Abstract
Serine-rich (Srr) proteins exposed at the surface of Gram-positive bacteria are a family of adhesins that contribute to the virulence of pathogenic staphylococci and streptococci. Lectin-binding experiments have previously shown that Srr proteins are heavily glycosylated. We report here the first mass-spectrometry analysis of the glycosylation of Streptococcus agalactiae Srr1. After Srr1 enrichment and trypsin digestion, potential glycopeptides were identified in collision induced dissociation spectra using X! Tandem. The approach was then refined using higher energy collisional dissociation fragmentation which led to the simultaneous loss of sugar residues, production of diagnostic oxonium ions and backbone fragmentation for glycopeptides. This feature was exploited in a new open source software tool (SpectrumFinder) developed for this work. By combining these approaches, 27 glycopeptides corresponding to six different segments of the N-terminal region of Srr1 [93–639] were identified. Our data unambiguously indicate that the same protein residue can be modified with different glycan combinations including N-acetylhexosamine, hexose, and a novel modification that was identified as O-acetylated-N-acetylhexosamine. Lectin binding and monosaccharide composition analysis strongly suggested that HexNAc and Hex correspond to N-acetylglucosamine and glucose, respectively. The same protein segment can be modified with a variety of glycans generating a wide structural diversity of Srr1. Electron transfer dissociation was used to assign glycosylation sites leading to the unambiguous identification of six serines and one threonine residues. Analysis of purified Srr1 produced in mutant strains lacking accessory glycosyltransferase encoding genes demonstrates that O-GlcNAcylation is an initial step in Srr1 glycosylation that is likely required for subsequent decoration with Hex. In summary, our data obtained by a combination of fragmentation mass spectrometry techniques associated to a new software tool, demonstrate glycosylation heterogeneity of Srr1, characterize a new protein modification, and identify six glycosylation sites located in the N-terminal region of the protein.
Protein glycosylation takes place in bacteria. A considerable amount of information on the biochemical pathways involved in protein glycosylation has been obtained in the genera Campylobacter and Neisseria (1–3). Hence, functional studies performed on Gram-negative bacteria have revealed important properties of protein glycosylation pathways. Like in eukaryotes, proteins can be glycosylated on asparagines (N-glycosylation) or serine/threonine residues (O-glycosylation). Two modes of glycan transfer onto proteins have been characterized in Gram-negative bacteria. In the first, the activated glycan is synthesized on a lipid carrier on the cytoplasmic side of the membrane. After membrane translocation, the glycan portion of the polyprenyl-phosphate-glycan intermediate is transferred en bloc to the protein by an oligosaccharyltransferase active in the periplasm. Such mechanism has been demonstrated for N-glycosylation in Campylobacter jejuni (4–6) and O-glycosylation in Neisseria gonorrheae (7–9). These pathways are responsible for the glycosylation of more than 65 proteins in C. jejuni (10) and 12 different proteins in N. gonorrheae (11). The second well-characterized type of transfer is sequential and arises in the cytoplasm: glycosyltransferases (GTs)1 catalyze the transfer of monosaccharide residues, under a nucleotide-diphosphate activated form, in a stepwise manner to the protein. This mechanism has long been considered to be restricted to O-glycosylation like in Campylobacter, Helicobacter, or Pseudomonas spp. for flagellar proteins (12–15). However, it was recently demonstrated that HMW1c, a GT of Haemophilus influenzae, was responsible for the cytoplasmic N-glycosylation of the adhesin HMW1 (16, 17).
Important families of glycoproteins have also been characterized in Gram-positive bacteria. The glycosylation of flagellins has been demonstrated in Clostridium spp. and in Listeria monocytogenes (13, 18); the modification is required for flagella assembly in C. difficile (13) and motility in L. monocytogenes (19, 20). In L. monocytogenes, the flagellin monomer is modified with N-acetylglucosamine (GlcNAc) residues at up to six different sites by GmaR, a cytoplasmic O-GlcNAc-transferase (OGT) homologous to eukaryotic OGTs (18). GmaR was the first prokaryotic enzyme of this type to be characterized (20). The type of sugar added to flagellins and the number of modified sites is different between species and is even strain-dependent in Clostridium difficile (13). Genomic analysis suggests that flagellin glycosylation in Clostridium spp. is performed on proteins through a sequential process in the cytoplasm (3). In mycobacteria and other actinomycetae, a general O-mannosylating system has been identified which involves a transmembrane O-mannosyltransferase similar to that found in eukaryotes (21–24). Importantly, in Mycobacterium tuberculosis the disruption of the pathway responsible of adhesins mannosylation was found to strongly attenuate its virulence (21). The glycosylation of surface (S-) layer proteins which cover the cell surface of a variety of bacterial species is a common modification of this class of proteins (25, 26). The extensive MS analysis performed in mutant backgrounds on the S-layer proteins of Geobacillus stearothermophilus and Paenibacillus alvei have made possible the reconstruction of the biosynthesis pathway for S-layer glycans, and also to propose a model for their export and transfer to S-layer proteins (27, 28). Interestingly, the P. alvei study also revealed that tyrosine residues can be modified by a bacterial protein glycosylation pathway (28). More recently cell wall autolysins expressed in different Lactobacilli spp. have been shown to be glycosylated (29–31); interestingly, the peptidoglycan hydrolytic activity of the autolysin Acm2 of Lactobacillus plantarum was found to be controlled by O-glycosylation (30). Although a large array of glycoproteins proteins have been characterized, it is notable that only O-type glycosylation has been described so far in Gram-positive bacteria.
Recently, a family of serine-rich (Srr) glycosylated proteins has been discovered in Streptococcus species and Staphylococcus aureus (32, 33). Well-studied members include the fimbriae-associated adhesin Fap1 of Streptococcus parasanguinis (34), the human-platelet binding protein GspB of Streptococcus gordonii (35), the SraP and PsrP adhesins of Staphylococcus aureus and Streptococcus pneumoniae, respectively (36, 37) and the Srr1 adhesin of S. agalactiae (36, 38–42). These proteins are exported to the cell surface through a dedicated transport system called SecA2 and are covalently anchored to peptidoglycan. Genome comparisons revealed striking similar genetic organization of srr loci characterized by the presence of a gene cluster encoding the Srr-dedicated secretion machinery (SecA2 secretion system) and a variable number of GTs (32, 33, 43). The role of Srr proteins as major bacterial adhesins was first demonstrated in the oral streptococci Streptococcus gordonii and Streptococcus parasanguinis (formerly S. sanguis) where they are required for full adhesion of bacterial cells to platelets or saliva-coated hydroxylapatite, respectively (34, 44). In the human pathogens S. aureus and S. pneumoniae, the Srr proteins corresponding to SraP and PrsP were shown to promote adhesion to platelets and lung cells, respectively (36, 37, 45). Importantly, Srr proteins have been associated with the virulence of all of these opportunistic pathogens (34, 36, 37, 45, 46).
Streptococcus agalactiae is the leading cause of sepsis and meningitis in neonates (47, 48). The species colonizes asymptomatically the mucosa of 20–30% of the human population and is an increasing cause of invasive infections in immunocompromised adults (49). S. agalactiae Srr1 is involved in adhesion to endothelial and epithelial cells (38–42,50). Such surface proteins are attractive vaccine candidates and S. agalactiae Srr protein has been shown to confer immunogenic protection in mice (51).
Srr1 binds both keratin 4 and fibrinogen through the same N-terminal binding region located between amino acid residues 303 and 641 (Fig. 1A) (39, 42, 52). Upstream of this binding domain, Srr1 displays two non-repeated regions (NR1 and NR2) showing no homology with known proteins which are separated by a first serine-rich region (SR1) where the proportion of the Ser/Thr residues is above 40%. The C-terminal part of Srr1 (SR2) contains more than 70% of Ser/Thr residues with 140 repetitions of the SASM/T motif that extends over 700 residues (Fig. 1A and supplemental Fig. S1). The C terminus also contains a LPXTG motif that is recognized by sortase for covalent attachment of Srr1 to the peptidoglycan (53). The Srr proteins synthesized in Streptococcus spp. and S. aureus exhibit a similar organization with variation in the functional binding domains and in the length of the C-terminal SR domain (32). We previously carried out an extensive functional analysis of the srr1-secA2 locus of S. agalactiae (38) (Fig. 1B). Genetic and biochemical data showed that the GTs GtfA and GtfB were essential for the addition of a N-acetylglucosamine (GlcNAc) to Srr1 and that six other GTs (GtfC to GtfH) were potentially involved in additional glycan modifications (38). It was also demonstrated for Srr1 and GspB that glycosylation takes place in the cytoplasm (38, 54).
Fig. 1.
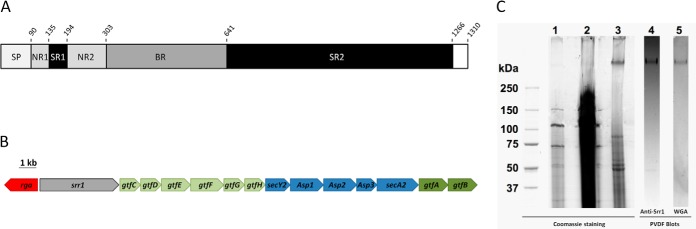
Modular organization of Srr1, scheme of srr1 locus and production of Srr1. A, The different domains of the 1310 residues in-length Srr1 protein of S. agalactiae are shown. SP: Signal peptide; NR: Non repeat region; SR: Serine rich region; BR: Binding region; LPXTG: domain containing the LPXTG-cell wall anchoring motif. Residues numbering is shown. B, Schematic view of the 23 kb srr1 locus. The genes encoding GTs are colored in green. Light Green for accessory GTs-encoding genes (gtfC to gtfH), dark green for essential GTs-encoding genes (gtfA and gtfB). In blue, the genes encoding proteins related to Srr1 secretion. In the opposite transcriptional orientation, in red, the rga gene encodes a positive transcriptional regulator of srr1 (gray). The embrace highlights the region deleted in the H36BSrtA*ΔgtfCH mutant strain. C, SDS-PAGE of Srr1 containing fractions. Lane 1: Supernatant of the medium after an over-night culture of H36B SrtA* strain. Lane 2: Supernatant concentrated 20-fold. Lane 3: Srr1 containing fraction after purification on WGA agarose column. Lane 4: Western blot analysis using polyclonal anti-Srr1 serum. Lane 5: Lectin blot analysis using biotinylated WGA. Arrowheads point to Srr1 band. The same proportion (5%) of the samples were loaded before (lane 2) and after (lane3) purification to highlight the efficiency of the purification procedure.
We report here the first detailed analysis of Srr1 glycosylation at the molecular level by means of mass spectrometry. Lectin affinity chromatography was used to enrich Srr1 before trypsin digestion Fig. 1C. Tryptic peptides were then analyzed by nanoLC-tandem MS (MS/MS) using various activation techniques. A dedicated software tool SpectrumFinder (SF), allowing the retrieval of glycopeptides after higher energy collisional dissociation (HCD) fragmentation was developed for this study. This combination of approaches led to the identification of 27 glycopeptides in the N-terminal part of the protein, outside the region containing the highest serine density. We demonstrated that besides glycosylation by N-acetylhexosamine (HexNAc) or hexose (Hex) residues, Srr1 can also be modified by an unexpected sugar characterized as O-acetylated–N-acetylhexosamine (O-AcHexNAc). To localize glycosylation sites, electron transfer dissociation (ETD) experiments were performed. We were able to identify six glycosylation sites that were associated to five serine and one threonine residues.
EXPERIMENTAL PROCEDURES
Chemicals
Cultures media were from BD (Beckton, Dickinson and Company, Sparks, MD). Water used was ultra-pure water obtained with a Milli-Q purifier (Millipore SAS, Molsheim, France). Lectins were from Vector laboratories (Vector Laboratories, Burlingame, CA, USA). The other chemicals were from Sigma Aldrich (Sigma-Aldrich, St. Louis, MO, USA) except when indicated. Antibodies were prepared as described (15).
Bacterial Strains, Culture Media and Growth Conditions
Streptococcus agalactiae H36B is a serotype Ib strain whose genome has been sequenced (29). This strain was chosen because of its high Srr1 production level, as compared with S. agalactiae NEM316 strain. To produce Srr1, we used the H36BSrtA* mutant strain where the catalytic residue Cys206 of the sortase A protein was changed into alanine by engineering the srtA gene (gbs0949) as described (55). As a consequence, the secreted proteins carrying a LPXTG motif like Srr1 were more efficiently released into the culture medium (53). This mutation does not affect Srr1 glycosylation as this post-translational modification is a cytoplasmic event taking place prior to the export process (38). To study the role of accessory GTs (GtfC-H) and the sequence of glycosylation events, the strain H36BSrtA*ΔgtfCH was constructed as previously described (38). S. agalactiae strains were cultured in Todd-Hewitt (TH) broth in standing filled flasks at 37 °C.
Srr1 Protein Production
Bacterial cell were removed from culture medium (1 liter) by centrifugation (10 min at 6000 × g) and the supernatant was filtered to 0.22 μm. The proteins released in medium during growth were precipitated overnight with ammonium sulfate (30%) at 4 °C under gentle agitation. Protein pellet obtained after centrifugation (15 min at 15,000 × g) was resuspended in 10 ml Wheat germ agglutinin (WGA) buffer (20 mm Tris, and 200 mm NaCl, pH 7.5) and dialyzed on Float-a-lyzer G2 10kDa (Spectrum lab, Breda, NL) overnight against WGA buffer. For lectin affinity chromatography, a 5/50 column (Amersham Bioscience) was packed with one ml WGA-agarose conjugate. The chromatographic steps were run on an Äkta purifier (Amersham Bioscience). The column was thoroughly washed with WGA buffer and protein sample was loaded at 0.1 ml/min at 4 °C. After a washing step with 10 ml WGA buffer, WGA bound proteins were eluted with 1 ml elution buffer (20 mm Tris, 200 mm NaCl, and 500 mm N-acetyl-d-glucosamine, pH 7.5) at 0.5 ml/min. Protein fractions were extensively dialyzed against 20 mm Tris, 200 mm NaCl, pH 7.5 on 10 KDa cut-off membrane Amicon Ultra, Millipore, Billerica, MA. Concentrated fractions were stored at −20 °C until further analysis. For the comparative analysis of Srr1 produced in H36BSrtA* and H36BSrtA*ΔgtfCH, the culture supernatants were concentrated 100-fold and loaded on SDS-PAGE (see next section). The Coomassie-stained protein band corresponding to Srr1 was excised and trypsin-treated as described below (Generation of Srr1 tryptic peptides).
SDS-PAGE, Blotting, Lectin Binding, and Immunodetection
Protein fractions were resolved on precast Criterion XT Bis Tris Gel 4–12% run in XT-MOPS buffer (Bio-Rad Life Sciences, Marnes-la-Coquette, France). Gels were stained using colloidal Coomassie dye G250 (GelCode, Thermo Fisher Scientific Inc, Rockford, IL, USA). For immunodetection or lectin binding, gels were washed in water and sunk in Bjerrum and Shaffer Nielsen buffer (BSN buffer): 48 mm Tris, 39 mm glycine, and 20% methanol, pH 9.2. Proteins were transferred on nitrocellulose or methanol activated PVDF membrane after their immersion in BSN buffer. Semi-dry transfer was done using Trans-blot S.D. apparatus (Bio-Rad) at 20 V for 30 min. Membrane were washed once in water and then in PBS Tween 0.1% (PBS-T). Membrane were blocked in PBS-T, 3% BSA (PBS-T-B) for 30 min. Anti-Srr1 polyclonal antibodies were used at 1:1000 dilution in PBS-T-B and membrane was incubated 1 h at room temperature or overnight at 4 °C. After three washes of 10 min in PBS-T, membranes were incubated with Alexa-488 (Invitrogen, Invitrogen SAS, St. Aubin, France) conjugated secondary antibody diluted 1:5000 in PBS-T-B. For lectin binding, succinylated WGA (sWGA) conjugated to fluorescein (Vector laboratories, Burlingame, CA) was used at a 1:2000 dilution. Membranes were scanned for fluorescence detection using a FLA-3000 instrument (Fuji photo film, Tokyo, Japan).
Compositional Analysis of Srr1-associated Monosaccharides
Monosaccharide composition of purified Srr1 was determined as trimethylsilyl derivatives by gas chromatography and mass spectrometry (GC-MS) after acid hydrolysis. Purified Srr1 was spotted on PVDF membrane and extensively washed in water. Hydrolysis of glycosidic bond was achieved by transferring the membrane in 100 μl of 4 m trifluoroacetic acid (TFA) for three hours at 110 °C.
The hydrolysate was dried under vacuum and resuspended in 100 μl of the derivatization mixture containing 1.34 m N-methyl-N-(trimethyl-silyl)-trifluoroacetamide (MSTFA) (Sigma-Aldrich) and 0.5 mm of xylose in pyridine as internal standard. The trimethylsilyl derivatives of carbohydrates were analyzed by GC-MS with an Agilent system (GC 6890+ and MS 5973N, Agilent Technologies, Santa Clara, CA). Samples were injected with an automatic injector (Gerstel PAL, Sursee, Switzerland). Gas chromatography was performed on a 30 m ZB-50 column with 0.25 mm inner diameter and 0.25 μm film thicknesses (Phenomenex, Torrence, CA). Helium was used as the carrier gas and set at a constant flow rate of 1.5 ml/min. The temperature program was 5 min isothermal heating at 80 °C, followed by a 20 °C/min oven temperature ramp to 300 °C, and a final 3 min heating at 300 °C. Compounds were identified by both their retention time and comparison of their electron ionization mass spectra profiles with those of the NIST 05 Mass spectral library (Scientific Instrument Services, Ringoes, NJ, USA). The quantification was done using an external standard calibration curves for each molecule (2.5–25 nmol injected) established with the peak area of specific ion and expressed in nmol/mg cell walls.
Srr1 Tryptic Digestion
Coomassie-stained Srr1 bands detected on gel were cut and rinsed in water. Tryptic digestion was performed in-gel by adding 200 ng sequencing grade modified trypsin (Promega France, Charbonnières, France) diluted in 25 mm NH4HCO3 for 18 h at 37 °C. Tryptic peptides were recovered by washing the gel pieces twice in 0.2% TFA-50% acetonitrile and once in 100% acetonitrile and the supernatant was evaporated to dryness. Srr1 alkylation was deliberately omitted before mass spectrometry analysis to avoid formation of S-carbamidomethylmethionine (CMM) on methionyl residues resulting from iodoacetamide treatment (there are 45 methionines in Srr1) (56). Upon collision induced dissociation (CID) fragmentation, CMM gives a typical neutral loss of 105 Da that is interfering with our manual research of glycan neutral loss. Srr1 contains only one cysteine residue.
LC-MS/MS (CID) of Srr1 Tryptic Digest
Liquid chromatography (LC) was performed on an Ultimate 3000 RSLC Nanosystem (Dionex, Thermo Scientific). The sample was loaded on a precolumn cartridge, desalted with 0.08% TFA and 2% acetonitrile, and peptides separation was achieved on a PepMap C18 column (stationary phase: C18 PepMap 100, 3 μm; column: 75 μm i.d., 150 mm; Dionex). Buffers were 0.1% formic acid, 2% acetonitrile (A) and 0.1% formic acid, and 80% acetonitrile (B). Peptide separation was achieved with a linear gradient from 0% to 38% B for 50 min at 300 nl/min at 40 °C.
Separated peptides were analyzed on-line with a LTQ-Orbitrap Discovery mass spectrometer (Thermo Electron SAS, Villebon, France) using a nanoelectrospray interface. Ionization (1.5 kV ionization potential) was performed in positive mode with liquid junction and a capillary probe (10 μm i.d.; New Objective). Peptide ions were analyzed using Xcalibur 2.07 with the following data-dependent acquisition steps: 1) full MS scan in orbitrap (mass-to-charge ratio (m/z) 600 to 2000, profil mode, resolution 30,000 at m/z 400); and 2) MS/MS in linear trap (qz 0.22, activation time 50 ms, and normalized collision energy (NCE) 35%; centroid mode). Step 2 was repeated for the four major ions detected in step 1. Singly charged ions were excluded and dynamic exclusion time was set to 60 s. To enhance mass accuracy in Orbitrap mass analyzer, the lock mass option was activated on dimethylcyclosiloxan (m/z 667.1764).
In Source Fragmentation (ISF) in LTQ-Orbitrap
ISF was used to form oxonium ions of interest by applying a capillary voltage of 80 V. Oxonium ions were further fragmented by CID in the LTQ using 40 NCE.
LC-MS/MS (HCD) of Srr1 Tryptic Digest
LC was performed with a NanoLC-Ultra Eksigent system. The sample was loaded at a flow rate of 7.5 μl/min onto a precolumn cartridge (Biosphere C18, 5 μm, 20 mm, 100 μm i.d.; NanoSeparations, Nieuwkoop, NL) with 0.1% (v/v) formic acid and 2% acetonitrile. After 3 min, the precolumn cartridge was connected to the separating column (Biosphere C18, 3 μm, 150 mm, 75 μm i.d.; NanoSeparations Nieuwkoop, NL). The buffers used were water (buffer A) and acetonitrile (buffer B) each containing 0.1% (v/v) formic acid. Peptides were separated using a linear gradient from 5% to 35% B for 37 min at 300 nl/min.
A Q Exactive mass spectrometer (ThermoFisher Scientific) with nanoelectrospray interface was used for peptide analysis. Ionization (1.5 kV ionization potential) was performed in positive mode using a liquid junction and a capillary probe (10 μm i.d.; New Objective). Peptide ions were analyzed using Xcalibur 2.2 with the following data-dependent acquisition steps: 1) full MS scan (mass-to-charge ratio (m/z) 600 to 2500, resolution 70,000 at m/z 400); and 2) MS/MS scan using Quadrupole selection (window of 3 Th) and HCD (NCE 30, resolution 17,500). Step 2 was repeated for the ten most intense ions (top 10) detected in step 1. Singly charged ions were excluded and dynamic exclusion time was set to 40 s. Lock mass option was activated using m/z 667.1764 of dimethylcyclosiloxan.
LC-MS/MS (ETD) of the Srr1 Tryptic Digest
LC was performed on an ultimate 3000 nanoLC (Dionex, Thermoscientific). Sample was loaded at 30 μl/min on a precolumn cartridge (stationary phase: C18 PepMap 100, 5 μm; column: 300 μm i.d., 5 mm; Dionex) and desalted with 0.1% TFA and 2% acetonitrile. After 3 min of loading, the precolumn cartridge was connected to an in-house separating C18 column (Reprosil C18-HD, 3 μm, column 75 μm i.d., 150 mm, Cluzeau France). Buffers were 0.1% formic acid, 2% acetonitrile (A) and 0.1% formic acid and 80% acetonitrile (B). Peptide separation was achieved with a linear gradient from 0% to 42% B for 35 min at 300 nl/min at 35 °C.
Two types of acquisition (using Xcalibur 2.1) were used. The first one was a combined CID/ETD run and is based on a i) full MS scan in the Orbitrap (m/z 300 to 2000) at resolution 15,000 (at m/z 400) and ii) CID and ETD MS/MS spectra (max 5 precursor ions) with analysis in the linear ion trap. In the second type of run, only ETD was used for MS/MS including all glycopeptide ions previously identified. The maximum trapping time, the activation time with fluoranthene and the NCE were respectively fixed at 200 ms, 180 ms, and 35%. All ETD spectra were manually inspected.
X! Tandem Analysis
The analysis of CID and HCD data was performed with X! Tandem CYCLONE (2011.12.01.1). The S. agalactiae H36B complete database (http://www.ncbi.nlm.nih.gov/genome/186?project_id = 54315) and a contaminant database (trypsin, keratins, and others classical contaminants) were used. Enzymatic cleavage was set as a trypsin digestion ([RK] (15)) with multi possible miscleavage. Precursor mass and fragment mass tolerance were set to 10 ppm and 0.5 Da, respectively for LTQ Orbitrap data and to 10 ppm for Q Exactive data. Methionine oxidation was taken into account as possible modification. A refinement search was performed that accepts semi-enzymatic cleavage of peptide and amino terminus acetylation (+42.0105). Only peptides with an E value smaller than 0.05 were considered for identification.
To identify glycopeptides in CID data, Ser/Thr/Tyr variable modification with HexNAc (+203.0793 Da), O-AcHexNAc (+245.0912 Da), and Hex (+162.0528 Da) were introduced in the search parameters. In spectra typical of neutral loss events, the intensity of b and y ions generated by peptide backbone fragmentation is much lower than the ion formed by the glycan loss. To take this effect into account, the dynamic range for scoring spectra was set to 1000.
Development of a New Software Tool for Spectra Comparison (SpectrumFinder)
The main goal of SpectrumFinder is to compare pairs of MS/MS spectra, taking into account the fact that the same peptide sequence carrying different glycans will lead to very similar fragmentation patterns in HCD after glycan loss.
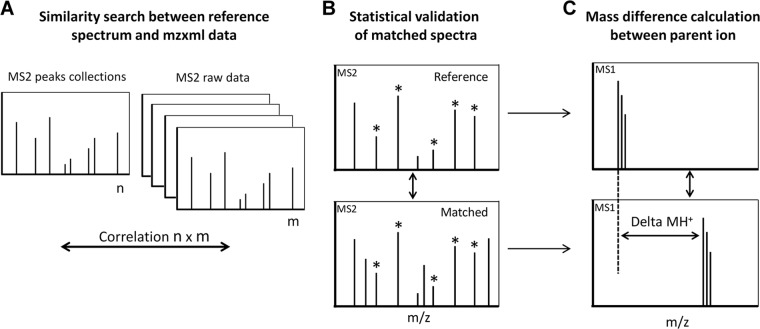
SpectrumFinder v1.0 is open source software developed for the need of this study (Fig. 2). It is distributed under the terms of the GNU Public License version 3 and available for Linux and Windows operating systems from http://pappso.inra.fr/bioinfo/sf. SpectrumFinder is written in C++. It uses a simple XML parameter file to build a collection of reference spectra and compare it to other spectra in mzXML files. The software requires MS/MS spectra recorded on the same instrument. After an initial spectral cleaning step based on background subtraction and low cut-off selection performed on a user-defined number of peaks, all MS2 spectra of a LC run are compared with a reference spectrum of interest (Fig. 2A) to assess their similarity. The similarity is established on the basis of different criteria Fig. 2B. First, a minimum number of shared peaks is required. Second, a “cosine similarity” is calculated between the two intensity vectors of paired-peaks. When a peak is absent, a null intensity is used. The cosine similarity measurement is sensitive to the orientation but not to magnitude. Consequently, it is a good indicator of common components between two vectors independently of their intensity. Third, a linear regression and a Pearson test are calculated between intensities of all shared peaks. The resulting p value estimates the similarity between the fragmentation patterns.
Fig. 2.
Workflow of SpectrumFinder processing. A, SpectrumFinder searches similar fragmentation pattern among MS2 spectra. Peaks are extracted from a reference spectrum and compared with an experimental data set. B, Similarities between MS2 spectra are statistically evaluated and a correlation score is calculated C, When a correlation is considered significant, the mass difference between the precursor ions of reference/computed spectra is calculated.
SpectrumFinder output is a text file Fig. 2C. Each match between the reference spectrum and a candidate spectrum is reported on a separate line. Columns contain information on the computed spectrum (scan number, sample name), cosine similarity, linear regression coefficient, and value of the Pearson test.
In this study, the reference library was deliberately restricted to spectra corresponding to potential glycopeptides (Fig. 3). In each case, the reference was manually chosen as the best quality spectrum (minimum six fragment ions with high signal to noise ratio). For each SpectrumFinder output, we systematically checked the corresponding MS2 spectra for the presence of diagnostic oxonium ions: m/z 163.0606 (Hex), m/z 204.0872 (HexNAc), m/z 366.1400 (HexHexNAc), and m/z 246.0977 (O-AcHexNAc). The mass difference between the parent ions in both the reference and computed spectra was calculated and manually fitted with an accuracy of ± 5 ppm as a combination of glycosylation (see above X! Tandem analysis) and other modifications: acetylation (+42.0105), formylation (+27.9949), methylation (+14.0156), water loss (-18.0153), oxidation of methionine residue (+15.9949), acetaldehyde (+26.01565), and ammonium (+17.0265) adducts.
Fig. 3.
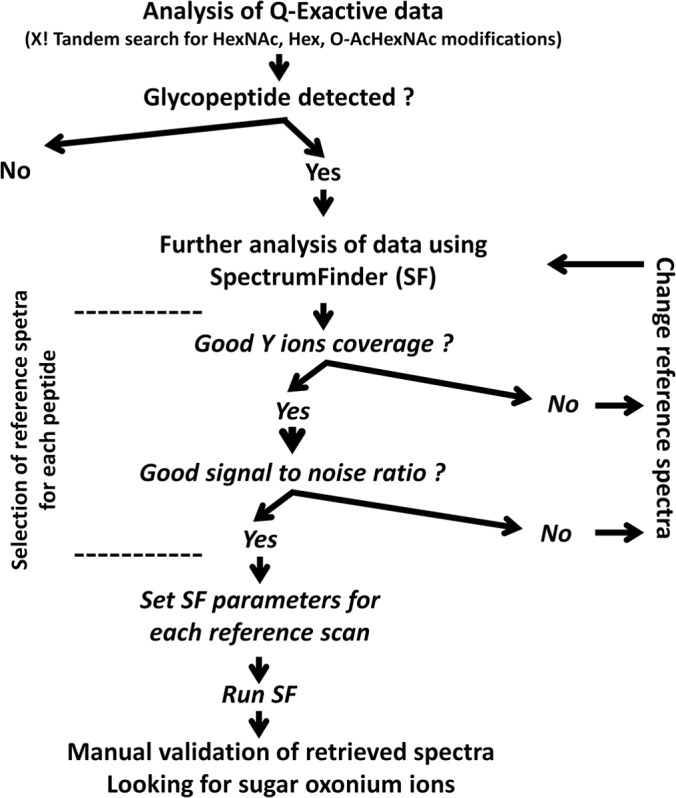
Schematic overview of Q Exactive data mining with X! Tandem and SpectrumFinder.
In contrast to other library search tools such as SpectraST (57) or X!Hunter (58), SpectrumFinder does not use any a priori knowledge on the mass of the precursor, because only fragment ions are compared.
Semi-quantitative Analysis of Glycopeptides
A glycoform is a characterized glycosylation variant of a peptide. The relative abundance of each glycoform of the [192–211] peptide of Srr1 produced in H36BSrtA* and H36BSrtA*ΔgtfCH strains was calculated by averaging the corresponding XIC peak area measured from three biological replicates. Normalization was done using the areas of non-glycosylated peptides ([335–345]; [373–387]; and [494–507]).
To perform semi-quantitative analysis of peptide glycoforms, the area of each peak corresponding to a parent ion was computed from an extracted ion chromatogram (XIC) with MassChroQ, a software that performs alignment, XIC extraction, peak detection and quantification on mzXML data (59) with a 10 ppm tolerance. The relative proportion of each glycopeptide is reported in Table II.
Table II. Report of Srr1 glycosylated peptides identified with Spectrum Finder after HCD fragmentation on Q Exactive.
| Peptide | Modification | Relative abundance (%)a | Supplemental figure number |
|---|---|---|---|
| S192TSISEVSNILETQASLTDK211 | None | 1.33% | Sup. Fig 5A |
| 2 HexNAc | 28.7% | Sup. Fig 5B | |
| 3 HexNAc | 26.51% | Sup. Fig 5C | |
| 1 HexNAc | 18.71% | Sup. Fig 5D | |
| 4 HexNAc | 7.00% | Sup. Fig 5E | |
| 1 O-AcHexNAc + 2 HexNAc | 5.29% | Sup. Fig 5F | |
| 4 HexNAc + 2 Hex | 4.41% | Sup. Fig 5G | |
| 1 O-AcHexNAc + 1 HexNAc | 2.32% | Sup. Fig 5H | |
| 1 O-AcHexNAc + 3 HexNAc | 2.22% | Sup. Fig 5I | |
| 1 O-AcHexNAc + 3 HexNAc + 2 Hex | 1.16% | Sup. Fig 5J | |
| 1 O-AcHexNAc | 1.11% | Sup. Fig 5K | |
| 4 HexNAc + 3 Hex | 0.74% | Sup. Fig 5L | |
| 5 HexNAc + 2 Hex | 0.14% | Sup. Fig 5M | |
| 2 O-AcHexNAc + 1 HexNAc | 0.12% | Sup. Fig 5N | |
| 4 HexNAc + 1 Hex | 0.1% | Sup. Fig 5O | |
| 2 O-AcHexNAc | 0.08% | Sup. Fig 5P | |
| 1 O-AcHexNAc + 4 HexNAc | 0.06% | Sup. Fig 5Q | |
| E214SFSANQIVTESSLVTDAGK233 | None | 77.84% | Sup. Fig 5R |
| 1 HexNAc | 17.06% | Sup. Fig 5S | |
| 1 O-AcHexNAc | 3.26% | Sup. Fig 5T | |
| 2 HexNAc | 0.84% | Sup. Fig 5U | |
| 1 O-AcHexNAc + 1 HexNAc | 0.29% | Sup. Fig 5V | |
| N234ASVSSLIEITKPK247 | None | 93.61% | Sup. Fig 5W |
| 1 HexNAc | 4.75% | Sup. Fig 5X | |
| 2 HexNAc | 4.01% | Sup. Fig 5Y | |
| M255SNESLITPEK265 | None | 99.3% | Sup. Fig 5Z |
| 1 HexNAc | 0.7% | Sup. Fig 5AA | |
| T275GNESLTPTIR285 | None | 5.29% | Sup. Fig 5AB |
| 1 HexNAc | 94% | Sup. Fig 5AC | |
| 1 O-AcHexNAc | 0.52% | Sup. Fig 5AD | |
| T451ATLNNQVASIEER464 | None | 72.12% | Sup. Fig 5AE |
| 1 O-AcHexNAc + 1 HexNAc | 27.44% | Sup. Fig 5AF | |
| 2 O-AcHexNAc | 0.45% | Sup. Fig 5AG |
a The relative abundance of each precursor was estimated using MassChroQ software.
RESULTS
Enrichment and Monosaccharide Composition of Full-length Srr1
S. agalactiae Srr1 is a 1310 amino acid protein with an expected molecular mass of 150 kDa. However upon SDS-PAGE Srr1 exhibits an apparent mass exceeding 250 kDa. This anomalous migration is likely to be a consequence of a high glycosylation level (Fig. 1C) combined to unusual amino acid content (50% of serine and threonine residues) which can alter the detergent binding capacity as compared with globular proteins (60, 61). We previously demonstrated a strong reactivity of Srr1 toward the sWGA lectin, which is specific to GlcNAc moieties (38). We thus took advantage of this property to devise a lectin-based enrichment step using agarose linked sWGA. The efficiency of the strategy is illustrated (Fig. 1C): the lectin-based enrichment was required to detect Srr1 by Coomassie Blue staining. Monosaccharide composition of purified Srr1 was determined after acid hydrolysis by GC-MS analysis of trimethylsilyl derivatives. Two monosaccharides were identified: GlcNAc and glucose (Glc) in a 1.8:1 ratio. Unlike what was reported for Srr Fap1 protein of S. parasanguinis, we did not find any evidence for the presence of galactose or N-acetylgalactosamine associated to Srr1 (34).
Glycopeptide Search in CID Spectra of Srr1 Tryptic Peptides
Srr1 tryptic digest was first analyzed by LC-MS/MS on a LTQ-Orbitrap instrument using CID. Because the highly repetitive C-terminal domain does not contain any lysine or arginine residues, the sequence coverage was found to be restricted to the N-terminal region [93–639] (see supplemental Fig. S1). As the first 90 residues of Srr1 corresponded to the atypical signal peptide cleaved by the SecA2 secretion machinery (62), only residues spanning from 90 to 435 of its mature form are potentially accessible to trypsin. We obtained 83% sequence coverage for this region (supplemental Fig. S1).
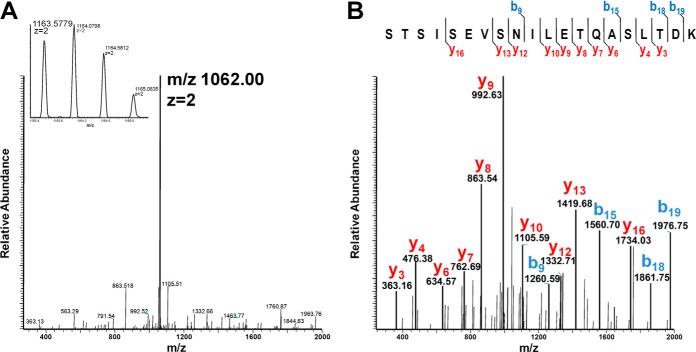
Because of the lability of the O-glycosidic bonds during CID experiments (63) glycan loss from the precursor ion is often the major fragmentation pathway observed in MS/MS spectra. The loss of HexNAc from the [192–211] tryptic peptide at m/z 1163.5779 leading to an abundant m/z 1062.00 is shown in Fig. 4A. A MS3 experiment performed on the deglycosylated form of the peptide at m/z 1062.00 confirmed its primary structure (Fig. 4B).
Fig. 4.
Mass spectrometric identification of Srr1 glycopeptides using CID MS/MS. CID-induced neutral loss of +1 HexNAc on the [192–211] peptide. Three successive MS cycles are shown: (inset A) full MS A, CID fragmentation of the most abundant ion (m/z 1163.5779 z = 2) corresponding to the [192–211] peptide modified with one HexNAc as demonstrated by B, the MS3 fragmentation of m/z 1062.00 z = 2.
In order to identify Srr1 glycopeptides, the set of CID spectra was first analyzed with X! Tandem. The automated analysis of 900 spectra (generated in one LC-MS/MS run), led to the identification of three peptide sequences [214–233], [234–247], and [275–285] carrying a monosaccharide (supplemental Fig. S2A–2D). MS/MS data were also manually inspected, looking for fragmentation spectra dominated by a single ion characteristic of a glycan loss event. This manual search led to the identification of 12 other glycopeptides (Table I). The highest diversity was observed for the [192–211]peptide found associated with ten different glycan combinations, a feature illustrating the heterogeneity of Srr1 glycosylation (Table I and supplemental Fig. S3A–3K). Glycosylation with a Hex was found only on peptides carrying at least four HexNAc (or O-AcHexNAc, see below), suggesting that the latter modification is a prerequisite for further Hex addition. The reactivity of Srr1 toward lectin and the monosaccharide composition analysis of full length protein showed that Srr1 was mainly modified with GlcNAc and Glc residues, strongly suggesting that HexNAc and Hex should correspond to these two sugars.
Table I. Report of Srr1 glycosylated peptides identified after neutral loss event in CID MS/MS experiment.
| Peptide sequence | Modification | (M+H)+ | MS/MS spectraa |
|---|---|---|---|
| S192TSISEVSNILETQASLTDK211 | None | 2123.0714 | Sup. Fig 3A |
| 1 HexNAc | 2326.1511 | Sup. Fig 3B | |
| 2 HexNAc | 2529.2308 | Sup. Fig 3C | |
| 3 HexNAc | 2732.3105 | Sup. Fig 3D | |
| 4 HexNAc | 2935.3888 | Sup. Fig 3E | |
| 4 HexNAc + 2 Hex | 3259.5066 | Sup. Fig 3F | |
| 4 HexNAc + 3 Hex | 3421.5648 | Sup. Fig 3G | |
| 1 O-AcHexNAc | 2368.1613 | Sup. Fig 3H | |
| 1 O-AcHexNAc + 1 HexNAc | 2571.2410 | Sup. Fig 3I | |
| 1 O-AcHexNAc + 2 HexNAc | 2774.3207 | Sup. Fig 3J | |
| 1 O-AcHexNAc + 3 HexNAc | 2977.4004 | Sup. Fig 3K | |
| E214SFSANQIVTESSLVTDAGK233 | None | 2083.0189 | Sup. Fig 3L |
| 1 HexNAc | 2286.0986 | Sup. Fig 3M | |
| 1 O-AcHexNAc | 2328.1088 | Sup. Fig 3N | |
| N234ASVSSLIEITKPK247 | None | 1486.8475 | Sup. Fig 3O |
| 1 HexNAc | 1689.9272 | Sup. Fig 3P | |
| 2 HexNAc | 1893.0069 | Sup. Fig 3Q | |
| T275GNESLTPTIR285 | None | 1188.6219 | NAb |
| 1 HexNAc | 1391.7016 | Sup. Fig 3R | |
| S266QVMIASDKTGNESLTPTIR285 | None | 2164.0914 | Sup. Fig 3S |
| 1 HexNAc | 2367.1711 | Sup. Fig 3T |
a The MS/MS spectra are reported in the corresponding supplemental figures.
b Not Available, MS/MS were selected between 600 and 2000 m/z.
An Unexpected Sugar Modification Identified on Srr1
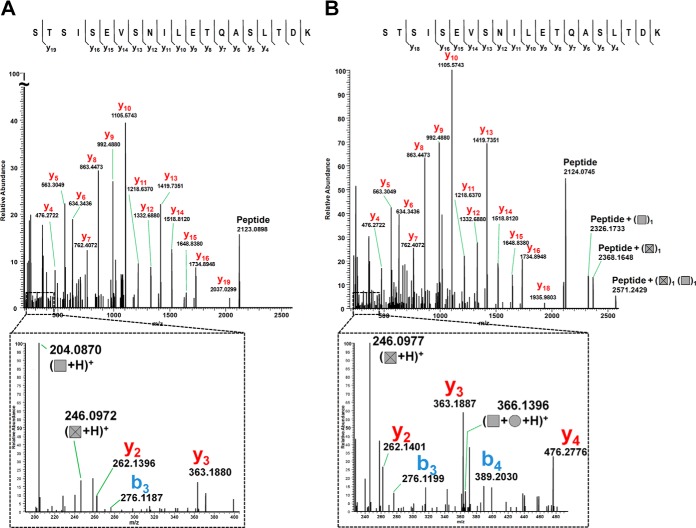
We recurrently observed a neutral loss of 245.09 Da in CID spectra (see for example supplemental Fig. S3H, 3N), that was found either alone or in association with HexNAc moieties on various peptides of Srr1. This mass did not correspond to any known post-translational protein modification but was recently associated to O-acetylated GlcNAc on Lactobacillus plantarum peptidoglycan (see proposed structure Fig. 5A) (64). To get more insight into the structure of this modification, CID spectra of oxonium ions at m/z 246.1 produced by ISF either from peptidoglycan fragments of L. plantarum (Fig. 5B) or Srr1 peptides (Fig. 5C) were generated and compared. The two fragmentation profiles matched perfectly and displayed the presence of ions m/z 210.1 and 228.0 characteristic of acetylated fragments. It was thus concluded that Srr1 can be modified by O-AcHexNAc molecule (Fig. 5). The additional acetylation cannot be precisely located from CID spectra alone, but the striking similarity between fragmentation profiles supports the hypothesis that the precursor ions share the same structure. The β-1,4-glycosidic linkage present in peptidoglycan makes the C4-position unlikely to be modified and we propose the C6-position as a putative site for the additional acetylation.
Fig. 5.
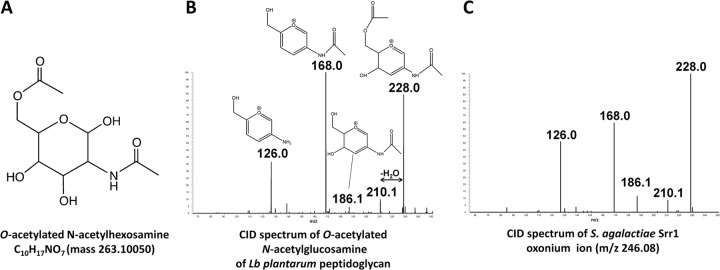
Identification of a novel Srr1 glycomodification by characterization of oxonium ions m/z 246. 0977. Oxonium ions were obtained by ISF of Lb. plantarum peptidoglycan and Srr1 glycopeptides as indicated under “Experimental Procedures.” A, Proposed structure of O-AcHexNAc (64). B, CID fragmentation of O-AcHexNAc oxonium ion from a Lactobacillus plantarum peptidoglycan sample. The characteristic diagnostic ions corresponding to acetylated fragments are: m/z 210.1 and m/z 228.0 as described in (64). C, CID fragmentation of the m/z 246.0977 oxonium ion detected from Srr1 sample.
Development of a New Software Tool to Analyze HCD MS/MS Data Obtained from Glycopeptides
As previously described, CID spectra of all glycopeptides are poorly informative because only the glycosidic bond cleavage is observed. To overcome this problem, we decided to use HCD fragmentation, which is slightly more energetic and allows for consecutive fragmentations. Digests previously analyzed on the LTQ were thus analyzed on a Q Exactive instrument (65). As expected, HCD MS/MS of glycopeptides was found to lead to different fragmentation patterns compared with CID. Interestingly, many y ions, characteristic of the backbone cleavage, could now be observed concomitantly with the formation of oxonium ions. Furthermore, various glycoforms of a given peptide were found to generate highly similar y-type ion profiles. Hence, glycopeptides sharing the same peptide backbone but varying in their glycosylation level display very similar fragmentation patterns while having different precursor masses. As an example, Fig. 6 shows the HCD fragmentation spectra of two different glycoforms of [192–211] peptide at m/z 925.4455 and m/z 1101.1749) illustrating the similarity of y ion profiles and detection of oxonium ions.
Fig. 6.
MS2 spectra of two glycoforms of [192–211] peptidefragmented in HCD illustrating the similar y ion profiles. A, Fragmentation of m/z 925.4455 z = 3, corresponding to the peptide +1 O-AcHexNAc and +2 HexNAc. B, Fragmentation of m/z 1101.1749 z = 3, ([192–211] + 1 O-AcHexNAc, 3 HexNAc, 2 Hex). Ions at m/z 204.0872, 246.0977, and m/z 366.1400 are oxonium ions. Glycan symbols: HexNAc (Square), Hex (Circle), O-AcHexNAc (Crossed square).
A software was developed to exploit the HCD spectra similarities between different glycoforms of a peptide. The software, coined Spectrum Finder, was dedicated to the automated search of specific peptide fragmentation pattern without a priori on the mass of the precursor (for details see experimental procedures and http://pappso.inra.fr/bioinfo/sf). As compared with X! Tandem, analysis of HCD data with SpectrumFinder revealed additional fragmentation spectra originating from the same peptide backbones. For each spectrum selected by SpectrumFinder, the mass difference between the precursor ion and the unmodified peptide was calculated and fitted to a glycan combination of Hex, HexNAc, and O-AcHexNAc (see experimental procedures and Fig. 2, Fig. 3). All the glycopeptides previously identified in the CID experiments (listed in Table I) could be retrieved, demonstrating the robustness of this new approach. Furthermore, 12 novel glycopeptides could be identified: nine corresponded to a novel sugar combination on a peptide sequence already described as glycosylated but two corresponded to peptide sequences that were not previously found as glycosylated ([255–265] and [451–464]) (Table II).
Using our strategy, 17 variants of the [192–211] peptide bearing one to seven sugar residues were identified. Importantly, the semi-quantitative analysis performed on all glycoforms of this peptide revealed that the non-glycosylated form represented less than 1%, whereas more than 80% of the glycosylated forms corresponded to modifications with one to four HexNAc residues (Table II). In this regard it should be stated that HexNAc peptide ion are considered to be suppressed in favor of non-glycosylated peptides (66). As noted above, we confirmed that a Hex was observed only on HexNAc containing peptides.
Two additional glycoforms carrying HexNAc or O-AcHexNAc were identified for the [214–233] peptide and one additional form for [275–285]. The comparison of the extracted ion chromatogram peak areas showed that the new glycopeptides retrieved by SpectrumFinder were of low abundance. These results highlight the gain of sensitivity provided by this approach. To check that none of the observed glycoforms were the result of ISF of larger ones, retention times measured for 16 different glycoforms of the [192–211] peptide were compared (supplemental Fig. S6). Except for the +1 HexNAc and +2 HexNAc glycoforms, all other glycoforms exhibited different retention times. This shows that ISF does not lead to a major bias in our measurements but has in some cases to be taken into account.
As expected an increase in the number of sugar residues leads to a shorter retention time. It is worth noting that in some cases two retention times are observed for the same glycopeptide, probably resulting in the presence of different isomers.
Searching HCD MS/MS spectra with SpectrumFinder led to the identification of a novel segment of Srr1 subject to glycosylation: the [451–464] peptide, which is located distant from the serine-rich region of the N-terminal part and belongs to the IgG domain of Srr1 bearing the fibrinogen (Fg) binding activity (39, 52).
Determination of Glycosylation Sites
ETD has been shown to be a valuable tool for the analysis of glycopeptides because the glycosidic bond can be preserved during the fragmentation process, allowing glycosylation sites to be accurately mapped (10, 67). Srr1 digest was therefore analyzed in both untargeted/targeted approaches using ETD as the fragmentation mode. All acquired ETD spectra were manually analyzed and annotated to locate the modification sites (supplemental Fig. S7). When combined, the different runs enabled the identification of five different peptide sequences bearing glycosylation (Table III).
Table III. Report of glycosylation site on Srr1 peptides identified after ETD fragmentation on LTQ-Orbitrap.
| Peptide | Modification | Parent ion observed (m/z), charge | Sugar position | Supplemental figure Number |
|---|---|---|---|---|
| S192TSISEVSNILETQASLTDK211 | 3 HexNAc | 911.4421, 3+ | (S192-S196) and S207 | Sup. Fig. 8A |
| 1 HexNAc | 776.0556, 3+ | S192-T193 | Sup. Fig. 8B | |
| 1 O-AcHexNAc | 790.0590, 3+ | S196 | Sup. Fig. 8C | |
| E214SFSANQIVTESSLVTDAGK233 | 1 HexNAc | 762.7047, 3+ | S226 | Sup. Fig. 8D |
| 1 O-AcHexNAc | 776.7081, 3+ | S226 | Sup. Fig. 8E | |
| N234ASVSSLIEITKPK247 | 1 HexNAc | 563.9810, 3+ | S239 | Sup. Fig. 8F |
| 2 HexNAc | 631.6742, 3+ | S236 and S239 | Sup. Fig. 8G | |
| T275GNESLTPTIR285 | 1 HexNAc | 696.3547, 2+ | S279 | Sup. Fig. 8H |
| 1 O-AcHexNAc | 717.3598, 2+ | S279 | Sup. Fig. 8I | |
| T451ATLNNQVASIEER464 | 1 O-AcHexNAc + 1 HexNAc | 665.6512, 3+ | T451 | Sup. Fig. 8J |
In the [192–211] +3 HexNAc glycopeptide (supplemental Fig. S8A, S8B), Ser207 was unambiguously identified with one HexNAc, and two other HexNAc that could be restricted to Ser192, Thr193, Ser194, or Ser196. The lack of fragments within the first five residues, which contain four potential modified amino acids, precludes any further assignment. The same peptide was also found with only +1 HexNAc, which can be assigned to either Ser192 or Thr193. The glycopeptide [214–233] was retrieved either with +1 HexNAc or +1 O-AcHexNAc and Ser226 was modified in both peptides. The [234–247] peptide was observed to carry either +1 or +2 HexNAc. In both cases Ser239 was found glycosylated and in the larger peptide an addition glycan is found on Ser236. In the [275–285] peptide, Ser279 was found to be modified either by a HexNAc or by a O-AcHexNAc. The last peptides that have been successfully fragmented by ETD were [451–464] +1 HexNAc or +1 O-AcHexNAc. Unexpectedly, only one residue (Thr451) was found to be modified with a HexNAc-O-AcHexNAc disaccharide. The presence in the ETD spectrum (supplemental Fig. S8J) of m/z 1748.80, which corresponded to a loss of O-AcHexNAc, indicated that it is the HexNAc moiety which is attached to the peptide backbone.
In total, six different glycosylation sites could be unambiguously assigned. These results confirm that Srr1 is O-glycosylated and suggest that serines are preferred over threonines. Interestingly, we noted that the four identified glycosylated serines were followed by a leucine, suggesting that the sequon SerLeu could be a preferred target of GtfAB. However, the threonine modification demonstrates that glycosylation sites are flexible.
Influence of GT Encoded in the srr1_locus on Srr1 Glycosylation
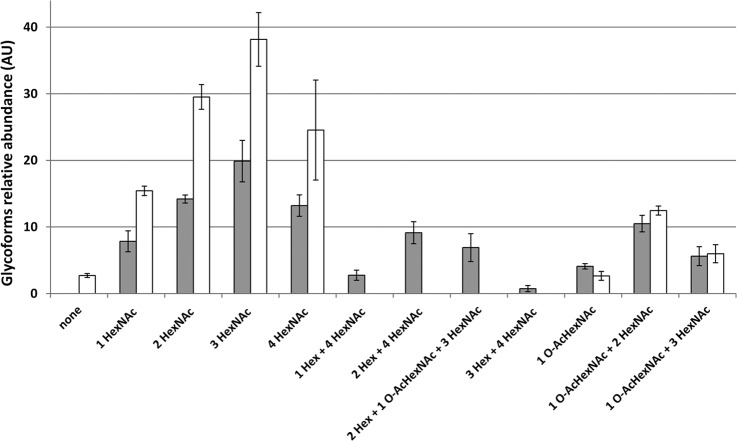
WGA lectin binds to all bacterial Srr adhesins tested so far, indicating that GlcNAc is the predominant sugar in this family of proteins (Fig. 1B) (35, 54). Our data show that all glycopeptides carried at least one HexNAc residues, suggesting that this addition could constitute an initial step in Srr1 glycosylation. We have previously shown that the GtfAB GTs encoded in the Srr1 locus are essential for Srr1 stability (38) as opposed to the six GtfC-H GTs (strain H36BSrtA*ΔgtfCH) (38). However, the protein produced in this genetic background displays a lower apparent molecular weight on SDS-PAGE that could be associated with a glycosylation defect (supplemental Fig. S8). To examine this hypothesis, we compared the abundance of selected Srr1 glycopeptides produced in strains possessing the full set of GT-encoding genes (H36BSrtA*) or where the so-called accessory GT-encoding genes were deleted (H36BSrtA*ΔgtfCH). The Srr1 protein bands were trypsin digested and analyzed on the LTQ-Orbitrap. The XICs corresponding to the glycoforms of the [192–211] peptide (Table I) were extracted from the two experiments. This peptide was chosen because of the large abundance and diversity of its glycoforms. As shown in Fig. 7, all the Hex-containing glycopeptides are now absent in the ΔgtfCH strain, suggesting that one or more of the corresponding GT-encoding genes is involved in its transfer onto GlcNAc-modified residues.
Fig. 7.
Impact of the GtfCH GTs on the abundance of the glycoforms of the [192–211] peptide. The glycoforms of the [192–211] peptide (Table II) issued from Srr1 synthesized in H36BSrtA* (gray bars) and H36BSrtA*ΔgtfCH (white bars) strains were quantified by calculating the area of each parent ions computed from XIC and normalized (see experimental procedures). LC-MS/MS experiments were performed in triplicates and standard deviation (S.D.) is reported for each quantified glycoform. All Hex containing glycoforms were absent in Srr1 produced in ΔgtfCH background.
DISCUSSION
Srr proteins constitute a widespread family of bacterial adhesins expressed in several Gram-positive bacterial species of human and veterinary clinical importance. Using MS-based approaches and dedicated software tool, we demonstrate that Srr1 produced in S. agalactiae displays a wide diversity of glycoforms attached to a protein segment located outside of SR1, the highest Ser/Thr density region of the N-terminal region (Fig. 8). Some glycopeptides are found with a various number of sugars, which highlights Srr1 glycosylation heterogeneity. The most striking example is associated to the [192–211] peptide that can be modified with at least 16 different combinations of HexNAc and Hex. Semi-quantitative analysis on this glycopeptide family shows that glycosylated forms would represent more than 98% of the total amount of the peptide (Table II). Among the glycoforms observed, some are not eluted as single and symmetrical peaks (see supplemental Fig. S6) suggesting the presence of different positional isomers that further increase the heterogeneity of the protein. Monosaccharide composition analysis of full-length Srr1 demonstrated that GlcNAc and Glc were the major sugar associated to the protein. Although, the present mass spectrometry analysis is restricted to the N-terminal part which contains only a fraction (28 Ser/Thr of the potential O-glycosylation sites (624 Ser/Thr) of Srr1, it is reasonable to assume that the HexNAc and Hex modifications correspond to GlcNAc and Glc residues. A definitive conclusion will require compositional analysis of the N-terminal region.
Fig. 8.
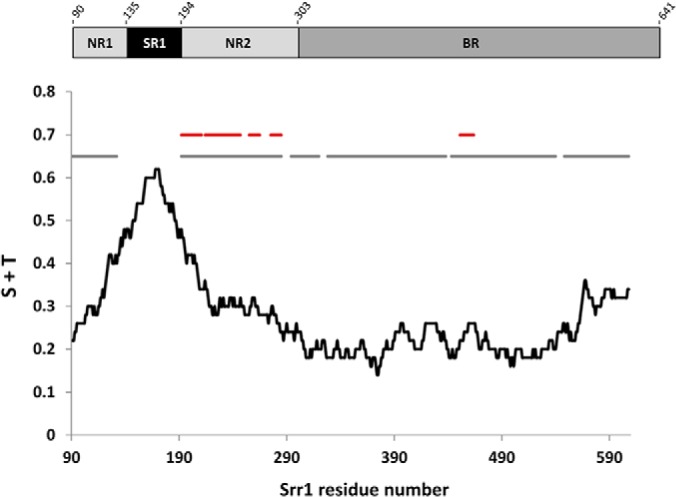
Serine and threonine content of the N-terminal region of Srr1 in relation with MS-identified peptide and glycopeptide. The black line displays the serine+threonine content of the sequence, calculated as a floating average with 50 residues in length sliding windows and expressed as proportion. The gray lines relate to the sequence numbering and correspond to region covered by the present mass spectrometry analysis. The red lines relate to the sequence numbering and correspond to glycosylated regions listed in Table II. The numbering starts at residue 91 corresponding to the secreted form of the protein.
An additional level of complexity is obtained by the optional and surprising acetylation of GlcNAc. The same chemical modification was recently described on the glycan backbone of L. plantarum peptidoglycan where it provides an increased resistance toward the muramidase activity of lysozyme (64). This modification is performed by OatB, a membrane-embedded acetyltransferase, which possesses 40% homology with Gbs0052, an ortholog present in S. agalactiae. It is possible that this enzyme also acetylates GlcNAc residues decorating Srr1. The capacity of an enzyme to act on different surface compounds is not uncommon; in several Gram-negative species it was demonstrated that enzymes belonging to the lipopolysaccharide biosynthesis machinery were required for O-glycosylation of flagellins (68–70). It was also found that a common pentasaccharide is used to O-glycosylate proteins and synthesize the capsule of the opportunistic pathogen Acinetobacter baumannii (71). Interestingly, a 245 Da component has recently been associated with glycan modifying surface proteins in Campylobacter hominis, the sole Campylobacter species considered as a human commensal (4). That the same post-translational modification of surface proteins was found in C. hominis and S. agalactiae, two unrelated species living in the same environment, could correspond to a niche-specific adaptation.
In the absence of the gtfCH genes encoding the GTs that belong to the GT2 and GT8 family (72), Hex-modified [192–211] glycopeptides are no more detected (Fig. 7). This suggests that the first glycosylation step of Srr1 is the addition of a GlcNAc residue through GtfAB activity, and is also a strong indication that the Hex transferring enzyme(s) is encoded by one of the six deleted genes. In line with this conclusion, it was recently demonstrated that Gtf3, the S. parasanguinis ortholog of GtfC (43% identity at the amino acid level), possesses a glucosyltransferase activity in vitro (73). Importantly, all O-AcHexNAc modifications were present on Srr1 produced in the ΔgtfCH strain (Fig. 7) demonstrating that the enzyme responsible of this modification was not encoded in the gtfCH locus.
As illustrated in Fig. 8, the [192–211] peptide is located near the region having the highest serine/threonine density (>40–60%) at the N terminus. A large part of that SR1 region [133–191] remained undetected in our experiments. This was not unexpected given the compositional bias (56% Ser/Thr), size and charge properties of the corresponding tryptic peptide. It is likely that the missing peptide is glycosylated but cannot be detected in our experimental conditions. In that respect, Srr proteins resemble eukaryotic O-glycosylated mucins, which are extremely difficult to analyze because of the heterogeneity and abundance of the glycomodifications (74–76). In S. gordonii, studies performed on truncated variants of GspB and based on the reactivity of lectin suggested that only SR1 region undergoes glycosylation (54). Our results demonstrate that residues located in the binding region of the protein such as [451–464] can also be glycosylated.
An important question is to understand what causes the high heterogeneity of Srr1 glycosylation. The biosynthesis of glycan and their attachment to proteins is dependent on a variety of biochemical parameters like the sugar nucleotide donor availability or the activity of glycosylating enzymes (77). Because glycosylation of proteins is a non-template process, variation of these parameters during cultivation could affect protein glycosylation as it has been shown for eukaryotic proteins (78). The consequences of varying S. agalactiae cultivation conditions (medium, carbon sources, temperature etc.) on Srr1 glycosylation characteristics (location, sugars added, and heterogeneity) could provide valuable clues on the mechanisms susceptible to influence the pathway.
The biological importance of protein glycosylation and how it can assist Gram-positive species in their commensal lifestyle is an important issue. Unfortunately, our current knowledge of the consequence of Srr1 glycosylation on its functional properties remains limited. Biochemical analysis of Srr proteins in GT expression mutants have established that GtfAB-dependent glycosylation is essential for Srr production, stability or export depending on the species studied (34, 38, 79, 80). When considering the biology of protein modification by GlcNAc, it is important to mention that all major pathways of central metabolism are required for UDP-GlcNAc synthesis (66). Thus, it can be hypothesized that O-GlcNAcylation of Srr1 could be indexed to nutrient availability because the sensor role of this modification is well established in eukaryotes (66).
Our data indicate that Srr1 glycosylation is not dependent on a strict consensus sequence, although we observed that glycosylation of serine was favored when it was followed by a leucine. There must be some additional structural elements of Srr1 that might control GT activity. The identification of these features will require to being able to explore the glycosylation of regions presenting the highest Ser/Thr density. The achievement of this objectives could be facilitated by the development of an in vitro glycosylation model similar to that developed for Fap1 (33).
Major efforts are underway to elucidate the mechanisms of bacterial pathogenesis for preventing and treating infections. A majority of the bacterial glycoproteins identified so far are localized at the cell-surface where they are involved in a variety of processes such as biofilm formation, adhesion, cell motility, or immune system escape (2, 79, 81). In Bacteroides fragilis, a Gram-negative human symbiont, the general O-glycosylation system is involved in the modification of tens of proteins, contributes to in vitro fitness of the species and is also required for the colonization of the mammalian gastro-intestinal gut (82, 83). These observations support the idea that protein glycosylation systems participate in the interaction of bacteria with mammalian host and contribute to commensalism or pathogenesis (1, 84).
Surface proteins represent obvious targets for the development of vaccines and Srr proteins of S. agalactiae have been proposed as good candidates (51). In this perspective it will be important to characterize the glycosylation dynamics of protein susceptible to be chosen as attractive antigens because this mechanism could circumvent antibody recognition.
Supplementary Material
Acknowledgments
We thank Prof. Colin Tinsley for his help in the design of Fig. 8.
Footnotes
Author contributions: T.C., A.G., and M.-Y.M. designed research; T.C., A.G., A.-M.d.G., and M.-Y.M. performed research; B.V., O.L., and S.D. contributed new reagents or analytic tools; T.C., A.G., J.C.-R., and M.-Y.M. analyzed data; T.C., J.C.-R., P.T.-C., and M.-Y.M. wrote the paper.
* We thank the French National Research Agency (ANR) for supporting the “GlycoPath” project. The DIM Malinf from the region Ile-de-France is also acknowledged for funding of the LTQ-Orbitrap Velos mass spectrometer.
 This article contains supplemental Figs. S1 to S8.
This article contains supplemental Figs. S1 to S8.
1 The abbreviations used are:
- GT
- glycosyltransferase
- BR
- binding region
- BSN
- Bjerrum and Shaffer Nielsen buffer
- CID
- collision induced dissociation
- ETD
- electron transfer dissociation
- Glc
- glucose
- GlcNAc
- N-acetylglucosamine
- HCD
- higher energy collisional dissociation
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- ISF
- in source fragmentation
- MS/MS
- tandem mass spectrometry
- NCE
- normalized collision energy
- NR
- non repeat region
- O-AcHexNAc
- O-acetylated-N-acetylhexosamine
- PAGE
- polyacrylamide gel electrophoresis
- SF
- SpectrumFinder
- SR
- serine-rich (region)
- Srr
- serine-rich (protein)
- sWGA
- succinylated wheat germ agglutinin
- TFA
- trifluoroacetic acid
- TH
- Todd-Hewitt broth
- Tris
- 2-amino-2-hydroxymethyl-1,3-propanediol
- WGA
- wheat germ agglutinin
- XIC
- extracted ion chromatogram.
REFERENCES
- 1. Nothaft H., Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 2. Nothaft H., Szymanski C. M. (2013) Bacterial protein N-glycosylation: new perspectives and applications. J. Biol. Chem. 288, 6912–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwashkiw J. A., Vozza N. F., Kinsella R. L., Feldman M. F. (2013) Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 89, 14–28 [DOI] [PubMed] [Google Scholar]
- 4. Nothaft H., Scott N. E., Vinogradov E., Liu X., Hu R., Beadle B., Fodor C., Miller W. G., Li J., Cordwell S. J., Szymanski C. M. (2012) Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol. Cell. Proteomics 11, 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glover K. J., Weerapana E., Imperiali B. (2005) In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U. S. A. 102, 14255–14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W., Aebi M. (2002) N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 [DOI] [PubMed] [Google Scholar]
- 7. Power P. M., Seib K. L., Jennings M. P. (2006) Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 347, 904–908 [DOI] [PubMed] [Google Scholar]
- 8. Hartley M. D., Morrison M. J., Aas F. E., Borud B., Koomey M., Imperiali B. (2011) Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aas F. E., Vik A., Vedde J., Koomey M., Egge-Jacobsen W. (2007) Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 65, 607–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott. N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Hojrup P., Packer N. H., Larsen M. R., Cordwell S. J. (2010) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031-MCP000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vik A., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W., Koomey M. (2009) Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 106, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thibault P., Logan S. M., Kelly J. F., Brisson J. R., Ewing C. P., Trust T. J., Guerry P. (2001) Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276, 34862–34870 [DOI] [PubMed] [Google Scholar]
- 13. Twine S. M., Reid C. W., Aubry A., McMullin D. R., Fulton K. M., Austin J., Logan S. M. (2009) Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191, 7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schirm M., Soo E. C., Aubry A. J., Austin J., Thibault P., Logan S. M. (2003) Structural, genetic, and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48, 1579–1592 [DOI] [PubMed] [Google Scholar]
- 15. Verma A., Schirm M., Arora S. K., Thibault P., Logan S. M., Ramphal R. (2006) Glycosylation of b-Type flagellin of Pseudomonas aeruginosa: structural and genetic basis. J. Bacteriol. 188, 4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grass S., Lichti C. F., Townsend R. R., Gross J., St. Geme J. W., 3rd (2010) The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6, e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gross J., Grass S., Davis A. E., Gilmore-Erdmann P., Townsend R. R., St. Geme J. W., 3rd (2008) The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 283, 26010–26015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schirm M., Kalmokoff M., Aubry A., Thibault P., Sandoz M., Logan S. M. (2004) Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J. Bacteriol. 186, 6721–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schirm M., Schoenhofen I. C., Logan S. M., Waldron K. C., Thibault P. (2005) Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem, 77, 7774–7782 [DOI] [PubMed] [Google Scholar]
- 20. Shen A., Kamp H. D., Grundling A., Higgins D. E. (2006) A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20, 3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu C. F., Tonini L., Malaga W., Beau M., Stella A., Bouyssie D., Jackson M. C., Nigou J., Puzo G., Guilhot C., Burlet-Schiltz O., Riviere M. (2013) Bacterial protein-O-mannosylating enzyme is crucial for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 110, 6560–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. VanderVen B. C., Harder J. D., Crick D. C., Belisle J. T. (2005) Export-mediated assembly of mycobacterial glycoproteins parallels eukaryotic pathways. Science 309, 941–943 [DOI] [PubMed] [Google Scholar]
- 23. Wehmeier S., Varghese A. S., Gurcha S. S., Tissot B., Panico M., Hitchen P., Morris H. R., Besra G. S., Dell A., Smith M. C. (2009) Glycosylation of the phosphate binding protein, PstS, in Streptomyces coelicolor by a pathway that resembles protein O-mannosylation in eukaryotes. Mol. Microbiol, 71, 421–433 [DOI] [PubMed] [Google Scholar]
- 24. Mahne M., Tauch A., Puhler A., Kalinowski J. (2006) The Corynebacterium glutamicum gene pmt encoding a glycosyltransferase related to eukaryotic protein-O-mannosyltransferases is essential for glycosylation of the resuscitation promoting factor (Rpf2) and other secreted proteins. FEMS Microbiol. Lett, 259, 226–233 [DOI] [PubMed] [Google Scholar]
- 25. Ristl R., Steiner K., Zarschler K., Zayni S., Messner P., Schaffer C. (2011) The s-layer glycome-adding to the sugar coat of bacteria. Int. J. Microbiol. 2011, epub ref: 20871840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fagan R. P., Fairweather N. F. (2014) Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222 [DOI] [PubMed] [Google Scholar]
- 27. Anzengruber J., Pabst M., Neumann L., Sekot G., Heinl S., Grabherr R., Altmann F., Messner P., Schaffer C. (2014) Protein O-glucosylation in Lactobacillus buchneri. Glycoconj. J. 31, 117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zarschler K., Janesch B., Pabst M., Altmann F., Messner P., Schaffer C. (2010) Protein tyrosine O-glycosylation-a rather unexplored prokaryotic glycosylation system. Glycobiology 20, 787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lebeer S., Claes I. J., Balog C. I., Schoofs G., Verhoeven T. L., Nys K., von Ossowski I., de Vos W. M., Tytgat H. L., Agostinis P., Palva A., Van Damme E. J., Deelder A. M., De Keersmaecker S. C., Wuhrer M., Vanderleyden J. (2012) The major secreted protein Msp1/p75 is O-glycosylated in Lactobacillus rhamnosus GG. Microb. Cell Fact. 11, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rolain T., Bernard E., Beaussart A., Degand H., Courtin P., Egge-Jacobsen W., Bron P. A., Morsomme P., Kleerebezem M., Chapot-Chartier M. P., Dufrene Y. F., Hols P. (2013) O-glycosylation as a novel control mechanism of peptidoglycan hydrolase activity. J. Biol. Chem. 288, 22233–22247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fredriksen L., Mathiesen G., Moen A., Bron P. A., Kleerebezem M., Eijsink V. G., Egge-Jacobsen W. (2011) The major autolysin Acm2 from Lactobacillus plantarum undergoes cytoplasmic O-glycosylation. J. Bacteriol. 194, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lizcano A., Sanchez C. J., Orihuela C. J. (2012) A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol. Oral Microbiol. 27, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou M., Wu H. (2009) Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155, 317–327 [DOI] [PubMed] [Google Scholar]
- 34. Stephenson A. E., Wu H., Novak J., Tomana M., Mintz K., Fives-Taylor P. (2002) The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43, 147–157 [DOI] [PubMed] [Google Scholar]
- 35. Takamatsu D., Bensing B. A., Sullam P. M. (2004) Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 52, 189–203 [DOI] [PubMed] [Google Scholar]
- 36. Rose L., Shivshankar P., Hinojosa E., Rodriguez A., Sanchez C. J., Orihuela C. J. (2008) Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J. Infect. Dis. 198, 375–383 [DOI] [PubMed] [Google Scholar]
- 37. Siboo I.R., Chambers H. F., Sullam P. M. (2005) Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 73, 2273–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mistou M. Y., Dramsi S., Brega S., Poyart C., Trieu-Cuot P. (2009) Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J. Bacteriol. 191, 4195–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seo H. S., Mu R., Kim B. J., Doran K. S., Sullam P. M. (2012) Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis. PLoS Pathog. 8, e1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seo H. S., Xiong Y. Q., Sullam P. M. (2013) Role of the serine-rich surface glycoprotein Srr1 of Streptococcus agalactiae in the pathogenesis of infective endocarditis. PLoS One 8, e64204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Sorge N. M., Quach D., Gurney M. A., Sullam P. M., Nizet V., Doran K. S. (2009) The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J. Infect. Dis. 199, 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Samen U., Eikmanns B. J., Reinscheid D. J., Borges F. (2007) The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect. Immun. 75, 5405–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Feltcher M. E., Braunstein M. (2012) Emerging themes in SecA2-mediated protein export. Nat. Rev. Microbiol. 10, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takamatsu D., Bensing B. A., Cheng H., Jarvis G. A., Siboo I. R., Lopez J. A., Griffiss J. M., Sullam P. M. (2005) Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol. Microbiol. 58, 380–392 [DOI] [PubMed] [Google Scholar]
- 45. Sanchez C. J., Shivshankar P., Stol K., Trakhtenbroit S., Sullam P. M., Sauer K., Hermans P. W., Orihuela C. J. (2010) The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 6, e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiong Y. Q., Bensing B. A., Bayer A. S., Chambers H. F., Sullam P. M. (2008) Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb. Pathog. 45, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Verani J. R., McGee L., Schrag S. J. (2010) Prevention of perinatal group B streptococcal disease–revised guidelines from CDC, 2010. MMWR Recomm. Rep. 59, 1–36 [PubMed] [Google Scholar]
- 48. Edmond K. M., Kortsalioudaki C., Scott S., Schrag S. J., Zaidi A. K., Cousens S., Heath P. T. (2012) Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet 379, 547–556 [DOI] [PubMed] [Google Scholar]
- 49. Phares C. R., Lynfield R., Farley M. M., Mohle-Boetani J., Harrison L. H., Petit S., Craig A. S., Schaffner W., Zansky S. M., Gershman K., Stefonek K. R., Albanese B. A., Zell E. R., Schuchat A., Schrag S. J. (2008) Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA 299, 2056–2065 [DOI] [PubMed] [Google Scholar]
- 50. Sheen T. R., Jimenez A., Wang N. Y., Banerjee A., van Sorge N. M., Doran K. S. (2011) Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J. Bacteriol. 193, 6834–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Doro F., Liberatori S., Rodriguez-Ortega M. J., Rinaudo C. D., Rosini R., Mora M., Scarselli M., Altindis E., D'Aurizio R., Stella M., Margarit I., Maione D., Telford J. L., Norais N., Grandi G. (2009) Surfome analysis as a fast track to vaccine discovery: identification of a novel protective antigen for Group B Streptococcus hypervirulent strain COH1. Mol. Cell. Proteomics 8, 1728–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seo H. S., Minasov G., Seepersaud R., Doran K. S., Dubrovska I., Shuvalova L., Anderson W. F., Iverson T. M., Sullam P. M. (2013) Characterization of fibrinogen binding by glycoproteins Srr1 and Srr2 of Streptococcus agalactiae. J. Biol. Chem. 288, 35982–35996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bierne H., Dramsi S. (2012) Spatial positioning of cell wall-anchored virulence factors in Gram-positive bacteria. Curr. Opin. Microbiol. 15, 715–723 [DOI] [PubMed] [Google Scholar]
- 54. Takamatsu D., Bensing B. A., Sullam P. M. (2004) Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J. Bacteriol. 186, 7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Konto-Ghiorghi Y., Mairey E., Mallet A., Dumenil G., Caliot E., Trieu-Cuot P., Dramsi S. (2009) Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5, e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krüger R., Hung C-W., Edelson-Averbukh M., Lehmann W. D. (2005) Iodoacetamide-alkylated methionine can mimic neutral loss of phosphoric acid from phosphopeptides as exemplified by nano-electrospray ionization quadrupole time-of-flight parent ion scanning. Rapid Commun. Mass Sp. 19, 1709–1716 [DOI] [PubMed] [Google Scholar]
- 57. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 58. Yen C. Y., Meyer-Arendt K., Eichelberger B., Sun S., Houel S., Old W. M., Knight R., Ahn N. G., Hunter L. E., Resing K. A. (2009) A simulated MS/MS library for spectrum-to-spectrum searching in large scale identification of proteins. Mol. Cell. Proteomics 8, 857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Valot B., Langella O., Nano E., Zivy M. (2011) MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics 11, 3572–3577 [DOI] [PubMed] [Google Scholar]
- 60. Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grefrath S. P., Reynolds J. A. (1974) The molecular weight of the major glycoprotein from the human erythrocyte membrane. Proc. Natl. Acad. Sci. U. S. A. 71, 3913–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rigel N. W., Braunstein M. (2008) A new twist on an old pathway-accessory Sec systems. Mol. Microbiol. 69, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wuhrer M., Catalina M. I., Deelder A. M., Hokke C. H. (2007) Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 849, 115–128 [DOI] [PubMed] [Google Scholar]
- 64. Bernard E., Rolain T., Courtin P., Guillot A., Langella P., Hols P., Chapot-Chartier M. P. (2011) Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286, 23950–23958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111 011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Trinidad J. C., Schoepfer R., Burlingame A. L., Medzihradszky K. F. (2013) N- and o-glycosylation in the murine synaptosome. Mol. Cell. Proteomics 12, 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tabei S. M., Hitchen P. G., Day-Williams M. J., Merino S., Vart R., Pang P. C., Horsburgh G. J., Viches S., Wilhelms M., Tomas J. M., Dell A., Shaw J. G. (2009) An Aeromonas caviae genomic island is required for both O-antigen lipopolysaccharide biosynthesis and flagellin glycosylation. J. Bacteriol. 191, 2851–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Miller W. L., Matewish M. J., McNally D. J., Ishiyama N., Anderson E. M., Brewer D., Brisson J. R., Berghuis A. M., Lam J. S. (2008) Flagellin glycosylation in Pseudomonas aeruginosa PAK requires the O-antigen biosynthesis enzyme WbpO. J. Biol. Chem. 283, 3507–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hug I., Feldman M. F. (2010) Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology 21, 138–151 [DOI] [PubMed] [Google Scholar]
- 71. Lees-Miller R. G., Iwashkiw J. A., Scott N. E., Seper A., Vinogradov E., Schild S., Feldman M. F. (2013) A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 89, 816–830 [DOI] [PubMed] [Google Scholar]
- 72. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2013) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhou M., Zhu F., Dong S., Pritchard D. G., Wu H. (2010) A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis. J. Biol. Chem. 285, 12140–12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Darula Z., Sherman J., Medzihradszky K. F. (2012) How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides. Mol. Cell. Proteomics 11, O111 016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tran D. T., Ten Hagen K. G. (2013) Mucin-type O-glycosylation during development. J. Biol. Chem. 288, 6921–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Christiansen M. N., Kolarich D., Nevalainen H., Packer N. H., Jensen P. H. (2010) Challenges of determining O-glycopeptide heterogeneity: a fungal glucanase model system. Anal. Chem. 82, 3500–3509 [DOI] [PubMed] [Google Scholar]
- 77. St. Amand M. M., Tran K., Radhakrishnan D., Robinson A. S., Ogunnaike B. A. (2014) Controllability analysis of protein glycosylation in CHO cells. PLoS One 9, e87973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jedrzejewski P. M., Del Val I. J., Constantinou A., Dell A., Haslam S. M., Polizzi K. M., Kontoravdi C. (2014) Towards controlling the glycoform: a model framework linking extracellular metabolites to antibody glycosylation. Int. J. Mol. Sci. 15, 4492–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Peng Z., Wu H., Ruiz T., Chen Q., Zhou M., Sun B., Fives-Taylor P. (2008) Role of gap3 in Fap1 glycosylation, stability, in vitro adhesion, and fimbrial and biofilm formation of Streptococcus parasanguinis. Oral Microbiol. Immunol. 23, 70–78 [DOI] [PubMed] [Google Scholar]
- 80. Wu R., Zhou M., Wu H. (2010) Purification and characterization of an active N-acetylglucosaminyltransferase enzyme complex from Streptococci. Appl. Environ. Microbiol. 76, 7966–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Iwashkiw J. A., Vozza N. F., Kinsella R. L., Feldman M. F. (2013) Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 89, 14–28 [DOI] [PubMed] [Google Scholar]
- 82. Fletcher C. M., Coyne M. J., Comstock L. E. (2010) Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J. Biol. Chem. 286, 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fletcher C. M., Coyne M. J., Villa O. F., Chatzidaki-Livanis M., Comstock L. E. (2009) A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Comstock L. E. (2009) Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell Host Microbe 5, 522–526 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.