Abstract
We elucidate the interaction between actin and specific membrane components, using real time live cell imaging, by delivering probes that enable access to components, that cannot be accessed genetically. We initially investigated the close interplay between Phosphatidylinositol 4,5-bisphosphate (PIP2) and the F-actin network. We show that, during the early stage of cell adhesion, PIP2 forms domains within the filopodia membrane. We studied these domains alongside cell spreading and observed that these very closely follow the actin tread-milling. We show that this mechanism is associated with an active transport of PIP2 rich organelles from the cell perinuclear area to the edge, along actin fibers. Finally, mapping other phospholipids and membrane components we observed that the PIP2 domains formation is correlated with sphingosine and cholesterol rafts.
Among all the different available techniques in molecular biology, fluorescence microscopy (FM) is by far one of the most used1. This technique, in fact, has the great advantage to obtain functional and structural information in a single experiment. Particularly, live cells imaging is becoming fundamental in the understanding of the dynamics of many biological processes. A typical strategy for carrying out live cell imaging include the use of fluorescent proteins (FP)2. The development of molecular genetics and engineering has allowed proteins manipulation and, consequently, the creation of fluorescent protein libraries. These proteins can be expressed within live cells in specific sub-cellular compartments. In addition to the intrinsic limitations associated with genetic manipulations, fluorescence proteins cannot be used for any cellular components that have a post-translational origin. This includes most glycols as well as phospholipids, sphingosines and sterols. The latter are essential molecules in the life of a cell, as they control compartmentalization, in cooperation with membrane proteins, leading to the formation of fluid membrane bounded structures3. It is now accepted that the plasma membrane has a specific regulatory role in several signaling pathways which is directly controlled by rearrangement into “raft” domains, that results from the fluctuations of local composition and membrane spontaneous curvature4,5,6,7,8. These micro- and nano- domains, composed of specific phospholipids and proteins9, have a central role in the regulation of many cellular functions such as signalling pathways, membrane shaping, cell motility and polarization4,8,10. Among the different membrane components, one of the most studied phospholipids is Phosphatidylinositol 4,5-bisphosphate (PIP2)11,12,13,14,15,16. PIP2 works as an anchoring points for several proteins whose function is to control membrane deformation.
These include several GTPases belonging to the Rho family (e.g., Rho, Rac and Cdc4215,17) as well as several actin and cytoskeleton regulators (e.g., ERM-proteins, Talin, WAVE/WASP, Gelsolin capping, ADF/Cofilin, Profilin and Twinfilin14,16,18). Although progress has been made to elucidate the different pathways and proteins involved in membrane/cytoskeleton interaction19, there are still lot of queries on (i) how PIP2 is transported along the F-actin polymerization sites, (ii) how the PIP2 membrane organization is related to the local lipid composition and (iii) how its function is controlled by other membrane components. Herein we show that we can access these important membrane components using effective intracellular delivery of fluorescently labelled phospholipids and actin probes. We have recently developed a nanotechnological platform to introduce probes within the cells without affecting their metabolic activity hence allowing live cell imaging. This is based on the use of synthetic vesicles (called polymersomes) formed by pH sensitive copolymers20,21,22,23. Here we use these to deliver, separately and simultaneously six different probes, two common phospholipids namely: 2-Decanoyl-1-(O-(11-(4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Propionyl)amino)Undecyl)-sn-Glycero-3-Phosphocholine (PC), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (PE), a cholesterol analogous: 22-(N-(7-Nitrobenz-2-Oxa-1,3-Diazol-4-yl)Amino)-23,24-Bisnor-5-Cholen-3β-Ol (CHOL), a sphingosine: N-((4-(4,4-difluoro-5-(2-thienyl)-4-bora-3a, 4a-diaza-s-indacene-3-yl)phenoxy)acetyl)sphingosine (S), as well as the inositol: TopFluor® phosphatidylinositol 4,5-bisphosphate (PIP2) and the peptide Phalloidin-ATTO647 (ACTIN) to stain F-actin. We first investigated the correlation between the cytoskeleton structures and the PIP2 membrane domains, to understand their role in cell adhesion and spreading processes. In particular, we explored the specific interactions between PIP2 membrane domains and the actin cytoskeleton, by means of real time imaging of living cells, during important processes such as cell adhesion and spreading. We complemented these studied by delivering three more membrane probes, in addition to PIP2 and ACTIN, specifically PC, CHOL and S. This has allowed us to study their specific distribution and reciprocal interconnections within the cell, and to explore their topological localization with the membrane, particularly at the cell edge where cell adhesion and spreading processes initiate.
Results
Validation of methodology
pH sensitive diblock copolymers poly(2-(methacryloyloxy)ethyl phosphorylcholine)-poly(2-(diisopropylamino)ethyl methacrylate) (PMPC-PDPA) are used to form polymersomes. The PMPC and the PDPA block convey two important functions to the final vector: (i) The hydrophilic PMPC enables the polymersome to interact with endocytosis related receptors and to facilitate the nanoscopic vesicle internalisation in several cell types24, (ii) the pH-sensitive (PDPA) (with a pKa of 6.5 under physiological conditions) allows a controlled release of the cargo (upon acidification) into the endosomes. Once internalised, the natural acidification in the endocytic pathway rapidly overcomes the pKa of the copolymer, forcing the polymersomes to dissociate into single chains. This causes an increase in the osmotic pressure within the early endosomes, which is compensated by the opening of temporary pores in their membrane, thus resulting in a local release of the cargo23,25. Polymersomes, being vesicles, have the ability to encapsulate hydrophobic and amphiphilic molecules within their membrane, as well as hydrophilic molecules within their aqueous lumen20,21,22. In addition, polymersomes possess intrinsic physicochemical characteristics, including high flexibility and stability, which make them ideal candidates for a broad range of bio-applications, ranging from drug delivery to diagnostics26,27. In order to establish the appropriate experimental methodology (see the material and methods section for more details) to introduce the relevant probes we have optimized the encapsulation of several fluorescent molecules (Figure S1) and we studied their effect on the cell metabolic activity using a MTT assay. As shown in Figure S2 all the formulations did not adversely affect cell viability at the concentration used.
In addition to the MTT assay, for the ACTIN probe, we have also tested whether the delivery of Phalloidin affect F-actin polymerization and consequently cell mobility. As shown in Figure S2D and Figure S3, the concentration used is sufficient to visualize F-actin but does not effect the cell motility as tested by a scratching assay.
In order to evaluate that the membrane distribution of the lipid probes is not affected by the fluorophore modification, we co-delivered two different PC lipids modified either by a 4-difluoro-4-bora-3a, 4a-diaza-s-indacene or by a pyrene. As shown in Figure S7, the two molecules have an almost identical cellular distribution, with overlap coefficient close to 1. This suggests that the actual fluorophore does not affect the bioactivity of the lipid and it consequent trafficking. Similarly, we studied the distribution and interaction in live cells, of the ACTIN probe and PIP2, in the presence of the compound CK-869. This is a blocker for F-actin assembly by inhibiting the actin-related Arp2/3 complex, that results in an actin accumulation at the cellular edges28. NIH-3T3 cells were first incubated, with polymersomes carrying PIP2 and ACTIN for 24 h. Confocal micrographs were processed and analyzed in detail to quantify the average percentage overlap signal values in both the central cell area and the periphery of each cell. In this case, the average overlap percentage value was found to be >85% for the peripheral and central areas of untreated cells, and >80% (again for both the areas) in the Arp 2/3 inhibited population (Figure S4 of supporting information). We confirmed this by repeating the same experiment with another lipid, PE which is known not to have specific interaction with the F-actin structures. The results are reported in Figure S5 of supporting information. In this case, non-inhibited cells displayed an average-overlap value, between the two channels (PE - ACTIN), less than 45% in the central area, and close to 30% at the periphery of cell. On the other hand, cells incubated with CK-869 presented a significant increase in the overlap value just in the central area (>65%). This result is probably due to an apoptotic process induced by the drug, which results in the intracellular vesicular accumulation of all the delivered probes. At the same time, the calculated overlap between ACTIN and PIP2 is similar to the values found in the previous analysis (see supporting information, Figure S4–5).
To further clarify any doubts regarding the natural distribution of the delivered PIP2, we performed another control experiment. Cells were first transfected with a CFP-PLCdelta-PH pDNA (see supporting information) and after that treated as previously described with PIP2 probe. The CFP-PLCdelta-PH construct encodes the pleckstrin homology (PH) domains of protein lipase C delta, fused with a fluorescent protein, that is reported to specifically interact and bind to PIP229,30. The results of this analysis are shown in Figure S6 and they present an average overlap value between CFP-PLCdelta-PH and PIP2 of >80%, thus further corroborating our findings. Most interestingly the majority of the overlap is localized at the plasma membrane where the lipase binds to PIP2. However, our lipid based approach allows also to visualize all the internal PIP2 rich organelles which are not accessible by the PH domain.
Moreover, to fully demonstrate that the cell membrane distribution of the delivered lipid probes was not driven by an excessive overexpression of the endogenous composition, a mathematical analysis was performed. The results of these analyses are reported in Table S1 of supporting information. From this calculation, we derived that the average contribution of the total concentration per singular lipid specie is less than 1%. We can thus assume that such a small increment should cause no effect on the cell lipid homeostasis.
In addition we also reported a 3D cellular reconstruction and graphical localization analysis between PIP2 and F-actin in Figure S8 of supporting information. We believe that this supports the evidence that PIP2 localizes naturally into clusters in the membrane. In fact, from this analysis it is possible to observe that PIP2 clusters are mostly localized and the bottom level of the cell membrane, and prevalently at the cell protrusions.
Finally, to verify the natural distribution of our polymersome-delivered probes, we investigated the sub-cellular localization of PC.
This phospholipid is well establish as dominant constituent of the endoplasmic reticulum (ER) membrane31. For this reason the evaluation of this compound after its polymersome-mediated intracellular delivery, can be adopted as a further validation regarding the natural distribution of the probes at the cell membrane level. Figure S9 of supporting information shows the obtained results. From this data, it is possible to observe a considerable accumulation of this lipid at the level of the intracellular reticular network. Further analysis identified this network as the ER compartment of the cell and proved also, in this way, the possibility to use our probes as reliable system to map out the different lipid compositions.
PIP2 and F-actin interactions during cell spreading
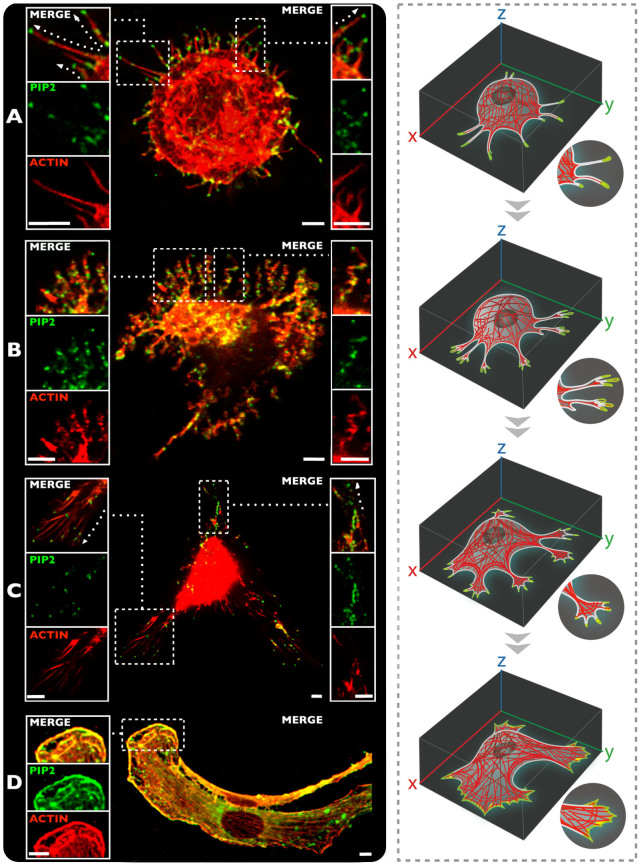
Figure 1 shows confocal micrographs of representative live cells labelled with PIP2 (green) and ACTIN (red) undergoing adhesion, showing: the early stages (Figure 1A–B), focal adhesion point formation (Figure 1C), and cellular spreading (Figure 1D). It is interesting to observe the different time-resolved PIP2 localization within the cell, and its interconnection within the actin structures. A detailed evaluation of this complex and dynamic process confirmed that PIP2 plays a pivotal role in driving and controlling the actin directionality. The representative picture of Figure 1A show a typical rounded cell with many filopodia distributing radially to sense the environment. In these, the PIP2 cluster forming an average of 2 to 3 domains per filopodia. As shown in the supporting movie (see movie SM1), these protrusions are still mobile and not committed to a specific substrate site and act as sensing pods. Our live cell imaging approach shows that PIP2 clusters are not only present on the filopodia tips but also alongside the tube in sites not yet attached to the substrate. As the cell undergoes more adhesion, (see Figure 1B) the filopodia stop the sensing and start to attach to the substrate (see movie SM2). As shown in Figure 1B, the PIP2 is still clustered into defined domains that appears to be associated with the filopodia kinks. Gradually the filopodia merge and start forming lamellapodia (Figure 1C) and likewise the PIP2 domains forming larger domains. These, ultimately, as the cell is fully spread, are almost homogeneously distributed alongside the actin strew fibers. These results suggest that PIP2, as expected, acts together with the cell sensing machinery, which feels the proximity of a surface, and guides the cell in that direction in order to approach the substrate and to develop the adhesion events. These results also strongly suggest an active transport and accumulation of this molecule toward the cellular protrusions (Figure 1B–C).
Figure 1. Live cell imaging of different phases during the cell adhesion and spreading process.
(A) Initial stage of cell adhesion and intracellular distribution and organization between ACTIN and PIP2 (10 min). (B) Translocation of PIP2 domains towards the cell protrusions (30 min). (C) Cell spreading process and PIP2 nano-domains accumulations in the focal adhesion points, followed by actin fibers (1 h). (D) PIP2 and ACTIN interactions during the cell spreading process (16 h). Scale bar: 5 μm.
PIP2 and actin: vesicular trafficking regulations
The data above suggests that PIP2 is somehow transported along the F-actin polymerization and the consequent cell spreading. This would suggest some sort of membrane composition control mechanism that orchestrates the cell adhesion and spreading process, in synergy with the actin tread-milling. In lower eukaryotes PIP2 transport is regulated by the vesicular trafficking from the cell interior to the cell protrusion to feed the extremity with relevant lipid signaling32. Exploiting our methodology to image live cells we investigated whether this vesicular trafficking mechanism is also present in high eukaryotes.
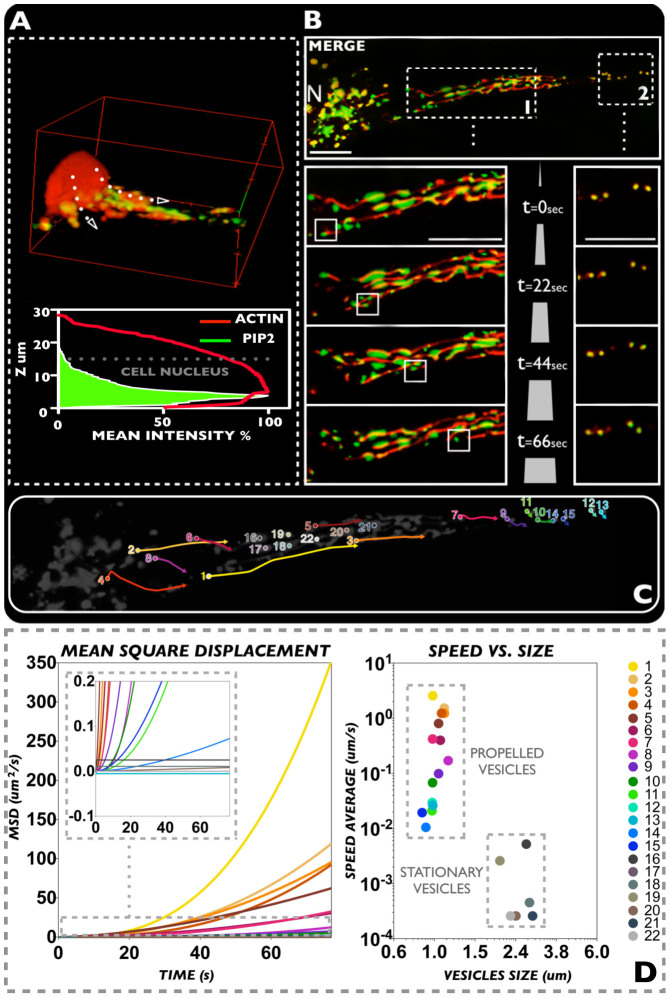
In Figure 2A we show a three dimensional reconstruction of a spreading fibroblast with labelled PIP2 (green) and ACTIN (red). The z-average distribution of the two probes shows that the lipid molecules distribute from the peri-nuclear area to the bottom and adhesive sections of the cell. We further investigated this distribution focusing on one single cell protrusion, tracking the movement of PIP2 and F-actin from the nucleus to the cell extremity. In Figure 2B a video rate imaging of a cell protrusion from the nuclear area to one extremity is imaged over 1.2 mins. As shown in all the time snapshots, the PIP2 is confined in vesicular organelles sitting on actin filaments that distribute along the cell protrusion. While a minority of PIP2 organelles remain still, most the others move along the filament and follow a specific direction from the nucleus to the cell edge (see Figure 2C). We further analyzed the time sequence plotting the mean square displacement (MSD) for 22 visible and trackable organelles. The MSD is the measure of the spatial extent of random motion and, when the tracked object moves according to a random walk, it changes linearly with time33. However, for anomalous diffusion processes such as sub-diffusional events and/or propelled objects, the MSD is no longer linear with time. If we use the generalized equation MSD = ktb, the exponent b can be used to classify the type of diffusion. As shown in figure 2D, most PIP2 organelles display non-linear MSD with exponent larger than 1, typical of propelled systems. This would suggest some form of active transport of the PIP2 organelles along the actin filament possibly involving myosin proteins34. Finally, we plotted the average speed for each organelle as a function of size showing two different distributions of small, motile vesicles and large, stationary organelles. Among the motile population, we measured speeds up to ~2 μm/s and within the range expected for actin/myosin propelled systems35.
Figure 2.
(A) Partial Z-stack cell reconstruction of PIP2 and ACTIN distribution. The latter has also been studied through a Z-stack versus mean intensity graphical reconstruction. (B) Live cell 2D imaging video (1.2 mins) of PIP2 and ACTIN distributions, showing the interaction between these two bio-molecules during vesicular trafficking process. (C) Scheme relative to the evaluated vesicles and the analyzed tracks. (D) Graphs expressing MSD and speed vs. size values resulting from the analyzed vesicles. Scale bar: 10 μm.
Membrane surface topology: lipids
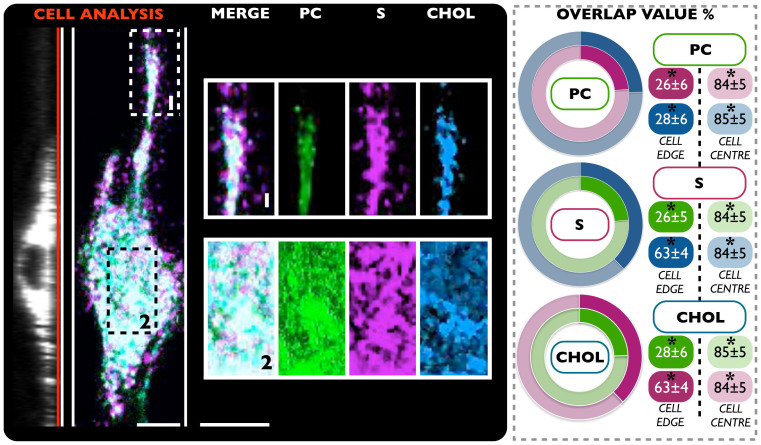
As previously shown in this work, the polymersome system enables delivery of more than one probe at the same time. This ability was therefore used to explore the relative distribution of multiple membrane components simultaneously, with a particular focus on lipid cluster domain organization and the interplay of these with the F-actin network. In this context, with the aim to visualize such processes, cells were initially treated with polymersomes loaded with three specific membrane probes: S (magenta), CHOL (blue) and PC (green) (Figure 3). This approach enabled the study of lipid raft distribution of these three compounds, within the cell membrane, by means of live cell imaging analysis. The result of fluorescence confocal laser scanning micrographs in live NIH-3T3 is shown in Figure 3. As easily recognizable, while the bottom nuclear region of cell mostly shows a homogenous lipids distributions, this organization appears radically changed at the peripheral area. On the cellular protrusion, in fact, the clusters of sphingosine appear displaced toward the edge. Here, a MATLAB®-based micro-domain overlap analysis has been carried out, to have a better indication about any possible differences regarding lipid-lipid interactions. The analysis between the three delivered compounds highlighted that cholesterol and sphingosine strongly interact (the overlap is 63 ± 4%) at the cellular edge, in contrast to the PC (overlap PC-CHOL = 28 ± 6% and PC-S = 26 ± 6%).
Figure 3. Confocal laser scanning micrograph.
The NIH-3T3 cell line were treated with three different polymersome samples: PC (green), S (magenta) and CHOL (blue). The results (average overlap value percent) show the dynamics and distribution of these biomolecules in a live environment. (*t-test, p-value > 0.05) No statistical differences between three independent experiments. Scale bar: 15 μm.
This data suggests the possibility to study the formation of rafts using labeled sphingosine and cholesterol. We then used this approach to study how these rafts and lipid clusters control the formation of PIP2 domains. We carried out an additional experiment where polymersomes loaded with CHOL, S and ACTIN were delivered. Moreover, cells were also treated with PIP2 probe to have a complete and simultaneous picture about the distribution and interconnection of PIP2 with both cholesterol and sphingosine membrane domains. Also in this case, the cells were analyzed with a bottom focus on both cell edge and centre. The obtained results in addition to the calculated overlap values of the signal of four molecules, are reported in Figure 4.
Figure 4. Confocal laser scanning micrograph.
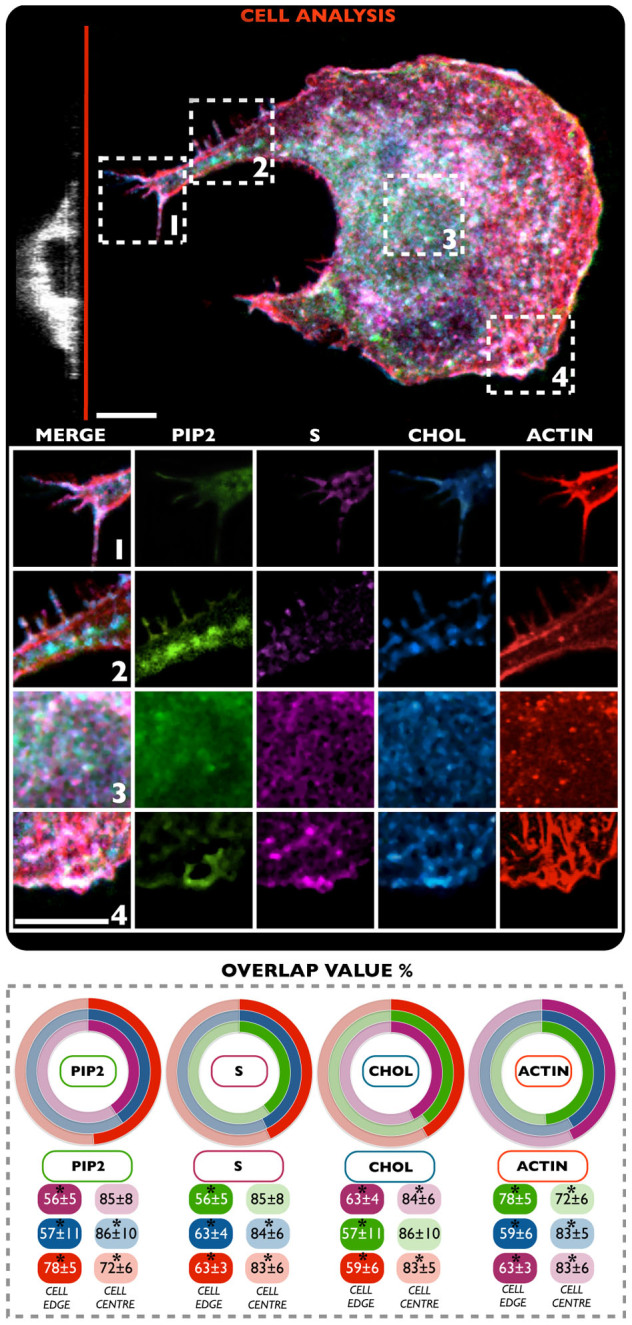
The NIH-3T3 cell line were treated with four different polymersome samples: S (magenta) and CHOL (blue), PIP2 (green) and ACTIN (red). The results (average overlap value percent) show the dynamics and distribution of four biomolecules in a live environment. (*t-test, p-value > 0.05) No statistical differences between three independent experiments. Scale bar: 10 μm.
As evident in Figure 4, this analysis further corroborates all the previous findings.
The percentage of overlap value per single channel, calculated with respect to the other three (graphs Figure 4), provides different information about the distribution of each delivered molecules. As reported before, the overlap value was calculated for each fluorescence channel, by using a MATLAB® -based micro-domain analysis. First, a strong overlap value between ACTIN and all the others channels: S (63 ± 3%), CHOL (59 ± 6%) and PIP2 (78 ± 5%) is evident at the cell edge. Second, and more importantly, the central area displayed an overlap value, for both S and CHOL, of 83 ± 5% with ACTIN. On the other hand, the overlap value of the PIP2 probe was slightly lower, i.e. 72 ± 6%. This is an additional confirmation of the importance of interactions occurring between PIP2 and F-actin structure, which are strongly promoted, especially at the cellular edge12,14. The same analysis was performed for the other delivered molecules, with the aim to understand the lipid raft distribution at the cell edge, and to correlate this with the active role played by actin structures in promoting/controlling it10. In this case, the cholesterol sample (CHOL) elicited a slightly higher interaction (about 63 ± 4% of signal overlap) with the sphingosine sample (S) at the cell edge, compared to the F-actin (ACTIN) (59 ± 6%) and PIP2 (57 ± 11%) probes. In the case of sphingosine, lower overlap values were found between S and the PIP2 channel (56 ± 5%). Interestingly, in this specific instance, an almost equal signal overlap between S and ACTIN, and S and CHOL was observed (about 63 ± 5% of overlap signal). This could be another indication on the importance and strong cooperation between this lipid with cholesterol and actin structures in stabilizing tubular structures, guiding cell adhesion and spreading processes10,36. Finally, the data on the delivered inositol reconfirms the strong and specific interaction with actin at the protrusions level (78 ± 5%), as well as with S (56 ± 5%) and CHOL (57 ± 11%) for the formation of lipid domains.
Discussion
A critical step towards shedding light on the complexity of cell function is the development of methodologies that enable access to important information with the minimal possible perturbation of the complex dynamics of cellular processes. Among all these, membrane dynamics is one the least studied, as most of its components cannot be genetically modified and thus studied by means of fluorescence proteins. Techniques based on small molecules can be applied to study these cell membranes constituents, but often their administration demands the use of cell-toxic solubilizing agents or highly detrimental membrane permeabilization and fixation protocols. In the proposed work, we deliver fluorescent probes for live cell imaging in a controlled and effective fashion using polymersomes20,23,25,26. Using this technology we demonstrated the potential to exploit these carriers to achieve an effective delivery, in live cells (NIH-3T3 cell line), of several membrane probes, namely: PC, PE, CHOL, S, PIP2, and Actin probes. The lack of toxicity to cells of our established delivery system enables the study of the dynamics of a live cell, with a particular focus on cell membrane dynamics. We initially focus on shedding light on the interaction between the PIP2 and F-actin network11,12,14. However, PIP2 localization has always been studied indirectly using either by immunofluorescence on fixed cells or using genetically induced fluorescent proteins containing pleckstrin homology (PH) domains29. Here we delivered a labeled and still functional PIP2 that allows us to directly track its exact localization within the membrane. The first observation that this method allows, it is that PIP2 forms domains that accumulate at the edge of the cells. As shown in Figure 1 these domains are associated with cell protrusions and follow them as they grow and sample the surrounding of the cell. The sequence shown in Figure 1 shows that at the early stages of adhesion, the cell edges have many small filopodia extending toward the cell surroundings for sampling its environment (see movie SM1). As the cell finds anchorage on the substrate, the protrusion evolves into more dendritic structures (see Figure 1B) and eventually into larger filopodia (see Figure 1C) and lamellapodias (see Figure 1D).
PIP2 form no more that two or three domains per protrusion then as the protrusion expands into lamellapodias, they start to merge along the actin stressed fibers. These observations suggest that PIP2 transport regulates the actin polymerization as well as the protrusion dynamics. Moreover, both actin polymerization driven membrane protrusion and its anchorage to the membrane require a continuous feeding of PIP2 to the cell extremity. As shown in Figure 2A, analysis of actin and PIP2 distribution within live cells shows a clear polarization of the latter towards the attached cell side, which in our experiments corresponds to the cell bottom. We, therefore, looked into the cell regions that goes from the peri-nuclear area to the cell extremity focusing on both PIP2 rich organelles and actin fibers. As shown in Figure 2B–C we tracked several of these organelles. Mean square displacement (MSD) analysis, average speed measurements and sizing of PIP2 rich organelles show that the majority of the small trafficking vesicles move towards the cell extremity, according to a precise directional propelled motion with speeds up to ~2 μm/s (Figure 2D). This suggests that vesicles are trafficked along the actin filaments, possibly involving molecular motors such as myosin. A similar mechanism was proposed for yeast and is known as the “Exocytic Signal Model” but to date, it has never been reported for higher eukaryotic cells32,37,38. According to our observations, we can also postulate that the actin/membrane interaction that is critical for controlling spreading and adhesion, is fed by PIP2 rich vesicles and that this transport follows closely actin polymerization/depolymerization cycles. Finally, exploiting our delivery systems we have also added other membrane probes to the live cells and studied their distribution at the cell edges.
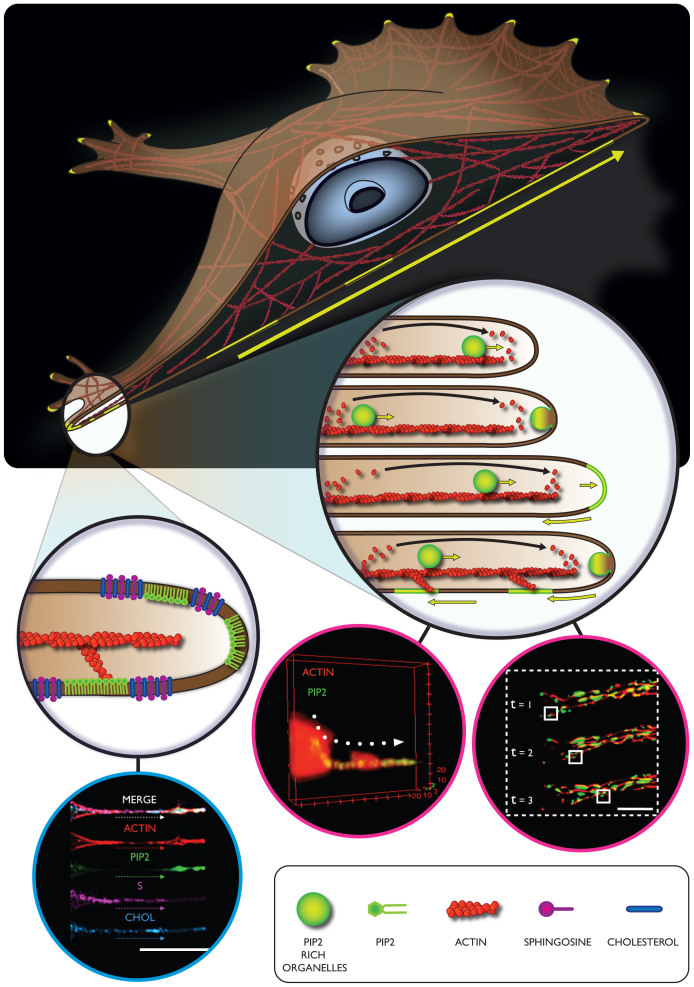
Two different experiments were set up, with the aim to understand, in a live cell model, the effective sub-membrane lipid distributions, and their possible interactions with actin. First, three different lipid probes were simultaneously delivered within cells (S, CHOL and PC), and confocal analysis was carried out at both, the centre and periphery of cells (Figure 3). While the results did not show relevant differences in the fluorescence signal in the central area of the cells, the peripheral sections displayed a significant overlap between S and CHOL. This could be explained by an enhanced cytoskeleton activity at the cellular edge, combined with a significant remodeling in the lipid domain arrangement39. This is an important proof indicating a strong cooperation between these two membrane constituents (cholesterol and sphingosine) during cell cytoplasmic membrane modifications, and cell protrusion formations, as also previously suggested by other work36. In particular, a specific regulation/interaction between cholesterol and sphingosine occurs, depending on specific cytoskeleton conformation40,41. According to previous findings, in fact, the presence and formation of cholesterol-sphingolipid domains is dependent on cytoskeleton integrity and its localization at the cell membrane level42. The proposed model depicts cholesterol as an active promoting factor able to regulate specific scaffold proteins and consequently enhance the formation of complex signaling43,44. These latter proteins, in turn, could be involved in the activations of other signaling factors implicated in the cytoskeleton modulation and, thus, in sphingolipid domains formations. All these assumptions have been further corroborated by our overlap experiments of lipid domains, depicted in Figure 4. This data suggests that cytoskeleton activity is significantly promoted at the cell periphery, as compared to the inner part of the cell39. As a consequence, specific cell membrane constituents and lipid domains arrangement should be organized with a precise and well-defined spatial distribution41,45. To further shed light on this hypothesis, both S and CHOL membrane constituents were simultaneously delivered with ACTIN, and PIP2 probes. The overlap of fluorescence signal percentage, per single channel, demonstrated that the F-actin strongly interacts (particularly at the peripheral area) with PIP2 (78 ± 5%) and with all the other molecules, S (63 ± 3%) and CHOL (59 ± 6%). On the other hand, this probe displayed a slightly minor interaction with PIP2 at the central area (72 ± 6%), as compared to the other two molecules, S and CHOL (83 ± 5%). This is in agreement with a strong cooperation occurring between PIP2 and actin molecules, especially at the cellular edge14. The analysis of all the other PM distributed probes, at the peripheral area of cells, revealed that sphingosine has a slightly higher interaction with ACTIN and CHOL (about 63 ± 5% of overlap signal), with respect to the PIP2 (56 ± 5%), although the slightly lower degree of interaction between CHOL and ACTIN (59 ± 6%) compared to S and ACTIN should be also mentioned. This shows that the formation of PIP2 domains is also regulated by other membrane components, particularly by cholesterol and sphingosine, already singled out in the study of lipid rafts. Particular relevant is the recent observation that these two membrane components play a critical role in stabilizing PIP2 domains29,46. It also worth noticing that both S and CHOL accumulate at the cell edges when these are deformed into protrusions. This would suggest that these two membrane components also control the spontaneous curvature of these highly deformed membranous structures36. On this basis we can thus propose the following mechanism, as in the schematic in Figure 5. As the cell samples the surroundings by forming cell protrusions, these gradually evolve into filopodia and lamellapodia and from our experiments we can see how PIP2 has a critical role in controlling both the protrusion dynamics, via controlling actin polymerization and the formation stress fibers and the consequent adhesion domains. Further we show that these protrusions formation and stabilization require the coordinated interaction of several membrane components, including sphingosine and cholesterol. Finally we also show, for the first time, strong evidence of transport from the central region of the cell to the extremity of critical adhesion components, via vesicular trafficking. As shown in Figure 5, we propose that this transport operates along the actin fibers and also serves as a membrane reservoir. Indeed, a single 200 nm trafficking vesicle moving into a 600 nm wide tube and fusing at its extremity will result in a growth of about 50% of its final length. The resulting PIP2 domains will then follow the actin tread-milling and possibly recycling back to the cell interior.
Figure 5. Schematic of a proposed mechanism involving different cellular components during important processes such as adhesion and spreading.
In the proposed mechanism, PIP2 has the central role to coordinate the actin polymerization and the formation of membrane adhesion domains. Furthermore we are showing a PIP2 rich organelles transport alongside the F-actin structures that possibly serve as membrane reservoir and finally culminate in cell protrusion elongation. Finally we are proposing sphingosine and cholesterol as important membrane constituents for the conformational stabilization of tubular cell fibers.
Methods
Cytoskeleton and phospholipid interactions
Mouse embryonic fibroblast cell line (NIH-3T3) were obtained from ATCC®, and used as a cellular model in all the experiments. Cell were cultured and maintained in Dulbecco's Modified Eagle Medium (DMEM) containing: 10 (v/v) fetal calf serum, 2 mM L-glutamine, 100 mg/ml streptomycin and 100 IU/ml penicillin at 37°C/95% air/5% CO2. Cell were periodically sub-cultured using Trypsin-EDTA solution 0.25% for the detachment process. For the evaluation of distribution and interactions between the two compounds TopFluor®PIP2 (PIP2) and Phalloidin-ATTO647 (ACTIN) during cell adhesion process, cells were initially seeded in a 24-well plate at a density of 4 × 104 cells per well, and grown for 24 h in complete DMEM medium. Then, cells were treated and incubated for 24 h with fresh medium containing PIP2 and ACTIN, all at 1 mg/ml final concentration of polymersomes. After this, fibroblast were washed three times with phosphate saline buffer (PBS), and detached from the plate surface using 0.25% Trypsin-EDTA solution.
Cells were then collected in a pellet through centrifugation at 1000 g × 5 minutes. Subsequently, cells were seeded again in glass bottom dishes (35 mm diameter) at a density of 1 × 104 in a DMEM imaging medium. The NIH-3T3 cells were then analyzed by means of confocal laser scanning microscopy, in living conditions, using a λex = 490 nm and a λem = 525 nm for the PIP2, and λex = 644 nm and λem = 670 nm for the ACTIN. We calculated an average of radiation dose applied to the scanning point (average scanning speed always less 1 s) of of 16.5 J/cm2 for the green channel (using 1.2% of the maximum output laser energy with physical length of 120.67 × 120.67 um), and of 0.5 J/cm2 for the far red channel(using in this case 1.5% of the maximum output laser energy at the equal physical length), both values are well below the reported dosage for cellular damage47.
The images were recorded, with a live cell imaging method in an appropriate environmental chambers (37°C/95% air/5% CO2), at different time points, i.e. 10 and 30 minutes, to analyze the early stage cell adhesion process. The first stage of cell adhesion process (10 minutes after cells seeding) was also recorded in a continuous two channels video, with 0.7 frames per second (see supporting information SM1). To evaluate the interactions between cytoskeleton and phospholipid, during the cell spreading process, the NIH-3T3 cells were first plated at a density of 4 × 103 cell/well for 24 h in complete DMEM medium. Then, fibroblasts were treated, as previously described, with fresh medium containing PIP2 and ACTIN (both at 1 mg/ml final polymersomes concentration). After 24 h of incubation time, the medium was removed, and cells were washed three times with Phosphate Buffer Saline (PBS), and incubated with fresh DMEM imaging medium. Subsequently, cells were analyzed by means of confocal live-cell imaging microscopy, with the same previously described parameters. Also for the vesicular trafficking analysis, the NIH-3T3 cells were treated again with the two polymersome preparations. For this experiment, cells were seeded at a density of 8 × 103 cells/well, and incubated with the polymersome formulations for 6 h. After this, the medium was removed, substituted with imaging DMEM, and samples were analyzed by means of live confocal microscopy, with the same parameters as described above.
Topological study
NIH-3T3 cells were cultured and maintained at the same conditions mentioned above. For this particular study two different experiments were performed. In the first one, the NIH-3T3 were seeded in glass bottom dishes (35 mm diameter) with a density of 4 × 103 cells/well, and incubated for 24 h in complete DMEM medium. Subsequently, the cells were treated with fresh DMEM medium containing three different polymersome formulations, namely BODIPY®Phosphocholine (PC), NBD-Cholesterol (CHOL), and BODIPY®TB-Sphingosine (S), all with a final polymersomes concentration of 1 mg/ml. After 24 h of incubation, this medium was removed and substituted (after three wash steps in PBS) with fresh imaging medium. Then, cells were analyzed by confocal laser scanning microscopy (in live cell mode) in an appropriate growth chamber (37°C/95% air/5% CO2). Cells were analyzed at λex/λem = 490 nm/525 nm for the PC, at λex/λem = 590 nm/621 nm for S, and finally at λex/λem = 428 nm/544 nm for the CHOL. In the second experiment, only the polymersome formulations have been modified. In particular, the NIH-3T3 cells were exposed to four different probes simultaneously, i.e. CHOL, S, PIP2 and ACTIN. Similar to the previous study, cells were seeded with a density of 4 × 103 cells/well, and incubated for 24 h in complete DMEM medium. After 24 h, the medium was replaced with fresh DMEM containing the four formulations (1 mg/ml final poplymersome concentration), and cells were then incubated for 24 h. After the incubation with the probes, the medium (containing the polymersomes) was replaced by fresh imaging DMEM, and cells were analyzed. The intracellular distribution of the cargo was characterized by means of a confocal analysis, with the following parameters: λex = 428 nm, λem = 544 nm for the CHOL, λex = 590 nm and λem = 621 nm for S, λex = 495 nm and λem = 503 nm for PIP2, and λex = 644 nm and λem = 670 nm for ACTIN.
Colocalisation coefficient calculation and microscopy data analysis
The quantification of colocalization between the two fluorophores was quantified using an in-house developed MATLAB® script (supporting information). The script provides the Pearson's coefficient, the derived overlap coefficient, and the Manders' coefficients. In fluorescence microscopy, the colocalization between two channels is a measure of the dependence of one channel to the other, which means how much an increase in the intensity in one channel is reflected by an increase in the intensity in the other channel. The Pearson's coefficient is a statistical tool, introduced by Karl Pearson, as a way to measure the strength of linear dependence between two variables48.
It is defined as:
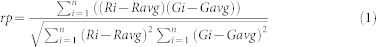 |
Where rp is the Pearson's coefficient, Gi and Ri are the intensity of the i pixel for the two channels R and G, Gavg and Ravg are the average intensity of the pixels in the whole image for both channels. The coefficient ranges from −1 (no correlation) to +1 (perfect correlation). The overlap coefficient is a simplified and easier coefficient to interpret derived from the Pearson's coefficient. Because it omits the average terms, this coefficient is dependent on the background, which should be uniform.
The overlap coefficient is calculated as:
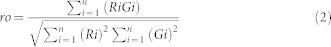 |
Finally, the imaging data analysis, to determine the mean distribution intensity regarding the PIP2 and ACTIN channels, with respect to the Z stack cell reconstruction (Figure 2A), was performed with using imageJ software. Additional specifications, in terms of reagents and materials used, polymersomes preparations and cargo encapsulations, physical and biological evaluations of different formulations, and majors clarifications about molecule-molecule specific interactions and distribution can be found in the SI Materials and Methods.
Author Contributions
L.C. performed the experiments and analysis. A.J. developed the MATLAB® script for the overlap and MSD calculations. A.L. and G.B. supervised the project. All authors contributed to and discussed the manuscript.
Supplementary Material
Supporting information
SI Video 1
SI Video 2
Acknowledgments
The authors thanks Miss Silvia Bianco for the design of the cartoons and Dr. Loris Rizzello for the critical proofing of the manuscript. L.C. thanks BTG/Biocompatibles for sponsoring his studentship and G.B. thanks the ERC for sponsoring part of his salary (ERC-STG-MEVIC).
References
- Stephens D. J. & Allan V. J. Light microscopy techniques for live cell imaging. Science 300, 82–86 (2003). [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. & Prasher D. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994). [DOI] [PubMed] [Google Scholar]
- Van Meer G., Voelker D. R. & Feigenson G. W. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K. & Ikonen E. Functional rafts in cell membranes. Nature 387, 569–572 (1997). [DOI] [PubMed] [Google Scholar]
- Stachowiak J. C., Brodsky F. M. & Miller E. A. A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat Cell Biol 15, 1019–1027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaby J. M., Momsen M., Kulkarni V. S. & Brown R. E. Cholesterol-induced interfacial area condensations of galactosylceramides and sphingomyelins with identical acyl chains. Biochemistry 35, 5696–5704 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- FM G. & A A. Biophysics of sphingolipids I. Membrane properties of sphingosine, ceramides and other simple sphingolipids. BBA-Biomembranes 1758, 1902–1921 (2006). [DOI] [PubMed] [Google Scholar]
- Dustin M. L. Shmoos, rafts, and uropods-the many facets of cell polarity. Cell 110, 13–18 (2002). [DOI] [PubMed] [Google Scholar]
- Simons K. & Gerl M. J. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol 11, 688–699 (2010). [DOI] [PubMed] [Google Scholar]
- Lingwood D. & Simons K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010). [DOI] [PubMed] [Google Scholar]
- Bohdanowicz M. & Grinstein S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol Rev 93, 69–106 (2013). [DOI] [PubMed] [Google Scholar]
- Di Paolo G. & De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 (2006). [DOI] [PubMed] [Google Scholar]
- Rozelle A. L. et al. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr Biol 10, 311–320 (2000). [DOI] [PubMed] [Google Scholar]
- Saarikangas J., Zhao H. & Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev 90, 259–289 (2010). [DOI] [PubMed] [Google Scholar]
- Logan M. R. & Mandato C. A. Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol Cell 98, 377–388 (2006). [DOI] [PubMed] [Google Scholar]
- Yin H. L. & Janmey P. A. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65, 761–789 (2003). [DOI] [PubMed] [Google Scholar]
- Bishop A. L. & Hall A. Rho GTPases and their effector proteins. Biochem. J. 348, 241–255 (2000). [PMC free article] [PubMed] [Google Scholar]
- Sechi A. S. & Wehland J. The actin cytoskeleton and plasma membrane connection: PtdIns(4,5)P(2) influences cytoskeletal protein activity at the plasma membrane. J Cell Sci 113, 3685–3695 (2000). [DOI] [PubMed] [Google Scholar]
- Iden S. & Collard J. G. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol 9, 846–859 (2008). [DOI] [PubMed] [Google Scholar]
- Massignani M. et al. Enhanced Fluorescence Imaging of Live Cells by Effective Cytosolic Delivery of Probes. Plos One 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton I. et al. Fully synthetic polymer vesicles for intracellular delivery of antibodies in live cells. FASEB J 27, 98–108 (2013). [DOI] [PubMed] [Google Scholar]
- Lomas H. et al. Biomimetic pH sensitive polymersomes for efficient DNA encapsulation and delivery. Adv Mater 19, 4238–4243 (2007). [Google Scholar]
- Du J., Tang Y., Lewis A. L. & Armes S. P. pH-Sensitive vesicles based on a biocompatible zwitterionic diblock copolymer. J Am Chem Soc 127, 17982–17983 (2005). [DOI] [PubMed] [Google Scholar]
- Massignani M. et al. Controlling cellular uptake by surface chemistry, size, and surface topology at the nanoscale. Small 5, 2424–2432 (2009). [DOI] [PubMed] [Google Scholar]
- LoPresti C., Lomas H., Massignani M., Smart T. & Battaglia G. Polymersomes: nature inspired nanometer sized compartments. J Mater Chem 19, 3576–3590 (2009). [Google Scholar]
- Discher D. E. et al. Emerging applications of polymersomes in delivery: From molecular dynamics to shrinkage of tumors. Prog Polym Sci 32, 838–857 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine D. H. et al. Polymersomes: a new multi-functional tool for cancer diagnosis and therapy. Methods 46, 25–32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- To C., Shilton B. H. & Di Guglielmo G. M. Synthetic triterpenoids target the Arp2/3 complex and inhibit branched actin polymerization. J Biol Chem 285, 27944–27957 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bogaart G. et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho R. J. et al. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol 151, 1353–1368 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meer G. & de Kroon A. I. P. M. Lipid map of the mammalian cell. J Cell Sci 124, 5–8 (2011). [DOI] [PubMed] [Google Scholar]
- Yakir-Tamang L. & Gerst J. E. Phosphoinositides, exocytosis and polarity in yeast: all about actin? Trends Cell Biol 19, 677–684 (2009). [DOI] [PubMed] [Google Scholar]
- Lemons D. S. & Gythiel A. Paul Langevin's 1908 paper “On the Theory of Brownian Motion”. Am J Phys 65, 1079–1081 (1997). [Google Scholar]
- Hammer J. A. & Sellers J. R. Walking to work: roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol 13, 13–26 (2012). [DOI] [PubMed] [Google Scholar]
- Pierobon P. et al. Velocity, processivity, and individual steps of single myosin V molecules in live cells. Biophys J 96, 4268–4275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A. et al. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J 24, 1537–1545 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheorghe D. M. et al. Interactions between the yeast SM22 homologue Scp1 and actin demonstrate the importance of actin bundling in endocytosis. J Biol Chem 283, 15037–15046 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein A. E. & Drubin D. G. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19, 287–332 (2003). [DOI] [PubMed] [Google Scholar]
- Pollard T. D. & Cooper J. A. Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. G. N. et al. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J Cell Biol 177, 717–730 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D. et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135, 1085–1097 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisz J. F. et al. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. P Natl Acad Sci USA (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.-Y., Weng J. & Anderson R. G. W. OSBP Is a Cholesterol-Regulated Scaffolding Protein in Control of ERK1/2 Activation. Science 307, 1472–1476 (2005). [DOI] [PubMed] [Google Scholar]
- Sheng R. et al. Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domain-containing scaffold proteins. Nat Commun 3, 1249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar K. et al. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 149, 1353–1367 (2012). [DOI] [PubMed] [Google Scholar]
- Eggeling C. et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162 (2009). [DOI] [PubMed] [Google Scholar]
- Wagner M. et al. Light dose is a limiting factor to maintain cell viability in fluorescence microscopy and single molecule detection. Int. J. Mol. Sci. 11, 956–966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K. Determination of the coefficient of correlation. Science 30, 23–25 (1909). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
SI Video 1
SI Video 2