Abstract
Cancer genome sequencing efforts have revealed the novel theme that chromatin modifiers are frequently mutated across a wide spectrum of cancers. Mutations in genes encoding subunits of SWI/SNF (BAF) chromatin remodeling complexes are particularly prevalent, occurring in 20% of all human cancers. As these are typically loss-of-function mutations and not directly therapeutically targetable, central goals have been to elucidate mechanism and identify vulnerabilities created by these mutations. Here we discuss emerging data that these mutations lead to the formation of aberrant residual SWI/SNF complexes that constitute a specific vulnerability and discuss the potential for exploiting these dependencies in SWI/SNF-mutant cancers.
Introduction
SWI/SNF complexes are evolutionarily conserved multi-subunit complexes that utilize the energy of ATP hydrolysis to mobilize nucleosomes and remodel chromatin (Kassabov et al., 2003; Phelan et al., 1999). These approximately 2 MDa complexes are made up of 12–15 subunits; they contain one of the two catalytic ATPase subunits, SMARCA4/BRG1 or SMARCA2/BRM, several core subunits including SMARCB1/SNF5/INI1/BAF47 and SMARCC1/BAF155 that are present in all SWI/SNF complexes, as well as subunits present in only some variants such as ARID1A and ARID1B, mutually exclusive subunits for BAF (BRG1-associated factor) varieties of the complexes, and PBRM1 and ARID2, specific for PBAF (polybromo BRG1-associated factor) varieties of the complexes (Wang et al., 1996; Wu et al., 2009). SWI/SNF complexes interact with transcription factors, co-activators and co-repressors, and are capable of mobilizing nucleosomes at target promoters and enhancers to modulate gene expression (Figure 1) (Hu et al., 2011; Tolstorukov et al., 2013; You et al., 2013) and have also been implicated in various types of DNA repair (Dykhuizen et al., 2013; Gong et al., 2006; Hara and Sancar, 2002; Park et al., 2006; Watanabe et al., 2014).
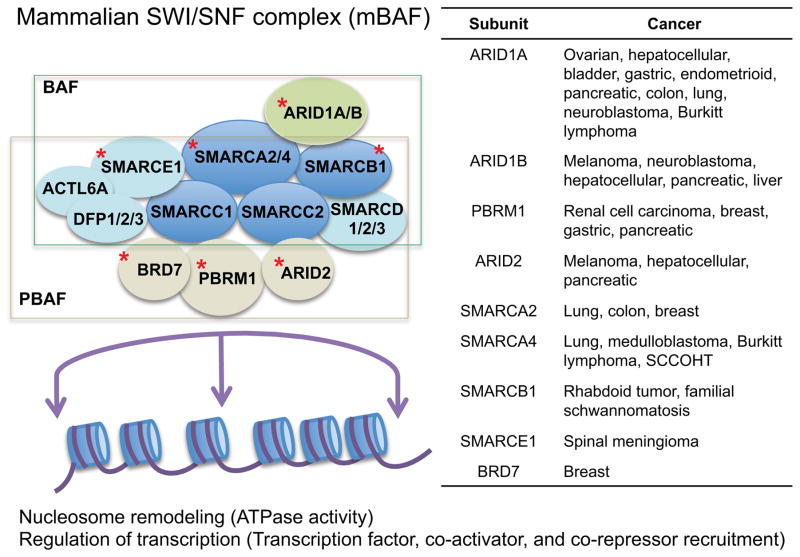
Figure 1. SWI/SNF complexes modulate transcription and genes encoding subunits of SWI/SNF complexes are mutated in cancer.
SWI/SNF complexes are found in two major subtypes, BAF and PBAF, and are comprised of multiple subunits (top left). SWI/SNF complexes contribute to transcription modulation by mobilizing nucleosomes and by interacting with transcription factors, coactivators, and corepressors on DNA. Subunits found mutated in cancer are denoted by a red star and are described in the table (top right).
With respect to a role in the control of gene expression, SWI/SNF complexes have been shown to serve roles in the transcriptional regulation of lineage specification and development in numerous model systems. For example, SWI/SNF complexes contribute to the development of T cells (Chi et al., 2002; Wang et al., 2011b), hepatocytes (Gresh et al., 2005), oligodendrocytes (Yu et al., 2013), and embryonic stem cell self-renewal and pluripotency (Gao et al., 2008; Ho et al., 2009). Specificity of SWI/SNF complexes in the control of these developmental programs is achieved in part through restricted expression and combinatorial assembly of variant SWI/SNF subunits. The SMARCD3 (BAF60C) subunit is expressed specifically in the embryonic heart, where it is essential for the control of cardiac development (Lickert et al., 2004). Similarly, a switch from the PHF10 (BAF45A) and ACTL6A (BAF53A) subunits, which are expressed in neural stem cells, to DPF1 (BAF45B), DPF3 (BAF45C), and ACTL6B (BAF53B) subunits is essential to control the transition of neural progenitors into post-mitotic mature neurons (Lessard et al., 2007; Wu et al., 2007). Such switching can modulate interaction with specific transcription factors (Kadam et al., 2000) and facilitates differential activation of transcriptional pathways. Ultimately, via combinatorial inclusion of variant subunits, several hundred versions of SWI/SNF complexes may exist (Wu et al., 2009) and serve instructive roles in the control of fate specification.
The first clue linking SWI/SNF complexes to cancer came in the late 1990s when mutations of the gene encoding the SMARCB1 (SNF5/INI1/BAF47) subunit were identified in rhabdoid tumors (RT), a rare but highly aggressive type of cancer that strikes young children (Biegel et al., 1999; Versteege et al., 1998). Smarcb1 was subsequently validated as a bona fide and potent tumor suppressor in genetically engineered mouse models (Guidi et al., 2001; Klochendler-Yeivin et al., 2000; Roberts et al., 2000, 2002). While this observation was first noted over a decade ago, it is only more recently via cancer genome sequencing studies that the high prevalence of SWI/SNF subunit mutations have been found in many types of cancer. At least eight genes encoding subunits of SWI/SNF complexes have been identified as recurrently mutated in cancers derived from nearly every tissue in the body, collectively occurring in 20% of all human cancers (Figure 1) (Kadoch et al., 2013; Shain and Pollack, 2013). For example, inactivating mutations of ARID1A are prevalent in a wide variety of cancers, including 45% of ovarian clear cell and endometrioid carcinomas (Jones et al., 2010; Wiegand et al., 2010), 19% of gastric cancers (Wang et al., 2011a), 19% of bladder cancers (Gui et al., 2011), 14% of hepatocellular cancers (Guichard et al., 2012), 12% of melanomas (Hodis et al., 2012), and also less frequently in colorectal, lung, breast, pancreas and several other cancer types (Kadoch et al., 2013; Shain and Pollack, 2013). SMARCA4 (BRG1), a catalytic ATPase and a core subunit of SWI/SNF complexes, is mutated in several cancer types including lung (Medina et al., 2008; Reisman et al., 2003), medulloblastoma (Parsons et al., 2011), pancreatic cancer (Wong et al., 2000), and most recently, small-cell carcinoma of the ovary, hypercalcemic type (SCCOHT) (Jelinic et al., 2014; Ramos et al., 2014; Witkowski et al., 2014). Other subunits have also been found to be mutated in cancer, such as PRBM1 (BAF180) in renal carcinoma (Varela et al., 2011) and ARID2 in melanoma (Hodis et al., 2012) and hepatocellular carcinoma (Li et al., 2011) (Figure 1). The mechanisms by which mutation of each individual subunit promotes oncogenesis and the function of mutated SWI/SNF complexes in cancer is now an active area of investigation.
Many studies have elucidated pathways that are regulated by SWI/SNF complexes, and how disruption of these gene expression programs by subunit mutation promotes cancer. For example, SWI/SNF can bind to RB and facilitate repression of RB target genes (Trouche et al., 1997). SWI/SNF also interacts with MYC, both as an activator and as a repressor (Cheng et al., 1999; Nagl et al., 2006). In part via disruption of RB function, inactivation of SMARCB1 leads to downregulation of p16INK4a and E2F targets, indicating that SWI/SNF plays a role in cell cycle regulation and differentiation (Betz et al., 2002; Isakoff et al., 2005; Oruetxebarria et al., 2004). Additionally, SWI/SNF complexes are required for specific regulation of interferon beta targets (Morozov et al., 2007; Ramirez-Carrozzi et al., 2009). It has been shown that SWI/SNF complexes can bind to the promoters of roughly one-third of all genes (Ho et al., 2009; Tolstorukov et al., 2013) and the above represent only a few of numerous pathways that have been shown to be SWI/SNF dependent.
With respect to the chromatin mechanisms that underlie regulation of targets, a largely antagonistic functional relationship between SWI/SNF and PRC2 complexes has been identified (Ho et al., 2009; Kennison and Tamkun, 1988; Kia et al., 2008; Wilson et al., 2010). Loss of SMARCB1 leads to upregulation of EZH2, as well as broad H3K27 trimethylation and repression of PRC2 targets, effects that are essential for cancer formation driven by SMARCB1 loss (Wilson et al., 2010). Targeted inhibition of EZH2 may represent a therapeutic opportunity for SMARCB1-mutant cancers (Knutson et al., 2013). While the mechanisms by which SWI/SNF mutations contribute to cancer are still being elucidated and the relative importance of contributions to transcriptional regulation vs. DNA repair are still in question, mutation of SWI/SNF subunits in cancer likely contributes to cancer at least in part by perturbing the regulation of transcriptional pathways involved in control of proliferation and fate specification (Eroglu et al., 2014).
It is interesting to note that while loss of function SWI/SNF subunit mutations seem most prevalent in cancer, point mutations have also been described, such as a small number of SMARCA4 missense mutations in medulloblastoma (Parsons et al., 2011). It is not yet understood whether these point mutations also result in loss of function of the protein, as in a classical tumor suppressor, or whether they result in partial loss, or even potentially oncogenic gain of function effects. Looking forward, elucidating the effects of these point mutations will likely provide further mechanistic understanding of the cancer promoting activity of SWI/SNF mutations. However, from a therapeutic standpoint, as mutations in genes encoding SWI/SNF complex subunits are often loss-of-function, including nonsense, frameshift, and large deletions (Lee et al., 2012; Versteege et al., 1998; Wang et al., 2014b; Wilson and Roberts, 2011), the products of the mutant genes themselves do not constitute obvious drug targets. Consequently, it is of great interest to identify specific vulnerabilities conferred by these mutations upon cancer cells that have the potential to provide new therapeutic opportunities.
The residual complex: a new class of vulnerability
One attractive hypothesis to account for many subunits of a single complex mutated is that all of the mutations are essentially equivalent and result in inactivation of SWI/SNF complexes. However, several findings seemed in conflict with such a possibility. First, the consequences of inactivation of genes encoding SWI/SNF subunits in mice are fairly distinct. For example, while inactivation of Smarcb1 and Smarca4 both result in early embryonic lethality at E3.5 (Guidi et al., 2001; Klochendler-Yeivin et al., 2000), knockout of Arid1a leads to the absence of mesoderm and arrest at E6.5 (Gao et al., 2008), silencing of Smarcd3 results in heart developmental defects (Lickert et al., 2004), and Smarca2-deficient mice are viable (Reyes et al., 1998). Consequently, subunit loss results in distinct developmental phenotypes. Second, loss of different subunits of the complexes is associated with different types of cancer. For example ARID1A is frequently mutated in ovarian cancer (Jones et al., 2010; Wiegand et al., 2010), SMARCA4 in lung cancer (Fukuoka et al., 2004; Medina et al., 2008; Reisman et al., 2003), PBRM1 in renal cancer (Varela et al., 2011), and SMARCB1 in rhabdoid tumors (Lee et al., 2012), with only modest overlap, suggesting distinct consequences for mutation of different subunits.
Consistent with this, conditional Smarcb1 deletion in mice results in formation of rhabdoid-like tumors and lymphomas (Roberts et al., 2002) while Smarca4 haploinsufficiency leads to mammary tumors (Bultman et al., 2008). Third, it has been shown that SWI/SNF complexes can assemble without SMARCB1 (Doan et al., 2004), SMARCA4 (Hoffman et al., 2014; Wilson et al., 2014) or ARID1A (Helming et al., 2014), indicating that residual complexes remain despite tumor suppressor subunit loss. Consequently, an alternate hypothesis was proposed: that loss of tumor suppressor subunits results in aberrant residual complexes that in turn actively drive oncogenesis (Wang et al., 2009). Essentially, oncogenesis was not due to tumor suppressor loss per se, but rather to gain of aberrantly functioning residual complexes. Consistent with this hypothesis was the demonstration of an essential role for the residual complex in driving cancer formation in SMARCB1-mutant cancers. Specifically, rather than accelerating cancer, or having no effect due to redundancy, the proliferation of SMARCB1-deficient human RT lines was blocked upon knockdown of SMARCA4, itself a tumor suppressor. In genetically engineered mouse models, inactivation of Smarca4 also blocked the in vivo tumor formation otherwise caused by Smarcb1 loss (Wang et al., 2009). These findings suggested that the functional activity of residual SWI/SNF complexes might be essential for cancer driven by SMARCB1 loss. However, SMARCB1-mutant cancers are quite rare and whether this concept was similarly true for cancers mutant in other SWI/SNF subunits was unknown. Additionally, since several SWI/SNF subunits serve important roles in various cell types, it remained unclear whether there was a differential requirement for these subunits between SWI/SNF-mutant cancers and normal cells, which would be necessary for a potential therapeutic approach based upon targeted inhibition of residual complexes.
Recently, data from three large scale screening publications (Helming et al., 2014; Hoffman et al., 2014; Wilson et al., 2014) have provided some insight to this question and suggest that an essential role for the residual complex extends to the wide spectrum of cancers harboring mutations in other subunits, and also suggests enhanced dependence upon at least some residual complex members occurs in SWI/SNF-mutant cancers. Project Achilles is a near-genome scale shRNA screen against 11,000 genes performed in over 200 human cancer cell lines (Cheung et al., 2011). Data from Project Achilles was used to search for dependencies created by SWI/SNF mutation. Within the cell lines in Project Achilles, genes encoding two SWI/SNF subunits were mutated at sufficient frequency to enable a search for vulnerabilities. ARID1A was mutant in 18 of 165 cell lines while SMARCA4 was mutant in 8 of 165 cell lines. Both of these comparisons resulted in novel insights – in both cases the number one dependence was upon a related SWI/SNF subunit.
In ARID1A-mutant cancer cells, ARID1B was identified as the number one dependency, suggesting that in the setting of ARID1A mutation, residual SWI/SNF complexes become specifically and differentially reliant upon ARID1B (Helming et al., 2014). ARID1A and ARID1B are 60% identical in protein sequence and are mutually exclusive since individual SWI/SNF chromatin remodeling complexes can contain either ARID1A or ARID1B but not both. Experiments aimed at validating results from the dependency screen showed that knocking down ARID1B specifically impaired the proliferation of ARID1A-mutant cancer cells but had minimal effect on ARID1A wildtype cancer cells. Mechanistically, while loss of ARID1B had no effect upon integrity of SWI/SNF complexes in wildtype cells, in the context of ARID1A mutation, the combined absence of ARID1A and ARID1B destabilized SWI/SNF complexes and resulted in dissociation of subunits, which was associated with loss of cell proliferation.
In SMARCA4-mutant cells in the Achilles screen, SMARCA2 was found to be specifically essential (Wilson et al., 2014), a relationship that was simultaneously and independently identified in another screen (Hoffman et al., 2014) and an earlier focused study in which the effects of SMARCA2 loss upon SMARCA4-mutant cancers were directly tested (Oike et al., 2013). Collectively, these reports suggest that the residual complexes created by SMARCA4 mutation rely on SMARCA2 as the remaining SWI/SNF ATPase subunit and thus cannot tolerate loss of SMARCA2. Notably, an aspect of the synthetic lethal relationship between SMARCA4 and SMARCA2 is distinct from that between ARID1A and ARID1B. While ARID1B loss destabilizes SWI/SNF complexes in ARID1A-mutant cancers (Helming et al., 2014), the residual complexes remain intact following SMARCA2 loss in SMARCA4-mutant cancers (Hoffman et al., 2014; Wilson et al., 2014). This finding suggests that even though SWI/SNF subunits can fully associate without a catalytic ATPase subunit, the ATPase activity is required for the proliferation of the cancer cells. Collectively, these recent findings indicate that residual complexes exist in a variety of SWI/SNF-mutant cancers and are essential for their growth.
While the dependency findings establish an essential role for residual SWI/SNF complexes in SWI/SNF-mutant cancers, the mechanism by which these residual complexes promote cancer remains poorly understood. One conceptual possibility is that the residual complexes essentially acquire neomorphic gain of function, which alters targeting and/or remodeling, and results in gene expression changes that facilitate transformation. Perhaps consistent with such a possibility, ARID1A and ARID1B have been reported to have opposing roles in regulation of proliferation in osteoblasts (Nagl et al., 2007). Consequently, loss of ARID1A may result in excessive incorporation of ARID1B and unbalanced regulation of proliferation versus differentiation. Similarly, SMARCA4 and SMARCA2 show differential expression patterns during development (Machida et al., 2001; Singh and Archer, 2014; Zheng et al., 2004) with SMARCA4 tending to be highly expressed in proliferating cells while SMARCA2 tends to be expressed in slowly cycling cells such as stem cells and in non-cycling differentiated cells (Reisman et al., 2009). SMARCA4 and SMARCA2 have also been shown to interact with different transcription factors (Kadam and Emerson, 2003), and have been implicated in the differential control of cell fate (Flowers et al., 2009; Zhang et al., 2011). In the neomorphic gain-of-function model, loss of function of SWI/SNF tumor suppressor subunits might be akin to oncogenic activation of residual SWI/SNF complexes. Accordingly, cancers become addicted to the residual complex resulting in differential dependency upon specific subunits compared to normal cells.
The precise mechanism by which residual mutant SWI/SNF complexes contribute to oncogenesis remains an active area of investigation. It will be of interest to understand whether mutation of SWI/SNF subunits in cancer results in mistargeting of the complexes to chromatin akin to the effects of altered methylation of the SMARCC1 subunit, which affect targeting of SWI/SNF complexes in breast cancer (Wang et al., 2014a). It will also be important to determine whether the oncogenic effect of SWI/SNF subunit mutations arises in part from altered balance of variant SWI/SNF complexes and further whether residual SWI/SNF complexes can properly remodel chromatin once they are bound. Additionally, as suggested by the fascinating finding that the SS18-SSX fusion functions primarily by ejecting SMARCB1 from SWI/SNF complexes (Kadoch and Crabtree, 2013), a key question is whether the cancer-associated mutations in part function by altering assembly of residual complexes. A major hope is that a deeper understanding of the mechanisms of mutant SWI/SNF complexes may facilitate development of targeted therapeutics.
A Surprise: Co-mutations
Several pieces of data suggest that such a model may be too simple. While ARID1A is perhaps the most frequently mutated SWI/SNF subunit, mutations of ARID1B have also been found in cancer (Kadoch et al., 2013; Sausen et al., 2013; Shain and Pollack, 2013). Given the finding of a synthetic lethal relationship between ARID1A inactivation and ARID1B knockdown, the prediction was that the mutation of ARID1A and ARID1B would not co-occur in the same cell line or tumor sample. Indeed, this hypothesis would seem to be a fundamental prediction of synthetic lethality: co-mutations should not occur. However, precisely the opposite was found: significant co-occurrence of ARID1A and ARID1B mutations, both in cancer cell lines and in primary cancers (Helming et al., 2014). What might account for both synthetic lethality and co-mutation? One possibility is that mutations in ARID1A or ARID1B, rather than leading to neomorphic gain of function, result in hypofunction of the residual complexes. In this scenario, ARID1A and ARID1B would have redundant functions with respect to tumor suppression and, akin to the concept of haploinsufficiency, reduced levels of ARID1, whether ARID1A or ARID1B, result in impaired control of gene expression and predispose to transformation. However, retaining some amount of ARID1 function may be essential for cell survival, and consequently these mutations also result in enhanced dependence upon ARID1B compared to normal cells. The mutational profile of ARID1A and ARID1B may provide support for such a model as cancer cell lines were identified that had biallelic mutations in ARID1A in which ARID1B was either wildtype or mutant on one allele; cancer cell lines with monoallelic mutations in both ARID1A and ARID1B; and rarely cancer cell lines in which a monoallelic mutation in ARID1A was accompanied by biallelic mutations in ARID1B. However, no cancer cell lines were identified in which ARID1A and ARID1B both contained biallelic mutations consistent with an essential role for some ARID1 function.
The situation is less clear for the relationship between SMARCA4 and SMARCA2. Mutations in SMARCA4 have been reported in several types of cancer (Fukuoka et al., 2004; Medina et al., 2008; Reisman et al., 2003) and mice haploinsufficient for SMARCA4 are predisposed to mammary tumors (Bultman et al., 2008). In contrast, SMARCA2 mutations are rare in primary tumors. Sequencing of SMARCA2 in non-melanomatous skin cancers identified a hotspot missense mutation in 3 of 16 cases (Moloney et al., 2009), a mutation class often associated with gain-of-function effects, although this mutation was not reported as significant in a subsequent exome study (Jayaraman et al., 2014). Lack of SMARCA2 expression has been noted in several cancer cell lines and primary cancers (Glaros et al., 2007), an effect challenging to interpret as in normal tissues SMARCA2 tends to be low in cells with high proliferative potential (Reisman et al., 2005). However, while Smarca2-deficient mice have not been reported prone to spontaneous tumors, they are 15% larger than control littermates, prone to prostate hyperplasia (Shen et al., 2008), and have increased susceptibility to tumor formation in an ethylcarbamate lung cancer model (Glaros et al., 2007) and a UV irradiation skin cancer model (Halliday et al., 2012). Ultimately, while not frequently mutated, it is possible that SMARCA2 may have tumor suppressor capabilities in human tissues. Interestingly, some cell lines and cancers have been reported, such as the SW-13 cancer cell line, in which SMARCA4 is mutated and SMARCA2 not expressed (Dunaief et al., 1994; Strobeck et al., 2002). Whether such cancers reflect emergence of resistance or whether the synthetic lethal relationship is in some way context dependent is unclear and an active area of investigation.
Therapeutic Potential
The synthetic lethal relationships described above raise potential opportunities for targeting of residual SWI/SNF complexes as a therapeutic approach for cancers with a SWI/SNF mutation. SMARCA2 contains two domains of particular note with respect to the potential for therapeutic targeting: a bromodomain and an ATPase domain (Wu et al., 2009). Substantial precedence has emerged for targeting of bomodomains, as the JQ1 BRD4 bromodomain inhibitor (Filippakopoulos et al., 2010) has shown promising effects in pre-clinical studies and clinical trials of BRD4 inhibition are now in progress (http://www.cancer.gov/clinicaltrials/search/view?cdrid=733416&version=HealthProfessional&protocolsearchid=12562170). A similar approach may be feasible for SMARCA2. While the crystal structures of the bromodomains of both SMARCA4 and SMARCA2 have been solved (Filippakopoulos et al., 2012), further studies, such as small molecule screening, will be necessary to determine if these domains are targetable. It is important to note, however, that it is not entirely clear whether the bromodomain is essential for the function of SMARCA2, as studies have shown that SMARCA2 activity is dependent on a high-mobility-group protein I/Y-like DNA binding domain (Bourachot et al., 1999). Alternatively, the ATPase domain of SMARCA2 could be targeted. While structures of yeast SWI/SNF complexes have been reported (Kasten et al., 2011), structure of the mammalian complexes and ATPase domains would be useful for evaluating the potential for targeted drug development. If targetable, SMARCA2 may be a promising target, as Smarca2 knockout mice are reported viable (Reyes et al., 1998). It is of interest to note that mutations of SMARCA2 have been found in some neural disorders (Ronan et al., 2013). A recurrent SMARCA2 missense mutation has been identified in schizophrenia where it is reported to reduce nuclear localization of SMARCA2 protein (Koga et al., 2009). Mutations of SMARCA2 have also been identified as the basis for the human developmental disorder Nicolaides-Baraitser syndrome (NBS) (Van Houdt et al., 2012). However, these mutations are either missense or small in frame deletions, and never inactivating, leading to the prediction that the SMARCA2 mutations in NBS are gain-of-function and thus likely not predictive of the effects of targeted inhibition.
One challenge in developing an inhibitor to SMARCA2 is its homology to SMARCA4, which may make it difficult to develop a compound that inhibits SMARCA2 but not SMARCA4. As noted above, SMARCA4 and SMARCA2 do display differential transcription factor interactions in part due to structural differences (Kadam and Emerson, 2003) but it remains to be determined whether such structural differences can be effectively exploited for targeting. It is also worthy of note that three recent publications identified SMARCA4 itself as a potential therapeutic target in small cell lung cancer (Romero et al., 2013), and in Myc-driven leukemias (Buscarlet et al., 2014; Shi et al., 2013). On the surface, this would seem a paradox – SMARCA4 having tumor suppressor activity in some contexts but specifically required for cancer maintenance in others. However, increasing data highlights the importance of context dependent control of gene expression in cancer. For example, gain-of-function mutations of EZH2 are found in lymphomas (McCabe et al., 2012) while loss of function mutations occur in myelodysplastic syndrome (Nikoloski et al., 2010). Even the canonical tumor suppressor p53, which can promote cancer via loss of function, is also associated with recurrent point mutations in cancer, some of which have been implicated as having gain-of-function oncogenic activity (van Oijen and Slootweg, 2000; Strano et al., 2007). Consequently, context is essential and whether therapeutic benefit might be derived from targeting SMARCA2, or SMARCA4, or both remains to be determined.
The relationship between ARID1A and ARID1B might also present therapeutic opportunity, as ARID1A is frequently mutated in many human cancers. A recent finding shows that stapled peptides can successfully disrupt the protein-protein interaction between EZH2 and the Polycomb PRC2 chromatin modifying complex (Kim et al., 2013) and an analogous strategy might be possible to disrupt the interaction between ARID1B and SWI/SNF complexes. ARID1B has also been associated with an E3 ubiquitin ligase that mediates the monoubiquitination of Histone 2B (Li et al., 2010). Targeting the E3 ligase-associated activity of ARID1B is an approach worth considering, although it is unknown whether the E3 ligase activity plays a role in the synthetic lethality.
Several potential limitations must be considered for therapeutic targeting of residual SWI/SNF complexes. One is therapeutic window. The results of recent screens suggest that at least in some cases there is differential dependence upon residual complex members between mutant and non-mutant cells. Whether this difference is great enough to constitute an effective therapeutic window, however, remains to be determined. There are also likely to be challenges in drug development, in both identifying essential domains and determining whether those domains can be feasibly targeted. An additional concern, given the tumor suppressor activity of several SWI/SNF subunit genes, is whether inhibition of SWI/SNF complexes might actually cause cancer. Even when considering ARID1B, in general, SWI/SNF-mutant cancer cell lines remain dependent upon absence of the missing subunit. For example, re-expression of SMARCB1 in SMARCB1-mutant cancers causes cell cycle arrest (Kuwahara et al., 2010) and re-expression of SMARCA4 in SMARCA4-mutant cancers results in reversion of malignant phenotype (Romero et al., 2012; Wong et al., 2000). Consequently, it seems likely that should treatment with a compound that inhibits a SWI/SNF subunit result in the formation of a cancer, such a cancer would likely remain dependent upon the absence of the subunit and that cessation of the inhibitor would be predicted to result in resolution of such a tumor.
Ultimately, for cancers driven by mutation of a gene encoding a SWI/SNF subunit, at least some of these mutations result in specific dependence upon residual complex members, which in turn may offer potential therapeutic targets. Given the large number of cancers harboring SWI/SNF mutations, investigation of the dependency mechanisms and of the potential to target these complexes has the potential for broad cancer relevance.
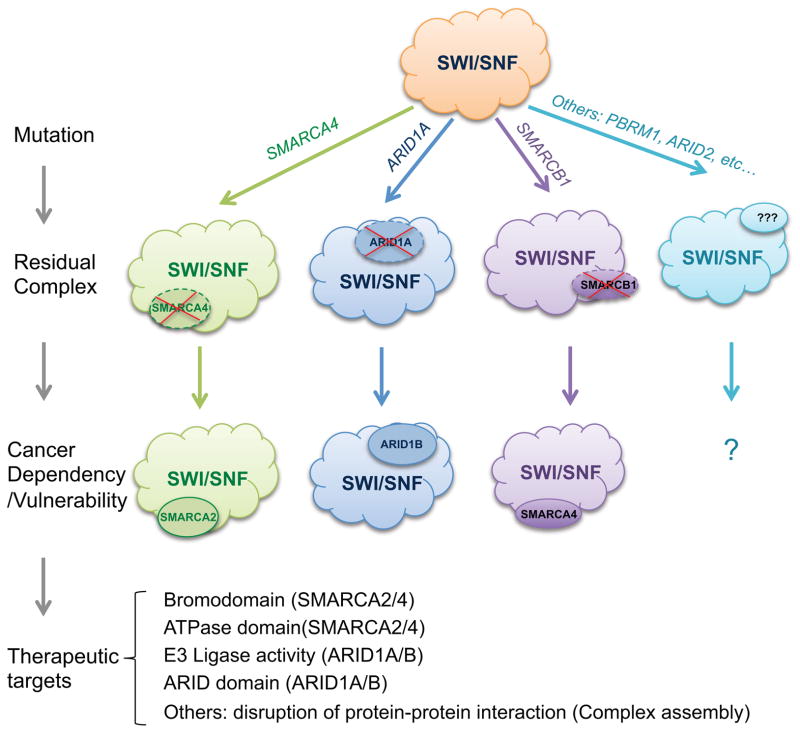
Figure 2. Residual SWI/SNF complexes are a vulnerability in cancers containing SWI/SNF subunit mutation.
Mutation of a gene encoding a SWI/SNF complex subunit results in the formation of a residual complex that is specifically dependent upon other subunits and essential for the growth of the cancer. Targeting subunits of this residual complex is a newly identified therapeutic opportunity.
Acknowledgments
The work in Dr. Roberts’ laboratory is partly supported by R01CA172152 (to C.W.M. Roberts) and R01CA113794 (to C.W.M. Roberts), and a U01NCI Mouse Models of Cancer Consortium Award (to C.W.M. Roberts). Alex’s Lemonade Stand Foundation, Hyundai Hope on Wheels, the Cure AT/RT Now fund, the Avalanna Fund, the Garrett B. Smith Foundation, Miles for Mary, the Ellison Foundation, and Cookies for Kids Cancer provided additional support. X. Wang is a recipient of a P.A.L.S.-St.Baldrick's Research Grant and is supported by postdoctoral fellowships from the Rally Foundation and David Abraham Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21:5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- Bourachot B, Yaniv M, Muchardt C. The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol Cell Biol. 1999;19:3931–3939. doi: 10.1128/mcb.19.6.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Buscarlet M, Krasteva V, Ho L, Simon C, Hébert J, Wilhelm B, Crabtree GR, Sauvageau G, Thibault P, Lessard JA. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood. 2014;123:1720–1728. doi: 10.1182/blood-2013-02-483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- Doan DN, Veal TM, Yan Z, Wang W, Jones SN, Imbalzano AN. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene. 2004;23:3462–3473. doi: 10.1038/sj.onc.1207472. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Ålin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, Pfister S, Cho YJ, Zhao K, Crabtree GR. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;497:624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu E, Burkard TR, Jiang Y, Saini N, Homem CCF, Reichert H, Knoblich JA. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers S, Nagl NG, Beck GR, Moran E. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J Biol Chem. 2009;284:10067–10075. doi: 10.1074/jbc.M808782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, Hong K, Settnek S, Gupta A, Buetow K, et al. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10:4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, Reisman D. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene. 2007;26:7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol. 2006;13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, Muchardt C, Hue L, Pontoglio M, Yaniv M, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad I, Ben Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, Smith TW, Imbalzano AN, Jones SN. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM, Zhou Y, Sou PW, Huang XXJ, Rana S, Bugeja MJ, Painter N, Scolyer RA, Muchardt C, Di Girolamo N, et al. The absence of Brm exacerbates photocarcinogenesis. Exp Dermatol. 2012;21:599–604. doi: 10.1111/j.1600-0625.2012.01522.x. [DOI] [PubMed] [Google Scholar]
- Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol. 2002;22:6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, Kim Y, Kryukov GV, Ghandi M, Aguirre AJ, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20:251–254. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, Bagdasarian L, Huber J, Lindeman A, Chen D, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci U S A. 2014;111:3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt JKJ, Nowakowska BA, Sousa SB, van Schaik BDC, Seuntjens E, Avonce N, Sifrim A, Abdul-Rahman OA, van den Boogaard M-JH, Bottani A, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–9. S1. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, Wang X, Biegel JA, Pomeroy SL, Mesirov JP, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. 2005;102 doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman SS, Rayhan DJ, Hazany S, Kolodney MS. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Invest Dermatol. 2014;134:213–220. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- Jelinic P, Mueller JJ, Olvera N, Dao F, Scott SN, Shah R, Gao J, Schultz N, Gonen M, Soslow RA, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46:424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih IM, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Vogelstein B, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Transcriptional Specificity of Human SWI/SNF BRG1 and BRM Chromatin Remodeling Complexes. Mol Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- Kasten MM, Clapier CR, Cairns BR. SnapShot: Chromatin remodeling: SWI/SNF. Cell. 2011;144:310, e1. doi: 10.1016/j.cell.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Porter Scott M, Chesworth R, Moyer MP, Copeland RA, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, Iritani S, Itokawa M, Inada T, Iwata N, et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet. 2009;18:2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- Kuwahara Y, Charboneau A, Knudsen ES, Weissman BE. Reexpression of hSNF5 in malignant rhabdoid tumor cell lines causes cell cycle arrest through a p21(CIP1/WAF1)-dependent mechanism. Cancer Res. 2010;70:1854–1865. doi: 10.1158/0008-5472.CAN-09-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. 2012:122. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJA, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. Mammalian SWI/SNF--a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol. 2010;30:1673–1688. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Machida Y, Murai K, Miyake K, Iijima S. Expression of chromatin remodeling factors during neural differentiation. J Biochem. 2001;129:43–49. doi: 10.1093/oxfordjournals.jbchem.a002834. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- Moloney FJ, Lyons JG, Bock VL, Huang XX, Bugeja MJ, Halliday GM. Hotspot mutation of Brahma in non-melanoma skin cancer. J Invest Dermatol. 2009;129:1012–1015. doi: 10.1038/jid.2008.319. [DOI] [PubMed] [Google Scholar]
- Morozov A, Lee SJ, Zhang Z-K, Cimica V, Zagzag D, Kalpana GV. INI1 induces interferon signaling and spindle checkpoint in rhabdoid tumors. Clin Cancer Res. 2007;13:4721–4730. doi: 10.1158/1078-0432.CCR-07-0054. [DOI] [PubMed] [Google Scholar]
- Nagl NG, Zweitzig DR, Thimmapaya B, Beck GR, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- Nagl NG, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SMC, Kuiper RP, Knops R, Massop M, Tönnissen ERLTM, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Van Oijen MGCT, Slootweg PJ. Gain-of-Function Mutations in the Tumor Suppressor Gene p53. Clin Cancer Res. 2000;6:2138–2145. [PubMed] [Google Scholar]
- Oike T, Ogiwara H, Tominaga Y, Ito K, Ando O, Tsuta K, Mizukami T, Shimada Y, Isomura H, Komachi M, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013;73:5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LMP, Mohd-Sarip A, Vries RGJ, Hoeben RC, Verrijzer CP. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- Park J-H, Park E-J, Lee H-S, Kim SJ, Hur S-K, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JCH, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P, Karnezis AN, Craig DW, Sekulic A, Russell ML, Hendricks WPD, Corneveaux JJ, Barrett MT, Shumansky K, Yang Y, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 2014;46:427–429. doi: 10.1038/ng.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson Ea. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in Human Lung Cancer Cell Lines and Primary Lung Cancers: Correlation with Poor Prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Bouldin TW, Weissman BE, Funkhouser WK. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl Immunohistochem Mol Morphol. 2005;13:66–74. doi: 10.1097/00129039-200503000-00011. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci U S A. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CWM, Leroux MM, Fleming MD, Orkin SH. Highly penetrant , rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- Romero OA, Setien F, John S, Gimenez-Xavier P, Gómez-López G, Pisano D, Condom E, Villanueva A, Hager GL, Sanchez-Cespedes M. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med. 2012;4:603–616. doi: 10.1002/emmm.201200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero OA, Torres-Diz M, Pros E, Savola S, Gomez A, Moran S, Saez C, Iwakawa R, Villanueva A, Montuenga LM, et al. MAX inactivation in small-cell lung cancer disrupts the MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0799. [DOI] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Papadopoulos N, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Powers N, Saini N, Comstock CES, Sharma A, Weaver K, Revelo MP, Gerald W, Williams E, Jessen WJ, et al. The SWI/SNF ATPase Brm is a gatekeeper of proliferative control in prostate cancer. Cancer Res. 2008;68:10154–10162. doi: 10.1158/0008-5472.CAN-08-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Whyte Wa, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe J-S, Minder JL, Mercan F, Wang E, Eckersley-Maslin Ma, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Archer TK. Analysis of the SWI/SNF chromatin-remodeling complex during early heart development and BAF250a repression cardiac gene transcription during P19 cell differentiation. Nucleic Acids Res. 2014;42:2958–2975. doi: 10.1093/nar/gkt1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Dell’Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G. Mutant p53: an oncogenic transcription factor. Oncogene. 2007;26:2212–2219. doi: 10.1038/sj.onc.1210296. [DOI] [PubMed] [Google Scholar]
- Strobeck MW, Reisman DN, Gunawardena RW, Betz BL, Angus SP, Knudsen KE, Kowalik TF, Weissman BE, Knudsen ES. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem. 2002;277:4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans Ja, Wilson BG, Park PJ, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci U S A. 2013;110:10165–10170. doi: 10.1073/pnas.1302209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci U S A. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sévenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan ASY, Tsui WY, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011a;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A, Wisinski KB, Huang W, Cai W, Pike JW, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014a;25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Côté J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sansam CG, Thom CS, Metzger D, Evans Ja, Nguyen PTL, Roberts CWM. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 2009;69:8094–8101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Werneck MBF, Wilson BG, Kim H-J, Kluk MJ, Thom CS, Wischhusen JW, Evans JA, Jesneck JL, Nguyen P, et al. TCR-dependent transformation of mature memory phenotype T cells in mice. J Clin Invest. 2011b;121:3834–3845. doi: 10.1172/JCI37210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Haswell JR, Roberts CWM. Molecular Pathways: SWI/SNF (BAF) Complexes Are Frequently Mutated in Cancer--Mechanisms and Potential Therapeutic Insights. Clin Cancer Res. 2014b;20:21–27. doi: 10.1158/1078-0432.CCR-13-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R, Ui A, Kanno S-I, Ogiwara H, Nagase T, Kohno T, Yasui A. SWI/SNF Factors Required for Cellular Resistance to DNA Damage Include ARID1A and ARID1B and Show Interdependent Protein Stability. Cancer Res. 2014;74:2465–2475. doi: 10.1158/0008-5472.CAN-13-3608. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CWM. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CWM. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Helming KC, Wang X, Kim Y, Vazquez F, Jagani Z, Hahn WC, Roberts CWM. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol. 2014 doi: 10.1128/MCB.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski L, Carrot-Zhang J, Albrecht S, Fahiminiya S, Hamel N, Tomiak E, Grynspan D, Saloustros E, Nadaf J, Rivera B, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–443. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- Wong AKC, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland A-M, et al. BRG1, a Component of the SWI-SNF Complex, Is Mutated in Multiple Human Tumor Cell Lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, De Carvalho DD, Dai C, Liu M, Pandiyan K, Zhou XJ, Liang G, Jones PA. SNF5 is an essential executor of epigenetic regulation during differentiation. PLoS Genet. 2013;9:e1003459. doi: 10.1371/journal.pgen.1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LMN, Mao M, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chen M, Kim J-R, Zhou J, Jones RE, Tune JD, Kassab GS, Metzger D, Ahlfeld S, Conway SJ, et al. SWI/SNF complexes containing Brahma or Brahma-related gene 1 play distinct roles in smooth muscle development. Mol Cell Biol. 2011;31:2618–2631. doi: 10.1128/MCB.01338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Patel B, McMenamin M, Paprocki AM, Schramm RD, Nagl NG, Wilsker D, Wang X, Moran E, Latham KE. Expression of genes encoding chromatin regulatory factors in developing rhesus monkey oocytes and preimplantation stage embryos: possible roles in genome activation. Biol Reprod. 2004;70:1419–1427. doi: 10.1095/biolreprod.103.023796. [DOI] [PubMed] [Google Scholar]