Abstract
The mammalian bladder urothelium classified as basal, intermediate, and terminally differentiated umbrella cells offers one of the most effective permeability barrier functions known to exist in nature because of the formation of apical uroplakin plaques and tight junctions. To improve our understanding of urothelial differentiation, we analyzed the microRNA (miRNA) expression profiles of mouse urinary tissues and by TaqMan miRNA analysis of microdissected urothelial layers and in situ miRNA-specific hybridization to determine the dependence of these miRNAs on the differentiation stage. Our in situ hybridization studies revealed that miR-205 was enriched in the undifferentiated basal and intermediate cell layers. We then used a quantitative proteomics approach to identify miR-205 target genes in primary cultured urothelial cells subjected to antagomir-mediated knockdown of specific miRNAs. Twenty-four genes were reproducibly regulated by miR-205; eleven of them were annotated as cell junction- and tight junction-related molecules. Western blot analysis demonstrated that antagomir-induced silencing of miR-205 in primary cultured urothelial cells elevated the expression levels of Tjp1, Cgnl1, and Cdc42. Ectopic expression of miR-205 in MDCK cells inhibited the expression of tight junction proteins and the formation of tight junctions. miR-205- knockdown urothelial cells showed alterations in keratin synthesis and increases of uroplakin Ia and Ib, which are the urothelial differentiation products. These results suggest that miR-205 may contribute a role in regulation of urothelial differentiation by modulating the expression of tight junction-related molecules.
The surfaces of the urinary bladder and the lower urinary tract are covered by urothelial cells; these are classified as basal, intermediate, and terminally differentiated superficial cells (umbrella cells) based on their localization relative to the basement membrane or lumen (for review, see (1)). The outermost umbrella cell layer is characterized by a highly specialized superstructure known as the plaques of asymmetric unit membrane (AUM)1 (2–4). These AUM plaques occupy ∼90% of the umbrella cell surface and are composed of uroplakins (a family of integral membrane proteins) (5, 6). Functionally, the urothelium contributes to forming an extensible barrier that accommodates the large changes in urine volume and offers highly specialized umbrella cells that have distinct apical and basolateral membrane domains demarcated by tight junctions (7–9).
The formation of tight junctions creates a tight seal and a high-resistance paracellular barrier between adjacent umbrella cells (7, 10). When the epithelial barrier is compromised, leakage of urine components into the underlying bladder layers results, with symptoms of interstitial cystitis (IC) (11, 12). The restoration of tight junction formations is one of the earliest cellular events in the bladder wound-healing process of IC (13, 14). The molecular constituents of the tight junction include transmembrane proteins, cytoplasmic proteins, and the cytoskeleton. The relevant transmembrane proteins (e.g. occludin, claudins, and JAMs) cross the junction and connect the membranes of adjacent cells to make a tight seal. Cytoplasmic tight junction-associated proteins form scaffolds that anchor tight junction membrane proteins and also attach to the cytoskeleton to further initiate cellular signaling pathways (e.g. to regulate polarity (15, 16) and proliferation (17–19). Among them the scaffolding protein, ZO-1 (tight junction protein 1, TJP1), directly interacts with transmembrane proteins and is required for the localization of occludin to the tight junction (20, 21). Cingulin (CGN) and paracingulin (cingulin-like 1; CGNL1) form links between tight junction proteins (TJP1, TJP2, TJP3, and JAM-A) and the actins cytoskeleton (22–24). CGN has been shown to inhibit RhoA activity and claudin-2 mediated cell proliferation by recruiting GEF-H1 (23, 24). CGNL1, also known as JACOP (junction-associated coiled-coil protein), appears to be structurally and functionally similar to CGN (22–24). In addition to down-regulating RhoA activity, CGNL1 reportedly up-regulates Rac1 activity via the Rac1-GEF Tiam1 interaction (24). Another scaffolding protein, angiomotin (Amot, a component of the Rich1/Amot complex), reportedly recruits the Cdc42 GTPase activating protein (i.e. Rich1) to tight junctions and helps maintain the stability of tight junction polarity complexes (25). The most differentiated, superficial cells of the urothelium develop tight junctions that form a seal between adjacent cells, thus preventing paracellular diffusion. As being an integral part of the urothelial differentiation program, the formation of tight junctions is generally known to be controlled at the transcriptional regulation and subcellular localization of the tight junction proteins (9, 26). However, little is known about the molecular mechanisms underlying the formation of tight junctions in the urothelium.
The small noncoding microRNAs (miRNAs) play critical roles in regulating gene expression via post-transcriptional gene silencing, and have emerged as a prominent class of gene regulators in a variety of biological pathways (27–29). In general, miRNAs are initially transcribed as longer primary transcripts, and then undergo a series of processing steps. These intermediate bands were thought to be the by-products from the RNA cleavage by two ribonuclease (RNase) III enzymes, Drosha and Dicer, also assembled into ribonucleoprotein (RNP) complexes before finally yield the single-stranded mature miRNAs (for review, see (30)). To date, the details of miRNA biogenesis and gene silencing are still unclear. The delicate miRNA system has created a possibility for a better understanding of urothelial differentiation by investigating miRNAs and their target genes. Therefore, we explored urothelial miRNA expression profiles and revealed that expression of miR-205 in a differentiation stage-specific manner in urothelium. Although several studies have elucidated that targets of miR-205 can modulate a wide spectrum of pathways in many cell types, including Akt signaling (31), PTEN (32), EMT program (33), and PI(3)K pathway (34). However, no study has systematically examined the role of miR-205 in normal urothelial differentiation. Recent advances in various “omics” approaches are employed to gain the knowledge of various cellular functions, pathways, and disease biomarker identification (35–38). In the present study, we used a multiple-omics approach to investigate the targets of miR-205 in primary cultured urothelial cells. Collectively, our results demonstrate a novel regulation wherein miR-205 modulates tight junction-related proteins during the urothelial cellular differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture, Antagomirs, and Pre-miR Precursors
Normal urothelial cells isolated from mouse bladder were primary cultured as described in previous studies (39). Briefly, after treating both with Dispase (2.5% w/v in DMEM; Invitrogen, Carlsbad, CA) at 37 °C for one hour and trypsin (0.25% w/v in PBS; Invitrogen) at 37 °C for 10 min, the detached urothelial cells were plated in the keratinocyte serum-free medium (KSFM; Invitrogen). Antagomirs directed against miR-205 (antagomiR-205; Ant-205), miR-709 (antagomiR-709; Ant-709), and scramble controls were synthesized by Dharmacon and were added to the culture medium to a final concentration of 1 mm for 72 h. The single-stranded RNAs consisted of a 19–22-nucleotide length with modifications as specified: Ant-205, 5′-CsAsGACUCCGGUGGAAUGAAsGsGsAs-Chol-3′; Ant-709, 5′-UsCsCUCCUGCCUCUGCCsUsCsCs-Chol-3′. Uppercase letters represent 2′-OMe-modified nucleotides, “s” represents a phosphorothioate backbone, and Chol represents cholesterol. For overexpression studies, Ambion Pre-miR precursors of miR-205 and Negative Control #1 were synthesized by Applied Biosystems (Foster City, CA).
RNA Isolation, miRNA Microarray, and mRNA Microarray Analyses
The total RNA isolation was carried out from inbred ICR mice tissues and primary cultured urothelial cells using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions for gene expression profiling and real-time RT-PCR analyses. The small RNA was enriched using a two-step procedure for miRNA microarray analysis. First, all high molecular weight RNA species over 300 nucleotides in length were removed by using the Millipore Microcon YM-100 Centrifugal Filter Unit. The flow-through containing small RNA was collected in the second step by using a Millipore Microcon YM-3 unit, which retains all RNA molecules over 10 nucleotides in length. The purified small RNA fraction was collected from the filter unit containing mature miRNA, tRNA, 5S rRNA, and 5.8S rRNA. These small mRNA fragments were processed by direct labeling with Cy5 for hybridizations of experimental, control, and degradation tests. Labeling and hybridization were performed as described previously (40). CombiMatrix Mouse MicroRNA CustomArray 4 × 2K slides with 537 miRNA probes were used for gene-expression profiling. Arrays were scanned using a Genepix 4000B Scanner (Axon) and raw pixel intensities were extracted with CombiMatrix Microarray Imager 5.8 software. Signal intensity for each spot was normalized by subtracting the local background that was set by the median signal from all mismatch probes of the area surrounding each spot from the total intensity. The probe signals that were more than two times the background signal were considered as “present.” After conducting the microarray analyses, the mean was computed over all miRNA signals from all samples, and the fold change relative to the mean for each miRNA was log2 transformed. Subsequently, a cluster analysis was completed on the transformed miRNA expression profiles of the samples. Hierarchical clustering with an uncentered correlation metric was performed by using a single linkage method of the software Cluster 3.0 (41), and the clustering tree and heat map were visualized by the software Java Treeview (42). In the heat map, the miRNAs with a value of more than +5 are shown as +5, and the miRNAs with a value of less than −5 are shown as −5. For mRNA microarrays, the total RNAs from primary cultured mouse urothelial cells subjected to antagomir treatment were prepared with TRIzol Reagent (Invitrogen) and labeled with Cy5 or Cy3 and then hybridized to the Agilent mouse whole genome Microarray Chip (4 × 44K). Fluorescence intensities were scanned with a GeneArray Scanner to obtain and scale quantitative information.
Northern blots and Real-time RT-PCR Analysis
The total RNA was fractionated in a 15% denaturing PAGE, then transferred to NC paper (Hybond-XL; Amersham Biosciences), before being immobilized by UV cross-linking. Anti-miRNA probes were labeled with γ-32P-ATP (5000 Ci/mmol, 20 mCi/ml; Pharmacia, New York, NY) by using T4 polynucleotide kinase (i.e. TnT) and then purified with a Sephadex G50 spin column (ProbeQuant G-50 Micro Column; Pharmacia). Next, the membrane was prehybridized in Ambion hybridization solution (ULTRAhyb-oligo hybridization buffer; Invitrogen) at 42 °C for one hour and then hybridized at 42 °C overnight. Following hybridization, the membranes were washed twice with 0.2 x SSC with 0.1% SDS at 65 °C for 30 min each time. The blot was normalized for RNA loading by hybridization with 5S rRNA. Real-time quantitative polymerase chain reaction (qRT-PCR) was carried out in the ABI 7500 Fast Real-time PCR system (Applied Biosystems) using Assay-on-Demand TaqMan probe sets (Applied Biosystems), according to the manufacturer's instruction.
LNA-anti-miRs and in situ Hybridization
Locked Nucleic Acid (LNA)-modified, digoxigenin (DIG)-labeled anti-miR-205 and anti-miR-709 probes were synthesized by Exiqon. Fresh-frozen mouse bladder tissue sections (16 μm) were fixed by immersion in 4% paraformaldehyde in PBS for 15 min and deproteinated with proteinase K (0.6 μg/ml) at 37 °C for 30 min. Sections were washed once with 0.2% glycine in PBS and then twice with PBS. Next, the sections were postfixed for 10 min in 4% PFA, followed by 10 min acetylation (0.1 m triethanolamine [pH 8], 0.25% acetic anhydride) and pre-hybridized in hybridization buffer (50% formamide, 1 × Denhardt's solution, 10 mm DTT, 5 × SSC, 0.1% Tween-20, 9.2 mm citric acid [pH 6], 50 mg/ml heparin, 1 mg/ml yeast RNA, 1 mg/ml salmon sperm DNA, and 10% dextran sulfate) at 45 °C for one hour. The slides were then hybridized overnight with 20 nm of DIG-labeled LNA probes in a humidified chamber at 45 °C and then rinsed in 5 × SSC and washed in 2.5 × SSC, 1.25 × SSC, 0.1 × SSC for 20 min each at room temperature. Immunological detection was carried out with the anti-DIG Fab conjugated to alkaline phosphatase (Roche), according to the manufacturer's recommendation. Methyl green was used for nuclear counterstaining.
iTRAQ Experiments
Equal amount of proteins from each treatment were digested with trypsin and labeled with iTRAQ reagents (Applied Biosystems) as described previously (43). Two four-plex iTRAQ comparisons were set up as follows. Comparison A: control-1 was labeled with iTRAQ reagent 114, control-2 with 115, Ant-205 with 116, Ant-709 with 117. Comparison B: Ant-709 was labeled with iTRAQ reagent 114, Ant-205 with 115, control-1 with 116, and control-2 with 117. The labeled peptides from each iTRAQ comparison were then pooled and fractionized on a reverse phase capillary column with an acetonitrile gradient in the presence of about 0.1% NH4OH (Gemini C18 column, 3 × 100 mm, 3 μm; Phenomenex, Torrence, CA) carried on a Breeze HPLC instrument (Waters, Milford, MA) (43). The effluents were collected and then pooled into ∼60 fractions dependent on the signal intensity of OD280. Each fraction was dried at vacuum and re-dissolved with 0.1% formic acid solution for the following LC-MS/MS analysis.
Liquid Chromatography-tandem Mass Spectrometry Analysis (LC-MS/MS Analysis)
LC-MS/MS analysis was performed on a Surveyor nanoflow system (Thermo Fisher, San Jose, CA) connected to the LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher, San Jose, CA) with a Finnigan Nanospray II ion source. Peptide mixtures were separated on a 75 μm × 100 mm PicoFrit BioBasic C18 capillary column (5 μm particle size; New Objective, Woburn, MA) using an acetonitrile gradient in the presence of 0.1% formic acid. The full-scan MS spectra (m/z 400–m/z 2000) were acquired on the Orbitrap mass analyzer with a resolution of 30,000 at m/z 400. The lock mass calibration feature was enabled to improve the accuracy of mass detection. Up to four of the most intense ions with a minimal signal intensity of 20,000 were sequentially isolated for MS/MS fragmentation in the order of intensity of precursor peaks detected in the linear ion trap using the pulsed Q dissociation (PQD) method with the normalized collision energy at 30%, Q activation at 0.7, the activation time set to 0.1 and the isolation width at 2.0. Four MS/MS microscans were averaged to create the proper signal intensity for peptide quantification using iTRAQ reporter ions. Ions with a singly charged state were excluded. The ions (MS ± 15 ppm) for MS/MS measurements were dynamically repeated once and excluded for 90 s.
MS-based Protein Identification and Quantification
Peptide samples were analyzed on a nano-LC-LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher) as described previously (43). The MS and MS/MS raw data were converted to MGF peak list files with DTASuperCharge 1.37 using SmartPicking with 200 segment size (Th) and a max search level of 8. The generated MGF files were searched against a SwissProt 56.0 mouse protein database (16078 sequences) using the MASCOT search engine (v.2.2, Matrix Science) with search parameters as follows: digestion with trypsin, variable modification of N-terminal acetylation of protein, conversion of N-terminal glutamine to pyro-glutamate, oxidation of methionine, methylthiolation at cysteine, and iTRAQ labeling at lysine residue and N terminus of peptides. One missed cleavage, mass accuracy of 6 ppm on the parent ions, and 0.5Da on fragment ions were allowed. Identified proteins were subjected to further filtering using open-source proteomic statistical analysis software (Trans-Proteomic Pipeline, TPP v4.2 JETSTREAM rev 1, http://www.proteomecenter.org/software.php), developed at the Institute for Systems Biology (Seattle, WA), with PeptideProphet and ProteinProphet probability cutoffs were set at 0.95. All of the predicted error rates were less than 0.01 in this study. The apportionment of identified peptides between protein groups were calculated by weighting and provided by the proteomic statistical analysis software. The maximum weight value of each peptide was one, by which the belonging of peptides between possible protein groups were evaluated and indicated. The maximum weight value was divided by the number of identified protein groups containing this peptide. The peptide with a weight value equal to “1” represented it only belonged to one identified protein group. A weight value equal to 0.5 indicated the peptide presented in two protein groups with the same identification confidence. On the one hand, the peptide with a higher value between 0.5 and 1 indicated being present in more than one protein groups identified with different confidence, and the one with a higher protein probability would own it with a higher weight value. Consequently, the proteins were identified by more than two distinct peptides. Quantitative information of proteins was simultaneously obtained using the default settings of the Libra quantitation tool built in the TPP platform as described previously (43). In this study, only the distinct peptides and their reporter ions with an intensity of higher than 20 were included for protein quantification. To minimize the variation of sample loading, the total ion intensities of each reporter ion channel were normalized to the average intensities of four channels. Then, the median value of the normalized spectrum ratios of peptides for a protein group was calculated and used as a protein ratio to minimize the effects of outliers.
Western Blot Analysis
For Western blotting, protein extracts of primary cultured urothelial cells treated with the antogomir against miR-205 (antagomiR-205 or Ant-205) or scramble control were prepared, and 50 μg of protein lysates were separated by 7.5% or 15% SDS-PAGE and then transferred to PVDF (Millipore). The membranes were blocked in 5% nonfat dry milk in PBS-T (0.05% Tween-20 in PBS) for 1 h. After blocking, the membranes were rinsed and incubated with anti-CGNL1 (anti-Paracingulin; Invitrogen mouse 39–8900, 1:1000), anti-Cdc42 (BD Transduction Laboratories mouse 610928, 1:500), anti-TJP1 (anti-ZO-1; Invitrogen rabbit 61–7300, 1:1000), anti-TXNRD1 (Abcam, Cambridge, MA; rabbit ab16840, 1:1000), anti-Krt5 (Covance, Princeton, NJ; rabbit PRB-160P, 1:500), anti-Krt8 (Abcam rabbit ab53280, 1:1000), anti-beta Actin (Novus Biologicals, Littleton, CO; mouse NB600–501, 1:10000), anti-GAPDH (Cell Signaling, Danvers, MA; rabbit 14C10, 1:1000), anti-UPIa (rabbit antiserum, 1:1000), or anti-UPIb (rabbit antiserum, 1:1000) (both antisera against uroplakins were from Dr. T.T. Sun, New York University). Binding of primary antibodies was revealed using HRP-coupled anti-mouse or anti-rabbit secondary antibodies (Millipore). Signals were developed by enhanced chemiluminescence (Immobilon Western; Millipore) and exposed to x-ray films (Fuji). Prestained molecular weight marker proteins were visualized with a LuminolPen HRP labeling (LH603; Visual Protein, Taiwan). The intensity values of the target protein signals were normalized with the intensity value of beta-actin.
Transepithelial Electrical Resistance (TER) Measurement and Lucifer Yellow Permeability Assay
MDCK cell monolayers were cultured on polycarbonate inserts in Costar Transwell 12-well plates (Corning Costar Corp) and transfected with the precursor miR-205 (or control RNA) to selected day intervals for TER measurement by using a voltohmmeter (EVOM; World Precision Instrument, Sarasota, FL), according to the manufacturer's instruction. The TER values were calculated by subtracting the contributions of the bare filter and medium. The measurements were repeated three times for each. The experiment was performed in triplicate. The same MDCK cells were also used for Lucifer Yellow permeability assays. The fluid-phase marker, Lucifer Yellow (Sigma-Aldrich, L0144) was dissolved in PBS (Sigma-Aldrich, L0144) and added to the top chamber of the Transwells® insert with MDCK monolayers that were grown in during the cells' confluence. Before each experiment, the culture media were removed and the cell monolayers were rinsed with PBS and allowed to equilibrate at 37 °C for one hour. At the end of the experiments, the insert plate was removed from the basolateral compartments, and the transported Lucifer Yellow dye in the opposite chamber was read with a fluorescent spectroscopy (SpectraMax M5, Molecular Devices, Sunnyvale, CA) at 485 nm excitation and 535 nm emission wavelengths. The percent of Lucifer Yellow that passed through the cell monolayer into the transport analysis plate was calculated by using the related fluorescent measured for an equilibrium dilution of Lucifer Yellow in a separate plate for reference. All time point values were measured in triplicate. Statistical analysis was performed by Student's t test.
RESULTS
Identification of miRNAs that are Associated with Differentiation of the Urothelium
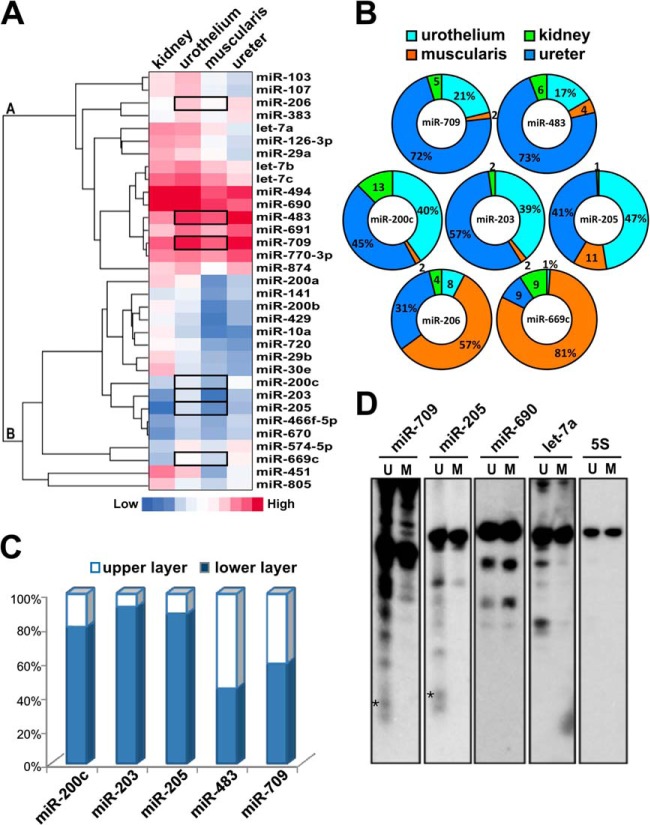
To explore which miRNAs may have potent roles in regulating urothelial differentiation, we first performed the exploratory microarray analyses of miRNA expression levels in RNA samples isolated from different urinary tissues including the mouse ureter, kidney, and the urothelial as well as muscularis layers of the bladder. By comparing the miRNA expression patterns from different urinary tissues, we found that 33 of the 170 detected miRNAs were differentially expressed in the bladder urothelium (Fig. 1A). Of these we chose to further validate several miRNAs that were expressed more abundantly in the bladder urothelial layer than in the muscularis layer: Group A (miR-709, miR-483, and miR-206) and Group B (miR-200c, miR-203, miR-205, and miR-669c) (see gene tree in Fig. 1A).
Fig. 1.
Identification of miRNAs that are associated with differentiation of the urothelium. A, Hierarchical clustering of miRNA expression in the mouse kidney, bladder urothelium, bladder muscularis, and ureter. The heat maps of 33 miRNAs are shown in horizontal rows. Each horizontal bar represents one probe set, and the color reflects its expression level (red and blue indicate high and low expression levels respectively). Clusters of genes with similar expression patterns are shown in the gene tree on the left with horizontal bars representing groups derived from the hierarchical clustering. B, Validation of miRNAs expression profiles of mouse urinary tissues. The threshold cycle (Ct) of each miRNA in TaqMan-based stem-loop PCR was recorded as a quantitative measure of the miRNA amount, and normalized against snoRNA 202/234 (calculated as 2[ΔCt]) and depicted as a relative percentage. C, Relative expression levels of miR-200c, miR-203, miR-205, miR-483, and miR-709 in undifferentiated basal cells and differentiated upper urothelial cells. Total RNA was collected from laser capture microdissected basal and upper urothelial cells to quantify miRNAs by TaqMan assays. D, The mature miR-205 and miR-709 were detected in urothelial (U) but not in muscular (M) layers of the bladder. Northern blot analyses of mouse urinary bladder using specific 32P-labeled oligonucleotide probes for miR-709, miR-205, miR-690, let-7a, and 5S. Only in the urothelium the mature miR-205 and miR-709 were visualized at ∼19-nt and ∼22-nt, denoted by dark stars. The larger molecule revealed the total transcripts of primary and pre-mRNA. 5S rRNA expression was used as a loading control.
By using independent RNA samples, we conducted the stem-loop-specific quantitative TaqMan miRNA assays to validate the microarray results. We confirmed five of these miRNAs (miR-709, miR-483, miR-200c, miR-203, and miR-205) were expressed by more than 4-fold in the urothelial layer than in the muscularis layer, whereas the other two miRNAs (miR-206 and miR-669c) did not show any enrichment in the urothelium (Fig. 1B). To determine the dependence of these five confirmed miRNAs on the urothelial differentiation stage, we further used laser-captured microdissection to dissect the upper well-differentiated urothelia and the lower undifferentiated cell layer fractions from the mesenchymal boundary, and then performed quantitative TaqMan assays on these two RNA pools. We found that the expression levels of miR-200c, miR-203, and miR-205 (Group B miRNAs) were higher in the lower undifferentiated fraction than in the upper well-differentiated fraction, whereas miR-483 and miR-709 (Group A miRNAs) were consistently expressed in both fractions (Fig. 1C).
Before performing miRNA in situ hybridization analysis to demonstrate the association between miRNAs and differentiation of urothelium we surveyed the expression level of several miRNAs with the Northern blot analysis. Comparison of two total RNAs isolated from mouse bladder urothelial or muscle tissues, we only found the signals corresponding to miR-205 and miR-709 in urothelial RNA, whereas all miRNA precursors were detectable (Fig. 1D). The longer exposure images of miR-690 and let-7a showed faint and very low abundant bands than the signals of miR-205 and miR-709 (data not shown). The results of miR-690 and miR-709 were found similar to an early report by Tang et al., that revealed the presence of a large amount of the precursors and faint broad bands corresponding to their mature miRNA (44).
Confirmed with miRNA in situ hybridization (Fig. 2A and supplemental Fig. S1) that miR-205 was dominantly expressed in the lower undifferentiated basal and intermediate cell layers, and only rarely in the terminally differentiated umbrella cells; well-differentiated cells were immunostained with the specific differentiation markers (Fig. 2B). In contrast, miR-709 was expressed in all cell layers of the urothelium, including the umbrella cells (Fig. 2A and supplemental Fig. S1). These results demonstrated the expression of miR-205 was associated with the different stages of urothelial differentiation and thus may be biologically significant in the development and function regulation of urothelium.
Fig. 2.
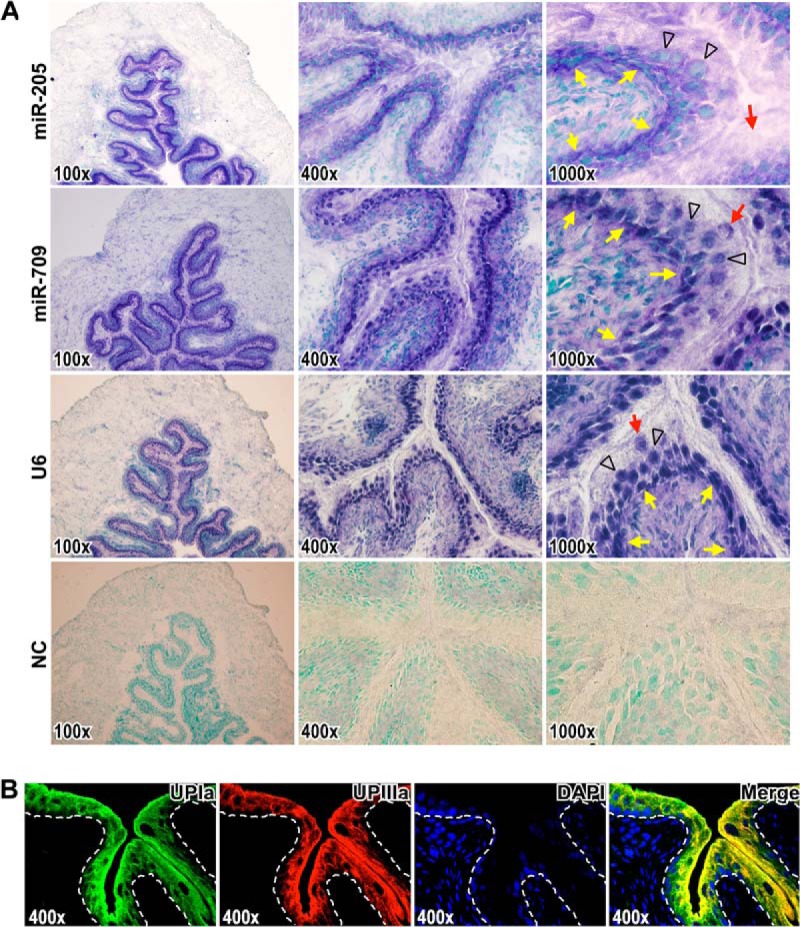
miR-205 is expressed in the lower undifferentiated layers of urothelium. A, In situ hybridization detection of miR-205 and miR-709 on mouse bladder sections. Images represent sections hybridized with antisense probes corresponding to miR-205, miR-709, positive control U6, and negative control (NC) in magnifications 100 x, 400 x, and 1000 x, respectively. The hybridization signals are in purple blue. Nuclei were counterstained with methyl green dye. The yellow arrows indicate the epithelium-mesenchyme boundary. The black arrowheads indicate the intermediate cells. The red arrows indicate the superficial umbrella cells. B, The urothelial differentiation was determined with the specific differentiation markers, uroplakin Ia (UPIa), and uroplakin IIIa (UPIIIa), in upper urothelial cell layers. Localization of UPIa (green) and UPIIIa (red) were observed by immunofluorescence (yellow in merged image). The dashed lines indicate the epithelium-mesenchyme boundary. DAPI-stained nuclei are shown in blue.
Identification of miR-205 Target Genes in the Urothelial Cells using Quantitative Proteomics
In this work, we conducted an iTRAQ-based quantitative analysis to further characterize miR-205 in normal urothelium, which typically showed a low rate of cell turnover in rodent bladder (45). By knocking down the expression of endogenous miR-205 in primary cultured mouse urothelial cells, we observed the expression changes to identify the potential target gene of miR-205. Antagomir treatment was used to knockdown endogenous miRNA expression in primary cultured cells where we observed 90% and 78% depletion of miR-205 and miR-709 respectively (supplemental Fig. S2). Protein extracts representing control and antagomir-treated urothelial cells were digested and labeled with different iTRAQ tags as described in the Experimental Procedure for relative quantitation by tandem mass spectrometry. Within two sets of comparisons, 1990 and 1771 proteins, as well as 1868 and 1668 proteins were identified and quantified in Comparison A (Exp-1) and B (Exp-2), respectively. A summary of proteomics results was shown in supplemental Table S1. In total, we identified 2169 nonredundant proteins, and quantified 2051 proteins in these two iTRAQ-based quantitative proteomics analyses (the details in supplemental Table S2 and S3). Because the in situ hybridization detection of miR-205, but not miR-709, was found in the lower, undifferentiated cell layers with a potentially differentiation-related expression, we therefore focused on identifying the targets of miR-205. The mean values and standard deviation (S.D.) of two sets of control comparison (C1/C2) were 1.04 ± 0.20 and 1.02 ± 0.12. Proteins that had a ratio greater than two in comparison (greater than 3SD from the mean of the control comparisons) were selected as candidate targets of miR-205. A total of 157 proteins increased more than 2-fold in response to antagomiR-205 treatment were considered as candidates (supplemental Table S2; 157 proteins marked in red).
By bioinformatic prediction assessing whether these genes contain a miR-205-binding site, all quantified proteins were assessed using the currently available and frequently used web-based miRNA target prediction programs (TargetScan 6.0, miRanda 2010, MicroCpsm Targets, and DIANA microT 3.0 programs). Because miRNAs regulate their target mRNAs predominantly by binding to the 3′ UTR, the annotated 3′ UTR of each transcript of a gene was used for target prediction. The method we chose for the predictive features combining characteristics in seed regions both the conserved and nonconserved miR-205 binding sites and also selected a loose score threshold including those imperfect seed matches to achieve the higher sensitivity (supplemental Table S4). By applying a more stringent strategy to identify the candidate target gene of miR-205 we further included the targets that must be predicted from more than two web-based prediction programs to enhance the specificity and minimize false positives.
We then assessed the 2051 quantified proteins with the lists of predicted miR-205 target gene by bioinformatics and found 265 genes (12.92%) containing the potential binding sites of miR-205. We also constructed the cumulative distribution plots of the log2-transformed protein expression fold-changes for those genes containing miR-205 target sites and all other expressed genes in response to antagomir-mediated knockdown of endogenous miR-205 in primary cultured urothelial cells. The cumulative distribution plots of the proteins with binding sites (265 proteins) represented a shift of the probability distribution to the right direction with higher values of protein ratio when compared with the distribution plots of the nontargeted proteins (1786) or to the total proteins (2.051), but there were no statistically significant differences (t test, p = 0.179 and 0.234, respectively) (upper panel of supplemental Fig. S3). Among the 157 differentially expressed proteins in response to antagomiR-205 knockdown treatment, 24 genes (15.29%) were identified from more than two miRNA target prediction programs, which was a more stringent strategy that we had applied to select the candidate targets to minimize false positives (Table I).
Table I. Summary of proteins identification and quantitation by iTRAQ analyses with more than a twofold change harboring potential target sites of miR-205 indicated from at least two of the web-based prediction programs.
| Gene Symbol | iTRAQ ratio (log2) Ant-205/Control |
TargetScan 6.0 | miRanda 2010 | MicroCosm Targets | DIANA microT3.0 | |
|---|---|---|---|---|---|---|
| Exp-1 | Exp-2 | |||||
| AMOT | 1.237 | ♦ |  |
• | ||
| ARHGDIA | 1.073 | ♦ | • | |||
| ATP6V1G1 | 1.475 | ♦ |  |
• | ||
| BTF3L4 | 1.502 | ♦ |  |
• | ||
| CALU | 1.028 | ♦ |  |
• | ||
| CDC42 | 1.145 | 1.116 | ♦ |  |
• | |
| CDS2 | 1.384 | ♦ | • | |||
| CGNL1 | 2.114 | 1.761 | ♦ |  |
• | |
| CHMP5 | 1.013 | ♦ | • | |||
| DDHD1 | 1.278 | ♦ | • | |||
| DNM2 | 1.005 | ♦ | • | |||
| HSPA14 | 1.078 | ♦ |  |
★ | • | |
| MAPK3 | 1.067 | ★ | • | |||
| MPP5 | 1.346 | ♦ | • | |||
| RCN2 | 1.740 | ♦ |  |
• | ||
| RDH10 | 1.142 | ♦ |  |
• | ||
| SEPT11 | 1.416 | ♦ |  |
• | ||
| SEPT2 | 1.076 | 1.204 | ♦ | • | ||
| SH3KBP1 | 1.023 | ♦ |  |
• | ||
| SLC25A13 | 1.218 | ♦ |  |
• | ||
| TCP11L1 | 1.109 | 1.214 | ♦ |  |
• | |
| TJP1 | 1.042 | ♦ |  |
• | ||
| VTI1B | 1.113 | ♦ |  |
• | ||
| ZADH2 | 1.401 | ♦ | • | |||
Note: All ratios are given in log2 values. Predicted mRNA targets of miR-205 were extracted from four web-based databases: TargetScan 6.0 (2707 targets, with a total of 332 conserved sites and 3004 poorly conserved sites), miRanda 2010 (3296 targets, with Good mirSVR score), MicroCosm Targets (881 targets, p value <0.05), and Diana microT 3.0 (3175 targets, miTG score >1).
To assess the roles of these 24 potential targets of miR-205, we analyzed their functional categories, gene ontologies, and related pathways using the Functional Annotation Clustering tool built in DAVID program (Database for Annotation, Visualization and Integrated Discovery, v6.7) (46, 47). Sixteen of the 24 potential targets were present in three significant function clusters with enrichment scores higher than 2 (p value < 0.01), in which those annotated categories can be related to the urothelial growth and differentiation (summarized in Table II). The first annotation cluster hit by eleven potential miR-205 targets is related to various junction types including tight-, cell-, occluding-, apical-, and cell-cell junctions and is summarized in Table II.
Table II. Functional annotation clustering of miR-205 target genes in urothelium.
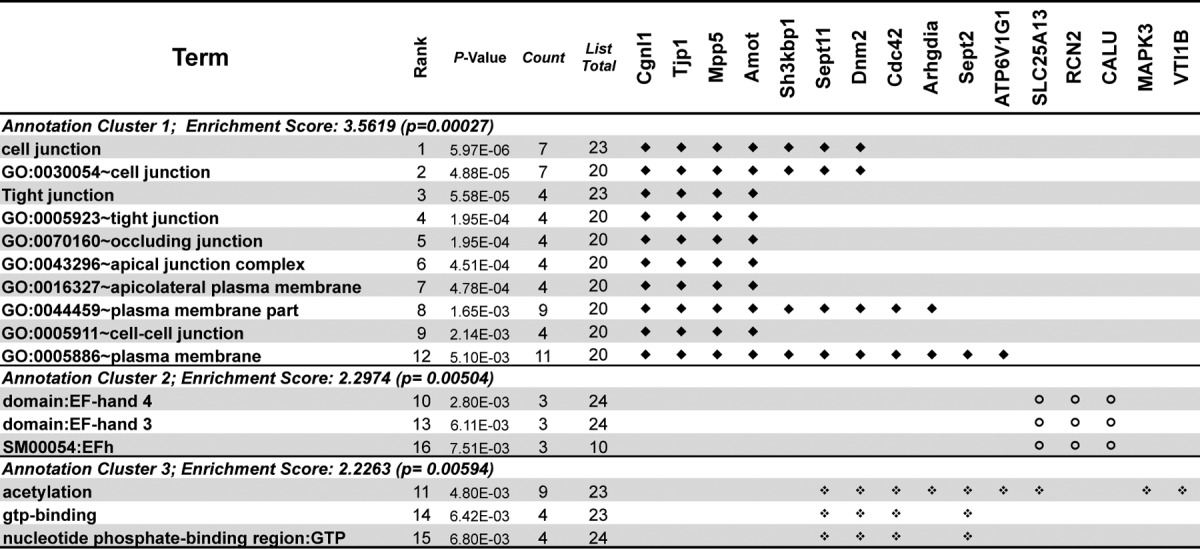
Note: Functional clusters were derived from gene-annotation enrichment analysis using DAVID with significant associations (p value cut-off < 0.01). Symbols are given for these target genes that are associated with a functional role of the junction regulation of the Annotation Cluster 1 (♦) or non-junction-related pathways of the Annotation Cluster 2 & 3 ( and
and 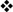 ).
).
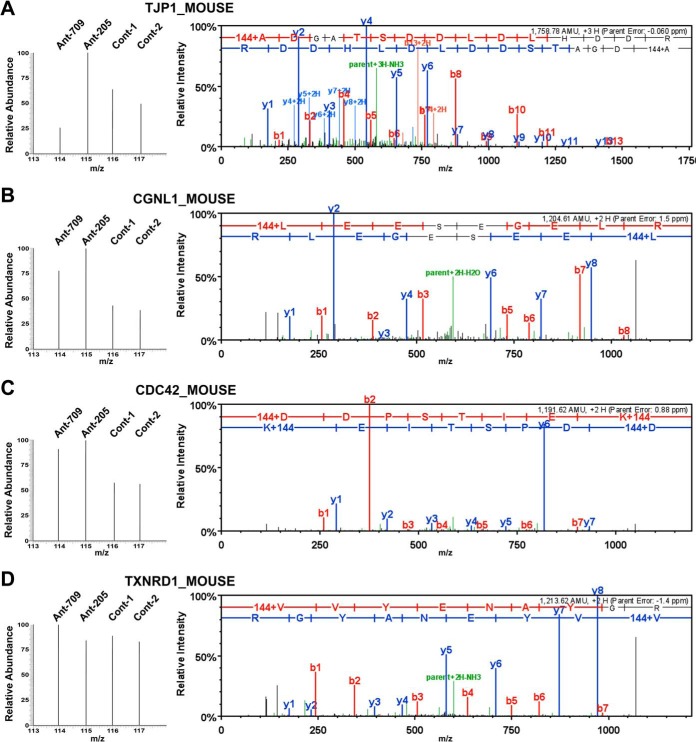
The iTRAQ-based identification and quantification of Tjp1, Cgnl1, Cdc42, and Txnrd1 are presented in Fig. 3, with representative peptide fragmentation spectra shown for each protein. The intensities of the zoom-in iTRAQ reporter ion spectra (m/z 114–117) of Tjp1, Cgnl1, and Cdc42 demonstrated that their protein expression levels increased as miR-205 expression decreased in the urothelium (Txnrd1 is shown as a nondifferentially expressed example). The correlations of these altered expression levels with that of miR-205 in urothelial cells was further verified by the Western blot analyses, which showed that the knockdown of endogenous miR-205 increased the expression levels of Tjp1, Cgnl1, and Cdc42, but not Txnrd1 (Fig. 4A).
Fig. 3.
iTRAQ-based identification and quantification of differentially expressed Tjp1, Cgnl1, and Cdc42 in response to antagomiR-205 knockdown treatment. Protein extracts representing the scramble control and antagomir-treated primary cultured urothelial cells were subjected to quantitative proteomic analyses. The representative peptide MS/MS fragmentation spectra used for identification and quantification of three identified miR-205 suppressed proteins are displayed: Tjp1 (A), Cgnl1 (B), and Cdc42 (C) are displayed. Txnrd1 (D) is shown as a nondifferential example. For each gene, the representative peptide sequence was deduced from the MS/MS spectrum (right) which contains peptide mass matched y-ions (marked in blue) and b-ions (marked in red). The peak intensities of iTRAQ reporter ions, m/z 114–117, located at the low mass range of MS/MS spectrum (left zoom-in spectra) were used to measure the relative abundance of the corresponding peptides in four samples as indicated.
Fig. 4.
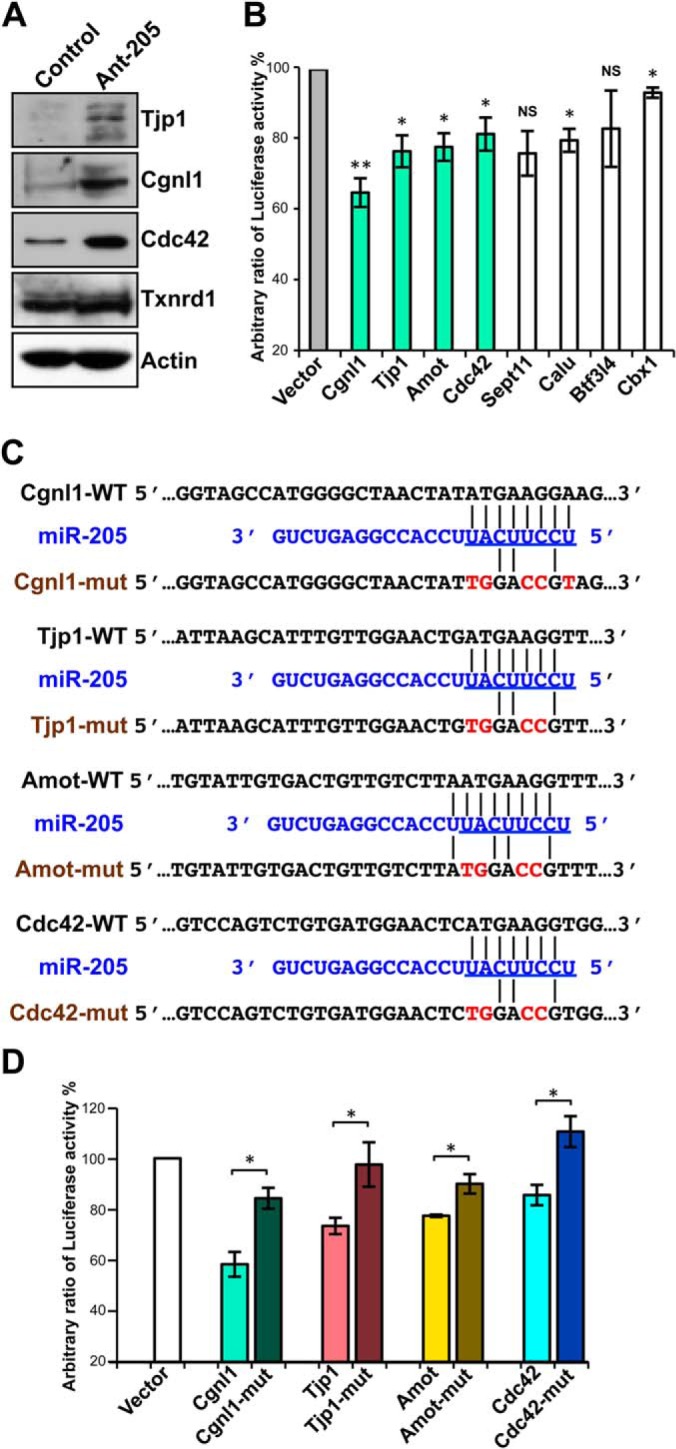
Several candidates are validated as target genes of miR-205. A, miR-205-mediated altered expression levels of target genes were verified by Western blot analyses showing that knockdown of endogenous miR-205 in primary cultured urothelial cells increased the expression levels of Tjp1, Cgnl1, and Cdc42, whereas the nonrelevant Txnrd1 and actin proteins were unaffected. B, Dual luciferase reporter assays show the influence of miRNA-target pairing to confirm direct targeting of miR-205 including Cgnl1, Tjp1, Amot, Cdc42, Sept11, Calu, and Btf3l4, but not Cbx1. 293T cells were co-transfected with 20 nm of either a precursor of miR-205 or a negative control dsRNA and the firefly luciferase reporter constructs bearing individual predicted target binding sites of miR-205 as indicated and a Renilla luciferase reporter plasmid as a transfection control. The cells were assayed for dual luciferase activities 24-h post-transfection. Compared with negative control-transfected cells, the reduction of firefly/Renilla luciferase expression ratios was revealed in the miRNA-transfected cells. The columns of junctions-related molecules are labeled in light green. Note the Cbx1 was not included in the candidate proteins listed and showed it was little affected. C, Sequence alignment of predicted putative and mutated miR-205-binding regions on the 3′ UTR of mRNAs were constructed and used in the luciferase reporter assay (mutations in red). D, The reductions of luciferase activity in the miRNA-transfected cells are displayed as the relative ratios of mutant and wild-type of four tight junctions-related molecules to vector control. The data are averages of triplicate transfections. For B and D, data are presented as means ± S.E. of three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001, by Student's t test.
Based on these results, we constructed luciferase reporters bearing individual target-gene 3′ UTR sequences containing the predicted miR-205 binding sites including Cgnl1, Tjp1, Amot, Cdc42, Sept11, Calu, and Btf3l4 to investigate the influence of miRNA-target pairings. Additionally, Cbx1 identified from only one prediction program was displayed as a parallel control in reporter assay. We then measured the miR-205-mediated repression regulation with the dual luciferase reporter assays in 293T cells. The sequences of the binding sites used in reporter construction are shown in supplemental Fig. S4. The miR-205-mediated reductions of luciferase activity are shown in Fig. 4B. The reporter harboring the 3′ UTR of Cgnl1 showed a large and statistically significant reduction (35.1% versus the control) in luciferase activity. The other tight junction-related molecules also experienced the significant drop following the expression of miR-205, except less significant on Sept11. We mutated the putative miR-mRNA interaction sequences of the tight junction-related molecules, Cgnl1, Tjp1, Amot, and Cdc42, to ensure specific interactions. Fig. 4C shows a miRNA:mRNA alignment and the putative and mutant binding sequences within the UTRs of the targets. As presented in Fig. 4D, the miR-205-mediated repressions of luciferase activity were not observed with the mutated constructs, indicating that this down-regulation was mediated by a direct interaction between miR-205 and the 3′ UTRs of targets.
miR-205 Down-regulates Cgnl1 at the Protein and mRNA Levels in the Urothelium
In addition to quantitative proteomic profiling, we conducted a transcriptomic analysis of antagomir-treated urothelial cells and revealed a total of 21,198 gene expression, which included 2154 genes (10.16%) containing the binding sites of miR-205 from more than two prediction programs. The cumulative distribution plots of the log2-transformed gene expression fold-changes determined by mRNA microarrays with the genes containing miR-205 target sites and all other expressed genes in response to knockdown of endogenous miR-205 in primary cultured urothelial cells were constructed. The cumulative distribution plots of the genes with binding sites (2154) represented a very minor shift of the probability distribution compared with the plots of the total proteins (21,198) or to the nontargeted proteins (19,044) (lower panel of supplemental Fig. S3). Our mRNA microarray analyses revealed that 859 genes were significantly up-regulated by more than 2-fold in response to antagomiR-205 treatment, 88 (10.24%) of them contain putative miR-205 binding sites identified from more than two prediction programs. Among them, five proteins were increased at both the protein (identified by iTRAQ assay) and mRNA (by mRNA microarray assays) levels following miR-205 knockdown by more than 2-fold; these included Cgnl1, Btf3l4, Ddhd1, Amot, and Rdh10 (supplemental Table S5). Interestingly, the mRNA level of Cgnl1 was significantly affected by antagomiR-205; this is consistent with our protein expression data showing an inverse correlation between the expression levels of Cgnl1 and miR-205 in primary cultured urothelial cells, and was verified by quantitative RT-PCR (supplemental Fig. S5). The Cgnl1 mRNA is known to be highly expressed in the mouse kidney (22), but its expression in bladder had not previously been reported. Here, we found that the expression of Cgnl1 mRNA in urothelium was comparable to that reported in the kidney, whereas only a minimal expression was observed in the muscularis layers of the bladder (supplemental Fig. S5). Taken together, our findings indicate that miR-205 down-regulates Cgnl1 at the RNA and protein levels in primary cultured urothelial cells, suggesting that Cgnl1 may play an important role in the regulatory signaling downstream of miR-205.
miR-205 Inhibits Tight Junction Formation
To examine the effects of miR-205 in modulating the formation of tight junctions, we ectopically transfected miR-205 in the MDCK cells, which are known to develop typical tight junctions at confluence. We found that overexpression of miR-205 interrupted the tight junction formation in MDCK cell. The contour immunofluorescence of ZO-1 became irregular and discontinuous staining at cell borders, which was visualized by standard immunofluorescence staining of the tight junction marker (anti-ZO-1) (Fig. 5A). The control group successfully developed the continuous network of tight junction at confluence, but the tight junction became irregular in the cells transfected with the miR-205 precursor; this was particularly notable at confluence, 72-h post-transfection (Fig. 5A). The Western blot analyses showed that ZO-1 protein expression decreased after miR-205 transfection (Fig. 5B, upper panels), whereas knockdown of miR-205 increased ZO-1 expression (Fig. 5B, lower panels). Similar results were found at 72-h post-transfection that protein expressions of CGNL1 and CDC42 decreased after miR-205 overexpression (supplemental Fig. S6).
Fig. 5.
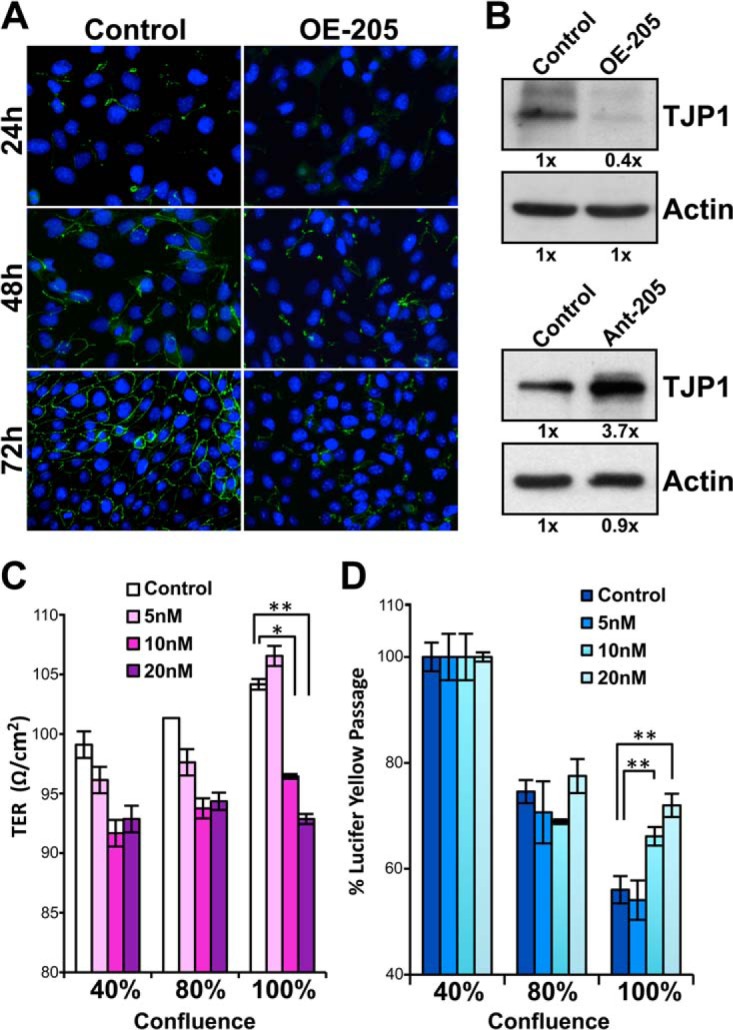
Negative effects of miR-205 on tight junction formation and membrane integrity in MDCK cells. A, Tight junction was visualized by immunofluorescence staining with anti-ZO-1 antibody (green) at 24-, 48-, and 72-h post-transfected with the miR-205 precursors (OE-205) or the control dsRNA. Nuclei were stained with DAPI (blue). Note the prominent inhibition of ZO-1 expression by miR-205 in the 72-h post-transfection samples. B, miR-205-mediated altered the expressions ZO-1 were harvested 72-h post-treatment and verified by Western blot analyses showing that overexpression of miR-205 suppressed ZO-1 expression (upper panels), whereas knockdown of miR-205 increased the expression of ZO-1 (lower panels). C and D, miR-205 overexpressed cells showed a decrease of transepithelial electrical resistance (TER) (C) and an increased paracellular permeability to Lucifer Yellow (D) in MDCK monolayer. The TER was measured to determine the integrity of the MDCK monolayer and found a decrease from 104.2 ± 2.1 Ω/cm2 (control) to 96.4 ± 0.9 Ω/cm2 (10 nm), and 92.9 ± 0.1 Ω/cm2 (20 nm) in confluent cells, which represent 7.5% and 10.8% decrease over control (C). The Lucifer Yellow flux was increased by 16% (10 nm) and 23% (20 nm) of the permeation rate in miR-205 transfected cells than the control cells (D). For C and D, data are presented as means ± S.E. of three independent experiments, *p < 0.05, **p < 0.01, by Student's t test.
The tight junction starts to form between epithelial cells after reaching confluence. The well-developed tight junction determines the integrity of an epithelial membrane and its selective paracellular permeability to ions and molecules. Therefore, we further assessed whether miR-205 affected the membrane integrity in MDCK monolayers by measuring their transepithelial electrical resistance (TER) and paracellular permeability to Lucifer Yellow. We found the results consistent with our immunostaining results that miR-205-overexpressing cells showed a significant decrease of the TER values (p < 0.01) in a dose-dependent manner (maximum in 20 nm; 10.8% decrease than controls in 100% confluence, 6.9% decrease in 80% confluence, and 6.3% decrease in 40% confluence (Fig. 5C)). In the same culture model, the permeability assays to Lucifer Yellow were conducted to determine the permeability coefficient. The percent of Lucifer Yellow dye (452 Da) transported the empty insert membrane was considered 100% to determine the transport rates in response to different treatments. The data showed that overexpression of miR-205 affected the permeability to Lucifer Yellow with a significant increase (p < 0.01) than controls after cells reached confluence (Fig. 5D). We conclude that miR-205 down-regulates the synthesis of tight junction-related molecules, the formation of tight junctions, and the integrity of the membrane, potentially suppressing cell polarity and differentiation.
Antagomir-mediated Knockdown of Endogenous miR-205 affects the Urothelial Cellular Differentiation Program
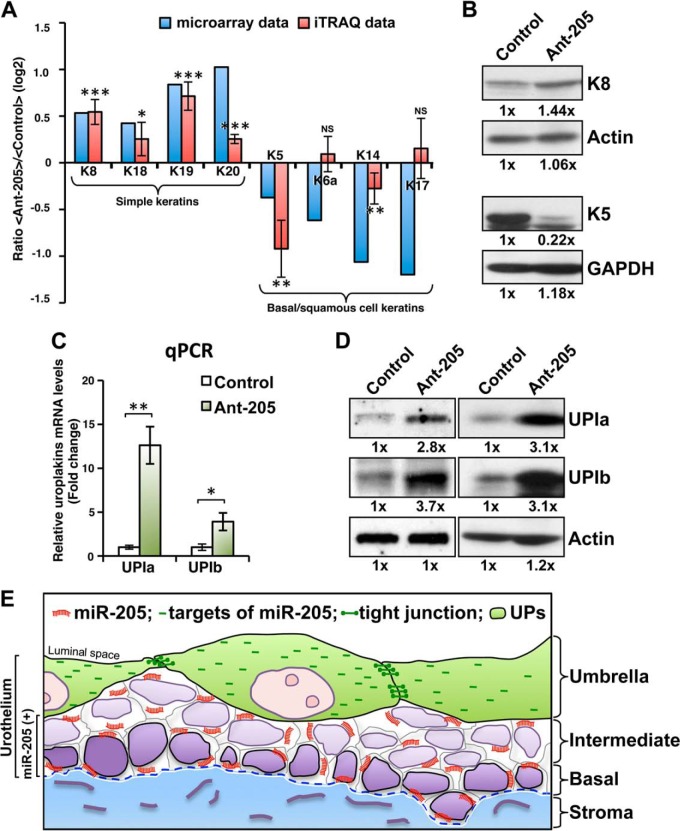
The multilayered urothelium is typically structured with differentiated surface cells and less matured basal cells. In general, the basal cell keratins K5 and K14 are regarded as markers for stratified squamous epithelia (48). The differentiation of most keratinocytes is accompanied by the general down-regulation of K5 and K14 and the induction of specific sets of keratins needed for the individual differentiation programs (48, 49). Because specific keratins are expressed differentially in various layers of urothelial cells (39, 50), we analyzed the keratin profiles of our mRNA microarray and proteomic data. Our iTRAQ-based quantitative proteomic analysis revealed that silencing of miR-205 in primary cultured urothelial cells altered the keratin protein pattern, down-regulated the expression levels of the basal and squamous keratins, K5 (47%) and K14 (17%) of the control level, and up-regulating the simple keratins (superficial cells), K8 (1.46-fold) and K19 (1.64-fold) (Fig. 6A). Similar to protein data, our microarray analysis showed that the mRNA expression levels of K8, K18, K19, and K20 were up-regulated, whereas those of K5, K6a, K14, and K17 were down-regulated (Fig. 6A). The altered protein levels of K8 and K5 were confirmed by the Western blot assays (Fig. 6B). These results show that miR-205 may play a role in the switching of urothelial differentiation programs because the expressions of the simple keratin were up-regulated and the basal and squamous keratins were down-regulated while knockdown of endogenous miR-205.
Fig. 6.
miR-205 suppresses the precocious urothelial differentiation programming. A, Features of keratins expression in primary cultured urothelial cells. Keratin profiles of our cDNA microarray data (blue columns) and iTRAQ data (red columns) are expressed as log2 ratios between miR-205-silenced and control cells. B, The altered protein levels of K8 and K5 were confirmed by Western blot analyses that showed silencing of miR-205 up-regulates the simple K8 (superficial) and down-regulates the K5 (basal). C and D, Effects of inducible expression of uroplakin Ia (UPIa) and uroplakin Ib (UPIb) in response to antogomor-205 treatment. UPIa and UPIb, two major differentiation markers of the urothelium, were found elevated at both mRNA (C) and protein (D) levels in miR-205 silenced cells compared with the control cells. Total RNA or proteins were collected 72 h after the initiation of treatment in primary cultured urothelial cells and subjected to qRT-PCR and Western blot analysis. E, Schematic representation of the composition of the urothelial cells and miR-205: the single strand miR-205 (shown in orange) is expressed in a differentiation-dependent manner and down-regulates its targets including the tight junction-related proteins (shown in green) in the intermediate and basal urothelial cells. The release of the negative regulation of miR-205 in umbrella cell may reprogram the formation of tight junctions occurs during terminal differentiation. For A, ratio of iTRAQ data is presented as means ± S.E. of two four-plex experiments. For C, data are presented as means ± S.E. of three independent experiments. Asterisks indicate statistically significance between control and miR-205 knockdown cells, *p < 0.05, **p < 0.01, ***p < 0.001, by Student's t test.
The differentiated superficial urothelium is characterized by the AUM plagues and differential antigen expression. Five highly conserved uroplakin proteins have been characterized from isolated AUM, including UPIa, UPIb, UPII, UPIIIa, and UPIIIb (1, 5, 6). In normal urothelium, the expression of uroplakins is used as specific markers of urothelial differentiation. Therefore, we measured the altered expression of UPIa and UPIb, two major products at terminal cytodifferentiation to help detect differentiation level to assess the potential importance of miR-205 on urothelial differentiation. Our study found both the mRNA (Fig. 6C) and protein (Fig. 6D) levels of UPIa and UPIb were increased when the expressions of miR-205 were reduced in the primary cultured urothelial cells. Notably, miR-205 did not directly target UPIa and UPIb because of a lack of predicted miR-205 binding site at their 3′ UTRs, nor for the detected keratins. Collectively, our results suggest that miR-205 may contribute to regulate urothelial differentiation program and uroplakins production in primary cultured urothelial cells by targeting the tight junction-related molecules.
DISCUSSION
The urothelium coating of the various surfaces of renal pelvis, ureters, bladder, and parts of urethra is a highly specialized transitional epithelium that is known to offer high elasticity and function as a high-resistance paracellular barrier. However, the molecular mechanisms underlying its differentiation are not well understood. In this study, we showed that by modulating the expression of tight junction-related proteins in primary cultured urothelial cells, miR-205 may contribute a role in governing the process of cellular differentiation of urothelium.
Mammalian bladder urothelium is composed of basal, intermediate, and terminally differentiated umbrella cell layers. It is generally accepted that umbrella cells gradually develop from basal and intermediate cells while they start to generate differentiation-related proteins that is, cytokeratins and uroplakins (4, 51, 52). The differentiated umbrella cells form the paracellular barrier are a single layer of highly polarized cells that have distinct apical and basolateral membrane domains demarcated by tight junctions (7–10). Recently in label-retaining cell (LRC) assay, which was estimated to identify the in vivo slow-cycling rat urothelial cells (possible stem/progenitor cells) have demonstrated that the basal LRCs were highly clonogenic in an in vitro stem cell assay (53). Other lineage tracing experiments in mice also revealed the basal cells gave rise to all layers of urothelium (54). Together these reports suggest the cellular differentiation most likely originates and resides in the basal layer of the urothelium, however, it is unclear what regulatory factors are involved in the urothelial differentiation.
miRNAs have been shown to regulate most biological processes, including cell growth, death, development, and differentiation. Here, we analyzed the miRNA expression signatures of the mouse bladder urothelium, and we report for the first time that miR-205 is expressed in a differentiation-dependent manner in the urothelium. Previous studies demonstrated that miR-205 is involved in both normal development and cancer growth. In corneal epithelial cells, miR-205 targets SHIP2 to trigger a coordinated damping of the Akt signaling pathway and helps confer the epithelial characteristics specific to eye function (31). In cooperation with members of the miR-200 family, miR-205 negatively regulates TGF-beta-induced epithelial-mesenchymal transition (EMT) by targeting the transcriptional factor, ZEB1 (33). miR-205 was also found to target and suppress ErbB3 and VEGF-A in tumor tissues (55); to promote apoptosis by targeting the HER3 receptor (56); and to increase cellular proliferation in prostate cancer by targeting the tumor suppressors, PTEN and SHIP2 (32). In a recent study, of genetic deletion of miR-205, Wang and colleagues found that the loss of miR-205 led to neonatal lethality and demonstrated a network of negative regulators of the PI(3)K pathway were directly targeted by miR-205 (34). All of the studies described above reveal the importance of miR-205; however, no report has offered a rigorous proteomics identification of the targets of miR-205 with diverse functions and properties.
General features of miRNA-mediated gene regulation include translational repression and miRNA-directed mRNA decay. Several reports have successfully used quantitative proteomic strategies to determine miRNAs-induced changes in protein expression for identifying miRNA targets (37, 57, 58). The stable isotope labeling by amino acids in cell culture (SILAC) is a widespread method for quantitative proteomics, and generally used in cultured cells to achieve complete labeling. Alternatively, the pulsed SILAC analyses have been used more delicately to determine the miRNA-mediated dynamic changes of cellular protein abundance (57, 59–61). In contrast, chemical labeling can be applied to all kinds of samples. Using iTRAQ labeling and LC-MS/MS analysis, HIF-1α induced under hypoxic conditions was identified as a direct target for miR-17–92 in lung cancer cell line (62). Several target proteins of miRNAs were also identified with iTRAQ proteomics in MCF-7 breast cancer cells in response to miRNA overexpression or endogenous knockdown (58, 63). Most recently, peripheral blood mononuclear cells from osteopetrosis patients were studied with iTRAQ proteomics and found the miRNA-target pair of miR-320a and Atf1 potentially associated with CLCN7 mutations in osteopetrosis (64). Therefore, in this study an iTRAQ-based quantitative analysis was used for global proteomic profiling to identify targets of miR-205 in primary cultured urothelial cells. By knocking down the expression of endogenous miR-205, we observed a significant increase in the relative abundance of 157 proteins by more than twofold in response to antagomir treatment. Of these, 24 target genes (15.29%) contained putative miR-205 binding sites in their 3′ UTRs including both perfect and imperfect seed match sequences from more than two miRNA target prediction programs, and were identified as potential direct targets of miR-205.
A parallel cDNA microarray analysis of the same primary cultured cells showed that miR-205 appeared to significantly alter the mRNA expression levels of 852 genes (by more than twofold) in primary cultured urothelial cells. Among them, 88 target genes (10.24%) contained putative miR-205 binding sites with both perfect and imperfect seed match sequences. Comparison of the 88 targets identified from the cDNA microarray data and 24 targets from quantitative proteomic data allowed us to identify five candidate targets that were significantly up-regulated at both the mRNA and protein levels following antagomiR-205 treatment. The sequence features on site efficacy of these five genes, including canonical seed-matches 8mer (Cgnl1) and 7mer-m8 (Amot, BTF3L4, DDHD1, and RDH10) sites in 3′ UTR sequences. In contrast, the other 19 targets were found altered in protein expressions, but no changes in their mRNA abundance in response to knockdown of endogenous miR-205, containing the canonical seed-match 7mer-A1 sites in 10 genes (52.3%), 7mer-m8 sites in 8 genes (42.1%), and 8mer site in one gene (5.3%) in their 3′ UTR, propelled toward a favorable prediction of mRNA degradation (8mer > 7mer-m8 > 7mer-A1 > 6mer >offset 6mer) (65–67). Because miRNAs regulate the expression of their target mRNAs predominantly by binding to the 3′ UTR, the target prediction programs we used in this study indicated the target sites only in their 3′ UTR. For those target sites located within their promoter sequences or open reading frame regions therefore would be neglected. In general, our findings conclusively suggest that combining bioinformatic analysis with iTRAQ-based quantitative proteomic analysis is considered to be an efficient method to detect functional targets of miRNA in the normal urothelial cells, which have a slow turnover rate.
Our advanced functional annotation revealed that miR-205 regulates mainly tight junction- and cell junction-related pathways in the intermediate and basal urothelial cells by targeting several junction-related genes (e.g. those encoding Tjp1, Cgnl1, Cdc42, Amot, and Mpp5). Tjp1, which is an intracellular component of tight and adherens junctions, regulates epithelial cell permeability and differentiation (68, 69). In the urothelium, Tjp1 is located at the apicolateral borders of the umbrella cells, where it participates in forming a tight seal between adjacent umbrella cells (7), and reportedly functions in urothelial differentiation program (7, 9). The other tight-junction-related molecules identified in this study have not previously been reported in the context of the normal urothelium. Among them, Cgnl1 is a tight-junction-associated plaque protein that links tight junction proteins and the cytoskeleton, and functions in cell proliferation via the recruitment of factors that regulate the activity of the small GTPases, Rac1, and RhoA (22, 24). Cdc42 is a member of the Rho GTPase family, and is known to govern cellular polarity, regulate tight junctions, and modulate tight junction-associated signaling (70, 71). Amot and Mpp5 are both scaffolding proteins; the former reportedly functions in tight junction maintenance and coordinates the regulation of Cdc42 (25), whereas the latter is believed to interact with Amot and is involved in tight junction biogenesis and the establishment of cell polarity (25). Thus, our novel findings highlight the potential role of miR-205 in the normal urothelium. Based on our data, we hypothesize that miR-205 contributes to regulating urothelial differentiation through its suppression of tight-junction-related proteins.
Knockdown the expression of endogenous miR-205 in primary cultured urothelial cells also led to an alteration of keratin synthesis. Normal urothelial differentiation is characterized by the expression of specialized uroplakin proteins and select keratin pairs, which predominate in an epithelial cell- and differentiation state-selective manner (48). Our data showed that primary cultured mouse urothelial cells mainly express the simple epithelial keratins, K8, K18, and K19, and the basal/squamous keratins, K5 and K14. The knockdown of endogenous miR-205 in primary cultured urothelial cells altered these expression patterns, decreasing the expression of the basal-cell keratins (K5 and K14) and increasing the expression of superficial-cell keratins (K8 and K19). Finally, knockdown of endogenous miR-205 increased the expression levels of the uroplakins, which are the major urothelial differentiation products.
Taken together, these results demonstrate that miR-205 is expressed in a differentiation-dependent manner in bladder urothelial cells, where it targets several tight junction-related molecules and associates with the specific cellular differentiation markers, suggesting that miR-205 may contribute to regulating the programming of urothelial cellular differentiation (Fig. 6E). The release of this miR-205-mediated negative regulation in umbrella cells may therefore be a critically important mechanism underlying the formation of tight junctions and urothelial plaques (two hallmarks of urothelial differentiation). We herein report a novel emerging role for miR-205 in regulating tight junction-related molecules during the urothelial cellular differentiation. In the future, improvements in the dynamic state alteration of miRNA targets related to human urothelial differentiation may facilitate strategies toward a better understanding of regulation networks of miRNA.
Supplementary Material
Acknowledgments
We thank Dr. Tung-Tien Sun (New York University School of Medicine) for providing antibodies against uroplakins and for helpful discussions; Dr. Ya-Chung Tian (Chang Gung Memorial Hospital) for assistance with the TER assay.
Footnotes
Author contributions: P.K.C. designed research; P.K.C., L.C., C. Liang, C. Lin, and Yu-Xun Chang performed research; P.K.C., L.C., Chien-Lun Chen, Chun-Hsien Chen, and Yu-Sun Chang analyzed data; P.K.C. and L.C. wrote the article.
* This study was supported by grants from the Ministry of Education (EMRPD190191 and EMRPD1C0021 to PK Chung), and grants from Chang Gung Medical Research Program (CMRPG392151 to CL Chen and PK Chung; CMRPG390801 to Dr. Yu-Hsiang Lin, CL Chen and PK Chung; CLRPD190012 and 190015 to Dr. Jau-Song Yu), Taiwan.
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S5.
1 The abbreviations used are:
- miRNA
- microRNA
- TJP
- tight junction proteins
- AUM
- asymmetric unit membrane
- JAMs
- junctional adhesion molecules
- EMT
- epithelial–mesenchymal transition
- LNA
- Locked Nucleic Acid
- iTRAQ
- isobaric tags for relative and absolute quantitation
- UTR
- untranslation region
- TER
- transepithelial electrical resistance.
REFERENCES
- 1. Wu X. R., Kong X. P., Pellicer A., Kreibich G., Sun T. T. (2009) Uroplakins in urothelial biology, function, and disease. Kidney Int. 75, 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koss L. G. (1969) The asymmetric unit membranes of the epithelium of the urinary bladder of the rat. An electron microscopic study of a mechanism of epithelial maturation and function. Lab. Invest. 21, 154–168 [PubMed] [Google Scholar]
- 3. Staehelin L. A., Chlapowski F. J., Bonneville M. A. (1972) Lumenal plasma membrane of the urinary bladder. I. Three-dimensional reconstruction from freeze-etch images. J. Cell Biol. 53, 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hicks R. M. (1975) The mammalian urinary bladder: an accommodating organ. Biol. Rev. Camb. Philos. Soc. 50, 215–246 [DOI] [PubMed] [Google Scholar]
- 5. Yu J., Manabe M., Wu X. R., Xu C., Surya B., Sun T. T. (1990) Uroplakin I: a 27-kD protein associated with the asymmetric unit membrane of mammalian urothelium. J. Cell Biol. 111, 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu X. R., Manabe M., Yu J., Sun T. T. (1990) Large scale purification and immunolocalization of bovine uroplakins I, II, and III. Molecular markers of urothelial differentiation. J. Biol. Chem. 265, 19170–19179 [PubMed] [Google Scholar]
- 7. Acharya P., Beckel J., Ruiz W. G., Wang E., Rojas R., Birder L., Apodaca G. (2004) Distribution of the tight junction proteins ZO-1, occludin, and claudin-4, -8, and -12 in bladder epithelium. Am. J. Physiol. Renal Physiol. 287, F305–F318 [DOI] [PubMed] [Google Scholar]
- 8. Lewis S. A., Eaton D. C., Diamond J. M. (1976) The mechanism of Na+ transport by rabbit urinary bladder. J. Membr. Biol. 28, 41–70 [DOI] [PubMed] [Google Scholar]
- 9. Varley C. L., Garthwaite M. A., Cross W., Hinley J., Trejdosiewicz L. K., Southgate J. (2006) PPARgamma-regulated tight junction development during human urothelial cytodifferentiation. J. Cell. Physiol. 208, 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kreplak L., Wang H., Aebi U., Kong X. P. (2007) Atomic force microscopy of mammalian urothelial surface. J. Mol. Biol. 374, 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parsons C. L., Lilly J. D., Stein P. (1991) Epithelial dysfunction in nonbacterial cystitis (interstitial cystitis). J. Urol. 145, 732–735 [DOI] [PubMed] [Google Scholar]
- 12. Lavelle J. P., Meyers S. A., Ruiz W. G., Buffington C. A., Zeidel M. L., Apodaca G. (2000) Urothelial pathophysiological changes in feline interstitial cystitis: a human model. Am. J. Physiol. Renal Physiol. 278, F540–F553 [DOI] [PubMed] [Google Scholar]
- 13. Kreft M. E., Sterle M., Veranic P., Jezernik K. (2005) Urothelial injuries and the early wound healing response: tight junctions and urothelial cytodifferentiation. Histochem. Cell Biol. 123, 529–539 [DOI] [PubMed] [Google Scholar]
- 14. Zhang C. O., Wang J. Y., Koch K. R., Keay S. (2005) Regulation of tight junction proteins and bladder epithelial paracellular permeability by an antiproliferative factor from patients with interstitial cystitis. J. Urol. 174, 2382–2387 [DOI] [PubMed] [Google Scholar]
- 15. Shin K., Straight S., Margolis B. (2005) PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J. Cell Biol. 168, 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fanning A. S., Van Itallie C. M., Anderson J. M. (2012) Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell 23, 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buse P., Woo P. L., Alexander D. B., Cha H. H., Reza A., Sirota N. D., Firestone G. L. (1995) Transforming growth factor-alpha abrogates glucocorticoid-stimulated tight junction formation and growth suppression in rat mammary epithelial tumor cells. J. Biol. Chem. 270, 6505–6514 [DOI] [PubMed] [Google Scholar]
- 18. Balda M. S., Garrett M. D., Matter K. (2003) The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitic L. L., Anderson J. M. (1998) Molecular architecture of tight junctions. Annu. Rev. Physiol. 60, 121–142 [DOI] [PubMed] [Google Scholar]
- 21. Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M. (2006) ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126, 741–754 [DOI] [PubMed] [Google Scholar]
- 22. Ohnishi H., Nakahara T., Furuse K., Sasaki H., Tsukita S., Furuse M. (2004) JACOP, a novel plaque protein localizing at the apical junctional complex with sequence similarity to cingulin. J. Biol. Chem. 279, 46014–46022 [DOI] [PubMed] [Google Scholar]
- 23. Aijaz S., D'Atri F., Citi S., Balda M. S., Matter K. (2005) Binding of GEF-H1 to the tight junction-associated adaptor cingulin results in inhibition of Rho signaling and G1/S phase transition. Dev. Cell 8, 777–786 [DOI] [PubMed] [Google Scholar]
- 24. Guillemot L., Paschoud S., Jond L., Foglia A., Citi S. (2008) Paracingulin regulates the activity of Rac1 and RhoA GTPases by recruiting Tiam1 and GEF-H1 to epithelial junctions. Mol. Biol. Cell 19, 4442–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wells C. D., Fawcett J. P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., Colwill K., Starostine A., Metalnikov P., Pawson T. (2006) A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell 125, 535–548 [DOI] [PubMed] [Google Scholar]
- 26. Rickard A., Dorokhov N., Ryerse J., Klumpp D. J., McHowat J. (2008) Characterization of tight junction proteins in cultured human urothelial cells. In vitro cellular & developmental biology. Animal 44, 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 28. Carrington J. C., Ambros V. (2003) Role of microRNAs in plant and animal development. Science 301, 336–338 [DOI] [PubMed] [Google Scholar]
- 29. He L., Hannon G. J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 30. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 31. Yu J., Ryan D. G., Getsios S., Oliveira-Fernandes M., Fatima A., Lavker R. M. (2008) MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. U. S. A. 105, 19300–19305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greene S. B., Gunaratne P. H., Hammond S. M., Rosen J. M. (2010) A putative role for microRNA-205 in mammary epithelial cell progenitors. J. Cell Sci. 123, 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 34. Wang D., Zhang Z., O'Loughlin E., Wang L., Fan X., Lai E. C., Yi R. (2013) MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol. 15, 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blanchet L., Smolinska A., Attali A., Stoop M. P., Ampt K. A., van Aken H., Suidgeest E., Tuinstra T., Wijmenga S. S., Luider T., Buydens L. M. (2011) Fusion of metabolomics and proteomics data for biomarkers discovery: case study on the experimental autoimmune encephalomyelitis. BMC Bioinformatics 12, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kurland I. J., Accili D., Burant C., Fischer S. M., Kahn B. B., Newgard C. B., Ramagiri S., Ronnett G. V., Ryals J. A., Sanders M., Shambaugh J., Shockcor J., Gross S. S. (2013) Application of combined omics platforms to accelerate biomedical discovery in diabesity. Ann. N.Y. Acad. Sci. 1287, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Q., Halvey P. J., Shyr Y., Slebos R. J., Liebler D. C., Zhang B. (2013) Integrative omics analysis reveals the importance and scope of translational repression in microRNA-mediated regulation. Mol. Cell. Proteomics 12, 1900–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y., Devescovi V., Chen S., Nardini C. (2013) Multilevel omic data integration in cancer cell lines: advanced annotation and emergent properties. BMC Syst Biol. 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang F. X., Bosland M. C., Huang H., Romih R., Baptiste S., Deng F. M., Wu X. R., Shapiro E., Sun T. T. (2005) Cellular basis of urothelial squamous metaplasia: roles of lineage heterogeneity and cell replacement. J. Cell Biol. 171, 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomson J. M., Parker J., Perou C. M., Hammond S. M. (2004) A custom microarray platform for analysis of microRNA gene expression. Nat. Methods 1, 47–53 [DOI] [PubMed] [Google Scholar]
- 41. de Hoon M. J., Imoto S., Nolan J., Miyano S. (2004) Open source clustering software. Bioinformatics 20, 1453–1454 [DOI] [PubMed] [Google Scholar]
- 42. Saldanha A. J. (2004) Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 43. Chang K. P., Yu J. S., Chien K. Y., Lee C. W., Liang Y., Liao C. T., Yen T. C., Lee L. Y., Huang L. L., Liu S. C., Chang Y. S., Chi L. M. (2011) Identification of PRDX4 and P4HA2 as metastasis-associated proteins in oral cavity squamous cell carcinoma by comparative tissue proteomics of microdissected specimens using iTRAQ technology. J. Proteome Res. 10, 4935–4947 [DOI] [PubMed] [Google Scholar]
- 44. Tang X., Gal J., Zhuang X., Wang W., Zhu H., Tang G. (2007) A simple array platform for microRNA analysis and its application in mouse tissues. Rna 13, 1803–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin B. F. (1972) Cell replacement and differentiation in transitional epithelium: a histological and autoradiographic study of the guinea-pig bladder and ureter. J. Anat. 112, 433–455 [PMC free article] [PubMed] [Google Scholar]
- 46. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 47. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moll R., Franke W. W., Schiller D. L., Geiger B., Krepler R. (1982) The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell 31, 11–24 [DOI] [PubMed] [Google Scholar]
- 49. Nelson W. G., Sun T. T. (1983) The 50- and 58-kdalton keratin classes as molecular markers for stratified squamous epithelia: cell culture studies. J. Cell Biol. 97, 244–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Surya B., Yu J., Manabe M., Sun T. T. (1990) Assessing the differentiation state of cultured bovine urothelial cells: elevated synthesis of stratification-related K5 and K6 keratins and persistent expression of uroplakin I. J. Cell Sci. 97 (Pt 3), 419–432 [DOI] [PubMed] [Google Scholar]
- 51. Veranic P., Romih R., Jezernik K. (2004) What determines differentiation of urothelial umbrella cells? Eur. J. Cell Biol. 83, 27–34 [DOI] [PubMed] [Google Scholar]
- 52. Romih R., Korosec P., de Mello W., Jr., Jezernik K. (2005) Differentiation of epithelial cells in the urinary tract. Cell Tissue Res. 320, 259–268 [DOI] [PubMed] [Google Scholar]
- 53. Kurzrock E. A., Lieu D. K., Degraffenried L. A., Chan C. W., Isseroff R. R. (2008) Label-retaining cells of the bladder: candidate urothelial stem cells. Am. J. Physiol. Renal Physiol. 294, F1415–F1421 [DOI] [PubMed] [Google Scholar]
- 54. Shin K., Lee J., Guo N., Kim J., Lim A., Qu L., Mysorekar I. U., Beachy P. A. (2011) Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature 472, 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu H., Zhu S., Mo Y. Y. (2009) Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 19, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Iorio M. V., Casalini P., Piovan C., Di Leva G., Merlo A., Triulzi T., Menard S., Croce C. M., Tagliabue E. (2009) microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 69, 2195–2200 [DOI] [PubMed] [Google Scholar]
- 57. Kaller M., Liffers S. T., Oeljeklaus S., Kuhlmann K., Roh S., Hoffmann R., Warscheid B., Hermeking H. (2011) Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol. Cell. Proteomics 10, M111 010462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leivonen S. K., Rokka A., Ostling P., Kohonen P., Corthals G. L., Kallioniemi O., Perala M. (2011) Identification of miR-193b targets in breast cancer cells and systems biological analysis of their functional impact. Mol. Cell. Proteomics 10, M110 005322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 60. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hausser J., Syed A. P., Selevsek N., van Nimwegen E., Jaskiewicz L., Aebersold R., Zavolan M. (2013) Timescales and bottlenecks in miRNA-dependent gene regulation. Mol. Syst. Biol. 9, 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taguchi A., Yanagisawa K., Tanaka M., Cao K., Matsuyama Y., Goto H., Takahashi T. (2008) Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res. 68, 5540–5545 [DOI] [PubMed] [Google Scholar]
- 63. Yang Y., Chaerkady R., Beer M. A., Mendell J. T., Pandey A. (2009) Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 9, 1374–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ou M., Zhang X., Dai Y., Gao J., Zhu M., Yang X., Li Y., Yang T., Ding M. (2014) Identification of potential microRNA-target pairs associated with osteopetrosis by deep sequencing, iTRAQ proteomics and bioinformatics. Eur. J. Hum. Genet. 22, 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nielsen C. B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C. B. (2007) Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. Rna 13, 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Anderson J. M., Van Itallie C. M. (1995) Tight junctions and the molecular basis for regulation of paracellular permeability. Am. J. Physiol. 269, G467–G475 [DOI] [PubMed] [Google Scholar]
- 69. Balda M. S., Matter K. (2000) The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 19, 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bruewer M., Hopkins A. M., Hobert M. E., Nusrat A., Madara J. L. (2004) RhoA, Rac1, and Cdc42 exert distinct effects on epithelial barrier via selective structural and biochemical modulation of junctional proteins and F-actin. Am. J. Physiol. Cell Physiol. 287, C327–335 [DOI] [PubMed] [Google Scholar]
- 71. Rojas R., Ruiz W. G., Leung S. M., Jou T. S., Apodaca G. (2001) Cdc42-dependent modulation of tight junctions and membrane protein traffic in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 12, 2257–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.