Summary
Cargo adaptor subunits of vesicle coat protein complexes sort transmembrane proteins to distinct membrane compartments in eukaryotic cells. The exomer complex is the only cargo adaptor known to sort proteins at the trans-Golgi network into secretory vesicles. Exomer function is regulated by the Arf1 GTPase, a master regulator of trafficking at the Golgi. We report the structure of exomer bound to two copies of Arf1. Exomer interacts with each Arf1 molecule via two surfaces; one is a non-canonical interface that regulates GTP hydrolysis. The structure uncovers an unexpected membrane-proximal hydrophobic element that exomer uses in cooperation with Arf1 to remodel membranes. Given the constrained motion of the exomer hinge region, we envision that exomer dynamically positions multiple membrane insertion elements to drive membrane fission. In contrast to other known cargo adaptors, exomer therefore couples two functions, cargo sorting and membrane fission, into a single complex.
Introduction
Transmembrane proteins are sorted into membrane vesicles and tubules for transport through the secretory and endocytic pathways of eukaryotic cells by the action of cargo adaptors (Schekman and Orci, 1996; Bonifacino and Glick, 2004; De Matteis and Luini, 2008). Many of these cargo adaptor complexes are recruited to the membrane surface by small GTPases of the Arf family (Donaldson and Jackson, 2011). In addition to packaging cargo into nascent vesicles, cargo adaptors perform a variety of other functions in vesicle biogenesis, including recruitment of accessory factors and triggering GTPase activity (Bonifacino and Lippincott-Schwartz, 2003). Our understanding of cargo sorting and vesicle formation has been enhanced by a number of structural studies of the clathrin-dependent cargo adaptors as well as the COPI and COPII coat protein complexes (Owen et al., 1999; Bi et al., 2002; Collins et al., 2002; Mossessova et al., 2003; Heldwein et al., 2004; Edeling et al., 2006; Bi et al., 2007; Jackson et al., 2010; Lee and Goldberg, 2010; Yu et al., 2012, Ren et al., 2013).
Remarkably, scant mechanistic information is available regarding cargo sorting in one of the most prominent eukaryotic trafficking pathways, in which cargo at the trans-Golgi network (TGN) is packaged into vesicles destined for the apical plasma membrane (PM) of polarized cells (Bard and Malhotra, 2006; Bonifacino, 2014). In the model organism S. cerevisiae, such secretory vesicles are the primary source of the lipids and proteins used to generate the plasma membrane of a daughter cell (Drubin and Nelson, 1996). The basic mechanisms underlying secretory vesicle cargo sorting and biogenesis remain elusive.
Arf1 is a conserved regulator of membrane trafficking, playing an important role in the recruitment of several cargo adaptors to the Golgi complex (D'Souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007). Arf1 possesses a membrane-inserting alpha-helix that can drive membrane curvature and potentially fission (McMahon and Gallop, 2005; Krauss et al., 2008; Lundmark et al., 2008; Drin and Antonny, 2010; Boucrot et al., 2012; Campelo and Malhotra, 2012). Exomer is an Arf1 GTPase-dependent cargo adaptor complex responsible for sorting a subset of cargo proteins at the TGN that are destined for the PM (Sanchajate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006). Although its homolog has not been identified in metazoans, exomer is the only known cargo adaptor responsible for sorting cargo at the TGN into secretory vesicles. Several other proteins have been identified to act in this pathway (von Blume et al., 2011; Malhotra and Campelo, 2011; Wakana et al., 2012; Gehart et al., 2012; Cruz-Garcia et al., 2013), but none of these act as cargo adaptors.
Exomer is a heterotetramer consisting of two copies of the core protein Chs5, and any two members of four paralogous proteins known as the ChAPs (Chs5 and Arf1 binding proteins: Chs6, Bud7, Bch1, and Bch2). The ChAPs determine cargo specificity by binding directly to the cytoplasmic tails of cargo proteins, but appear to be structurally interchangeable within the complex (Trautwein et al., 2006; Wang et al., 2006; Sanchajate and Schekman, 2006; Barfield et al., 2009; Rockenbauch et al., 2012; Paczkowski et al., 2012). The two Chs5 molecules dimerize through a β-sandwich that functions as a molecular hinge (Richardson and Fromme, 2013), and the ChAPs bind to Chs5 through a single long alpha-helix immediately C-terminal to the dimerization motif (Paczkowski et al., 2012). The FBE (FN3 and BRCT of exomer) domain of Chs5 was found to bind directly to Arf1, although the strength of this interaction appeared insensitive to the nucleotide-bound state of Arf1 (Paczkowski et al., 2012). We previously reported that multiple interactions between exomer, Arf1, and the membrane surface cooperate to correctly target exomer to the TGN (Paczkowski et al., 2012), but the structural basis for this cooperation is unknown.
To gain insight into how Arf1 regulates the function of exomer, we determined the structure of an Arf1/exomer complex. The structure reveals two interfaces between exomer and Arf1; we show both are important for membrane recruitment and exomer function. The symmetry of the structure allows us to define unambiguously the membrane binding interface and positioning of both exomer and Arf1 relative to the membrane. We demonstrate that exomer amplifies the ability of Arf1 to remodel membranes. This activity depends on a conserved hydrophobic element within exomer that is juxtaposed to the membrane surface. We propose that exomer drives vesicle biogenesis by directly participating in both cargo sorting and membrane remodeling, with fission arising from membrane insertion by both exomer and exomer-bound Arf1.
Results
Structure of the Exomer-Arf1 Complex
We determined the structure of the Arf1/exomer complex using crystals grown from a mixture of the Bch1 subunit, the Chs5 subunit (residues 1-299, which are sufficient for exomer function (Paczkowski et al., 2012)), and a version of Arf1 lacking its membrane-inserting amphipathic N-terminal helix (ΔN17-Arf1) locked in its active state through the combined effects of a GTPase-defective Q71L mutation and binding to the non-hydrolyzable GTP analog, GMPPNP. The structure of the complex was determined by molecular replacement using available structures of ΔN17-Arf1 (Shiba et al., 2003) and the Chs5 (residues 1-77, lacking FBE domain)/Bch1 exomer complex (Richardson and Fromme, 2013). The structure was refined using a 3.4 Å native dataset to yield a model with appropriate statistics (Table S1). Although the resolution of the data used to refine the complex is modest, our analysis of the structure benefited from available higher resolution structures of all of the components (1.50 Å for ΔN17-Arf1, 2.9 Å for Chs5(1-77)/Bch1, and 2.75 Å for Chs5(1-299)/Chs6).
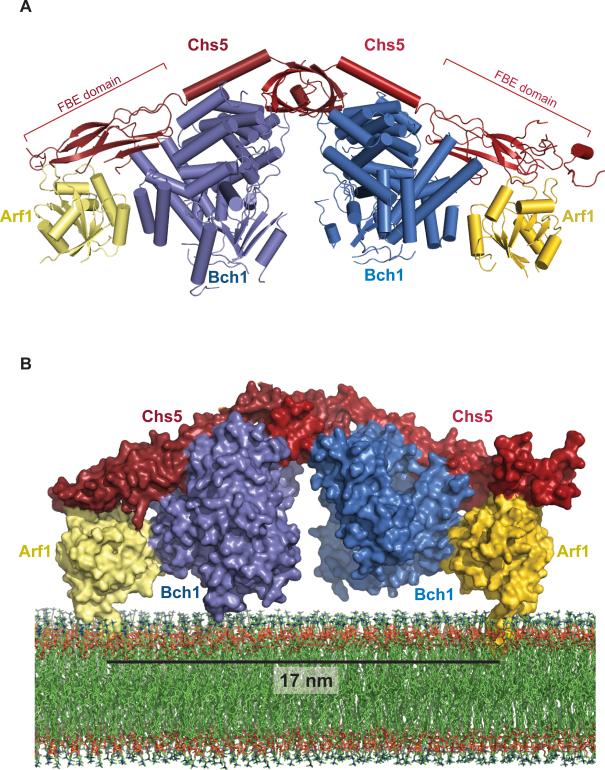
The asymmetric unit contains two copies of a hexameric complex, with each hexamer containing two copies each of ΔN17-Arf1, Bch1, and Chs5 (Figures 1A, S1A-C, and Movie S1). Regions of the asymmetric unit display varying degrees of disorder, consistent with the known flexibility of the exomer complex (Richardson and Fromme, 2013). The C-terminal portions of the FBE domains exhibited the largest degree of disorder, and were modeled by superposition of the structure of the FBE domain from the previously determined Chs5/Chs6 structure (Paczkowski et al., 2013) onto the well-ordered portions of the FBE domains of the Arf1/exomer complex (Figure S1D).
Figure 1. Crystal Structure of the Exomer-Arf1 complex.
A) The crystal structure of exomer bound to Arf1 shown as a ribbon diagram. Chs5 is shown in red, Bch1 in blue, and ΔN17-Arf1-GMPPNP in gold. Residues 1-274 of Chs5 constitutes a functional fragment (Paczkowski et al., 2012). Chs5 residues 170-299 were not resolved in the crystal structure.
B) Space-filling model of exomer and Arf1 bound to membranes, generated by superimposing the structure of full-length Arf1-GTP bound to lipids (Liu et al., 2010) onto ΔN17-Arf1-GMPPNP in the complex. The lipid bilayer was derived from a previous study (Heller et al., 1993).
Arf1 possesses an N-terminal amphipathic helix that inserts into the outer leaflet of the lipid bilayer. We superimposed the previously determined NMR structure of intact Arf1-GTP bound to a lipid micelle (Liu et al., 2010) onto the Arf1 molecules in the structure of the Arf1/exomer complex. Given the symmetry of the complex and using the N-terminal helix of Arf1 as an anchoring point, we identified the orientation of the complex relative to the membrane surface unambiguously (Figure 1B). This orientation was confirmed experimentally as described below.
The Bch1 subunit and the N-terminal dimerization domain of Chs5 adopt a similar conformation in the Arf1-bound exomer complex compared to the isolated Chs5/Bch1 exomer complex. However, the orientation of the FBE domain in the Arf1-bound complex is significantly different relative to its positioning in the Chs5/Chs6 complex, in which the FBE domain was constrained via a crystal contact with a neighboring complex. The FBE domain has undergone an approximately 180o rotation about the flexible linker connecting it to the long alpha-helix of Chs5, as well as a translation of approximately 24 Å, measured from the distal end of the FBE domain (Figure S1E). The reason for this new orientation is readily apparent – the FBE domain makes direct contact with Arf1 (Figures 1A and 2A), confirming the interaction observed previously by biochemical analysis (Paczkowski et al., 2012).
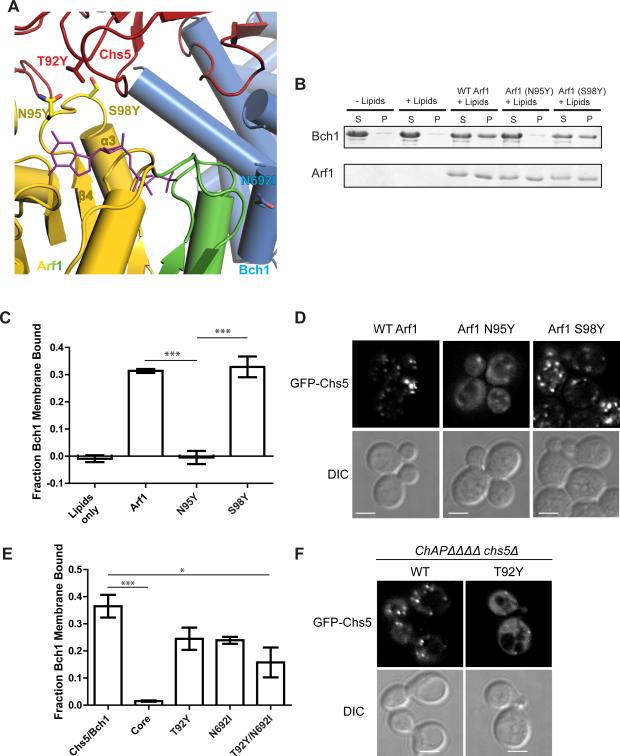
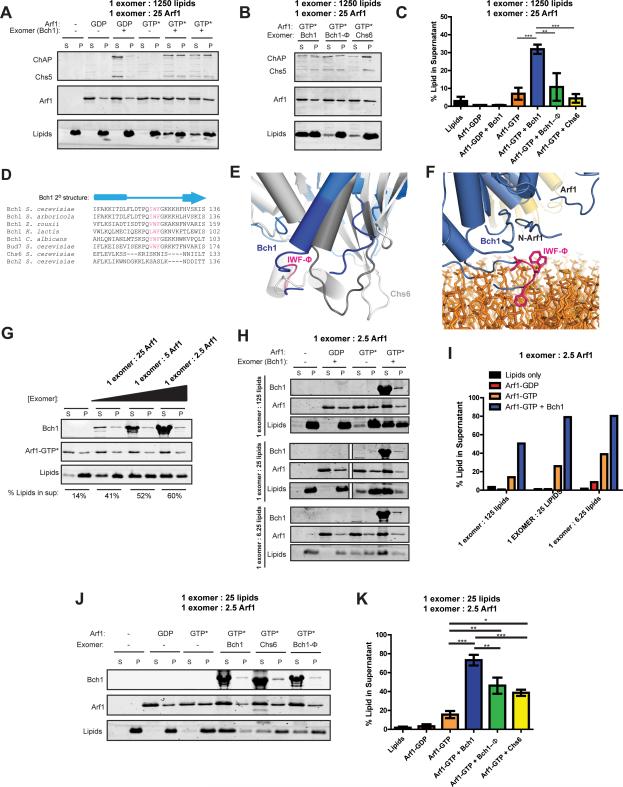
Figure 2. Two interfaces mediate the Arf1-exomer interaction.
A) Close-up view of the two interfaces Arf1 makes with the exomer complex shown in a ribbon diagram. Chs5 is shown in red, Bch1 in blue, and Arf1 in gold, with its switch residues highlighted in green. The nucleotide, GMPPNP, is shown in purple. Mutated residues are shown as sticks.
B) Liposome pelleting assay to assess membrane recruitment of the Chs5(1-299)/Bch1 exomer complex by myristoylated Arf1. N95Y and S98Y are Arf1 mutants. S, supernatant. P, pellet. The appearance of protein in the pellet fraction indicates membrane binding. For simplicity, only the Bch1 bands are shown for exomer complexes. See also Figures S2 and S3.
C) Quantification of the data shown in panel (B). Error bars represent SEM, n = 4, with significance determined by one-way ANOVA with post-processing to correct for multiple comparisons. Only significant comparisons are noted.
D) Images of plasmid-borne GFP-Chs5 in strains harboring wild-type or mutant Arf1. A single deconvolved focal plane is shown at equivalent light levels for each experiment. Scale bar, 2 μm.
E) Quantification of membrane recruitment of Chs5(1-299)/Bch1 mutants. “Core” is the Chs5(1-77)/Bch1 complex which lacks the FBE domain. T92Y is Chs5-T92Y, N692I is Bch1-N692I.
F) Plasmids expressing GFP-Chs5(1-299), wild-type or T92Y mutant, were introduced into a strain lacking exomer (chs5Δ chs6Δ bud7Δ bch1Δ bch2Δ), and imaged. Scale bar 2 μm.
Arf1 recruits exomer to membranes via two separate interfaces
In addition to the interaction of Arf1 with the Chs5 FBE domain, the structure reveals that exomer makes a previously unidentified contact with Arf1 through the Bch1 subunit. Like all small GTPases, the GTP-bound state of Arf1 is known to bind effectors through its “switch” regions (Khan and Ménétrey, 2013). Bch1, through a highly conserved helix, makes contact with the switch II region of Arf1 (Figures 2A and S1F) consistent with GTP-dependent recruitment of the complex to the membrane surface.
The FBE domain makes a non-canonical contact with Arf1 in a non-switch region near the nucleotide-binding pocket (Figure 2A). The FN3 subdomain of the Chs5 FBE domain forms an interface with Arf1 in a loop between β4 and α3, burying approximately 290 Å2 of solvent accessible surface. No other Arf1 effectors are known to bind to this region of Arf1 (Khan and Ménétrey, 2013). We previously found that, in contrast to the Arf1 effector COPI, which stimulates ArfGAP (GTPase-activating protein) activity (Yu et al., 2012), exomer inhibits the activity of the yeast ArfGAP Age2, likely by competing for binding to the Arf1 substrate (Paczkowski et al., 2012). Comparing the structure of the Arf1/exomer complex with that of the ArfGAP ASAP3 bound to Arf6 (Ismail et al., 2010) reveals that the FBE domain binding site does indeed significantly overlap with the binding site of the GAP domain (Figure S1G). It is possible, given the large amount of displacement of the FBE domain we observed between different crystal structures, that the FBE domain could be displaced during or after vesicle budding to allow the GAP access to the Arf, as previously proposed (Paczkowski et al., 2012).
The relevance of the two distinct exomer-Arf1 interfaces observed in the crystal structure was tested by mutational analysis. We employed both liposome flotation and quantitative liposome pelleting assays to monitor the Arf1-dependent recruitment of exomer to membranes in vitro. Arf1 bound to membranes in a GTP dependent manner, while the Chs5/Bch1 exomer complex was able to bind membranes only in the presence of membrane-anchored Arf1-GTP (Figures 2B and S2A). Introducing a single mutation, N95Y, in the observed exomer-binding loop of Arf1 (Figure 2A) resulted in a complete loss of exomer binding. Mutating a nearby residue in the loop, S98Y, which does not make clear contact with the Chs5 FBE domain, resulted in wild type levels of binding (Figure 2B,C). As a test of the specificity of this Arf1 mutation for the exomer interface, we performed a similar experiment using the clathrin-dependent cargo adaptor, Gga1. The N95Y mutant version of Arf1 recruited the Gga1 VHS-GAT domain to membranes at a level identical to that of wild-type Arf1 (Figure S2B,C). Thus, exomer binding to this non-switch region of Arf1 appears to be a novel and specific binding mode for cargo adaptor recruitment, but is consistent with previous observations that effectors often use more than one interface to interact with Arf1 (Khan and Ménétrey, 2013).
We assessed the consequence of disrupting the Arf1/FBE domain interaction in vivo. We first examined the effects on exomer localization to the TGN. In yeast strains harboring the Arf1-N95Y mutation, GFP-Chs5 was significantly mislocalized to the cytoplasm and was observed at fewer TGN punctae in comparison to cells with wild-type Arf1 (Figures 2D and S2D). The localization of Sec7, used as a marker for the TGN, was unaffected by this mutation in Arf1 (Figure S2E). These results indicate the Arf1-N95Y mutant disrupts Chs5 localization without perturbing the overall number of TGN compartments.
The Arf1-N95Y mutant displayed a significant growth phenotype on rich media, which is likely independent of its effects on exomer function, as cells lacking exomer have no detectable growth phenotype on rich media (Figure S2F). To monitor the effect of the Arf1-N95Y mutant on exomer function, we used a growth assay that monitors trafficking of the model exomer cargo, Chs3 (Valdivieso et al., 1991). Chs3 is a chitin synthase that is responsible for incorporating chitin into the cell wall (Shaw et al., 1991). The toxin calcofluor white is used to assay for exomer function based upon its ability to bind chitin in the cell wall and cause cell wall stress and death (Herth and Schnepf, 1980). Consequently, cells are more resistant to the toxin when exomer function is disrupted. We observed that the Arf1-N95Y mutant cells grew marginally better than wild-type cells in the presence of the toxin (Figure S2F). As the growth of the Arf1-N95Y mutant is slower than wild-type cells in the absence of the toxin, the subtly increased growth on calcofluor white suggests that exomer function was indeed perturbed by disruption of the Arf1-FBE domain interaction, as would be expected based on the Chs5 mislocalization phenotype.
To determine the relative contributions of the two different Arf1-exomer interaction interfaces, we generated exomer mutants using residues at each interface. Deletion of the entire FBE domain resulted in a complete loss of exomer binding (Figure 2E), as seen previously (Paczkowski et al., 2012). A single point mutant in the FBE domain, T92Y, resulted in a modest decrease in exomer binding (Figures 2E and S3A,B). To test the effect of this mutation in vivo, we observed the localization of GFP-Chs5. Both the wild-type and T92Y mutant Chs5 proteins showed proper TGN localization and function (Figure S3C,D). However, deleting all four ChAPs led to a complete mislocalization of GFP-Chs5-T92Y, whereas wild-type GFP-Chs5 was still properly localized to the TGN through its direct interaction with Arf1 (Figure 2F).
Mutation of a conserved asparagine residue (N692I) at the interface between Bch1 and the switch region of Arf1 resulted in a partial loss of exomer binding (Figures 2E, S1F, and S3B). Combination of the two Arf1-binding mutations (Chs5-T92Y/Bch1-N692I) resulted in an even greater reduction in exomer membrane recruitment compared to either of the single mutations (Figures 2E and S3B). Thus, both interfaces play an important role in binding Arf1.
Different ChAP subunits bind membranes with different affinities
Based upon sequence homology, the four yeast ChAP proteins can be separated into two distinct subclasses, with Bch1 in one class and Chs6 in the other (Trautwein et al., 2006). In vivo, a given exomer complex appears to consist of either two identical ChAPs or two different ChAPs (Sanchajate and Schekman, 2006; Paczkowski et al., 2012). The Chs5/Chs6 complex binds to liposomes independent of recruitment by Arf1, indicating a high affinity for membranes (Paczkowski et al., 2012) (Figure 3A,B). In contrast, we could not detect binding of the Chs5/Bch1 complex to liposomes unless membrane-bound Arf1 was also present (Figure 3A,B). This result indicates that Bch1 has a lower affinity for membranes compared to Chs6.
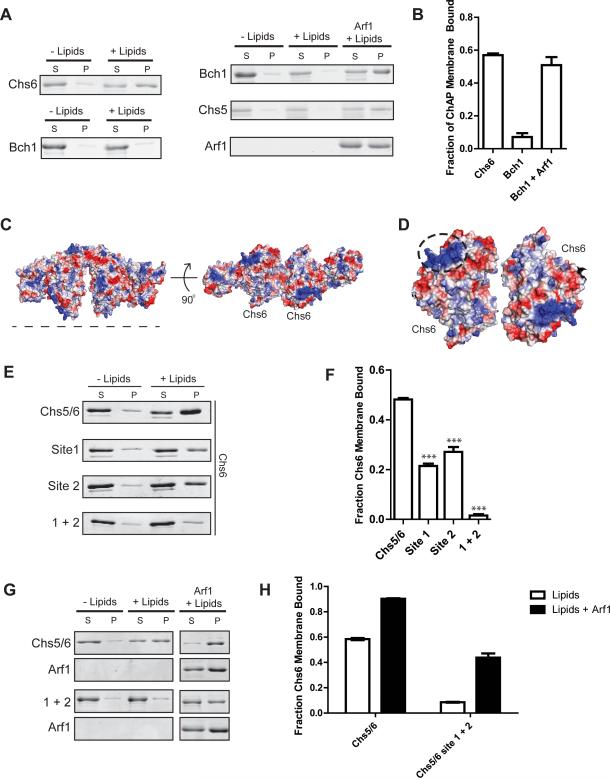
Figure 3. ChAP subunits bind membranes with different affinities.
A) Liposome pelleting assays comparing membrane binding affinity of Chs5(1-299)/Chs6 to that of Chs5(1-299)/Bch1 (left side). Chs5/Bch1 binding to membranes is detectable only when Arf1 is present (right side).
B) Quantification of the data shown in panel (A), n = 3.
C) Electrostatic surface potential of the modeled Chs5/Chs6/Arf1 complex, generated by superimposing the structure of Chs6 (Paczkowski et al., 2012) onto Bch1. Blue indicates positive potential, red indicates negative potential. The dotted line represents the proposed membrane-binding interface. A similar analysis of Bch1 was not practical, due to a significant number of residues in disordered loops on this surface.
D) View of the membrane binding surface of two Chs6 subunits, with the dashed circle highlighting the locations of site 1 and 2 comprising the positively charged cluster.
E) Liposome pelleting assay testing Chs5(1-299)/Chs6 membrane binding surface mutants. Site 1, K86A/H87A/K89A. Site 2, R446A/K449A. 1+2 is a combination of both sites.
F) Quantification of the data from panel (E), n = 3.
G) Liposome pelleting assay to test Arf1-dependent membrane binding of Chs5(1-299)/Chs6 mutants.
H) Quantification of the data from panel (A), n = 3.
We generated mutants to test whether the proposed membrane-binding surface is important for interacting with membranes. Examination of this surface of Chs6 revealed a cluster of positively charged residues (Figure 3C,D). We removed the positive charge from two different sites within this cluster, K86/H87/K89 (“site 1”) and R446/K449 (“site 2”), by substitution with alanine residues, and tested membrane binding of the resulting Chs5/Chs6 complexes. When the site mutants were mutated individually, we saw a ~50% decrease in membrane binding of each mutant (Figure 3E,F). When the two sites were mutated together, membrane binding of the Chs5/Chs6 complex was completely lost (Figure 3E,F).
In order to determine more precisely the contributions of membrane binding and Arf1 binding to exomer membrane recruitment, we assessed Arf1-dependent recruitment of a Chs5/Chs6 exomer complex harboring a mutant membrane-binding surface (“site 1+2” mutant). The presence of Arf1 increased the membrane-bound fraction of both the wild-type and mutant Chs5/Chs6 complexes (Figure 3G,H).
We also tested whether similar mutations in the proposed membrane-binding surface of Bch1 would reduce the amount of the complex recruited to membranes by Arf1. We were unable to accurately visualize the corresponding electrostatic surface of Bch1 due to existence of many disordered loops on this surface that we were unable to model. We therefore chose mutants by structural analogy to Chs6 and by considering sequence conservation. Neutralizing two positively charged sites on this surface in Bch1, H79/H81/K83/K85 (“site 1”) and K126/K127/K128 (“site 2”), resulted in a ~60% decrease in Chs5/Bch1 membrane recruitment by Arf1 (Figure 4A,B). Taken together, these results indicate that we have identified the membrane-binding surface on both the Chs6 and Bch1 exomer subunits, confirming the model presented in Figure 1B.
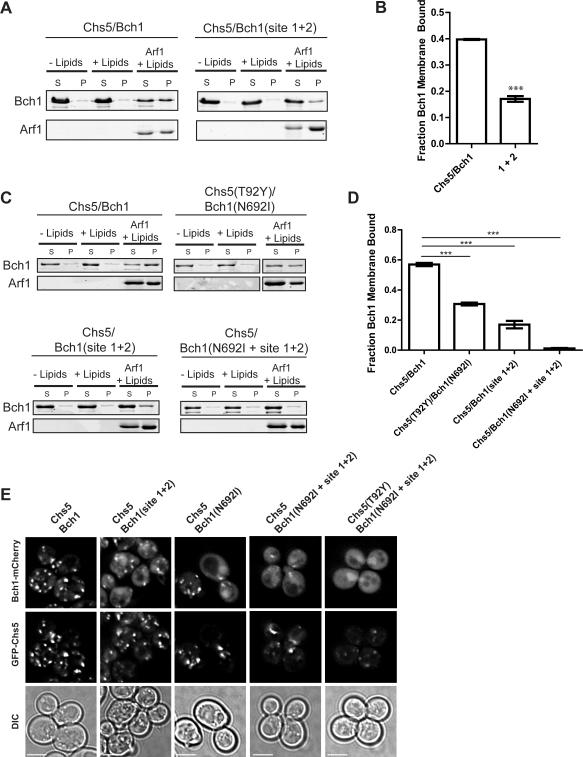
Figure 4. Exomer membrane recruitment occurs through multiple interactions with Arf1 and membranes.
A) Liposome pelleting assay to test Arf1-dependent membrane binding of Chs5(1-299)/Bch1 mutants.
B) Quantification of panel (G), n = 3, with significance determined by an unpaired t-test.
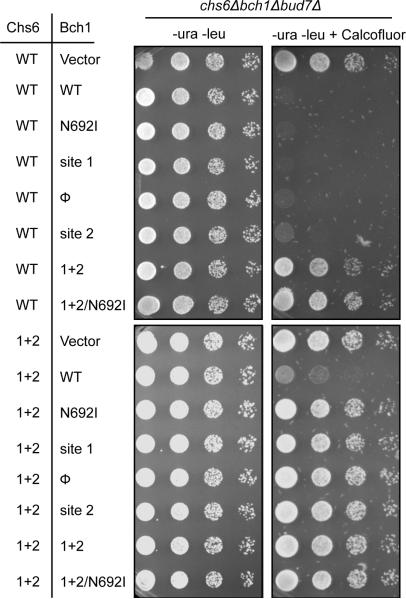
C) Liposome pelleting assay to determine the contributions of multiple exomer interfaces. Chs5-T92Y and Bch1-N692I are Arf1 interface mutations, site 1+2 is the membrane interface mutation.
D) Quantification of the data shown in panel (C), n = 3.
E) Plasmids expressing GFP-Chs5 and Bch1-mCherry constructs were introduced into a chs5Δchs6Δbud7Δbch1Δ strain and imaged. Scale bar, 2 μm.
We reasoned that combining mutations disrupting both the Arf1 interaction and the membrane interface should result in a complete loss of membrane-binding. Mutations disrupting the Arf1 interaction (Chs5-T92Y and Bch1-N692I) reduced the levels of exomer binding by approximately 50% (Figure 4C,D). Similarly, the site 1+2 mutation in Bch1 also resulted in a marked decrease in membrane binding (Figure 4C,D). However, when the site 1+2 mutation was combined with a single Arf1-interface mutation (Bch1-N692I), binding to membranes was completely abrogated (Figure 4C,D).
Strength of interactions in vitro correlates with function in vivo
In order to determine the impact of these mutations on recruitment of exomer to the TGN in vivo, we monitored the localization of both GFP-Chs5 and Bch1-mCherry in a strain lacking all of the exomer subunits except for the Bch2 ChAP (chs5Δ bch1Δ bud7Δ chs6Δ). Both Chs5 and Bch1 localized to the TGN in this strain, as expected (Figure 4E) (Trautwein et al., 2006). Mutation of either the membrane-binding interface (site 1+2) or the Bch1-Arf1 interface (N692I) caused a slight mislocalization of Bch1 to the cytoplasm, which was exacerbated by combining these two mutations (Figure 4E). GFP-Chs5 localization remained normal in these mutants, likely due to its ability to bind Arf1. However, combining mutations in the Chs5 FBE domain (T92Y), the Bch1-Arf1 interface (N692I) and the membrane binding interface (site 1+2) resulted in complete mislocalization of Bch1 to the cytoplasm and significant mislocalization of Chs5 (Figure 4E). These results correlate well with the in vitro biochemical data and provide further support for the role of multiple interfaces in recruitment of exomer to the TGN.
To determine the effect of disrupting each interaction interface on exomer function, we monitored growth of mutant cells on media containing calcofluor white. We used a yeast strain (chs6Δ bch1Δ bud7Δ cells) that requires wild-type copies of both Chs6 and Bch1 to be added on plasmids in order to assemble a functional exomer complex capable of trafficking Chs3. Alone, neither single site mutation (site 1 or site 2) nor the Arf1-interface mutation (N692I) in Bch1 disrupted exomer function (Figure 5, top panel). However, the Bch1 site 1+2 mutant exhibited resistance to growth on the toxin, indicating disruption of Chs3 trafficking. The site 1+2 mutant version of Chs6 exhibited only modest resistance, but robust resistance was observed when this membrane binding mutation of Chs6 was combined with any of the single Bch1 mutants (Figure 5, bottom panel), indicative of additive effects. Thus, the exomer interfaces identified by the Arf1-exomer crystal structure are relevant in vivo, and the extent of exomer function correlates well with its ability to be recruited to membranes in vitro.
Figure 5. Exomer function correlates with its ability to be recruited to membranes in vitro.
Plasmids expressing Chs6-GFP and Bch1-mCherry constructs were introduced into a chs6Δbch1Δbud7Δ strain. Bch1 interface mutations were introduced together with either wild-type Chs6 or the Chs6 site 1+2 mutant. Serial dilutions were spotted on the indicated media and imaged after 2 days at 30°C. Calcofluor was used at 100 μg/ml. Cells with disrupted Chs3 trafficking grow in the presence of calcofluor white.
Exomer amplifies Arf1 membrane remodeling activity
Small GTPases of the Arf family have been previously implicated in directly inducing membrane deformation through the action of an N-terminal amphipathic helix that inserts into the membrane (Lee et al., 2005; Lundmark et al. 2008; Krauss et al., 2008). Other proteins involved in vesicle biogenesis are known to influence membrane architecture and induce fission by inserting elements into the membrane (Wang et al., 2009; Campelo et al., 2010; Boucrot et al., 2012). In light of these observations, we were interested in using our newfound knowledge regarding how exomer interacts with membranes to determine what role, if any, exomer plays in membrane remodeling (defined as any activity that shapes membranes or results in fission).
We employed a previously reported membrane vesiculation assay that uses differential sedimentation to observe protein-induced changes in liposome size due to fission (Boucrot et al., 2012). Using approximately physiological protein concentrations and liposomes derived from Folch lipids, we found that activated Arf1 increased the amount of lipids remaining in the supernatant, consistent with a reduction in liposome size due to vesiculation (Figure 6A,C). Strikingly, combining activated Arf1 with the Chs5/Bch1 exomer complex induced significantly more lipids to remain in the supernatant (Figure 6A,C). This effect was nucleotide-dependent, as the amount of lipids in the supernatant was not significant in reactions containing GDP-bound Arf1, with or without exomer. These results indicate the Arf1/exomer complex remodels membranes in vitro.
Figure 6. Exomer amplifies Arf1 membrane remodeling activity.
A) Liposome vesiculation assay to examine membrane remodeling by Arf1 and exomer. Substrate liposomes sediment to the pellet (P). Smaller liposomes generated by membrane fission remain in the supernatant (S). The Chs5(1-299)/Bch1 exomer complex was used in these experiments. GTP* = GMPPNP. Molar ratios of components are indicated.
B) Similar to (A), but comparing Chs5(1-299)/Bch1, the hydrophobic mutation in Chs5(1-299)/Bch1 (Φ), and Chs5(1-299)/Chs6.
C) Quantification of vesiculation in (A) and (B). n = 3.
D) Multiple sequence alignment showing conservation and secondary structure of the region surrounding the Bch1 hydrophobic element. The IWF motif sequence is conserved in Bch1 across multiple species as well as with the homologous ChAP Bud7. The more distantly related ChAPs (Bch2 and Chs6) do not possess this motif.
E) Superposition of the membrane proximal regions of Chs6 (gray) and Bch1 (blue). Residues included in the multiple sequence alignment in (E) are darkened. The hydrophobic membrane insertion motif is colored pink. This loop in Bch1 inhabits a position occupied by two helices that are present in Chs6 but absent from the Bch1 protein. This loop is flexible in solution, as its electron density is visible in only one of four copies in the ASU.
F) Structural view of the Bch1 hydrophobic element modeled adjacent to a membrane. Bch1 is colored blue, with the IWF sequence colored pink. Lipids are colored orange and Arf1 is yellow. The N-terminus of ΔN17-Arf1 is noted.
G) Titration of Chs5/Bch1 exomer complex in the vesiculation reactions. At the highest exomer concentration, the exomer:lipid ratio is 1:25. Quantification is noted at the bottom of the gel.
H) Titration of lipids in the vesiculation reactions.
I) Quantification of (H).
J) Similar to (A) and (B), with higher protein:lipid ratio.
K) Quantification of (J).
See also Figure S4.
We found that the Chs5/Chs6 exomer complex, in contrast to the Chs5/Bch1 complex, had no observable effect on the amount of lipids in the supernatant of vesiculation reactions when combined with activated Arf1 (Figure 6B,C). We hypothesized that differences between the Bch1 and Chs6 membrane-binding surfaces might account for this behavior. Comparison of these surfaces revealed the existence of a conserved hydrophobic patch of residues present in Bch1 and the related ChAP Bud7, but absent from Chs6 and its related ChAP Bch2 (Figure 6D,E, S4A). Given its hydrophobicity and proximity to the membrane surface (Figure 6F, S4A), we hypothesized that this region can insert into the outer leaflet of the lipid bilayer, acting as a membrane insertion element, or “wedge” to remodel membranes. Reducing the hydrophobicity of this element in Bch1 by an alanine substitution mutation (I122A, W123A, F124A: Φ) had no significant effect on the membrane affinity of the Chs5/Bch1 exomer complex (Figure S4B,C). However, this mutation significantly decreased membrane-remodeling activity (Figure 6B,C). These results indicate the hydrophobic element in Bch1 plays a critical role in deforming membranes, likely via membrane insertion. This may explain why either Bch1 or Bud7 is required, in addition to Chs6, for Chs3 trafficking (Sanchajate and Schekman, 2006, Trautwein et al., 2006).
By titrating the amount of liposomes and exomer in the reactions, we found that vesiculation was more efficient at higher concentrations of exomer (Figure 6G), and if the lipid to exomer molar ratio was 25:1 (Figures 6H,I, and S4D). Under these conditions, the surface of liposomes should be densely packed with the exomer/Arf1 complex, implying that efficient vesiculation requires protein coating of the membrane surface. At high protein density, the hydrophobic element played a significant role in vesiculation, but was no longer essential for activity (Figure 6J,K). We note that a significant amount of exomer-induced vesiculation was observed at much lower protein density (Figure 6B-C), suggesting the potential for local cooperativity of protein complexes.
Under basal conditions, the hydrophobic element was dispensable for exomer function in vivo (Figure 5, upper panel). However, when this element was disrupted in a sensitized background (Chs6 site 1+2 mutant), exomer function was completely abolished (Figure 5, lower panel). As the in vitro binding experiments indicate that the hydrophobic element is not required for membrane binding, this element is likely to play an important role in membrane remodeling in vivo.
We tested the importance of lipid composition by comparing reactions with membranes derived from Folch lipids to reactions with membranes derived from a TGN-like lipid mix. The Folch membranes, which are commonly used in studies examining membrane-remodeling activity, supported a more robust remodeling activity than did the TGN-like membranes (Figure S4E). This result highlights the importance of lipid-modifying enzymes acting at sites of TGN vesicle budding (Malhotra & Campelo, 2011; Xu et al., 2013), which may create a localized lipid environment that is distinct from the bulk of the TGN. This idea is supported by the finding that sphingolipids and sterols are enriched in secretory vesicles relative to the TGN (Klemm et al., 2009).
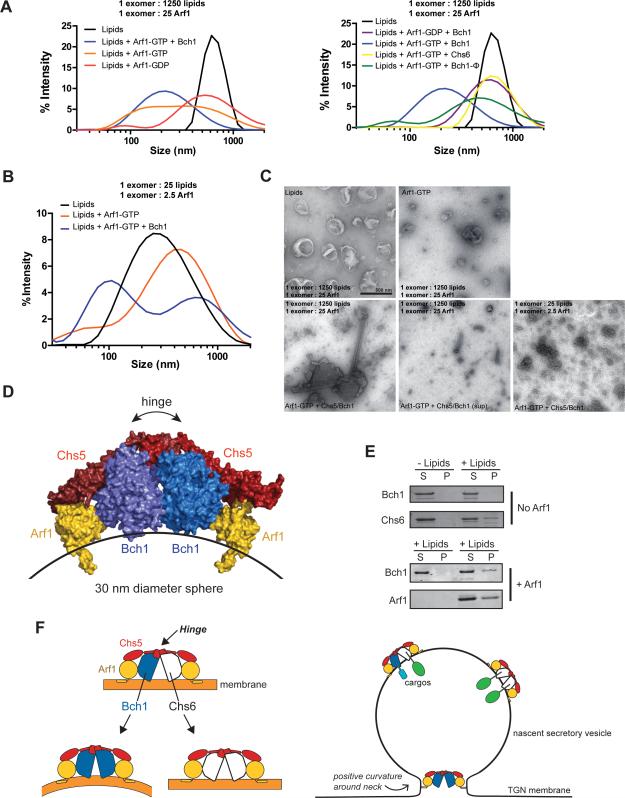
To further characterize the membrane remodeling activity of exomer, we performed dynamic light scattering (DLS) to quantitatively measure the sizes of liposomes derived from the vesiculation reactions performed under both physiological and high protein density conditions. Incubation of liposomes with Arf1-GDP, with or without the Chs5/Bch1 complex, resulted in liposome populations with a size range similar to the untreated control, albeit with a broader size distribution (Figure 7A). Activated Arf1 remodeled membranes to generate a subpopulation of smaller liposomes, consistent with our observations by SDS-PAGE. Combining activated Arf1 and the Chs5/Bch1 complex resulted in a greater percentage of these small liposomes (Figure 7A), indicative of enhanced vesiculation activity. At high protein density, this effect was even more pronounced (Figure 7B).
Figure 7. Characterization of exomer membrane remodeling activity.
A) Vesiculation reactions were performed as in Fig. 6A,B, but analyzed by DLS. Samples were separated into two different graphs for simplicity; they constitute an example of a single experiment.
B) DLS analysis of reactions similar to Fig. 6J.
C) Representative negative stain transmission electron micrographs of vesiculation reactions. Scale bar, 500 nm.
D) Structural model of the exomer/Arf1 complex generated by normal model analysis. The Chs5 hinge would allow the complex to bind to a highly curved membrane.
E) Pelleting of sucrose-laden 30 nm liposomes demonstrates exomer complexes bind to highly curved membranes.
F) Model for the dual functions of exomer in cargo sorting and membrane remodeling. Due to differences in membrane-remodeling capacity, we envision distinct exomer complexes may be responsible for cargo sorting (bound to cargo within the nascent vesicle) and vesicle fission (positioned at the constrained neck).
See also Figure S5.
Neither the Chs5/Chs6 complex nor the Chs5/Bch1 hydrophobic mutant enhanced the Arf1 vesiculation effect, as analyzed by DLS (Figure 7A). These results correlate well with the SDS gel analysis, and further support the notion that the hydrophobic element in Bch1 plays an important role in exomer-dependent membrane remodeling.
To gain insight into the nature of the products generated by exomer membrane remodeling, we imaged the vesiculation reactions by negative-stain electron microscopy. Strikingly, in reactions with activated Arf1 and Chs5/Bch1 we observed lipid tubules (Figures 7C and S5A,B). Although somewhat rare, such tubules were not observed in reactions containing activated Arf1 without exomer. At higher protein densities, clusters of small liposomes were observed. Quantification of electron micrographs of the supernatant of Arf1/Chs5/Bch1 vesiculation reactions correlates well with the DLS analysis (Figure S5C). Thus, the cooperative membrane remodeling activity of Arf1 and the Chs5/Bch1 exomer complex can produce liposomes of the same approximate size as endogenous secretory vesicles (Novick and Schekman, 1979).
Fission reactions require a transition through a highly curved (~20 nm diameter) tubular intermediate, which is the “neck” of a budding vesicle (Kozlovsky and Kozlov, 2003; Agrawal et al., 2010). Although the static Arf1/exomer crystal structure appears unsuited for this geometry, the Chs5 N-terminus acts as a molecular hinge, allowing for a constrained range of flexible motion (Richardson and Fromme, 2013). We used normal mode analysis to determine a plausible structural model of the Arf1/exomer complex bound to a highly curved membrane. Without any dramatic rearrangements, flexing of the hinge would allow the complex to fit comfortably on the surface of a 30 nm diameter membrane tubule (Figure 7D). We further confirmed that exomer binds to 30 nm diameter liposomes in vitro (Figures 7E, S5D). Therefore, we propose that the exomer/Arf1 complex directly catalyzes fission by organizing membrane insertion elements in close proximity to a budding vesicle neck (Figure 7F).
Discussion
Understanding how cargos are sorted into secretory vesicles is a fundamental question in cell biology. Although several proteins have been identified in mammalian cells as acting in TGN-to-PM transport (von Blume et al., 2011; Malhotra and Campelo, 2011; Wakana et al., 2012; Gehart et al., 2012; Cruz-Garcia et al., 2013), the only known cargo adaptor for secretory vesicle cargos in any eukaryotic cell is the Arf1-dependent exomer complex, which traffics approximately 1-5% of PM proteins in yeast. Vesicle formation involves two distinct processes that must be coordinated: cargo sorting and membrane remodeling. Exomer binding to cargo has been demonstrated, but the mechanism by which exomer binds and sculpts the membrane is unknown. In this study, we determined the molecular mechanism for exomer membrane recruitment. Arf1 interacts with the ChAP subunit via its switch regions, and interacts with the FBE domain of Chs5 through a non-switch region near the nucleotide-binding pocket. We envision the Arf1-ChAP interaction acting as the initial sensor for activated Arf1, while the Chs5 FBE domain acts as a stabilizer, to maintain the complex on the membrane surface in order to facilitate interactions of the ChAP subunits with cargo.
We provide evidence that exomer plays a direct role in remodeling membranes in cooperation with Arf1, therefore enabling exomer to couple cargo sorting with vesicle fission. Previous studies have implicated Arf1 in membrane remodeling (Lundmark et al., 2008; Krauss et al. 2008; Boucrot et al. 2012), but did not investigate the role of cargo sorting machinery. Our findings suggest that exomer, both by providing its own membrane insertion element and by coordinating the membrane insertion elements of two Arf1 molecules, can drive fission. Exomer may enforce orientations of these insertion elements, thereby amplifiying the intrinsic membrane remodeling activity of Arf1.
The protein dynamin drives fission of endocytic vesicles (Kosaka and Ikeda, 1983; Hinshaw and Schmid, 1995; Ferguson and De Camilli, 2012), and a recent report proposed that the flexibility of the dynamin PH domain relative to the membrane surface was a criticial component of dynamin-induced fission (Shnyrova et al., 2013). We speculate that the exomer hinge motion is also critical for exomer function in secretory vesicle fission, allowing for dynamic positioning of membrane insertion elements during vesicle biogenesis. Given the difficulty in generating mutants that specifically affect the flexibility of the hinge motion, future work will be needed to test this idea.
A previous study found that Arf1 and exomer were not sufficient to induce membrane curvature, but a heterogeneous mixture of ChAP subunits was tested at different concentrations than we utilized (Wang et al., 2006). We found that exomer membrane remodeling activity strongly depends upon a membrane-proximal hydrophobic element that is found in only a subset of the ChAPs (i.e. Bch1), as well as the lipid composition and lipid/protein ratios used. In vivo, exomer likely consists of heterotetramers with mixed, but defined, ChAP composition (Sanchajate and Schekman, 2006; Trautwein et al., 2006; Paczkowski et al., 2012). We were unable to test the membrane remodeling activity of mixed heterotetramers because we found that subunits within an exomer complex are highly dynamic: a purified Chs52/Bch1/Chs6 complex equilibrates into a mixed population consisting of Chs52/Chs62, Chs52/Bch12, and Chs52/Bch1/Chs6 complexes (data not shown). Perhaps the membrane remodeling activity of exomer in vivo is regulated in part by the particular composition of exomer complexes engaged in vesicle biogenesis (Figure 7F).
Analysis of the Bch1 mutant with a mutated hydrophobic element suggested this element was dispensable in vivo. However, this mutant retained a reduced capacity to remodel membranes with Arf1. Furthermore, the element was required for exomer function in a sensitized background in which the membrane-binding interface of exomer was weakened. Our interpretation of these results is that the hydrophobic element contributes to membrane remodeling in vivo. This interpretation may explain a previous observation that proper Chs3 trafficking requires either Bch1 or Bud7 (Sanchajate and Schekman, 2006, Trautwein et al., 2006). The distinct properties of the Bch1/Bud7 ChAPs may be generally required for exomer trafficking.
In conclusion, we propose that exomer directs membrane remodeling both through the action of its own novel membrane insertion motif and by precisely positioning Arf1 membrane insertion elements in a particular geometry. Exomer and Arf1 therefore represent a minimal machinery for vesicle biogenesis.
Experimental Procedures
Plasmids and strains
See Supplemental Experimental Procedures for a list of plasmids and yeast strains used.
Microscopy
Cells were grown in synthetic dropout media and imaged in log phase (OD600 ~ 0.5). Live cells were imaged at room temperature on a DeltaVision RT wide-field deconvolution microscope (Applied Precision).
Protein expression and purification
Recombinant exomer was purified as described (Paczkowski et al., 2012). ΔN17-Arf1-Q71L Arf1 was purified as a TEV protease-cleavable 6xHis-tag fusion. Recombinant myristolylated yeast Arf1 was purified as described (Ha et al., 2005).
Crystallization and structure determination
The Arf1-Chs5/Bch1 complex was crystallized by the hanging drop vapor diffusion method. The crystals belong to the P21 space group with unit cell dimensions a = 115 Å, b = 208 Å and c = 155 Å. Diffraction data were processed using HKL-2000 (Otwinowski and Minor, 1997). The structure was solved using Phaser in Phenix (Adams et al., 2010) by molecular replacement with the structures of Bch1 and ΔN17-Arf1-Q71L as search models. Due to the modest resolution of the data, we used density modification to reduce bias in electron density maps used for model building. Model building was performed using Coot (Emsley et al., 2010). Refinement was performed in Phenix. Structural images were generated with PyMOL. Throughout this work, when describing specific interactions we provide images of the best-resolved example present in the asymmetric unit. Specific chains used for each figure are reported in Supplemental Experimental Procedures.
Normal mode analysis
Calculation of the normal modes of the exomer/Arf1 complex was performed similar to our previous report (Richardson and Fromme, 2013), using the elNemo server (Suhre and Sanejouand, 2004). The lowest frequency non-trivial normal mode was used for the model shown in Figure 7D.
Preparation of synthetic liposomes
Liposomes were prepared as described (Paczkowski et al., 2012, Richardson et al., 2012). Each figure panel and corresponding quantification represents experiments done with a single batch of liposomes for consistency.
Liposome pelleting and vesiculation assays
Liposome pelleting assays and quantification of results were performed essentially as previously described, using TGN-like lipids (Paczkowski et al., 2012). The vesiculation assays were performed similarly to a previous report, using Folch lipids (Boucrot et al., 2012). Samples were analyzed by SDS-PAGE or DLS. Due to differences in lipid composition and protein concentration, no detectable vesiculation occurs under the conditions of the pelleting assays.
Membrane vesiculation assay
This assay was performed similarly to a previous report (Boucrot et al., 2012). For “physiological” conditions, 10 μg Folch liposomes (250 μM) were incubated with 4 μg myristoylated Arf1 (5 μM), 625 μM EDTA, and 125 μM GMPPNP or GDP for 10 min at room temperature. We note that previous studies demonstrating a more drastic effect of Arf1 utilized higher concentrations (10 μM) of the protein. MgCl2 was added to 2.5 mM and incubation continued for 5 min at room temperature. 1 μg exomer was added (200 nM), followed by a 10 min incubation. For the higher protein density conditions, 2 μg Folch liposomes (50 μM) and 10 μg exomer (2 μM) were used, and the amounts of other components were unchanged. Samples were spun at 20°C for 15 min at 132000 g. The supernatant and pellet fractions were subjected to SDS-PAGE. The gel was stained with a Coomassie solution (lacking methanol) before scanning on an Odyssey imager (Li-COR). Lipid band intensities were measured using ImageJ to determine the relative amounts of lipids in the supernatant and pellet.
Dynamic light scattering (DLS)
Samples were taken from the vesiculation assays prior to centrifugation, or from the supernatant fraction as noted. The average liposome diameter was determined by DLS using a Malvern Zetasizer Nano ZPS using a He-Ne laser (633nm) at 20° C. Three rounds of measurements were taken of each sample to ensure accuracy. Data was processed by Zetasizer software and the percent intensity of the different size populations was determined.
Statistical tests
Significance was determined by one-way ANOVA with Tukey's test for multiple comparisons (when comparing three or more groups), or by an unpaired T-test (when comparing two groups).
Supplementary Material
Acknowledgements
We thank the laboratories of S. Emr and B. Brown for use of their microscopes, as well as to C. Stefan, A. Manford, M. Henne, and N. Buchkovich for assistance with microscopy. We thank J. Bonifacino for the VHS-GAT-Gga1 plasmid. We thank B. Richardson for plasmids and expertise. We thank J. Chang for strain and plasmid construction. R. Kahn created the E. coli strain harboring Arf1/Nmt1. We thank D. Costello and the lab of S. Daniel for assistance with DLS. We thank H. Sondermann, J. Chappie, M. Henne, and B. Richardson for critical reading of the manuscript. We received expert advice from M. Szebenyi at CHESS, which is supported under NSF award DMR0936384, and NIH award GM103485. This work was supported by startup funds to JCF from Cornell University, by a NIH Training Grant (T32GM07273) award to JEP, and by NIH grant R01GM098621.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
Coordinates and structure factors for the Arf1/Exomer complex have been deposited in the Protein Data Bank (www.rcsb.org) under accession number 4Q66.
See also Supplemental Experimental Procedures.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Ricardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal NJ, Nukpezah J, Radhakrishnan R. Minimal mesoscale model for protein-mediate vesiculation in clathrin-dependent endocytosis. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000926. doi: 10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F, Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol. 2006;22:439–455. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]
- Barfield RM, Fromme JC, Schekman R. The exomer coat complex transports Fus1p to the plasma membrane via a novel plasma membrane sorting signal in yeast. Mol Biol Cell. 2009;20:4985–4996. doi: 10.1091/mbc.E09-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of the Sec23-Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPIII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Lippincott-Schwartz J. Coat proteins: shaping membrane transport. Nat Rev Mol Cell Biol. 2003;4:409–414. doi: 10.1038/nrm1099. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS. Adaptor proteins involved in polarized sorting. J. Cell Biol. 2014;204:7–17. doi: 10.1083/jcb.201310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Çamdere G, Liska N, Evergren E, McMahon HT, Kozlov MM. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F, Fabrikant G, McMahon HT, Kozlov MM. Modeling membrane shaping by proteins: focus on EHD2 and N-BAR domains. FEBS Letters. 2010;584:1830–1839. doi: 10.1016/j.febslet.2009.10.023. [DOI] [PubMed] [Google Scholar]
- Campelo F, Malhotra V. Membrane fission: the biogenesis of transport carriers. Annu Rev Biochem. 2012;81:407–427. doi: 10.1146/annurev-biochem-051710-094912. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP 2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia D, Ortega-Bellido M, Scarpa M, Villeneuve J, Jovic M, Porzner M, Balla T, Seufferlein T, Malhotra V. Recruitment of artaptins to the trans-Golgi network by PI(4)P and their involvement in cargo export. EMBO J. 2013;32:1717–1729. doi: 10.1038/emboj.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Letters. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev Cell. 2006;10:329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. Dynamin, a membrane-remodeling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehart H, Goginashvill A, Beck R, Morvan J, Erbs E, Formentini I, De Matteis MA, Schwab Y, Wieland FT, Ricci R. The BAR domain protein Arfaptin-1 controls secretory granule biogenesis at the trans-Golgi network. Dev Cell. 2012;23:756–768. doi: 10.1016/j.devcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Ha VL, Thomas GM, Stauffer S, Randazzo PA. Preparation of myristoylated Arf1 and Arf6. Methods Enzymol. 2005;404:164–174. doi: 10.1016/S0076-6879(05)04016-4. [DOI] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller H, Schaefer M, Schulten K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal phases. J Phys Chem. 1993;97:8343–8360. [Google Scholar]
- Herth W, Schnepf E. The fluorochrome, calcofluor white, binds to structural polysaccharide fibrils. Protoplasma. 1980;105:129–133. [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Ismail SA, Vetter IR, Sot B, Wittinghofer A. The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism. Cell. 2010;141:812–821. doi: 10.1016/j.cell.2010.03.051. [DOI] [PubMed] [Google Scholar]
- Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James KC, Collins BM, Honing S, Evans PR, Owen DJ. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Ménétrey J. Structural biology of Arf and Rab GTPases’ effector recruitment and specificity. Structure. 2013;21:1284–1297. doi: 10.1016/j.str.2013.06.016. [DOI] [PubMed] [Google Scholar]
- Klemm RW, Ejsing CS, Surma MA, Kaiser HJ, Gerl MJ, Sampaio JL, de Robillard Q, Ferguson C, Proszynski TJ, Shevchenko A, Simons K. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Reversible blockage of membrane retrieval and endocytosis in the garland cell of temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol. 1983;93:499–507. doi: 10.1083/jcb.97.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky Y, Kozlov MM. Membrane fission: model for intermediate structures. Biophys J. 2003;85:85–96. doi: 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, Jia JY, Roux A, Beck R, Wieland FT, De Camilli P, Haucke V. Arf1-GTP-induced tubule formation suggests a function of rf1 family proteins in curvature acquisition at sites of vesicle budding. J Biol Chem. 2008;41:27717–23. doi: 10.1074/jbc.M804528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf-GTP. Nat Struc Mol Biol. 2010;17:876–881. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun Y, Oster GF, Drubin DG. Mechanochemical crosstalk during endocytic vesicle formation. Curr Opin Cell Biol. 2010;22:36–43. doi: 10.1016/j.ceb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R, Dohert GJ, Vallis Y, Peter BJ, McMahon HT. Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J. 2008;414:189–194. doi: 10.1042/BJ20081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Campelo F. PKD regulates membrane fission to generate TGN to cell surface transport carriers. Cold Spring Harb Perspect Biol. 2011;3:1–9. doi: 10.1101/cshperspect.a005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature sensitive mutant of Saccharomyces cerevisiae. PNAS. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Noble ME, Hunter JB, Dafforn TR, Evans PR, McMahon HT. A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell. 1999;97:805–815. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- Paczkowski JE, Richardson BC, Strassner AM, Fromme JC. The exomer cargo adaptor structure reveals a novel GTPase-binding domain. EMBO J. 2012;31:4191–4203. doi: 10.1038/emboj.2012.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Faria GG, Canagarajah BJ, Bonifacino JS, Hurley JH. Stuctural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell. 2013;152:755–767. doi: 10.1016/j.cell.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BC, McDonold CM, Fromme JC. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev Cell. 2012;22:799–810. doi: 10.1016/j.devcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BC, Fromme JC. The exomer cargo adaptor features a flexible hinge domain. Structure. 2013;21:486–492. doi: 10.1016/j.str.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockenbauch U, Ritz AM, Sacristan C, Roncero C, Spang A. The complex interactions of Chs5p, the ChAPs, and the cargo Chs3p. Mol Biol Cell. 2012;23:4402–4415. doi: 10.1091/mbc.E11-12-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchatjate S, Schekman R. Chs5/6 complex: a multiprotein complex that interacts with and conveys chitin synthase III from the trans-Golgi network to the cell surface. Mol Biol Cell. 2006;17:4157–4166. doi: 10.1091/mbc.E06-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdiviseo MH, Durán A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, Wakatsuki S. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol. 2003;10:386–393. doi: 10.1038/nsb920. [DOI] [PubMed] [Google Scholar]
- Shnyrova AV, Bashkirov PV, Akimov SA, Pucadyil TJ, Zimmerberg J, Schmid SL, Frolov VA. Geometric catalysis of membrane fission driven by flexible dynamin rings. Science. 2013;339:1433–1436. doi: 10.1126/science.1233920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Sanejouand Y-H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32(Web Server issue):W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein M, Schindler C, Gauss R, Dengjel J, Hartmann E, Spang A. Arf1p, Chs5p, and ther ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 2006;25:943–954. doi: 10.1038/sj.emboj.7601007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdiviseo MH, Mol PC, Shaw JA, Cabib E, Durán A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyce cerevisiae. J Cell Biol. 1991;114:101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J, Alleaume AM, Cantero-Recasens G, Curwin A, Carreras-Sureda A, Zimmermann T, van Galen J, Wakana Y, Valverde MA, Malhotra V. ADF/cofilin regulates secretory cargo sorting at the TGN via the Ca2+ ATPase SPCA1. Dev Cell. 2011;17:652–662. doi: 10.1016/j.devcel.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Wakana Y, van Galen J, Meissner F, Scarpa M, Polishchuk RS, Mann M, Malhotra V. A new class of carriers that transport selective cargo from the trans Golgi network to the cell surface. EMBO J. 2012;31:3976–3990. doi: 10.1038/emboj.2012.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Hamamoto S, Orci L, Schekman R. Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin.Syndapin. PNAS. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR. Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol. 2013;202:875–886. doi: 10.1083/jcb.201305094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Breitman M, Goldberg J. A structure-based mechanism for Arf1-dependent recruitment of coatomer to membranes. Cell. 2012;148:530–542. doi: 10.1016/j.cell.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.