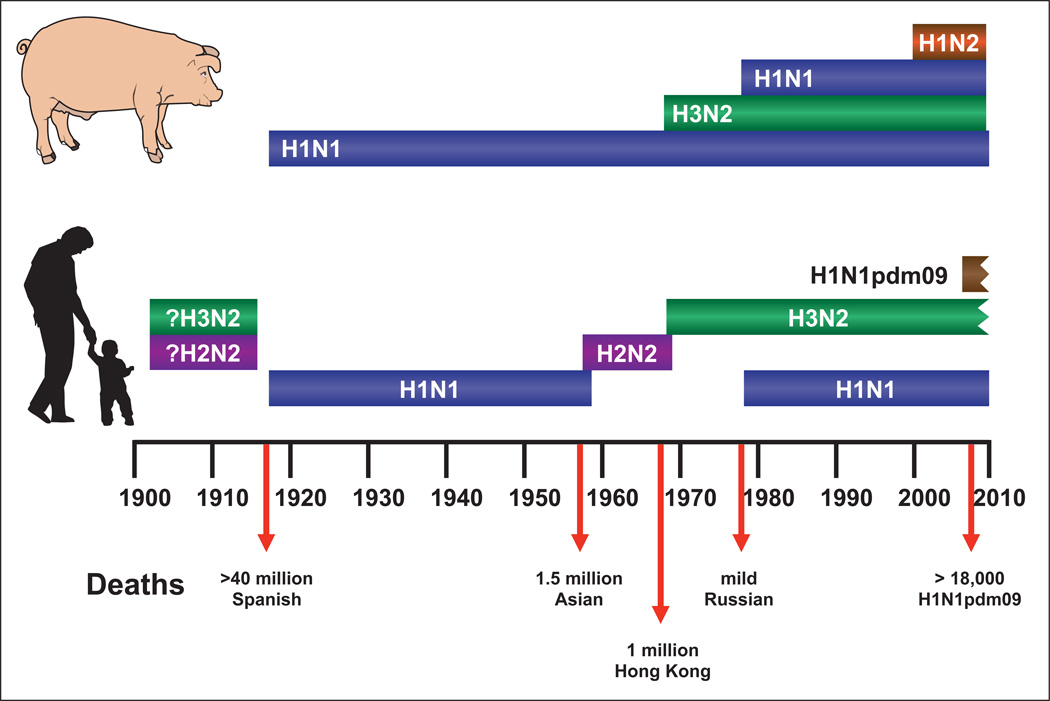
In the past century, four influenza pandemics have occurred in humans (Fig. 2), and seroarcheology studies suggests that they were preceded by outbreaks of H2N2 and H3N2 in avian species that subsequently re-emerged in humans. The H1N1 pandemic of 1918 (Spanish) was the most catastrophic, followed by the H2N2 pandemic of 1957 (Asian), and the H3N2 pandemic of 1968 (Hong Kong). The H1N1 virus that disappeared from humans in 1957 re-emerged in 1977, and in 2009 an H1N1 pandemic occurred while the descendants of the 1977 virus were still circulating. Each pandemic of influenza acquired gene segments from the aquatic bird reservoir by reassortment, which resulted in viral expression of novel HA and NA surface glycoproteins to which the majority of the human population had little or no immunity.
Figure 2.
Influenza pandemics in humans and pigs in the past century. Only three subtypes of influenza A viruses have caused pandemics in humans: H1N1, H2N2, and H3N2. H1N1 and H3N2 have also caused outbreaks in swine during the same period. On the basis of seroarcheological studies, it is proposed that H2N2 and H3N2 subtypes have previously infected humans. The most severe pandemic, which was caused by the Spanish influenza of 1918, resulted in more than 40 million deaths worldwide, many of which resulted from co-infection with bacteria. Subsequent pandemics have been less severe. Because H2N2 viruses have not circulated in humans for nearly 50 years, most people are immunologically naive. The H2N2 viruses, however, continue to circulate in aquatic birds. The jagged edges on the ends of the H1N1pdm09 and H3N2 bars indicate that these viruses continue to circulate.
The major challenge in the influenza community is planning for a pandemic while making recommendations on strain selection to combat antigenic drift in the circulating influenza subtypes. The influenza network established by the World Health Organization (WHO), which is known as the Global Influenza Surveillance and Response System (GISRS), consists of more than 120 national laboratories that monitor influenza globally. Antigenic analysis, sequence analysis, and population susceptibility based on antibody levels in human sera are used to make recommendations on vaccine content for the Northern and Southern Hemispheres.
Recommendations on vaccine strains have to be made 6–8 months before the vaccine can be produced; thus, the dominant strain may change during the interim owing to antigenic drift. The WHO has a remarkable 45-year record of success predicting the dominant circulating strain. On only two occasions has the recommended strain not matched the strain that emerged. In 1997–1998, the A/Wuhan/359/1995-like (H3N2) virus was the recommended vaccine strain for the H3N2 subtype, but the U.S. manufacturers chose to use A/Nanchang/933/1995 (H3N2) virus instead because of its high-yield growth properties. The antigenically drifted A/Sydney/5/1997 (H3N2) emerged and caused infection of vaccinated persons.8 In 2003–2004, the A/Panama/2007/1999 (H3N2) virus was the recommended vaccine strain, but A/Fujian/411/2002 (H3N2) became dominant.9
Antigenic drift
Antigenic drift results from the interplay between the influenza virus and the immune response of the host. Amino acid substitution in the HA and NA surface glycoproteins are of primary importance; if a substitution permits the virus to escape from immune control and provides a fitness advantage, then that virus has the opportunity to emerge as a novel epidemic strain, replacing the circulating strain.10 Antigenic mapping and sequence analysis of the escape mutants selected in the presence of monoclonal antibodies (mAbs) against the HA and NA surface glycoproteins defined four antigenic sites (epitopes) on the globular head of the HA,11 and further work increased that number to five antigenic sites (A–E).12 HA glycoprotein is considered the most important antigen in antigenic drift and has received the most attention, which may be a continuing flaw in influenza preparedness. Here, we will use the A/Hong Kong (H3N2) family of viruses to illustrate this phenomenon.
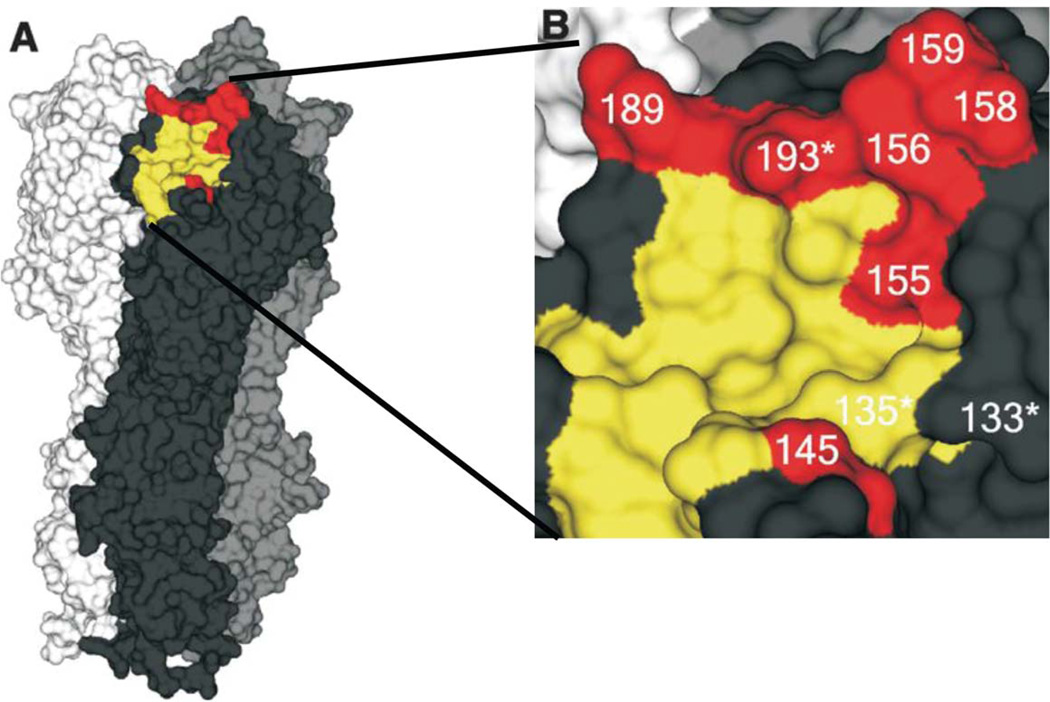
Since the emergence of A/Hong Kong/1/1968 (H3N2) virus 46 years ago, the WHO has recommended 29 changes in vaccine composition.13 Molecular analysis of the HAs showed that amino acid substitutions causing antigenic drift were largely confined to epitopes A–E.12 The working concept was that four amino acid changes in at least two antigenic sites were required for antigenic drift to occur. Current analysis of antigenic drift in H3N2 viruses indicates that epitopes A and B are key and that single amino acid changes in these sites have determined the major antigenic changes in the H3N2 subtype since 1968.14 Thus, the substitution of Q189K in the HA of A/England/42/1972 (H3N2) virus was crucial in changing the viruses’ antigenicity to that found in the A/Victoria/3/1975 (H3N2) virus. This amino acid substitution borders the receptor-binding site, and the key amino acid changes responsible for the major antigenic changes in the H1N1, H3N2, and influenza B viruses occur in that ridge (Fig. 3). The role(s) of the other compensatory mutations in determining antigenicity and fitness remains unresolved.
Figure 3.
The hemagglutinin (HA) molecule of A/Aichi/2/68 (H3N2) influenza virus in which the amino acid residues that are crucial in antigenic drift are indicated. (A) The trimeric model of the HA molecule. One monomer is shown in black; key amino acids in antigenic drift are shown in red; and the receptor-binding site is shown in yellow. (B) Residue 145 is located in antigenic site A, and residues 155, 156, 158, 159, and 193 are in antigenic site B. These are considered “cluster-transition” amino acids and correspond with amino acids in the HA of H1N1 and influenza B viruses. An asterisk indicates accessory substitutions. Reprinted from Ref. 14 with permission from the AAAS.
Antigenic shift
Antigenic shifts occur by reassortment and introduction of a novel HA, NA, or both gene segments from the aquatic bird reservoir into currently circulating human influenza viruses. The virus subtypes of concern in aquatic birds can be divided into three tiers: those that have caused pandemics in humans (H1, H2, H3) make up tier 1; those that have sporadically infected humans (H5, H7, H9) make up tier 2; and the remaining subtypes make up tier 3.15 Because H6N1 and H10N8 have caused isolated human infections,16,17 additional subtypes will probably be reclassified as tier 2. Here we will consider the continuing challenges presented by tier 1 (H2N2 and H2N3) and tier 2 viruses.
H2N2 influenza viruses
In 1957, an H2N2 influenza virus caused an Asian pandemic with an estimated global mortality of approximately 1.5 million.18 The virus circulated for a decade and then disappeared from humans in 1968. However, the H2N2 subtype of influenza virus continues to circulate in the aquatic birds of the world, and is a potential pandemic threat because there is precedence for recycling of the H1, H2, and H3 subtypes, and because the majority of the world’s population is immunologically naive to the H2N2 subtype.
Surveillance of aquatic birds in the Americas and Europe has indicated that H2N2 influenza viruses occur relatively infrequently. No H2N2 influenza viruses have been detected among the 1011 influenza isolates from shorebirds and gulls at Delaware Bay or from migratory ducks in Alberta, Canada in the past decade. In comparison, nine H2N2 influenza virus isolates were found in wild ducks by the Minnesota Center of Excellence for Influenza Research and Surveillance (CEIRS) in the Central Flyway in North America in the past decade. Surveillance in domestic poultry, on the other hand, has detected the continuing circulation of low-pathogenic H2N2 influenza in chickens and guinea fowl in New York and New Jersey live poultry markets from 1994 to 1997, and in chickens in Pennsylvania live-poultry markets in 2004. Surveillance in wild aquatic birds in Europe and Asia has reported sporadic isolations of H2N2 over the past decade, with 10 influenza H2N2 viruses reported in GenBank.
To determine the risk of human infections with H2N2 influenza viruses, 22 avian H2N2 influenza viruses from the past 6 decades were assayed for their potential to replicate in human and swine tissue culture and in mice and ferrets.19 The avian H2N2 viruses had a low rate of genetic and antigenic evolution and remained similar to isolates circulating near the time of the 1957 pandemic. Most isolates replicated in mice and differentiated human bronchial epithelial cells, but replication was low or undetectable in swine trachea explants. Multiple isolates replicated in ferrets, and three viruses were transmitted to direct-contact cage mates (Table 1). Markers of mammalian adaptation in HA and PB2 proteins were absent from all isolates, and the viruses retained a preference for avian-like α2,3-linked sialic acid receptors. Most isolates remained antigenically similar to prototype A/Singapore/1/1957 (H2N2) virus, suggesting they could be controlled by the 1957 pandemic vaccine candidate. The H2N2 viruses tested were all susceptible to anti-influenza drugs (NAIs and adamantanes). Like the 1957 pandemic H2N2 virus,20 the H2N2 influenza virus with the highest risk (i.e., A/duck/Hong Kong/319/1979 (H2N2)) was from the Eurasian avian lineage.19 Although infrequently isolated from aquatic birds, the currently isolated avian H2N2 viruses merit continued global surveillance and risk assessment, especially the Eurasian viruses.
Table 1.
Risk assessment of avian H2N2 influenza viruses
| Influenza A virus |
Receptor specificitya |
NHBE cellsb |
Swine explantsc |
Miced | Ferretse | HA 226f |
PB2 627g |
M2 inhibitorsh |
NAIsi | Similarity to pandemic vaccinej |
Risk score |
Risk classificationk |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mal/MT/61 | 0 | 3 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 8 | Intermediate |
| Dk/HK273/78 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | Low |
| Dk/HK319/79 | 0 | 3 | 1 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 10 | High |
| Mal/Pot177/83 | 0 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 6 | Intermediate |
| Mal/Pot179/83 | 0 | 3 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 8 | Intermediate |
| Ck/Jena/84 | 0 | 3 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 8 | Intermediate |
| Dk/NJ/95 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | Low |
| GF/NY/95 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | Low |
| Ck/NJ/95 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | Low |
| Sw/MO/06 (H2N3) | 1 | 3 | 1 | 3 | 3 | 1 | 1 | 0 | 0 | 0 | 13 | High |
| VN/1203 (H5N1)l | 0 | 3 | 1 | 3 | 1 | 0 | 1 | 1 | 0 | N/A | 10 | High |
Polymer binding: > 2-fold preference for SA α2,3 = score (0); > 2-fold preference for SA α2,6 = (1)
Replication kinetics: none = score (0); low = (1); intermediate/delayed = (2); high/rapid = (3)
No replication = score (0); productive replication = (1)
Morbidity: low/none = score (0); intermediate = (1); high = (2); high with mortality = (3)
No replication in donors = score (0); replication in donors, no transmission = (1); replication in one contact = (2); replication in both contacts = (3); aerosol transmission = (4)
Consensus at HA residue 226: avian consensus (Q) = score (0); mammalian consensus (L) = (1)
Consensus at PB2 residue 627: avian consensus (E) = score (0); mammalian consensus (K) = (1)
Adamantane-sensitive residues in M2 = score (0); one or more resistant residues in M2 = (1)
Inhibition by NAI: < 1 µM dose = score (0); > 1 µM dose = (1)
Antigenically similar, as determined by HI assay: virus inhibited by A/Singapore/1/57 sera ≥ 1:40 = score (0); not inhibited = (1)
Risk classification: low risk (< 5); intermediate risk (5–9); high risk (> 10)
Replication of VN/1203 (H5N1) in swine was determined in vivo rather than in tissue explants. The NAI score met the criteria for susceptibility to oseltamivir but not zanamivir
H2N3 influenza virus
During the past decade, H2N3 has been the most prevalent H2 influenza subtype in migratory aquatic birds in the Americas and Europe. In 2006, H2N3 spread from avian species to swine in the United States and caused an outbreak of respiratory disease.21 The virus, with an L226 residue of the H2 glycoprotein, acquired mammalian-like characteristics in receptor binding and an ability to replicate without adaptation in mice and ferrets. Surprisingly, this H2N3 virus did not establish a lineage in swine, and has since disappeared from that species.
H5N1, H7N9, and H9N2 influenza viruses
Since the late 1990s, H5N1 and H7N9 viruses have emerged in domestic poultry: H5N1 virus emerged in 1997 and H7N9 virus in 2013. Both have the ability to sporadically transmit to humans and cause high mortality. As of early 2014, there have been 650 human cases of H5N1 with 386 deaths and 259 cases of H7N9 with 71 deaths. Many similarities and some differences are apparent between the novel H5N1 and H7N9 influenza viruses that arose in China and sporadically transmit with high mortality to humans (Table 2). Both viruses were triple reassortants, with six gene segments from H9N2 influenza viruses; H5N1 is highly pathogenic in gallinaceous poultry (chicken and quail), whereas H7N9 causes inapparent infections in poultry. H7N9 is more transmissible to humans than is H5N1, but the latter causes higher mortality (>60%) than the former (~30%). Both cause similar symptoms, specifically acute respiratory distress syndrome and multiorgan failure.
Table 2.
Similarities and differences between H5N1 and H7N9 influenza viruses
| Property | H5N1 influenza virus | H7N9 influenza virus |
|---|---|---|
| Source | Domestic poultry | Domestic poultry |
| Pathogenicity in poultry | Highly pathogenic; systemic replication | Low pathogenic; respiratory tract replication |
| Pathogenicity in humans | High lethality; ≤ 60% mortality | High lethality; ≤ 30% mortality |
| Median age and sex of infected humans | Young adult (20–30 years); slight female bias | Older men (62 years) |
| Symptoms in humans | Respiratory distress syndrome; multiorgan failure | Respiratory distress syndrome; multiorgan failure |
| Time to first 100 cases | > 3 years | 57 days |
| Properties of virus | Triple reassortant Surface genes from domestic poultry Internal genes from H9N2–domestic poultry |
Triple reassortant Surface genes from wild birds Internal genes from H9N2–domestic poultry |
| Seasonality | Peak activity in poultry and humans during cooler months | Peak activity in humans during winter months |
| Influence of closure of live bird market | Interrupted initial outbreak in Hong Kong | Interrupted initial outbreak in Northeast China |
| Sensitivity to antivirals | Resistant to adamantanes Sensitive to NAIs |
Resistant to adamantanes Sensitive to NAIs |
| Resistance to NAIs | Human isolates ~ 60%, avian isolates ~ 30% | Human strains have NAI-resistant substitutions |
| Control strategies in domestic poultry | Stamping out successful with potential for reintroduction | Closure of live bird market |
| Vaccination | Reduces disease in poultry and humans but not at the cost of endemicity | Not currently used |
| Replication and transmissibility in swine | Replicates without disease signs but fails to transmit | Replicates without disease signs but fails to transmit |
| Replication and transmissibility in ferrets | Some early strains are lethal and spread systemically Transmits to in-cage contacts but not by aerosol |
Mild respiratory infections Transmits to in-cage contacts and limited aerosol transmission |
| Molecular properties | ||
| HA receptor binding | Predominantly SA α2,3 binding | Heterogenous with SA α2,3 and SA α2,6 binding |
| NA stalk deletion | No | Yes, associated with replication in domestic poultry |
| PB2-E627K | No | Yes, associated with mammalian adaptation |
| Replication in swans and geese | Lethal | No disease signs |
| Replication in ducks | Some breeds die; others show no disease signs | No disease signs |
NOTE: NIAs, neuraminidase inhibitors
Despite the exposure of many humans to these viruses in the live poultry markets, relatively few are infected, as evidenced by serological studies for H5N1 viruses.22 The high mortality has been associated with a cytokine storm in persons with a defect in their interferon-induced transmembrane protein 3 (IFITM3).23 The continuing challenge with avian influenza viruses is to be prepared if they acquire the mutations that will permit them to transmit between humans. Highly pathogenic H5N1 influenza viruses have circulated for more than 17 years and have become endemic in multiple countries, including China, Vietnam, Indonesia, and Egypt. As a result, there was a sense of complacency that, despite continued evolution, these viruses may not have the potential to acquire the ability to transmit between humans. Using the ferret model of influenza infection, gain-of-function studies in three separate laboratories have shown that as few as three amino acid substitutions in the HA can generate mammalian-transmissible viruses by indirect aerosol droplets.24–26 Despite controversy about accidental release of these viruses or the malicious use of this information for bioterrorism, these studies established that mammalian-transmissible H5N1 virus can arise in nature.
The common denominator in the genesis of H5N1 and H7N9 influenza viruses is reassortment between a precursor virus, either H5N1 or H7N9, and H9N2 influenza virus. The resultant virus possesses the surface glycoproteins from either H5N1 or H7N9 influenza viruses and the six internal genes from H9N2 viruses. Before 1997, no low- or high-pathogenic H5 influenza viruses were detected in humans; but H7 influenza viruses had caused modest numbers of conjunctivitis cases in humans27 and occasional respiratory infections, including one death and infection in 89 persons in the Netherlands.28 Reassortment with H9N2 led to a marked increase in human infections with both H5N1 and H7N9 viruses; hence, H9N2 can be considered an “enabler” of infection. The lineage of H9N2 influenza viruses that donated the internal gene segments to H5N1 and H7N9 were first detected in chickens in Asia in 1997.29,30 These H9N2 viruses have evolved into multiple lineages and sublineages, and are endemic in chickens and quail in most countries in Eurasia.31,32 In domestic poultry, H9N2 viruses replicate primarily in the upper respiratory tract and cause inapparent infection, unless the birds are co-infected with bacterial, parasitic, or other viral agents. Co-infection can result in decreased egg production and respiratory disease. H9N2 viruses have acquired a preference for human-like receptors (i.e., α2,6 sialic acid)33–35 and sporadically cause mild infections in humans and swine.36,37
Since the emergence of the H5N1 viruses, multiple genotypes have evolved with replacements of the internal gene segments originally supplied by H9N2. Therefore, although H9N2 is hypothesized to be the initial enabler, further reassortment events can occur. The emergence of H7N9 influenza virus in February 2013 was as a triple reassortant possessing six internal gene segments from H9N2.38 The report of a lethal human infection in China in late 2013 with an H10N8 influenza virus possessing internal gene segments from H9N2 adds support to the notion that H9N2 is an enabler virus. The challenge is to determine the molecular basis for this enabling ability of H9N2 and focus efforts on surveillance and risk assessment of H9N2 influenza viruses.
The challenges presented by the novel H5N1 and H7N9 influenza viruses differ depending on an agricultural or a public health perspective. The novel H7N9 reassortant virus poses limited or no threat to the poultry or pork industry, and the absence of overt disease signs provides no warning for transmission to humans; H7N9 is therefore more insidious than H5N1. The H5N1 viruses are highly pathogenic in unvaccinated poultry and can cause up to 100% mortality; hence, they are of great concern both to veterinary and public health officials, as this high mortality provides a warning system for potential human infection. In vaccinated poultry, H5N1 virus can shed without disease signs, which presents a conundrum in control strategies. Live poultry markets serve as the ultimate mixing and breeding site for influenza from the perspective of reassortment and transmission to humans. The closure of live poultry markets in Hong Kong in 1997 illustrated the importance of such markets: during the initial H5N1 outbreak in Hong Kong, there were 18 human cases with six deaths, and the epidemiological curve was rising sharply. After closure of the live poultry markets, the number of human cases fell to zero, and the initial H5N1 reassortant was eradicated. In 2013, the closure of the live poultry markets in Northeast China contributed to the cessation of human cases over the summer months. Reopening of poultry markets and cooler weather has contributed to a gradual increase in human H7N9 cases, with spread to the southern provinces of China. As of early 2014, the number of people infected during the second wave of H7N9 has exceeded that of the first wave.
Permanent closure of all live poultry markets is the ultimate challenge, and may be an important step toward reducing the risk of pandemic influenza. In the interim, the introduction of rest days and the banning of poultry carryover from one day to the next could dramatically reduce the risk of transmission of influenza from poultry to people.39
Emergence of the 2009 H1N1 pandemic virus
In 2009, the pandemic H1N1 influenza virus (H1N1pdm09) emerged and spread from swine to humans in Mexico. The lessons learned during that outbreak have provided valuable insight into dealing with the continued challenges of influenza, in terms of its unpredictability and severity.
Unpredictability
At the time of the 2009 pandemic, the influenza community was preparing for the spread of H5N1 in humans, with the possibility of a pandemic and plans to use government-stockpiled antivirals to ameliorate the first wave of infections while vaccines were being prepared and amplified. The 2009 H1N1 pandemic was completely unexpected. Although the pandemic plans and antiviral stockpiles provided strategies for dealing with an H1N1 pandemic in 2009, the H1N1 seed stocks did not match antigenically. The emergence of this H1N1 with pandemic potential while the descendants of the 1918 H1N1 Spanish pandemic were still causing seasonal influenza was unprecedented. It served to re-emphasize the recycling of the H1, H2, and H3 subtypes in humans and the need for continued risk assessment of H2N2 influenza viruses.
Severity
Another unpredictable aspect of the 2009 H1N1 pandemic was the severity of the infections. Initial reports on antigenic and structural similarity to the HA of the 1918 Spanish influenza H1N1 virus40 and the percentage of young people hospitalized (6.5%)41 raised the possibility of a severe pandemic. Although severe in young immunologically naive persons, pregnant women, and obese people, when considered over the global population, the 2009 H1N1 pandemic was relatively mild with a mortality of approximately 100,000 worldwide (Fig. 2). It is important to stress that “mild” is a relative term applied to the overall H1N1pdm09 pandemic; over 100,000 persons died, amounting to the loss of many years of life of ultimate severity. The absence of virulence markers in the PB2 gene (no lysine at residue 627), the nonfunctional PB1-F2, and the absence of a PDZ domain in NS1 may, in part, explain the lower pathogenicity of H1N1pdm09 virus compared with the Spanish 1918 H1N1 virus.
The lower-than-predicted severity of the 2009 H1N1 influenza caused a problem in perception, because no criteria in the WHO’s six-point pandemic-phasing advice system were in place to alert public health officials about severity. How to best alert the world about a pandemic spread and its severity without causing global panic remains a challenge.
Surveillance in swine
Another challenge raised by the 2009 H1N1 pandemic was the inadequate global surveillance of swine for influenza, despite proposed role for swine as an intermediate host.42 Recognizing the importance of systematic swine surveillance in Hong Kong after the emergence of H5N1, the CEIRS program found that while the H1N1pdm09 virus was not detected in swine in Asia, precursors of the H1N1pdm09 virus had been circulating in swine for as long as 17 years before 2009 without detection.43
The importance of swine surveillance was subsequently recognized, and the OFFLU network, with expertise in animal surveillance, was developed by the World Organization for Animal Health (OIE) and the Food and Agriculture Organization of the United Nations (FAO). These networks have worked with the WHO to greatly improve swine surveillance for influenza and are now continuously monitoring populations with the recognition that influenza viruses in swine are very dynamic in reassorting.44 Although the H1N1pdm09 influenza virus in humans has not reassorted with the co-circulating H3N2 viruses, virus in swine has mated with the H3N2 triple reassortant to produce H3N2v (variant H3N2), which has the M gene from H1N1pdm09 virus and seven genes from the H3N2 triple-reassortant virus that has circulated in pigs since the late 1990s. These H3N2v viruses are highly transmissible to children from show pigs at U.S. agricultural fairs.45 However, characterization of 306 influenza H3N2v human isolates demonstrated limited person-to-person transmission. These studies illustrate the importance of animal surveillance for influenza and the growing awareness of the importance of joining the veterinary and human surveillance in the one world–one health concept.
One lesson learned from the 2009 H1N1 pandemic was that, despite improvements in preparation of influenza vaccines, the current strategies of preparing either inactivated or live-attenuated influenza vaccines takes more than 6 months. Consequently, vaccines were not available for controlling the first wave of the pandemic.46 Antivirals, therefore, are the first line of defense in the face of an influenza pandemic. The continuing challenges include being prepared by producing vaccines, seed stocks, universal vaccines, and stockpiles of different classes of antiviral drugs that target multiple pathways.
Challenges of vaccination
Vaccines remain a cornerstone for controlling influenza virus infections in humans. Vaccination raises neutralizing antibodies that predominately target the HA glycoprotein, a major antigenic determinant of influenza virus. Current trivalent influenza vaccines induce the production of neutralizing antibodies that are specific to the H1 and H3 subtypes of human influenza A virus and to a single HA phylogenetic lineage (Yamagata or Victoria) of influenza B virus.47 Beginning with the 2013–2014 influenza season, quadrivalent, live-attenuated (FluMist Quadrivalent, MedImmune) and inactivated (Fluarix Quadrivalent, GlaxoSmithKline, and Fluzone, Sanofi Pasteur) influenza vaccines are also available. These new vaccines will most likely replace the earlier trivalent formulations.48
Because the neutralizing antibodies in annual influenza vaccines predominantly recognize specific antigenic determinants on the globular head of the HA surface glycoprotein, their effectiveness relies on the antigenic similarities between the HA of the vaccine strain and that of the seasonal viruses. Appropriate vaccine strains are selected based on worldwide influenza surveillance data and identification of a strain most likely to circulate during the impending influenza season.
Selecting a vaccine strain that antigenically matches a currently circulating influenza strain has many challenges and can result in suboptimal protection. These challenges include continuous antigenic changes in the HA molecule, which can lead to the escape of recognition by immune response and inability to predict the emergence of a novel antigenic variant. Including representative viruses from two influenza B lineages in the vaccine will reduce its potential for mismatch with the circulating strain. The Centers for Disease Control (CDC) estimates that 40,000 to 275,000 illnesses per year could be prevented by reducing influenza B virus infections.49 Thus, alternative strategies for controlling emerging influenza viruses are greatly needed, and developing an influenza vaccine that confers broad-spectrum, long-term protection is desirable.
Universal vaccines
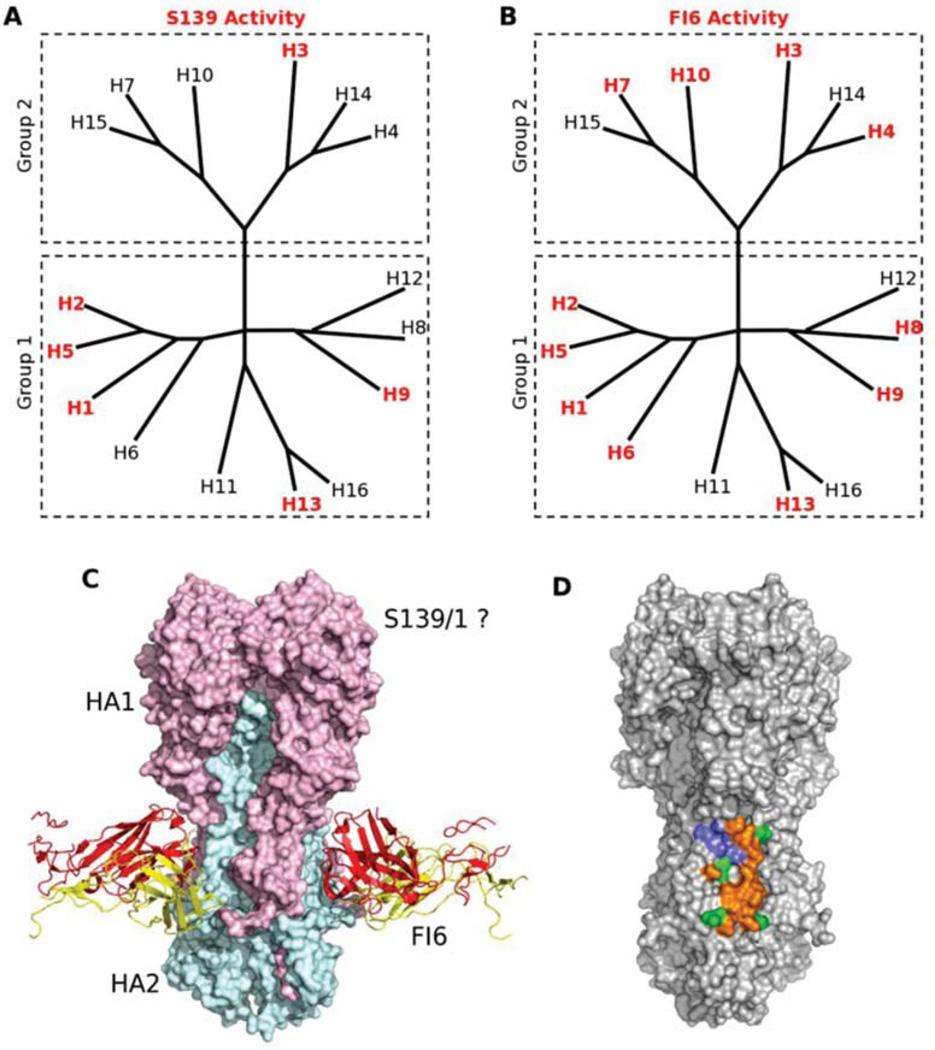
Research focused on broadly neutralizing antibodies against the HA of influenza viruses has greatly expanded in recent years. Identifying mAbs that can neutralize a wide range of influenza viruses has brought new perspectives to antiviral therapy and antibody-guided rational design of a universal vaccine. On the basis of their reactivity patterns with influenza A viruses, the identified antibodies can be divided into three groups: (1) cross-reactive with group 1 HAs, (2) cross-reactive with group 2 HAs, and (3) reactive against both groups 1 and 2 HAs (Fig. 4). The C179 mAb binds to the HA stem region and cross-neutralizes influenza viruses of the H1, H2, H5, H6, and H9 subtypes, which are representatives of group 1 HAs.50 Human CR6261 and F10 mAbs are encoded in the immunoglobulin VH1–69 germline segment and have broad activity against group 1 HAs of influenza viruses.51–53 These antibodies do not block virus attachment but rather inhibit the conformational changes in the HA required for fusion of viral and endosomal membranes.54 Few antibodies have been reported with broad activity against group 2 HAs of influenza viruses.
Figure 4.
Monoclonal antibodies are cross-reactive with hemagglutinins (HAs) of influenza A viruses of groups 1 and 2. (A–B) Phylogenetic trees of the 16 HA influenza A virus subtypes classified into groups 1 and 2 are shown. Phylogenetic trees with HA subtypes bound or neutralized by S139/1 (A) or FI6 (B) are highlighted in red. Note that both antibodies neutralize subtypes from both major virus groups. (C) Like CR6261 and CR8020, FI6 also recognizes an epitope on the HA stem. HA is depicted as a molecular surface, with the HA1 and HA2 subunits from one protomer highlighted in pink and cyan, respectively. The heavy and light chains of FI6 are shown as red and yellow ribbons, respectively. Although the precise location of its epitope is unknown, escape mutations suggest that S139/1 recognizes the HA1 head region. (D) The epitopes of FI6 and CR6261/F10 overlap extensively (orange surface), and only a small number of HA residues are recognized by FI6 only (green surface) or CR6261 only (blue surface). Reprinted from Ref. 54 with permission from Elsevier.
Human CR8020 mAb binds to the stem region of H3, H4, H7, H10, H14, and H15 HAs and neutralizes influenza viruses of H3, H7, and H10 subtypes.55 CR8020 mAb directly binds the fusion peptide and prevents its release in the endosome, thereby inhibiting the conformational changes in HA that are necessary for fusion. Additionally, CR8020 binds to HA0 and inhibits its maturation to HA1 or HA2, sterically preventing cleavage by host proteases.55 Recently, Friensen and colleagues56 identified a CR8043 mAb that binds a conserved epitope in the HA stem of group 2 HAs. CR8043 has in vitro neutralizing activity against H3 and H10 viruses and protects mice against challenge with a lethal dose of either H3N2 or H7N7 virus.56 A murine mAb S139/1 has activity against both group 1 and 2 HAs and binds the conserved region close to the receptor-binding site on the head of the HA molecule.57 Additionally, the two human antibodies, FI6v3 and CR9114 neutralize all viruses tested containing group 1 and 2 HAs.58,59 These mAbs bind the epitope that is highly similar to the epitopes of CR6261 and F10 antibodies but with minor important differences, allowing them to bind group 1 and 2 viruses.
Structural analyses by X-ray crystallography and electron microscopy have shown that all isolated broadly neutralizing antibodies target at least two distinct sites on the stem region of the HA molecule: CR6261, F10, and F16 mAbs target one site; CR8020 mAb targets another and a single site on the HA1 globular head.54 CR8033 and CR8071 neutralize influenza B viruses from both Yamagata and Victoria lineages.59 These mAbs recognize two distinct, conserved epitopes on the HA of influenza B viruses and appear to neutralize them by preventing virus progeny release.59 Currently, broadly neutralizing antibodies are in development as mAb therapies and undergoing clinical trials in the United States.60
Identifying weak spots in influenza virus defense can also lead to the development of promising drug candidates, such as proteins and small molecules that mimic the antibody interactions and thus compete for receptor binding.61 However, the ultimate goal is to produce a universal influenza vaccine that re-elicits antibodies to the conserved epitopes of HA and, thus, provides long-term protection against a broad range of influenza virus subtypes. To achieve this goal, use of novel immunization strategies is needed that include either immunization with a series of antigenically distinct HAs or a two-stage prime-boost strategy. Indeed, broadly neutralizing antibodies against the HA-stem determinants have been recently generated by priming with a DNA vaccine followed by boosting with a seasonal vaccine.62,63
Universal vaccines can be produced by engineering vaccine candidates that lack strain-specific, immunodominant epitopes on the HA head and instead carry cross-reactive, non-immunodominant epitopes on the HA stem. The challenge of this approach is making the conserved HA determinants more accessible to the immune system. To date, several headless HA immunogens have been generated by deleting most of the HA1, but for reasons not fully understood, these immunogens do not generate a broadly cross-reactive neutralizing antibody response in animal models.64–66
The highly variable HA-head region has been the focus of influenza vaccine development for the past 60 years, and the immune response generated in the course of vaccination with traditional influenza vaccine resembles natural infection with influenza virus. Generating an immune response to the highly conserved regions of the HA protein requires further studies and better understanding of the molecular basis for neutralization of influenza viruses caused by neutralizing and non-neutralizing antibodies. Recent data on vaccine-associated enhanced respiratory disease were reported after vaccinating pigs with whole inactivated H1N2 (human-like) virus and then challenging with heterologous H1N1pdm09 virus.67 The authors suggested that vaccination with whole inactivated H1N2 induced antibodies that cross reacted with the H1N1pdm09 virus. Those antibodies bind to the HA-stem region, which is close to the fusion peptide, and induce a more efficient conformational conversion to the fusogenic form of HA. This leads to enhanced fusion of H1N1pdm09 virus to target cells, and hence enhanced disease.67 The following two mechanisms also have potential for causing antibody-enhanced disease: (1) non-neutralizing HA stem–specific antibodies may bind to HA and direct the virus to Fc receptor–bearing cells, which might then become infected through Fc receptor–mediated endocytosis, or (2) antibodies against HA on the surface of influenza virus–infected cells may bind and induce antibody-mediated complement activation, which could lead to inflammation and cell lysis.68 Therefore, although available scientific data on broadly neutralizing antibodies is very promising, more research is required regarding rational design of safe universal influenza vaccines and the induction of a focused, broad, and long-lasting immune response.
Adjuvants
Adding adjuvants to influenza vaccines has the potential to promote antigen immunogenicity and both humoral and cell-mediated immune responses, reduce the minimal antigen dose, promote antigen sparing and the global availability of influenza vaccine, induce a broader immune response against drifted variants not present in the vaccine, and induce added levels of protection in the elderly.69
Earlier adjuvants, which were relatively quite effective, included water in a mineral oil emulsion and an emulsifier with or without Mycobacterium.70 However, mineral oil emulsions (without Mycobacterium) induced nodules in humans,71 so their use was discontinued. These early adjuvants are still the ones of choice for use in inactivated-influenza vaccines for poultry. Among the early adjuvants, the only approved one for use with human vaccines was alum, which is a relatively poor adjuvant for potentiating the immune response to subtype-specific influenza vaccines.
Adjuvants currently under development are based on the biodegradable oil squalene, which is found in shark liver, plants, and animals, including humans. Two squalene-based adjuvants have been approved for use with influenza vaccines in humans in Europe: MF59 by Novartis and ASO3 by GlaxoSmithKline.69 The main difference between the two is that ASO3 contains α-tocophenol (vitamin E) in addition to squalene. The importance of these squalene-based adjuvants is clearly demonstrated with H5N1 influenza–subunit vaccine; without adjuvant, two 90-µg doses of antigen were required to induce a modest HA inhibition titer (1/40) in 54% of human subjects;72 with a squalene-based adjuvant, as little as 1.9 to 3.75 µg antigen was sufficient to induce a similar response.73
The mechanism of action of adjuvants in potentiating immune responses is complex and not yet fully elucidated. The original idea of a depot that slowly releases antigen is an oversimplification. The current concept is that adjuvants induce cytokines, which recruit immune cells to the site of injection, potentiate antigen uptake by monocytes, and stimulate differentiation of monocytes into dendritic cells. The dendritic cells then prime naive T cells, which in turn migrate to draining lymph nodes, where they trigger an augmented immune response to the antigen.69
If, or when, the highly pathogenic H5N1 or low-pathogenic H7N9 influenza viruses acquire mutations to enable efficient human-to-human transmissibility, there will obviously be an urgent need for use of adjuvants that provide for antigen sparing and immune protection. H5N1 virus is a relatively weak immunogen; H7N9 is better, though it is not a strong antigen. Thus, one of the immunological challenges here is to understand the molecular basis of poor antigenicity and how to circumvent the problem of variable immunogenticity.
Although both currently available squalene-based adjuvants hold great promise for augmenting the immune response to influenza, AS03 has been associated with narcolepsy—a rare syndrome characterized by uncontrollable sleepiness—when administered with subunit-specific vaccine against H1N1pdm09 influenza virus.74 This associated narcolepsy affects approximately 1 in 3000 children or young adolescents;74 sleepiness is caused by a lack of the neuropeptide hypocretin, which is secreted by the neurons that control wakefulness. Remarkably, the HA of H1N1pdm09 virus contains a short peptide (residues 275–287) that is identical to a 13–amino acid peptide in hypocretin.75 When immunized with H1N1pdm09 vaccine in ASO3, persons susceptible to the induction of narcolepsy produce CD4+ T cells expressing human leukocyte antigen (HLA)-DQ0602 that become activated by the 13 amino acid peptide of hypocretin and then destroy the hypocretin-secreting cells in the hypothalamus. Thus, a rare combination of specific HLA expression and immunogenicity produced by H1N1pdm09 peptide when augmented with ASO3 provides an explanation for the induction of narcolepsy. Why narcolepsy has been associated with one squalene-based vaccine and not the other remains to be explained. Vaccines have enormous value in protecting the vast majority of persons, though rare disorders can occur. The challenge is to define and resolve these risk factors, because the benefits of vaccines far outweigh the risks.
Challenges of meeting the need for new antiviral drugs
Therapeutic interventions can be an important line of defense against influenza, especially when vaccine efficacy is low or vaccine is unavailable. However, several factors limit the effectiveness of antiviral therapy against influenza. The major challenge in antiviral influenza therapy is that only one class of drugs, NAIs, is currently available. Another FDA-approved class of anti-influenza drugs, the adamantanes (amantadine and rimantadine), are not recommended by the CDC for prophylaxis or treatment of influenza owing to the widespread resistance among human seasonal H1N1 and H3N2 influenza viruses. Moreover, adamantanes are ineffective against influenza B viruses.76
NAIs are virus-specific drugs that bind the enzymatically active site of the NA of influenza A or B, thereby interfering with the functions of the protein. A major function of NA is to cleave terminal sialic acid residues from the cell surface, promoting the release of newly formed virions.77 Therefore, NAIs inhibit the release of budding viral particles from infected host cells and virus spread from infected cells to uninfected cells. Unlike antibiotics that can eliminate or greatly reduce bacterial burden, NAIs merely pause the progression of disease by preventing new host cells from being infected with influenza viruses. To effectively treat influenza infection, NAIs should be administered within 48 hours of the onset of symptoms, which is a significant challenge in many countries.78 Although, some data indicated that the benefit may be obtained later in the course of disease for patients in underdeveloped regions of the world and those requiring hospitalization. If administered early in the clinical course, NAIs alter the tempo of infection, allowing normal immune-clearance mechanisms to rapidly eliminate the influenza virus. Therefore, in addition to having only a single class, the drugs must be administered during a narrow window of therapeutic efficacy, which presents a serious challenge for physicians prescribing them.
Neuraminidase inhibitors
Among the four existing NAIs, two have been available worldwide since the 1999–2000 influenza season: oral oseltamivir (Tamiflu®, Roche) and inhaled zanamivir (Relenza®, GlaxoSmithKline). Additionally, intravenous (i.v.) peramivir (Rapiacta®, Peramiflu®, BioCryst Pharmaceuticals) has been approved for the treatment of influenza in adults and children in Japan, South Korea, and China; and inhaled laninamivir octanoate (Inavir®, Daiichi Sankyo) has been approved for the treatment of influenza in adults and children in Japan.79 Peramivir has unique structural properties that differ from those of oseltamivir and zanamivir;80 it is characterized by tight binding to the NA site for more than 24 hours and slow dissociation.81 Laninamivir octanoate has demonstrated clinical efficacy against seasonal influenza A and B and the H1N1pdm09 viruses.82 The high concentration of the active form laninamivir persists for at least for 5 days in the lung, exceeding the level required to inhibit the replication of influenza virus. Therefore, influenza can be treated with a single administration of laninamivir octanoate, which is a major clinical benefit.
Resistance to neuraminidase inhibitors
The emergence of NAI-resistant influenza viruses is a substantial challenge to the effectiveness of antiviral therapy. The occurrence of NAI resistance–associated amino acid substitution(s) affects the binding affinity between the NA enzyme and the inhibitor and allows influenza viruses to evade the action of NAIs.83 The molecular markers of NAI resistance were identified either at the catalytic or framework residues of the NA glycoprotein and vary for oseltamivir, zanamivir, and peramivir. No laninamivir-resistant substitutions have been reported yet.
Because oseltamivir has a large hydrophobic side chain, the influenza NA must undergo rearrangements to enable drug binding. To form a suitable receptive pocket in NA, the E276 residue must rotate and bond with R224;84 thus any mutation that affects this rearrangement (e.g., inhibition of the rotation of the E276 residue and prevention of pocket formation) may reduce the binding affinity of oseltamivir, thereby lowering its efficacy. Amino acid changes in NA commonly associated with oseltamivir resistance vary for different NA types and subtypes. Oseltamivir-resistant influenza A viruses of N1 subtype most frequently carry H274Y or N294S amino acid substitutions in NA (N2 numbering); viruses of the N2 subtype carry E119V or R292K substitutions; and NAI-resistant variants of influenza B viruses harbor R152K or D198N.85
The molecular structure of zanamivir includes a guanidino group, which interacts with the conserved E119 residue in the NA-active center pocket.86 The higher structural homology to the NA natural substrate (sialic acid) and the lower zanamivir use (compared to oseltamivir) are the most probable factors that account for the infrequent isolation of zanamivir-resistant variants in the clinical setting.87 Peramivir possesses a hydrophobic group similar to that of oseltamivir, and a guanidino group similar to that of zanamivir. Therefore, like oseltamivir, peramivir requires structural rearrangements to accommodate its hydrophobic side chain,80 and substitutions that affect the activity of oseltamivir and zanamivir can also affect that of peramivir.
NAI-resistant viruses can emerge either under drug-selection pressure or naturally (without drug intervention). Before 2007, the incidence of oseltamivir resistance in clinical trial samples was 0.3% in adults and 4% in children.88,89 The high prevalence of oseltamivir-resistant A/Brisbane/59/2007(H1N1)-like viruses (subclade 2B) carrying the NA H274Y–resistance marker was reported worldwide during the 2007–2009 influenza seasons. Epidemiologic studies did not show evidence of an association between the development of resistance and oseltamivir use.90–92 Thus, H274Y NA amino acid substitution occurred naturally, and viruses with this mutation acquired remarkable transmissibility and superior fitness compared to their drug-sensitive counterparts.
Several changes in the NA can counteract the adverse effects of the H274Y mutation. D344N,93,94 Q222R, and V234M95 substitutions counteract the reduced NA affinity and surface accumulation; therefore, these substitutions are permissive for the H274Y NA change. Since the first wave of the 2009 pandemic and with the disappearance of the A/Brisbane/59/2007 (H1N1) strain, the overall level of oseltamivir resistance amongst H1N1pdm09 variants has remained relatively low (~ 1%).96,97 A few clusters of oseltamivir-resistant cases not associated with treatment and most likely involving transmission of H1N1pdm09 mutant strains have been reported in the United Kingdom,98 Australia,99 and Vietnam.100 The complexity of the NAI-resistance pattern underlies the challenges for antiviral surveillance among influenza viruses, which is an essential risk-assessment component for newly emerging viruses or viruses isolated from the avian/human interface.
Parenteral formulations of antiviral drugs
The lack of widely available parenteral formulations of antiviral drugs significantly limits virus-specific therapies for use in severe and complicated influenza. To date in the United States, only emergency use of intravenous peramivir is approved by the FDA; in Japan, South Korea, and China, intravenous peramivir is approved for the treatment of influenza. New parenteral formulations of zanamivir are also advancing to clinical practice.
The avian influenza H5N1 and H7N9 viruses and the H1N1pdm09 viruses with D222G HA mutation are particularly virulent.38,101 Critically ill patients with H5N1, H1N1pdm09, or H7N9 virus infections have been difficult to treat until the recent availability of parenteral formulations of peramivir and zanamivir. Most patients with severe infection have risk factors including old age, pregnancy, underlying diseases, and differences in host genetic background, which make it difficult to determine the optimal dose and duration of NAI therapy. In addition to the correction of hypoxemia and critical care support, effective antiviral therapy, with either increased dose of oral oseltamivir or, preferably, intravenous peramivir or zanamivir, is the foundation for specific treatment of influenza-associated pneumonia.102 Although earlier antiviral treatment resulted in a greater clinical benefit, observational studies support the use of systemic NAIs in hospitalized patients with influenza, even when begun 48 hours after onset of illness.
What is in the drug pipeline?
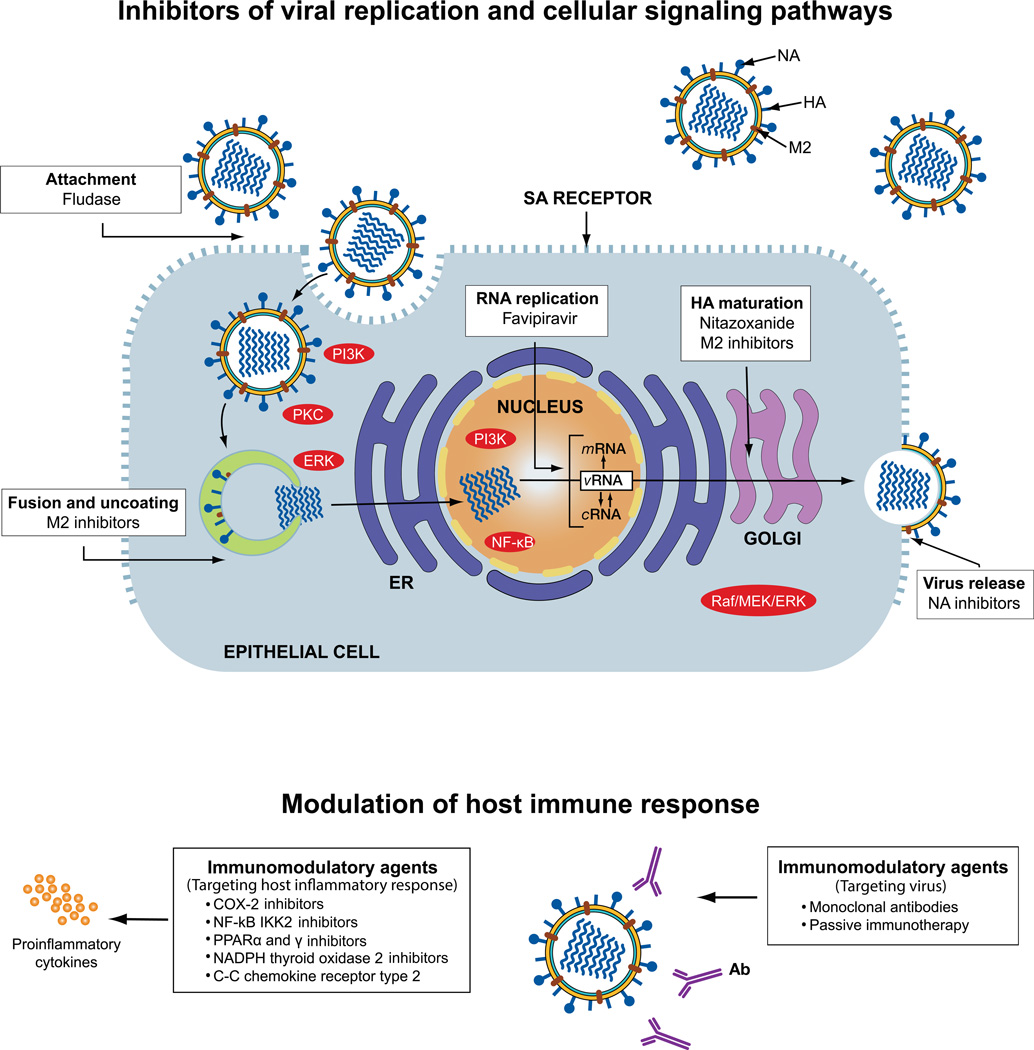
The limitations of the currently available therapeutics used to control influenza are the driving forces for the discovery and development of novel drugs and approaches. Such drugs may act as a first line of defense against the spread of influenza infection and buy time for vaccine development, particularly in a human pandemic setting. Anti-influenza drugs in pipeline can be divided into two major groups: drugs undergoing clinical trials and investigational drugs at different stages of preclinical development (Fig. 5).
Figure 5.
Schematic representation of influenza virus replication cycle and drugs currently used for prophylaxis and/or treatment of influenza, drugs in clinical trials, and those in preclinical development. Antiviral therapy predominately relies on virus-specific drugs such as the neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir, which bind the enzymatically active site of the NA of influenza A or B viruses and inhibit the release of budding viral particles from infected host cells. M2 inhibitors (amantadine and rimantadine) block the ion channel activity of the M2 protein of influenza A virus at the stage of virus fusion and uncoating. Drugs undergoing clinical trials in the U.S. for the prevention and treatment of influenza include Fludase (DAS181), Favipiravir (T-705), and Nitazoxanide. Sialidase inhibitor Fludase cleaves sialic acid (SA) α2,3- and SA α2,6-linked cellular receptors and inhibits virus attachment to them. Prodrug favipiravir is converted intracellularly to its active metabolite, T-705 ribofuranosyltriphosphate, which inhibits RNA polymerase activity. Nitazoxanide affects the terminal glycosylation of the hemaggluttinin (HA) glycoprotein, thereby impairing HA trafficking between the endoplasmic reticulum (ER) and the Golgi complex. The host’s immune response can be modulated by inhibitors of various cellular signaling pathways and cascades during the virus’ replication cycle. Although several inhibitors are currently in preclinical development, these investigational immunomodulatory agents can be categorized into two groups based on their target: those that target the host’s inflammatory response (left) and those that target the virus (right). Ab, antibody; COX2, cyclooxygenase-2; cRNA, complementary RNA; ERK, extracellular signaling–regulated kinase; HA, hemagglutinin; NA, neuraminidase; M2, matrix protein; MEK, MAP/ERK kinase; mRNA, viral messenger RNA; PI3K, phosphatidylinositol-3 kinase; PPAR, peroxisome proliferator-activated receptor; PKC, protein kinase C; vRNA, viral RNA.
Drugs in clinical trials
Currently, Fludase® (DAS181), favipiravir (T-705), and nitazoxanide are being evaluated in clinical trials in the United States for the prevention and treatment of influenza. Fludase consists of a bacterial sialidase catalytic domain derived from Actinomyces viscous linked to a respiratory epithelium growth factor–like anchoring domain (amphiregulin tag), which is required for effective targeting of epithelial cells.103 The sialidase activity of Fludase cleaves α2,3- and α2,6-linked sialic acid receptors on the cell surface, and their removal prevents influenza virus attachment. Fludase possesses antiviral activity against a broad range of influenza A and B viruses in cell culture and animal models.104 A phase II study among patients with uncomplicated cases of influenza infection demonstrated significantly reduced viral load and significantly shorter time to sustained decreased viral shedding in the group that received multiple doses of Fludase compared with a placebo group.105 The multidose schedule of Fludase was more effective than the single-dose regimen.105 Thus, Fludase shows promise for prevention and treatment of influenza virus infection, but potential concerns about using sialidase therapy still need to be addressed, including systemic dissemination, development of anti-DAS181 antibodies, and the role of secondary bacterial infection.
Favipiravir has a novel mechanism of action; it selectively inhibits influenza viral RNA polymerase by phosphoribosylated favipiravir–RTP (favipiravir ribofuranosyl-5′-triphosphate), an active form of the drug.106 This activity competes with purine nucleic acids; thus, viral RNA polymerase mistakenly recognizes favipiravir–RTP as a purine nucleotide. Favipiravir also shows antiviral activity against a wide range of types and subtypes of influenza viruses, including NAI-resistant strains.107 Favipiravir showed potent efficacy in murine lethal-infection models, including a highly pathogenic influenza H5N1 virus, a model with high virus titer, and a delayed-treatment model.108 Favipiravir is undergoing phase II clinical trials in the United States and is now approved for the treatment of influenza in Japan. This drug may be a leading candidate for use in controlled studies of antiviral combinations.
Nitazoxanide is a thiazolide antiparasitic agent that is approved in the United States for treatment of Cryptosporidium parvum or Giardia intestinalis infections in non-immunodeficient children and adults.109 Nitazoxanide is active against a broad range of DNA and RNA viruses, including hepatitis B and C and influenza in vitro.110,111 The mechanism of nitazoxanide action has not been fully elucidated for influenza viruses; however, some data suggest that it blocks terminal glycosylation of the HA glycoprotein. If so, such an activity would impair trafficking of the HA between the endoplasmic reticulum and the Golgi complex, thereby preventing its transport to the cell surface.112 Nitazoxanide, which is characterized by a favorable safety profile and has been confirmed in clinical trials and post-marketing experience, is currently undergoing clinical development for influenza treatment.
Drugs in preclinical development
The novel strategies and approaches toward developing drugs that can contain influenza are broad; however, they can be summarized as follows: (1) drugs that target a specific influenza viral protein, (2) drugs that target the cellular host factors essential for viral replication, and (3) drugs that modulate the host’s immune response (Fig. 5).
Viral targets
One favorable target for drug development is inhibiting influenza virus entry into host cells. Blocking this first step of infection should efficiently block virus propagation. This category of compounds consists of anti-HA mAbs; single-domain antibody fragments (also referred to as nanobodies) derived from camelid immunoglobulins; cyanovirin-N, a lectin isolated from Escherichia coli; sialyl-containing macromolecules (gangliosides sialylparagloboside and GM3); Fludase; and others.113 Viral fusion can be prevented by inhibiting acidification in endosomes. One of the most potent inhibitors for this purpose is bafilomycin A1 and concanamycin A. Bafilomycins belong to the family of macrolide antibiotics that inhibit vacuolar-type proton pumps involved in viral entry.114 Small molecules that bind to the stem region of HA and thereby hinder the conformational changes required for fusion represent another class of fusion inhibitors. The first to be discovered was tert-butyl hydroquinone, which has been crystallized in complex with HA.4 Subsequently, several compounds that act in a similar way have been described, but resistant variants can develop rapidly within a few passages in cell culture and thus do not have promise for future development.113
Using structural information and classical medicinal chemistry approaches, scientists recently discovered novel M2 inhibitors that are active against both adamantane-sensitive and adamantane-resistant influenza A viruses.115 Several functions of NS1 have been described that form the basis of its anti-interferon activity, and most therapeutic targeting strategies include disruption of these functions to relieve viral inhibition of the innate immune response. Only a few compounds discovered to target virus proteins and interfere with specific events in the virus-replication cycle have shown high potency and a lack of emergence of resistant variants and low toxicity. Thus, only a few reach clinical trials.
Cellular targets
The cellular factors or pathways essential for influenza virus replication are under development as new antiviral targets, and compounds targeting the Raf/MEK/ERK signaling pathway, NF-κB signaling, the PI3K/Akt pathway, or PKC signaling cascades might be promising.116 The advantage of these compounds is an extremely low risk of emergence of drug-resistant variants. Inhibitors of intracellular signaling cascades that are essential for virus replication have been identified as the most promising candidates. In the near future, we may gain insight into the cell biology of influenza virus and develop antivirals that target host factors rather than viral proteins.
Immunomodulatory therapy
The future of influenza control most likely involves improved means of preventing infection, coupled with combined strategies to slow the virus’ entry and mitigate the immunopathologic consequences of infection. Hyperinduction of proinflammatory cytokine production (or “cytokine storm”) has been observed in patients with severe influenza, such as those infected with avian H5N1 and H7N9 and H1N1pdm09 viruses, and in macaques infected with reconstituted 1918 Spanish H1N1.117–119 The investigational immunomodulatory agents targeting the host inflammatory response include compounds that target the cyclooxygenase (COX) pathway, and in particular COX2 (acetaminophen, mesalazine, celecoxib, zaltoprofen, and rofecoxib), inhibit NF-κB IKK2 (acetylsalicylic acid), peroxisome proliferator-activated receptors α and γ (gemfibrozil, ibuprofen, rosiglitazone, and pioglitazone), NADPH thyroid oxidase 2 (apocynin), C-C chemokine receptor type 2, high-mobility group protein B1 (statins), sphingosine-1-phosphate receptors (AAL-R, RP-002, and CYM-5442), and several other inhibitors.120 Several anti-inflammatory compounds have already been tested in patients with severe influenza infections, though the role of immunomodulatory therapy is still unclear owing to a lack of data from randomized, controlled clinical trials.
Another strategy for immunomodulatory therapy is boosting innate immunity. One way to enhance innate immunity is to stimulate natural killer (NK) cell activity. Influenza virus evades NK cell immunity by directly killing NK cells and inhibiting cell cytotoxicity.121,122 The γδ T cells have similar characteristics to NK cells and antigen-presenting cells and function in innate and adaptive immune responses to infectious agents and tumors. However, γδ T cells constitute only 1–5% of T cells in the blood and peripheral organs of adult humans.123,124 Human Vγ9Vδ2T cells expanded by the compound aminobisphosphonate pamidronate reduce disease severity and mortality caused by human seasonal H1N1 and avian H5N1 influenza viruses and control lung inflammation and viral replication.125 However, a better understanding of common cell-signaling pathways associated with acute lung injury caused by influenza virus is necessary to understand the causes of immunopathologic changes. This knowledge will help determine which immunomodulatory interventions are useful and predict the appropriate timing and consequences of their use.
Passive immunotherapy with convalescent plasma or with hyperimmune globulin prepared from convalescent plasma appears to be useful as an adjunct therapy for H5N1 and H1N1pdm09 infections. Recent clinical trials have shown that convalescent plasma effectively reduced mortality, respiratory tract viral load, and serum cytokine levels in 20 patients with severe H1N1pdm09 infection requiring intensive care support.126 Patients with severe H1N1pdm09 influenza who were treated with intravenous hyperimmune γ-globulin from persons who had survived the same disease had a lower peak viral load and lower mortality than controls, if treatment was begun within 5 days of symptom onset.127
Combination therapy prevents drug resistance
To counter the challenge of drug resistance, combination therapy can be designed that targets different viral proteins or host cellular factors with a non-overlapping resistance profile.128,129 Such combinations will most likely enhance the effectiveness of antiviral therapy. Moreover, it may reduce either the likelihood of developing single-drug resistance or the adverse effect(s) of any drug resistance that develops.
One challenge to designing combination therapy for treating influenza is the lack of approved compounds with different mechanisms of action. Therefore, studies have been conducted to evaluate the effects of double and triple combinations of FDA-approved NAIs, adamantanes (FDA approved but currently not recommended for treatment of influenza), and ribavirin (FDA-approved for the treatment of hepatitis C but not approved for influenza) in vitro and in vivo.130–133 These combinations generally produce additive-to-synergistic inhibition of influenza virus in vitro and provide greater protection than monotherapy in influenza-infected mice. Combination therapy prevents the selection of drug-resistant viruses.132 However, most of these studies evaluated the efficacy of combination therapy against influenza viruses that were susceptible to all of the drugs in the combination.
Elucidating the efficacy of combination regimens against drug-resistant viruses is essential. A triple-drug regimen consisting of oseltamivir, amantadine, and ribavirin (known as TCAD) has shown activity against viruses resistant to adamantanes or oseltamivir in vitro and in murine models. Therapeutic outcomes of the TCAD regimen were comparable to those of oseltamivir monotherapy in critically ill adult patients infected with H1N1pdm09 influenza virus.134 Currently, the National Institute of Allergy and Infectious Diseases (NIAID) is conducting a multisite, double-blinded, randomized phase II study to evaluate the efficacy of TCAD therapy versus oseltamivir monotherapy in patients infected with influenza and at risk of complications.60 In addition, the NIAID is conducting a companion study evaluating oseltamivir monotherapy versus placebo in patients with low-risk influenza infection.60 Finally, the value of non-pharmaceutical interventions (wearing face masks, washing hands) should be widely used during seasonal influenza epidemics.
Challenges of variable host susceptibility
A major challenge in understanding the transmissibility of seasonal influenza, and especially novel influenza viruses such as H5N1, H7N9, and H9N2, is variability in host susceptibility. Hundreds of thousands of people must have been exposed to H5N1 and H9N2 viruses since their emergence in the late 1990s, and already thousands must have been exposed to H7N9, yet a surprisingly small number have been infected. Why? Some animal species (e.g., rabbits and sheep) are completely resistant to influenza. Others (e.g., mallard ducks) permit replication of highly pathogenic H5N1 influenza viruses but do not demonstrate disease signs, whereas gallinaceous poultry (i.e., chicken, quail) are susceptible.
Insight into the genetic basis of host susceptibility is in its infancy; however, with the resolution of the genomes of multiple influenza viruses and those of multiple hosts, including humans, swine, and avian species, invaluable information is coming to light. The difference in susceptibility between mallard ducks and domestic chickens has been linked, at least in part, to the absence of the dsRNA helicase enzyme retinoic acid–inducible gene I (RIG-I) in chickens and its retention in ducks.135 During evolutionary development, chickens lost RIG-I, which is required for interferon synthesis and part of the immune response capacity to infection.
The relatively small number of humans infected with the novel H5N1 influenza virus suggests a genotypic predisposition; the extremely limited number of human-to-human clusters of H5N1 transmission has occurred among family members, which suggests genetic susceptibility.136 The fatal outcome of H5N1 in humans has been associated with high viral loads and high levels of cytokine induction.117 High levels of cytokines in the lungs of humans infected with the novel H7N9 influenza virus have been associated with genomic variation in the gene IFITM3 (interferon induced transmembrane protein 3).23 These findings support those of severe disease and increased mortality with aberrant innate immune responses of macaques to the 1918 Spanish influenza virus.118 The differences in susceptibility of mouse strains to H5N1 influenza virus has been also associated with high virus loads and the production of proinflammatory cytokines in highly susceptible strains like DBA 2J mice.137,138 The challenge is to determine which defects (e.g., IFITM3) are markers of influenza susceptibility.
Challenges of controlling influenza via poultry vaccination
Vaccination is considered the optimal strategy for controlling influenza in humans and is recommended by the WHO; however, vaccination of domestic poultry for the control of influenza is more controversial. Countries that want to eradicate influenza viruses forbid vaccination because antibodies induced by vaccination interfere with serologic monitoring to detect natural influenza infection. However, vaccination is recommended by the OIE as part of the control strategy for avian influenza.139
Poultry vaccination can be efficacious and has served to reduce disease signs in birds and the frequency of human cases of H5N1 in countries that have adopted vaccination as their primary control strategy (China, Vietnam, Indonesia, and Egypt). The use of poultry vaccination in Vietnam in 2006 dramatically reduced the incidence of H5N1 infection in people and poultry from 61 human cases with 19 deaths and 1068 outbreaks in poultry in 2005 to zero human cases and 36 outbreaks in poultry in 2006. However, poultry vaccination does not induce sterilizing immunity in a flock; and some birds can be infected and shed virus with no disease signs. Added to this has been the reluctance of duck farmers to vaccinate because most domestic duck species show limited or no disease signs from H5N1 infection.
Poultry vaccination can be part of a strategy to eradicate influenza when used to ring vaccinate infected flocks, with the goal of stamping out the virus. This strategy has been successful when used with improved biosecurity and compensation of farmers. Unfortunately, it has not eradicated H5N1 in any country that adopted poultry vaccination as their principal control strategy. Poultry farmers tend to view vaccination as a means to reduce disease in their flocks and not as a step toward eradicating the virus. Consequently, poultry vaccination has contributed to establishing endemicity in each of the countries that adopted it without a strong commitment to eradication. When vaccination in poultry is used only to reduce disease impact, it promotes antigenic drift, evolution of distinguishable clades, and development of variants with human transmissibility.
Another consequence of continued vaccine use in poultry is the need to change vaccine seed stocks. Although influenza strain changes can be made easily in human vaccines, the same is not necessarily true in poultry vaccines. Full-efficiency trials of vaccines are required to revalidate the product. Because inactivated influenza viruses are delivered in water in oil adjuvants, they induce cross-protection between variants, such that vaccine strain changes are needed less frequently.139 The downside of poultry vaccines is that they are not standardized for antigen content like human influenza vaccines, but rely on efficacy testing that is not necessarily done on each vaccine lot. Thus, poultry vaccination is a paradoxical practice that can save the lives of poultry and people, but when it is not used as part of an eradication plan serves to drive antigenic drift and perpetuate virus.
Conclusions
Here we have discussed the challenges faced by veterinary and public health officials as they attempt to control influenza. Among those many challenges, we consider the following ones to be potential game changers.
Development of a universal vaccine for influenza
Fundamental knowledge of the structure and immunogenicity of the HA of influenza virus has revealed a common antigenic domain in the region of the fusion peptide. A vaccine that is effective against all subtypes of influenza, is safe, induces long-lasting immunity, and is efficacious in lower animals and humans would be the holy grail of influenza vaccinology. The potential of universal vaccination for influenza merits the fullest support. The development of universal mAbs for use in severe cases of influenza and the possible development of small molecule antivirals in addition to a universal vaccine are exciting possibilities that could revolutionize the control of influenza. However, vaccination is not risk free, and we must be aware that augmenting immune responses to a minor determinant near the fusion peptide might, in some instances, potentiate virus replication.
Closure of live poultry markets
The zoonotic nature of influenza offers the potential for reducing the introduction of novel strains of influenza to humans at the human–animal interface. Closure of the live poultry markets in China has dramatically decreased human infections with H5N1 and H7N9 influenza viruses. Because live poultry markets are an optimal site for genetic mixing of influenza viruses from different hosts, their global closure would have the potential to reduce the genesis of novel influenza viruses and their transmission to humans. However, abrupt worldwide closure of live poultry markets would greatly disrupt the poultry industry and traditional lifestyles. A more practical approach is to improve sanitation and biosecurity in the live markets by instituting a no-carryover policy of live poultry from one day to the next.39 In the long term, eventual closure of live poultry markets worldwide is desirable.
Development of new antiviral drugs that target multiple pathways
Our current reliance on NAIs has the same risk associated with all monotherapies—the development of drug resistance. During the 2007–2009 influenza seasons, the emergence and spread of human H1N1 influenza viruses that were naturally NAI resistant illustrates the urgent need for novel drugs and combination therapy. Novel treatments may specifically target influenza viral proteins or cellular factors essential for virus replication. Alternatively, they may modulate the immune response of the host. A number of drugs in the pipeline target the polymerase (favipiravir), HA (nitazoxanide), and sialic acid receptors (Fludase). Compounds involved in influenza-mediated inflammation are available, and they lack the risk of facilitating antiviral resistance. Their potential role in improving clinical outcome in severe cases of influenza should not be ignored, but explored as an adjunct therapy or part of combination therapy. Other approaches to be considered are to modulate the host’s innate immune response or use immunotherapy with antibodies against the universal epitopes on the HA protein.
Elucidation of the genomics of influenza viruses
Which influenza viruses in the natural reservoirs in aquatic birds and bats have the ability to transmit to mammals including humans? Which viruses have the genetic information that will permit them to transmit to humans and spread between humans? Although large databases continue to accumulate the sequences of human H1 and H3 influenza viruses, similar genome information on wild bird H1 and H3 viruses is lacking, as is that for H5, H7, and H9 viruses. It is time to fill these gaps in knowledge and determine whether we can exploit genomic information to establish which influenza viruses have pandemic potential.
Longer-term influenza-resistant animals
Eradicating influenza viruses from the zoonotic reservoir is currently not feasible; thus, an alternative solution would be to develop influenza-resistant animals. If intermediate hosts (i.e., pigs, poultry, and horses) were engineered to be resistant to influenza, then transfer between the avian reservoir and humans would be reduced. As we identify genes that confer resistance to influenza, they could be inserted into the susceptible hosts. This approach would probably involve multiple genes. Thus, the insertion of the RIG-I gene from ducks into chickens could reduce disease signs but may not reduce the level of virus replication. As the genomics of those species that are completely resistant to influenza are established fully, influenza virus–resistant animals may be developed.
It is apparent that there are still many challenges and unanswered questions about the evolution of influenza viruses and the genesis of pandemic strains. Ultimately, much more information on the virus from multiple hosts needs to be gained to determine if all influenza viruses have pandemic potential.
Acknowledgments
This work was supported by Contract No. HHSN266200700005C from the National Institute of Allergy and Infectious Diseases, and by the National Institutes of Health, the Department of Health and Human Services, and the American Lebanese Syrian Associated Charities (ALSAC). We thank Angela McArthur for excellent scientific editing and James Knowles for assistance in preparing the manuscript.
References
- 1.Shaw ML, Palese P. Orthomyxoviradae: the viruses and their replication. In: Knipe D, Howley P, editors. Fields’ Virology, Sixth Edition. Philadelphia, PA: Lippin Williams & Wilkins; 2011. pp. 1647–1689. [Google Scholar]
- 2.Webster RG, Bean WJ, Gorman OT, et al. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1994;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jernigan DB, Cox NJ. Human influenza: One Health, One World. In: Webster RG, Monto AS, Braciale TJ, Lamb RA, editors. Textbook of Influenza, Second Edition. West Sussex: John Wiley and Sons; 2013. pp. 3–19. [Google Scholar]
- 4.Russell RJ, Kerry PS, Stevens DJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perdue ML. Molecular determinants of pathogenicity for avian influenza viruses. In: Swayne DE, editor. Avian Influenza. Ames, Iowa: Blackwell Publishing; 2008. pp. 23–41. [Google Scholar]
- 7.Klenk HD, Garten W, Matrosovich M. Pathogenesis. In: Webster RG, Monto AS, Braciale TJ, Lamb RA, editors. Textbook of Influenza, Second Edition. West Sussex: John Wiley and Sons; 2013. pp. 157–171. [Google Scholar]
- 8.CDC. Surveillance for Influenza-United States, 1997–98, 1998–99, and 1999–00 Seasons. MMWR Surveillance Summaries. 2002;51(SS07):1–10. [PubMed] [Google Scholar]
- 9.CDC. Update: Influenza Activity-United States, 2003–04 Season. MMWR Weekly. 2003;49:1197–1202. [PubMed] [Google Scholar]
- 10.Treanor J. Influenza vaccine--outmaneuvering antigenic shift and drift. N. Engl. J. Med. 2004;350:218–220. doi: 10.1056/NEJMp038238. [DOI] [PubMed] [Google Scholar]
- 11.Laver WG, Air GM, Webster RG, et al. Antigenic drift in type A influenza virus: sequence differences in the hemagglutinin of Hong Kong (H3N2) variants selected with monoclonal hybridoma antibodies. Virology. 1979;98:226–237. doi: 10.1016/0042-6822(79)90540-3. [DOI] [PubMed] [Google Scholar]
- 12.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Ann. Rev. Immunol. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization (WHO) WHO Report on Global Surveillance of Epidemic-prone Infectious Diseases WHO/CDS/CSR/ISR2000.1. [Accessed February 17, 2014]; http://www.shimclinic.com/singapore/influenza-vaccine-history.
- 14.Koel BF, Burke DF, Bestebroer TM, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 15.Nabel GJ, Fauci AS. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat. Med. 2010;16:1389–1391. doi: 10.1038/nm1210-1389. [DOI] [PubMed] [Google Scholar]
- 16.Yuan J, Zhang L, Kan X, et al. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin. Infect. Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 17.ProMED-mail. Avian Influenza, Human (158): China (Ziangxi) H10N8, Fatal. [cited 2014 Feb 17];2013 Dec 17; http://www.promedmail.org, archive number 20131217.2121718. [Google Scholar]
- 18.Simonsen L, Clarke MJ, Schonberger LB, et al. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J. Infect. Dis. 1998;178:53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- 19.Jones JC, Baranovich T, Marathe BM, et al. Risk assessment of H2N2 influenza viruses from the avian reservoir. J. Virol. 2013;88:1175–1188. doi: 10.1128/JVI.02526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schäfer JR, Kawaoka Y, Bean WJ, et al. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 21.Ma W, Vincent AL, Gramer MR, et al. Identification of H2N3 influenza A viruses from swine in the United States. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20949–20954. doi: 10.1073/pnas.0710286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Kerkhove MD, Riley S, Lipsitch M, et al. Comment on "Seroevidence for H5N1 influenza infections in humans: meta-analysis". Science. 2012;336:1506. doi: 10.1126/science.1221434. author reply 1506. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Zhang A, Wan Y, et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. U. S. A. 2013;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhang Q, Kong H, et al. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science. 2013;340:1459–1463. doi: 10.1126/science.1229455. [DOI] [PubMed] [Google Scholar]
- 27.Belser JA, Davis CT, Balish A, et al. Pathogenesis, transmissibility, and ocular tropism of a highly pathogenic avian influenza A (H7N3) virus associated with human conjunctivitis. J. Virol. 2013;87:5746–5754. doi: 10.1128/JVI.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koopmans M, Fouchier R, Wilbrink B, et al. Update on human infections with highly pathogenic avian influenza virus A/H7N7 during an outbreak in poultry in The Netherlands. Euro Surveill. 2003;7:2217. [Google Scholar]
- 29.Mo IP, Song CS, Kim KS, et al. An occurrence of non-highly pathogenic avian influenza in Korea. In: Swayne D, Slemons R, editors. Proceedings of the Fourth International Symposium on Avian Influenza; Tallahassee. United States Animal Health Association, Rose Printing Company; 1997. pp. 379–383. [Google Scholar]
- 30.Guo YJ, Krauss S, Senne DA, et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 31.Alexander DJ. Report on avian influenza in the Eastern Hemisphere during 1997–2002. Avian Dis. 2003;47(3 Suppl):792–797. doi: 10.1637/0005-2086-47.s3.792. [DOI] [PubMed] [Google Scholar]
- 32.Fusaro A, Monne I, Salviato A, et al. Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J. Virol. 2011;85:8413–8421. doi: 10.1128/JVI.00219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YP, Shaw M, Gregory V, et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 35.Shanmuganatham K, Feeroz MM, Jones-Engel L, et al. Antigenic and molecular characterization of avian influenza A(H9N2) viruses, Bangladesh. Emerg. Infect. Dis. 2013;19:1393–1402. doi: 10.3201/eid1909.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butt KM, Smith GJ, Chen H, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Zhou YJ, Li GX, et al. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet. Microbiol. 2011;149:254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl.J. Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 39.Leung YH, Lau EH, Zhang LJ, et al. Avian influenza and ban on overnight poultry storage in live poultry markets, Hong Kong. Emerg. Infect. Dis. 2012;18:1339–1341. doi: 10.3201/eid1808.111879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu R, Ekiert DC, Krause JC, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domínguez-Cherit G, Lapinsky SE, Macias AE, et al. Critically Ill patients with 2009 influenza A(H1N1) in Mexico. JAMA. 2009;302:1880–1887. doi: 10.1001/jama.2009.1536. [DOI] [PubMed] [Google Scholar]
- 42.Scholtissek C. Pigs as “mixing vessels” for the creation of new pandemic influenza A viruses. Med. Principles. Pract. 1990/1991;2:65–71. [Google Scholar]
- 43.Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 44.Ducatez MF, Hause B, Stigger-Rosser E, et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis. 2011;17:1624–1629. doi: 10.3201/1709.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jhung MA, Epperson S, Biggerstaff M, et al. Outbreak of variant influenza A(H3N2) virus in the United States. Clin. Infect. Dis. 2013;57:1703–1712. doi: 10.1093/cid/cit649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monto AS, Webster RG. Influenza pandemics: History and lessons learned”. In: Webster RG, Monto AS, Braciale TJ, Lamb RA, editors. Textbook of Influenza, Second Edition. West Sussex: John Wiley and Sons; pp. 20–33. [Google Scholar]
- 47.Fiore AE, Uyeki TM, Broder K, et al. Prevention and control of influenza with vaccines: recommendations of the advisory committee on immunization practices (ACIP) 2010. MMWR Recomm. Rep. 2010;59:1–62. [PubMed] [Google Scholar]
- 48.CDC. Summary Recommendations: Prevention and Control of Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices—(ACIP)—United States, 2013–14. [Accessed April 23, 2014]; http://www.cdc.gov/flu/professionals/acip/2013-summary-recommendations.htm.
- 49.Reed C, Meltzer MI, Finelli L, et al. Public health impact of including two lineages of influenza B in a quadrivalent seasonal influenza vaccine. Vaccine. 2012;30:1993–1998. doi: 10.1016/j.vaccine.2011.12.098. [DOI] [PubMed] [Google Scholar]
- 50.Okuno Y, Isegawa Y, Sasao F, et al. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Throsby M, van den Brink E, Jongeneelen M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr. Opin. Virol. 2012;2:134–141. doi: 10.1016/j.coviro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ekiert DC, Friesen RH, Bhabha G, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friensen RH, Lee PS, Stoop EJ, et al. A common solution to group 2 influenza virus neutralization. Proc. Natl. Acad. Sci. U. S. A. 2013;111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida R, Igarashi M, Ozaki H, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 59.Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Clinical Trials.gov. [Accessed March 5, 2014];2010 http://www.ClinicalTrials.gov. http://clinicaltrials.gov.
- 61.Xu R, Krause JC, McBride R, et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat. Struct. Mol. Biol. 2013;20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei CJ, Boyington JC, McTamney PM, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 63.Wei CJ, Yassine HM, McTamney PM, et al. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci. Transl. Med. 2012;4:147ra114. doi: 10.1126/scitranslmed.3004273. [DOI] [PubMed] [Google Scholar]
- 64.Bommakanti G, Citron MP, Hepler RW, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13701–13706. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steel J, Lowen AC, Wang TT, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1:e00018–e00110. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang TT, Tan GS, Hai R, et al. Vaccination with asynthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khurana S, Loving CL, Manischewitz J, et al. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013;5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- 68.Crowe JE., Jr Universal flu vaccines: primum non nocere. Sci. Transl. Med. 2013;28:200fs34. doi: 10.1126/scitranslmed.3007118. [DOI] [PubMed] [Google Scholar]
- 69.O’Hagan DT, Tsai T, Reed S. Emulsion-based adjuvants for improved influenza vaccines. In: Rappuoli R, Del Giudice G, editors. Influenza vaccines for the future. Second edition. Basel: Springer; 2011. pp. 327–357. [Google Scholar]
- 70.Freund J, Casals J, Hosmer EP. Sensitization and antibody formation after injection of turbecle bacilli and paraffin oil. Proc. Exp. Biol. Med. 1937;37:509–513. [Google Scholar]
- 71.Stuart-Harris CH. Adjuvant influenza vaccines. Bull World Health Organ. 1969;41:617–621. [PMC free article] [PubMed] [Google Scholar]
- 72.Treanor JJ, Campbell JD, Zangwill KM, et al. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N. Engl. J. Med. 2006;354:1343–1351. doi: 10.1056/NEJMoa055778. [DOI] [PubMed] [Google Scholar]
- 73.Levie K, Leroux-Roels I, Hoppenbrouwers K, et al. An adjuvanted, low-dose, pandemic influenza A (H5N1) vaccine candidate is safe, immunogenic, and induces cross-reactive immune responses in healthy adults. J. Infect. Dis. 2008;198:642–649. doi: 10.1086/590913. [DOI] [PubMed] [Google Scholar]
- 74.Luca G, Haba-Rubio J, Dauvilliers Y, et al. Clinical, polysomnographic and genome-wide association analyses of narcolepsy with cataplexy: a European Narcolepsy Network study. J. Sleep. Res. 2013;22:482–495. doi: 10.1111/jsr.12044. [DOI] [PubMed] [Google Scholar]
- 75.De la Herrán-Arita AK, Kornum BR, Mahlios J, et al. CD4+ T cell autoimmunity to hypocretin/orexin and cross-reactivity to a 2009 H1N1 influenza A epitope in narcolepsy. Sci. Transl. Med. 2013;5:216ra176. doi: 10.1126/scitranslmed.3007762. [DOI] [PubMed] [Google Scholar]
- 76.Davies WL, Grunert RR, Haff RF, et al. Antiviral activity of 1-adamantanamine (Amantadine) Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- 77.Air GM. Influenza neuraminidase. Influenza Other Respir. Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aoki FY, Macleod MD, MD, Paggiaro P, et al. Early administration of oral oseltamivir increases the benefits of influenza treatment. J. Antimicrob. Chemother. 2003;51:123–129. doi: 10.1093/jac/dkg007. [DOI] [PubMed] [Google Scholar]
- 79.Wathen MW, Barro M, Bright RA. Antivirals in seasonal and pandemic influenza-future perspectives. Influenza and Other Respir. Viruses. 2013;7:76–80. doi: 10.1111/irv.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Babu YS, Chand P, Bantia S, et al. BCX-1812-a novel, highly potent, orally active and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 2000;43:3482–3486. doi: 10.1021/jm0002679. [DOI] [PubMed] [Google Scholar]
- 81.Bantia S, Arnold CS, Parker CD, et al. Anti-influenza virus activity of peramivir in mice with single intramuscular injection. Antiviral Res. 2006;69:39–45. doi: 10.1016/j.antiviral.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 82.Ikematsu H, Kawai N. Laninamivir octanoate: a new long-acting neuraminidase inhibitor for the treatment of influenza. Expert. Rev. Anti. Infect. Ther. 2011;9:851–857. doi: 10.1586/eri.11.112. [DOI] [PubMed] [Google Scholar]
- 83.Gubareva LV. Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors. Virus Res. 2004;103:199–203. doi: 10.1016/j.virusres.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 84.Collins PJ, Haire LF, Lin YP, et al. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature. 2008;453:1258–1261. doi: 10.1038/nature06956. [DOI] [PubMed] [Google Scholar]
- 85.Samson M, Pizzorno A, Abed Y, et al. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res. 2013;98:174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 86.Zürcher T, Yates PJ, Daly J, et al. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J. Antimicrob. Chemother. 2006;58:723–732. doi: 10.1093/jac/dkl321. [DOI] [PubMed] [Google Scholar]
- 87.Thorlund K, Awad T, Boivin G, et al. Systematic review of influenza resistance to the neuraminidase inhibitors. BMC Infect. Dis. 2011;11:134. doi: 10.1186/1471-2334-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monto AS, McKimm-Breschkin JL, Macken C, et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 2006;50:2395–2402. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ward P, Small I, Smith J, et al. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J Antimicrob. Chemother. 2005;55(Suppl 1):i5–i21. doi: 10.1093/jac/dki018. [DOI] [PubMed] [Google Scholar]
- 90.Dharan NJ, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–1041. doi: 10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- 91.Hauge SH, Dudman S, Borgen K, et al. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007–08. Emerg. Infect. Dis. 2009;15:155–162. doi: 10.3201/eid1502.081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe 2007–08 season. Emerg. Infect. Dis. 2009;15:552–560. doi: 10.3201/eid1504.081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Collins PJ, Haire LF, Lin YP, et al. Structural basis for oseltamivir resistance of influenza viruses. Vaccine. 2009;27:6317–6323. doi: 10.1016/j.vaccine.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 94.Rameix-Welti MA, Munier S, Le Gal S, et al. Neuraminidase of 2007–2008 influenza A(H1N1) viruses shows increased affinity for sialic acids due to the D344N substitution. Antivir. Ther. 2011;16:597–603. doi: 10.3851/IMP1804. [DOI] [PubMed] [Google Scholar]
- 95.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Renaud C, Kuypers J, Englund JA. Emerging oseltamivir resistance in seasonal and pandemic influenza A/H1N1. J. Clin. Virol. 2011;52:70–78. doi: 10.1016/j.jcv.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 97.Storms AD, Gubareva LV, Su S, et al. Oseltamivir-resistant pandemic (H1N1) 2009 virus infections, United States,2010–11. Emerg. Infect. Dis. 2012;18:308–311. doi: 10.3201/eid1802.111466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lackenby A, Moran Gilad J, Pebody R, et al. Continued emergence and changing epidemiology of oseltamivir-resistant influenza A(H1N1)2009 virus, United Kingdom, winter 2010/11. Euro Surveill. 2011;16:19784. [PubMed] [Google Scholar]
- 99.Hurt AC, Hardie K, Wilson NJ, et al. Community transmission of oseltamivir resistant A(H1N1)pdm09 influenza. N. Engl. J. Med. 2011;365:2541–2542. doi: 10.1056/NEJMc1111078. [DOI] [PubMed] [Google Scholar]
- 100.Mai LQ, Wertheim HF, Duong TN, et al. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N. Engl. J. Med. 2010;362:86–87. doi: 10.1056/NEJMc0910448. [DOI] [PubMed] [Google Scholar]
- 101.Houng HS, Garner J, Zhou Y, et al. Emergent 2009 influenza A(H1N1) viruses containing HA D222N mutation associated with severe clinical outcomes in the Americas. J. Clin. Virol. 2012;53:12–15. doi: 10.1016/j.jcv.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 102.World Health Organization. WHO guidelines for pharmacological management of pandemic influenza A/H1N1 2009 and other influenza viruses 2010. [Accessed February 17, 2014];2010 http://www.who.int/csr/resources/publications/swineflu/h1n1_guidelines_pharmaceutical_mngt.pdf. [PubMed]
- 103.Malakhov MP, Aschenbrenner LM, Smee DF, et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob. Agents Chemother. 2006;50:1470–1479. doi: 10.1128/AAC.50.4.1470-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicholls JM, Moss RB, Haslam SM. The use of sialidase therapy for respiratory viral infections. Antiviral Res. 2013;98:401–409. doi: 10.1016/j.antiviral.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moss RB, Hansen C, Sanders RL, et al. A Phase II study of DAS181, a novel host directed antiviral for the treatment of influenza infection. J. Infect. Dis. 2012;206:1844–1851. doi: 10.1093/infdis/jis622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Furuta Y, Takahashi K, Kuno-Maekawa M, et al. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sleeman K, Mishin VP, Deyde VM, et al. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob. Agents Chemother. 2010;54:2517–2524. doi: 10.1128/AAC.01739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kiso M, Takahashi K, Sakai-Tagawa Y, et al. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 2010;107:882–887. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fox L, Saravolatz LD. Nitazoxanide a new thiazolide antiparasitic agent. Clin. Infect. Dis. 2006;40:1173–1180. doi: 10.1086/428839. [DOI] [PubMed] [Google Scholar]
- 110.Korba BE, Montero AB, Farrar K, et al. Nitazoxanide, tizoxanide and other thiazolides are potent inhibitors of hepatitis B virus and hepatitis C virus replication. Antivir. Res. 2008;77:56–63. doi: 10.1016/j.antiviral.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 111.Stachulski AV, Pidathala C, Row EC, et al. Thiazolides as novel antiviral agents. 2. Inhibition of hepatitis C virus replication. J. Med. Chem. 2011;54:8670–8680. doi: 10.1021/jm201264t. [DOI] [PubMed] [Google Scholar]
- 112.Rossignol JF, La Frazia S, Chiappa L, et al. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J. Biol Chem. 2009;284:29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Edinger TO, Pohl MO, Stertz S. Entry of influenza A virus: host factors and antiviral targets. J. Gen. Virol. 2014;95(Pt 2):263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 114.Ochiai H, Sakai S, Hirabayashi T, et al. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza and B viruses in MDCK cells. Antiviral Res. 1995;27:425–430. doi: 10.1016/0166-3542(95)00040-s. [DOI] [PubMed] [Google Scholar]
- 115.Wang J, Ma C, Jo H, et al. Discovery of novel dual inhibitors of the wild-type and the most prevalent drug resistant mutant, S31N, of the M2 proton channel from influenza A virus. J. Med. Chem. 2013;56:2804–2812. doi: 10.1021/jm301538e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Planz O. Development of cellular signaling pathway inhibitors as new antivirals against influenza. Antiviral Res. 2013;98:457–468. doi: 10.1016/j.antiviral.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 117.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kobasa D, Jones SM, Shinya K, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 119.WHO Writing Committee. Clinical Aspects of Pandemic Influenza A (H1N1) 2009. N. Engl. J. Med. 2010;362:1708–1719. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 120.Darwish I, Mubareka S, Liles WC. Immunomodulatory therapy for severe influenza. Expert Review of Anti-infective Therapy. 2011;9:807–822. doi: 10.1586/eri.11.56. [DOI] [PubMed] [Google Scholar]
- 121.Mao H, Tu W, Qin G, et al. Influenza virus directly infects human natural killer cells and induces cell apoptosis. J. Virol. 2009;83:9215–9222. doi: 10.1128/JVI.00805-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mao H, Tu W, Liu Y, et al. Inhibition of human natural killer cell activity by influenza virions and hemagglutinin. J. Virol. 2010;84:4148–4157. doi: 10.1128/JVI.02340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat. Rev. Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 124.Bonneville M, O’Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- 125.Tu W, Zheng J, Liu Y, et al. The aminobisphosphonate pamidronate controls influenza pathogenesis by expanding a gammadelta T cell population in humanized mice. J Exp. Med. 2011;208:1511–1522. doi: 10.1084/jem.20110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hung IF, To KK, Lee CK, et al. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin. Infect. Dis. 2011;52:510–519. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hung IF, To KK, Lee CK, et al. Hyperimmune intravenous immunoglobulin treatment: a multicentre double-blind randomized controlled trial for patients with severe A(H1N1)pdm09 infection. Chest. 2013;44:464–473. doi: 10.1378/chest.12-2907. [DOI] [PubMed] [Google Scholar]
- 128.Govorkova EA, Webster RG. Combination chemotherapy for influenza. Viruses. 2010;2:1510–1529. doi: 10.3390/v2081510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoopes JD, Driebe EM, Kelley E, et al. Triple combination antiviral drug (TCAD) composed of amantadine, oseltamivir, and ribavirin impedes the selection of drug-resistant influenza A virus. PLoS One. 2011;6:e29778. doi: 10.1371/journal.pone.0029778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ilyushina NA, Hoffmann E, Salomon R, et al. Amantadine-oseltamivir combination therapy for H5N1 influenza virus infection in mice. Antiviral Therapy. 2007;12:363–370. [PubMed] [Google Scholar]
- 131.Ilyushina NA, Hay A, Yilmaz N, et al. Oseltamivir-ribavirin combination therapy for highly pathogenic H5N1 influenza virus infection in mice. Antimicrob. Agents and Chemother. 2008;52:3889–3897. doi: 10.1128/AAC.01579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nguyen JT, Smee DF, Barnard DL, et al. Efficacy of combined therapy with amantadine, oseltamivir, and ribavirin in vivo against susceptible and amantadine-resistant influenza A viruses. PLoS One. 2012;7:e31006. doi: 10.1371/journal.pone.0031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smee DF, Hurst BL, Wong MH, et al. Effects of double combinations of amantadine, oseltamivir, and ribavirin on influenza A (H5N1) virus infections in cell culture and in mice. Antimicrob. Agents Chemother. 2009;53:2120–2128. doi: 10.1128/AAC.01012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim WY, Young SG, Huh JW, et al. Triple-combination antiviral drug for pandemic H1N1 influenza virus infection in critically ill patients on mechanical ventilation. Antimicrob. Agents Chemother. 2011;55:5703–5709. doi: 10.1128/AAC.05529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barber MR, Aldridge JR, Jr, Webster RG, et al. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Olsen SJ, Ungchusak K, Sovann L, et al. Family clustering of avian influenza A (H5N1) Emerg. Infect. Dis. 2005;11:1799–1801. doi: 10.3201/eid1111.050646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boon AC, deBeauchamp J, Hollmann A, et al. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J. Virol. 2009;83:10417–10426. doi: 10.1128/JVI.00514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boon AC, Finkelstein D, Zheng M, et al. H5N1 influenza virus pathogenesis in genetically diverse mice is mediated at the level of viral load. MBio. 2011;2:e00171–e00211. doi: 10.1128/mBio.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Swayne DE, Kapczynski DR. Vaccines, vaccination, and immunology for avian influenza viruses in poultry. In: Swayne DE, editor. Avian Influenza. Ames: Blackwell Publishing; 2008. pp. 407–451. [Google Scholar]