Abstract
Quantitative phosphoproteomics workflows traditionally involve additional sample labeling and fractionation steps for accurate and in-depth analysis. Here we report a high-throughput, straightforward, and comprehensive label-free phosphoproteomics approach using the highly selective, reproducible, and sensitive Ti4+-IMAC phosphopeptide enrichment method. We demonstrate the applicability of this approach by monitoring the phosphoproteome dynamics of Jurkat T cells stimulated by prostaglandin E2 (PGE2) over six different time points, measuring in total 108 snapshots of the phosphoproteome. In total, we quantitatively monitored 12,799 unique phosphosites over all time points with very high quantitative reproducibility (average r > 0.9 over 100 measurements and a median cv < 0.2). PGE2 is known to increase cellular cAMP levels, thereby activating PKA. The in-depth analysis revealed temporal regulation of a wide variety of phosphosites associated not only with PKA, but also with a variety of other classes of kinases. Following PGE2 stimulation, several pathways became only transiently activated, revealing that in-depth dynamic profiling requires techniques with high temporal resolution. Moreover, the large publicly available dataset provides a valuable resource for downstream PGE2 signaling dynamics in T cells, and cAMP-mediated signaling in particular. More generally, our method enables in-depth, quantitative, high-throughput phosphoproteome screening on any system, requiring very little sample, sample preparation, and analysis time.
In cancer, alterations in signal transduction networks can give rise to uncontrolled cell growth (1). For example, aberrant T-cell signaling can result in the manifestation of leukemia (2). The immune regulatory actions of T-cells are mediated by autocrine and paracrine prostaglandin E2 (PGE2),1 secreted by many different cell types, including tumor cells (3, 4). When PGE2 binds to its G-protein-coupled receptors, the secondary messenger cyclic adenosine monophosphate (cAMP) is produced, leading to the activation of intracellular signal transduction cascades primarily involving protein kinase A (PKA).
The dynamics of protein phosphorylation can nowadays be studied via mass spectrometry (MS)-based proteomics (5–7), which enables the confident identification of many thousands of phosphorylation sites in a single cellular system or tissue. As phosphoproteins, and their corresponding peptides after enzymatic digestion, are present at much lower abundances than their unmodified counterparts, enrichment of the phosphopeptide population prior to LC-MS/MS is a necessity. Methodologies for the global analysis of the phosphoproteome are most frequently based on immobilized metal ion affinity chromatography (IMAC) (8, 9) or titanium dioxide (TiO2) (10, 11) chromatography (12). Recently, we described titanium-IMAC (Ti4+-IMAC)-based enrichment as a new affinity matrix (13, 14), providing even greater selectivity and sensitivity than earlier methods (15).
Quantification of phosphorylation dynamics poses an additional analytical challenge typically requiring stable isotope labeling of samples (16, 17). A more straightforward approach would be so-called label-free quantification, which is cost-effective, alleviates the restriction on the number of samples that can be quantitatively monitored, and can reach a very high dynamic range in quantification. Label-free quantification requires, however, careful experimental design to prevent the introduction of sample biases (7). This is especially true for the analysis of post-translational modifications, which requires that the enrichment steps be highly reproducible both qualitatively and quantitatively. In this study we assessed the qualitative and quantitative reproducibility of Ti4+-IMAC phosphopeptide enrichment and demonstrated its applicability in a large-scale dynamic phosphoproteomics study in Jurkat T cells stimulated with PGE2.
EXPERIMENTAL PROCEDURES
Cell Culture and Digest Preparation
HeLa cells were grown to confluence in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (Lonza, Basel, Switzerland), after which cells were washed twice with PBS and harvested. Jurkat T lymphoma cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (Lonza). Before PGE2 stimulation, cells were centrifuged for 1 min at 1500g, growth medium was removed, and the cells were resuspended at a final concentration of 1 × 106 to 2 × 106 cells/ml in RPMI. Next, cells were supplemented with 0 (control) or 10 μm PGE2 and incubated for 5, 10, 20, 30, or 60 min. After treatment, Jurkat cells were washed twice with PBS and harvested. Cell lysis was performed on ice by sonication in buffer containing 50 mm ammonium bicarbonate (pH 8.0), 8 m urea, 1 mm sodium orthovanadate, complete EDTA-free protease inhibitor mixture (Roche), and phosSTOP phosphatase inhibitor mixture (Roche). Cell debris was then removed by centrifugation at 20,000g for 15 min at 4 °C. The total protein concentration was measured using a Bradford assay (Bio-Rad) and then split into 1-mg aliquots for enzymatic digestion. Proteins were reduced with DTT at a final concentration of 4 mm at 56 °C for 25 min; subsequently samples were alkylated with iodoacetamide at a final concentration of 8 mm at room temperature for 30 min in the dark. Proteins were then digested using Lys-C (1 μg of Lys-C per 75 μg of protein) and incubated for 4 h at 37 °C. The solution was then diluted to a final urea concentration of 2 m with 50 mm ammonium bicarbonate, trypsin was added (1 μg of trypsin per 100 μg of protein), and the solution was incubated at 37 °C overnight. The digestion was quenched by acidification to 5% formic acid. The digests were desalted using Sep-Pak C18 cartridges, dried in vacuo, and stored at −80 °C for further use.
Phosphopeptide Enrichment via Ti4+-IMAC
IMAC material was prepared and used essentially as previously described (14). Briefly, the Ti4+-IMAC beads (500 μg of beads/200 μl pipette tip) were loaded onto GELoader tips (Eppendorf, Hamburg, Germany) using a C8 plug. To reduce variations of enrichment processes, in-parallel spin tip enrichment was used. The Ti4+-IMAC columns were conditioned using 50 μl of loading buffer consisting of 80% acetonitrile (ACN)/6% trifluoroacetic acid (TFA) and centrifugation at 200g for 10 min. The protein digests were dissolved in 80% ACN/6% TFA and split in aliquots corresponding to ∼100 μg and ∼250 μg for the HeLa and Jurkat cell lysates, respectively. The aliquots were transferred to the spin tips and centrifuged at 100g for 30 min. Then, the columns were sequentially washed with 50 μl of washing buffer 1 (50% ACN, 0.5% TFA containing 200 mm NaCl) and 50 μl of washing buffer 2 (50% ACN/0.1% TFA) and centrifuged at 170g for 15 min. The bound peptides were eluted into a new tube (already containing 35 μl of 10% formic acid) with 20 μl of 10% ammonia by centrifugation at 100g for 20 min. A final elution was performed with 5 μl of 80% ACN/2% formic acid for 10 min. The collected eluate was further acidified by the addition of 3 μl of 100% formic acid prior to nano-LC-MS/MS analysis.
Reverse Phase Chromatography and Mass Spectrometry
Peptides were subjected to reverse phase nano-LC-MS/MS analysis using a Proxeon EASY-nLC 1000 (Thermo Scientific, Odense, Denmark) with an analytical column heater (40 °C) and an LTQ-Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany). Peptides were first trapped (Reprosil C18, Dr Maisch, GmbH, Ammerbuch, Germany, 3 μm, 2 cm × 100 μm) at a maximum pressure of 800 bar with 100% solvent A (0.1% formic acid in water) before being separated on the analytical column (either Agilent Poroshell 120 EC-C18, 2.7 μm, 40 cm × 50 μm for the HeLa cell samples or Agilent Zorbax SB-C18, 1.8 μm, 40 cm × 75 μm for the Jurkat T cell samples) (Agilent, Santa Clara, CA). Peptides were chromatographically separated by a 90-min gradient from 7% to 30% solvent B (0.1% formic acid in ACN) at a flow rate of 150 or 100 nL/min. The total measurement time for each sample was 110 min. The eluent was sprayed via a distal coated fused silica emitter (360-μm outer diameter, 20-μm inner diameter, 10-μm tip inner diameter; constructed in-house) butt-connected to the analytical column. The electrospray voltage was set at 1.7 kV. The mass spectrometer was operated in a data-dependent mode to automatically switch between MS and MS/MS. Briefly, survey full-scan MS spectra were acquired in the Orbitrap analyzer, scanning from m/z 350 to m/z 1500 at a resolution of 60,000 at m/z 400 using an automatic gain control setting of 1e6 ions. Charge state screening was enabled, and precursors with either unknown or 1+ charge states were excluded. After the survey scan, the 20 most intense precursors were selected for subsequent decision-tree-based ion trap CID or ETD fragmentation (18). The normalized collision energy for CID was set at 35%, and supplemental activation for ETD and dynamic exclusion were enabled (exclusion size list: 500; exclusion duration: 40 s).
Data Processing
Out of 108 nano-LC-MS/MS runs, 8 runs were not consistent with the other replicates and were discarded prior to data analysis. Raw data were processed with MaxQuant version 1.3.0.5 (19). MS and MS/MS spectra were searched against concatenated forward-decoy Swiss-Prot Homo sapiens database version 2012 09 (40,992 sequences) using the Andromeda search engine. The database search was performed with the following parameters: an initial mass tolerance of ±20 ppm and a final mass tolerance of ±6 ppm for precursor masses, ±0.6 Da for CID and ETD ion trap fragment ions, and two missed cleavages allowed. Cysteine carbamidomethylation was used as a fixed modification, and methionine oxidation, protein N-terminal acetylation, and serine, threonine, and tyrosine phosphorylation were included as variable modifications. The false discovery rate was set at 0.01 for peptides, proteins, and phosphosites; the minimum peptide length allowed was six amino acids; and a minimum Andromeda peptide score of 60 was required. The match-between-runs feature was enabled. A site localization probability of at least 0.75 and a score difference of at least 5 were used as thresholds for the localization of phosphoresidues. Normalization was performed by subtracting the median of log transformed intensities for each nano-LC-MS/MS run. To identify significantly regulated phosphorylation sites, a two-sample t test was performed with a permutation-based false discovery rate of 0.005 (randomizations were set at 500, and s0 was tuned to achieve a minimum 2-fold regulation and varied between 0.35 and 0.4). Phosphorylation sites regulated in at least 1 of all 15 possible comparisons were included for soft clustering analysis using GProX version 1.1.9 (20). Log base 2 intensities were Z-scored prior to fuzzy clustering using a fuzzification value of 2, 100 iterations, and a minimum membership of 0.35. The number of clusters was empirically derived by varying the number between 4 and 10, where 8 clusters returned the most distinctive clusters. The high-confidence localized phosphorylation sites were further analyzed using iceLogo (21) using a p value of 0.01 with the entire human proteome (Swiss-Prot human) as a background. Protein Gene Ontology biological process, Gene Ontology molecular function, Gene Ontology cellular component, and class were queried from the Panther database (22), and enrichment analysis was performed using a Fisher's exact test in Perseus with a 2-fold enrichment cutoff and a Benjamin–Hochberg false discovery rate of 0.02. All clustered phosphorylation sites were subjected to the NetworKIN 3.0 kinase prediction algorithm (23) to predict possible upstream kinases. A score threshold of 0.5 was applied, and only the top 10 predictions were used. To reveal kinases differentially active in each cluster, kinase activity enrichment was calculated as the ratio between the number of sites predicted by a specific kinase and the total number sites in each cluster. Predicted kinase substrates in each cluster were searched in String 9.05 (24) for interactors using the standard settings, and 10 new nodes were added to build protein networks. Signaling networks were then visualized in Cytoscape.
All raw mass spectrometry data files and MaxQuant output files have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (25) with the dataset identifier PXD000293. All peptide spectral matches and their MS/MS spectra can be viewed in the freely available Maxquant Viewer.
Antibodies Used for Western Blotting
For Western blot analysis, the following antibodies were used: rabbit anti-phospho-Filamin A (Ser2152, #4761), rabbit anti-phospho-Cofilin (Ser3, #3311), rabbit anti-phospho-ATF2 (Thr71, #5112), anti-rabbit IgG HRP (#7074), and anti-mouse IgG HRP (#7076) from Cell Signaling Technology (Danvers, MA) and mouse anti-α-Tubulin (#T9026) from Sigma.
RESULTS
We first assessed the feasibility of the highly selective and sensitive Ti4+-IMAC phosphopeptide enrichment strategy (13–15) for large-scale and high-throughput label-free quantification of phosphorylation dynamics. Therefore, we first interrogated its specificity and reproducibility for consecutive enrichments in HeLa cells. The data from three independent Ti4+-IMAC enrichments clearly show high specificity and qualitative reproducibility (Figs. 1A and 1B, respectively).
Fig. 1.
High specificity and reproducibility of Ti4+-IMAC phosphopeptide enrichment assessed in HeLa cells. A, the high percentage of phosphopeptides in three separate enrichments indicates the high specificity of the Ti4+-IMAC enrichment. B, the overlap of identified phosphopeptides in three MS replicates from a single enrichment (MS 1–3, upper panel) is similar to the overlap of three separate enrichments (E1–E3, lower panel), indicating qualitatively highly reproducible phosphopeptide enrichment. C, phosphopeptide intensity (log base 2) comparisons for MS replicates (upper panel) and enrichment replicates (lower panel), showing highly accurate quantitative reproducibility of the Ti4+-IMAC enrichment method.
Virtually all peptides observed were phosphopeptides (>98% out of ∼5000 peptide spectral matches), and the overlap of observed phosphopeptides between independent Ti4+-IMAC phosphopeptide enrichments was similar to that in technical replicate nano-LC-MS runs. Importantly, the quantitative reproducibility between the different enrichment replicates was high (Fig. 1C, supplemental Fig. S1), with an average Pearson correlation of 0.92 and a median cv of 0.2, evidently meeting the above-described constraint for label-free quantification of post-translational modifications.
In-depth Temporal Label-free Phosphoproteome Quantification
Satisfied with the high reproducibility and depth of analysis, we next monitored temporal phosphorylation signaling events in Jurkat T cells at six time points following stimulation of their G-protein-coupled receptors, EP1 to EP4, with the ligand PGE2. This stimulation affects several intracellular signaling cascades, involving primarily the cAMP/PKA and the PI3K/AKT pathways (26).
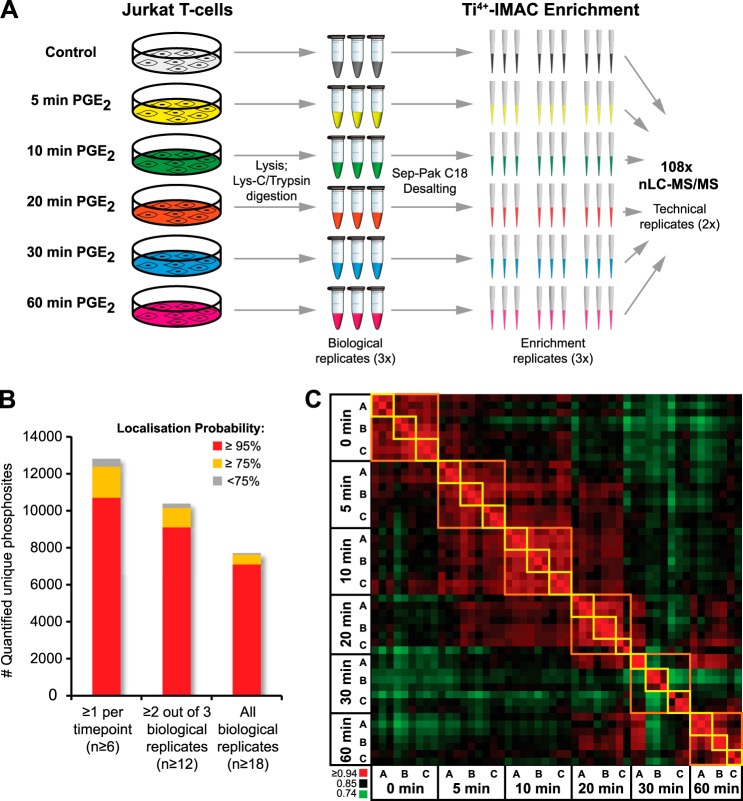
Jurkat cells were harvested after 0, 5, 10, 20, 30, and 60 min of PGE2 stimulation. For every time point, three biological replicates were harvested, cells were lysed, and proteins were digested with Lys-C and trypsin. Phosphopeptides were enriched using for every biological replicate three independent Ti4+-IMAC enrichment columns, requiring only 200 μg of protein digest as input material each. All 54 samples (six time points, three biological replicates, three Ti4+-IMAC enrichments; Fig. 2A) were analyzed twice via nano-LC-MS/MS using 2-h gradients and decision-tree guided peptide fragmentation, switching between CID and ETD based on peptide physiochemical properties (18, 27). In total this resulted in 108 nano-LC-MS/MS runs, of which 95% were successful, amounting to 216 h of MS analysis time and identifying cumulatively 21,443 unique phosphosites from 50 individual Ti4+-IMAC phosphopeptide enrichments. Of these 21,443 unique phosphosites, we could quantitatively follow the presence of 12,799 over all time points and 10,375 in at least two out of three biological replicates (Fig. 2B). To evaluate the quality of the complete experiment, we assessed the correlation and median cv between all Ti4+-IMAC enrichments of the different biological replicates and performed a Pearson's correlation of all phosphopeptide intensities of the successful nano-LC-MS/MS runs, plotted in a heat map (Table I, Fig. 2C). The heat map illustrates the reproducibility in phosphosite intensities between the different enrichment replicates (yellow squares) and the similarity within the three biological replicates (A, B, and C, orange squares). The time point showing the greatest biological similarity was 10 min after activation, and the greatest biological variability was observed after 30 min. As the enrichment reproducibility remained very high, the variation observed at the 30-min time point should be attributed mostly to biological variability rather than sample/enrichment handling variability.
Fig. 2.
High-throughput label-free phosphoproteomics. A, three biological replicates of Jurkat T lymphocyte cells stimulated with prostaglandin E2 (PGE2) and harvested at six different time points were lysed, digested, and desalted prior to phosphopeptide enrichment. Each biological replicate was divided over three Ti4+-IMAC phosphopeptide enrichment columns, resulting in 54 samples that were analyzed twice via 2-h nano-LC-MS/MS with CID/ETD fragmentation, amounting to a total of 108 phosphoproteomic datasets. B, number of unique phosphosites quantified using different criteria. The minimum number of quantification events is reported in brackets. C, Pearson intensity correlation heat map of all runs showing high reproducibility in phosphosite intensity for the phosphopeptide enrichment replicates (yellow squares) and high similarity between biological replicates A–C (orange squares).
Table I. Enrichment triplicate reproducibility.
| Sample | Average Pearson intensity correlation |
Median coefficient of variation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 min | 5 min | 10 min | 20 min | 30 min | 60 min | 0 min | 5 min | 10 min | 20 min | 30 min | 60 min | |
| BioA | 0.92 | 0.92 | 0.93 | 0.9 | 0.94 | 0.94 | 0.23 | 0.20 | 0.21 | 0.18 | 0.14 | 0.18 |
| BioB | 0.92 | 0.92 | 0.93 | 0.94 | 0.91 | 0.94 | 0.21 | 0.21 | 0.21 | 0.17 | 0.18 | 0.14 |
| BioC | 0.94 | 0.92 | 0.93 | 0.9 | 0.9 | 0.93 | 0.19 | 0.21 | 0.18 | 0.16 | 0.16 | 0.18 |
Jurkat Phosphoproteome Dynamics
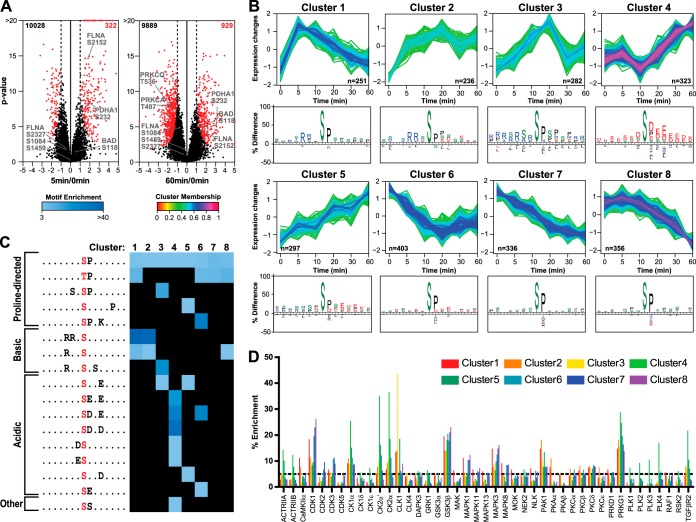
Next, significantly regulated phosphosites were strictly elected using two-sample t tests, comparing all time points using at least three measurements per phosphosite (Fig. 3A; 5 min/0 min and 60 min/0 min). In total, 2983 phosphosites were significantly regulated using the rather stringent criteria, present in all six time points in two out of three biological replicates at a false discovery rate of 0.005. Although a large number of phosphosites were regulated directly upon stimulation with PGE2 (5 min), a greater number of phosphosites were regulated at later time points, indicating downstream phosphorylation amplification in activated signaling cascades (Fig. 3A, supplemental Fig. S2).
Fig. 3.
Analysis of differentially regulated phosphosites. A, volcano plots of treatment versus control comparisons. p values (−log base 10, two-sample t test) are plotted as a function of the phosphosite ratio (log base 2) for 5 and 60 min versus 0 min of PGE2 treatment. Regulated sites are colored in red (permutation based false discovery rate = 0.005, s0 adjusted for 2-fold regulation). B, soft clustering analysis of significantly regulated phosphosites resulted in five differentially up-regulated and three differentially down-regulated clusters. For each cluster, the phosphosite sequence logos is plotted, showing amino acid frequencies that are significantly different compared to the entire human proteome (p < 0.01). Early up-regulated sites contain basophilic motifs, late up-regulated sites contain acidophilic motifs, and down-regulated sites contain only proline directed consensus sites. C, linear kinase motifs enriched in each cluster by Motif-X analysis. Significantly overrepresented kinase motifs were determined by querying the data against the IPI human database using a p value of 1E-6, a minimum number of occurrences of 20, and a minimum 3.0-fold enrichment relative to background. D, kinases predicted to be active upon PGE2 stimulation according to the NetworKIN algorithm. The y-axis represents the percentage of phosphorylated substrates predicted for each kinase in each cluster.
All regulated phosphosites were analyzed for common temporal regulation patterns using unsupervised clustering. This resulted in eight clusters of distinct temporal dynamics, with five clusters displaying patterns related to up-regulated phosphosite abundance and three clusters displaying primarily down-regulation (Fig. 3B). Gene ontology analysis revealed enrichment of endocytosis and mRNA-related terms in early-intermediate up-regulated clusters 2 and 3, whereas cluster 4, corresponding to late up-regulation, showed predominantly DNA-related ontology terms (supplemental Fig. S3).
Interestingly, the different clusters also showed a clear distinction in enriched sequence motifs, portrayed by IceLogo (28) (Figs. 3B and 3C), related directly to distinct kinase activity. Clusters 1 and 2, which showed strong up-regulation after 5 min of PGE2 stimulation, represent a clear basophilic kinase motif complemented by proline directed motifs. When we look in more detail at the sequence motifs present in this cluster, using motif-X (29), we can distinguish several distinct submotifs that are significantly enriched and mutually exclusive (Fig. 3C). Proline directed kinase substrate motifs are present in all clusters and therefore do not display clear temporal changes. In contrast, the extracted basophilic domains were mostly phosphorylated shortly after PGE2 stimulation (5, 10 min), whereas the acidophilic substrate motifs became more enriched at later times (≥20 min), likely as a secondary response to PGE2.
To predict the kinases responsible for the observed changes in phosphorylation and to gain more detailed information about the PGE2-induced signaling network, we subjected all regulated phosphosites to NetworKIN (23) (Fig. 3D). This analysis revealed an early increase in phosphorylation of CDK1, MAPK1, MAPK3, and MAPK8 substrates that decreased over time (highly enriched in clusters 1, 6, 7, and 8), possibly involving a negative feedback loop. Kinase substrates activated immediately after PGE2 stimulation, gradually damping out over time (cluster 1) or remaining up-regulated (cluster 2), were predominantly linked to CAMKIIa, PAK1, PKAα, PKAβ, and PKCε. Interestingly, the kinases CLK1 and CLK4 showed a clear, distinct activity profile corresponding to a delayed maximum activity at 20 min followed by a strong reduction for the proceeding time points (cluster 3). CLK1 is known to phosphorylate proteins of the mRNA spliceosomal complex and is likely the kinase responsible for the highly enriched mRNA processing gene ontology term in cluster 3 (supplemental Fig. S3).
The set of kinases predicted to be specifically activated after a prolonged PGE2 treatment (≥20 min; cluster 4) included the activin receptors (ACTRIIA, ACTRIIB), casein kinases (CK1α, -δ, and -ε; CK2α), Polo-like kinases (PLK1, -2, -3, and -4), and TGF-β receptor type 2. These (receptor) kinases are activated at a later stage and therefore probably represent downstream targets of the primary kinases affected at earlier times. Finally, the clusters showing mainly down-regulation, such as cluster 8, primarily contained cyclin-dependent and mitogen-activated kinases.
DISCUSSION
Here, we describe the application of Ti4+-IMAC-based phosphopeptide enrichment in combination with label-free quantification used to investigate in-depth temporal phosphorylation dynamics, enabling the reproducible quantification of more than 12,000 unique phosphorylation sites over six time points, including both biological and analytical replicates. The reported label-free approach is generic and nonrestrictive in the amount of sample and MS instrument time required. Our large dataset also provides specific new insights into the intricacy of PGE2-induced phosphorylation dynamics in T cells.
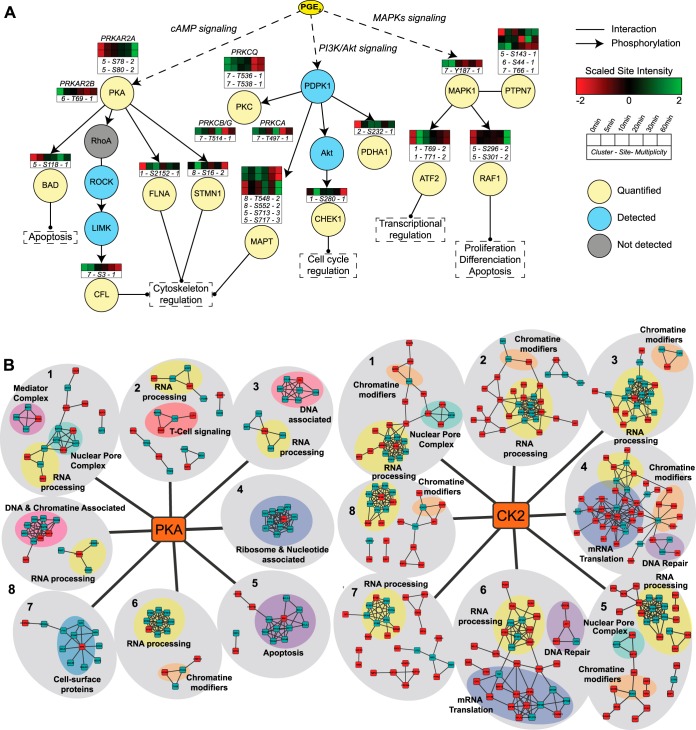
PGE2 is known to induce the increase of cellular cAMP levels, thereby activating primarily PKA. PKA subsequently initiates phosphorylation of downstream substrates. Details on PGE2/PKA-specific phosphorylation sites are still scarce; however, we can define early, intermediate, and late responders to PGE2 stimulation using a manually curated network, extracted from the online UniProt Knowledgebase and PhosphoSitePlus database (30) (Fig. 4A). PKA activation is evident in our data through several examples, such as the early up-regulation of S2152 on FLNA and S16 on STMN1, which are direct substrates of PKA and have functional implications in cytoskeleton reorganization. More downstream substrates were also identified, such as S3 on CFL, which showed decreased phosphosite abundance at the late time points, probably because of the inactivation of intermediate substrates RhoA, ROCK, and LIMK. These dynamic changes in phosphorylation can also be investigated via quantitative Western blotting, albeit in a low-throughput manner and only for sites for which specific phospho-antibodies are available. Therefore, we compared our MS-based method to quantitative Western blotting of the substrates FLNA (S2152), CFL (S3), and ATF2 (T71) (supplemental Fig. S4). The quantitative data for both methods showed good agreement with the MS-based quantitative data, and this gives credence to our MS-based quantitative data on all the other thousands of phosphorylation sites.
Fig. 4.
Downstream signaling of PGE2 stimulation reveals early, intermediate, and late responders. A, network manually curated using the reference databases UniProt and PhosphoSitePlus. Arrows represent known phosphorylation events between upstream kinases and their substrates. B, protein complexes and/or networks of predicted PKA (left panel) and CK2 (right panel) substrates. Significantly regulated substrates for each cluster and additional proteins belonging to the same protein complex are shown in red and green, respectively. Interaction data and common protein features were retrieved from the STRING and UniProt databases, respectively. See supplemental Figs. S5 and S6 for a more detailed version.
Interestingly, the abundance of the phosphosite S118 on BAD, a direct PKA substrate, was gradually up-regulated at later time points, indicating additional indirect regulatory mechanisms that block the pro-apoptotic activity of BAD (31). The MAPK and PI3K/AKT signaling pathways were also represented among the regulated phosphosites. MAPK1/ERK2 can be activated upon an increase in cAMP levels through its phosphorylation by the upstream kinase activators (MEK1 and MEK2) and the dissociation of MAPK1 from phosphorylated PTPN7, a protein phosphatase that acts preferentially on tyrosine-phosphorylated MAPK1. This results in the activation of MAPK1 and phosphorylation of its downstream substrates such as ATF2 T71, which was found to be up-regulated in both our in-depth phosphoproteomics dataset and the quantitative Western blot analysis (supplemental Fig. S4). Moreover, negative feedback phosphorylation of MAPK1 on RAF1 (S296 and S301), yielding an inactive, desensitized kinase, was also observed in our data. The PI3K/AKT pathway is activated by PGE2 stimulation through the EP4 receptor (32). This results in the activation of AKT signaling, which was apparent from the presence of its downstream substrate CHEK1 S280 in cluster 1.
In addition, differentially regulated protein complexes/networks for each cluster were identified using the predicted kinase substrates of PKA and CK2 from NetworKIN and the interaction data from STRING (Fig. 4B, supplemental Figs. S5 and S6). PGE2 stimulation has a clear effect on RNA processing (mRNA splicing, mRNA decapping, RNA binding) and DNA/chromatin binding, as many regulated phosphosites predicted to be regulated by PKA and CK2 were involved in these categories in multiple clusters. More specifically for PKA, a T cell signaling protein network was only identified in the set of phosphosites up-regulated for the entire PGE2 stimulation duration (cluster 2), suggesting a strong dependence of these PKA-dependent signaling proteins on PGE2 presence. Casein kinase 2 was highly enriched for substrates in clusters 4, 5, and 6, corresponding to a delayed regulation to PGE2 treatment (Fig. 3D). Interestingly, CK2-specific substrate interaction networks were mainly involved in DNA repair and mRNA translation (initiation, elongation, rRNA, tRNA binding factors) (clusters 4 and 6).
Overall, our work demonstrates the high potential of combining Ti4+-IMAC phosphopeptide enrichment, high-resolution nano-LC-MS/MS, and label-free quantification to interrogate in a comprehensive manner phosphosite abundance regulation in a large sample set. Previous studies using label-free phosphoproteomics resulted in lower coverage, identifying up to ∼1500 phosphosites in single runs and about 700 phosphosites reproducibly over four replicates (33–35). To increase the depth of their analysis, Kettenbach and Gerber used a two-dimensional enrichment and separation approach and were able to identify over 10,000 sites (35). However, because an additional fractionation step was introduced, the total analysis and instrument time had to be increased, and such a two-dimensional approach, with samples separated over different LC-MS runs, also compromises quantitative accuracy. Using the single-run-per-sample strategy reported here, the phosphoproteome could be analyzed in depth (>24,000 phosphosites in total) in a quantitatively reproducible manner (>10,000 quantified in at least 12 replicates, r > 0.9). Moreover, the presented methodology enabled the comprehensive analysis of the phosphoproteome from a large number of different samples (>100) using relatively small sample amounts. As a result of our high enrichment reproducibility (Fig. 2C, yellow squares), for future experiments we suggest that a single enrichment (i.e. 200 μg) per biological replicate will be sufficient, minimizing even further the instrument time and sample amount needed. The quality of the data and strength of the approach could be validated by previously curated phosphosite regulations and Western blotting, when antibodies are available. Finally, the presented data also constitute a comprehensive resource for downstream PGE2 signaling dynamics in T cells, and PKA signaling dynamics in general.
Supplementary Material
Acknowledgments
We thank Shabaz Mohammed, Houjiang Zhou, and Serena Di Palma for fruitful discussion and help with the Ti4+-IMAC enrichment protocol.
Footnotes
Author contributions: A.M.A. and A.J.H. designed research; E.L.d. and P.G. performed research; E.L.d. and P.G. analyzed data; E.L.d., P.G., A.M.A., and A.J.H. wrote the paper.
* This work was supported by The Netherlands Proteomics Centre and The Netherlands Organization for Scientific Research (NWO)–supported large-scale proteomics facility Proteins@Work (Project 184.032.201). NWO is also acknowledged for the VIDI grant of AFMA (723.012.102). Additionally, the PRIME-XS Project (Grant Agreement 262067), funded by the European Union Seventh Framework Program, is kindly acknowledged for financial support.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PGE2
- prostaglandin E2
- ACN
- acetonitrile
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- IMAC
- immobilized metal ion affinity chromatography
- Ti4+-IMAC
- immobilized titanium (IV) ion affinity chromatography
- CID
- collision-induced dissociation
- ETD
- electron transfer dissociation.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 2. Van Vlierberghe P., Ferrando A. (2012) The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Invest. 122, 3398–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kalinski P. (2012) Regulation of immune responses by prostaglandin E2. J. Immunol. 188, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chemnitz J. M., Driesen J., Classen S., Riley J. L., Debey S., Beyer M., Popov A., Zander T., Schultze J. L. (2006) Prostaglandin E2 impairs CD4+ T cell activation by inhibition of lck: implications in Hodgkin's lymphoma. Cancer Res. 66, 1114–1122 [DOI] [PubMed] [Google Scholar]
- 5. Altelaar A. F., Munoz J., Heck A. J. (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 [DOI] [PubMed] [Google Scholar]
- 6. Cox J., Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Fonslow B. R., Shan B., Baek M. C., Yates J. R., 3rd. (2013) Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 113, 2343–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Villen J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villen J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 11. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., Colman-Lerner A., Nolan G. P., Nesvizhskii A. I., Peter M., Loewith R., von Mering C., Aebersold R. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang P. H., White F. M. (2008) Phosphoproteomics: unraveling the signaling web. Mol. Cell 31, 777–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou H., Ye M., Dong J., Han G., Jiang X., Wu R., Zou H. (2008) Specific phosphopeptide enrichment with immobilized titanium ion affinity chromatography adsorbent for phosphoproteome analysis. J. Proteome Res. 7, 3957–3967 [DOI] [PubMed] [Google Scholar]
- 14. Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A. J., Zou H., Mohammed S. (2013) Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480 [DOI] [PubMed] [Google Scholar]
- 15. Zhou H., Low T. Y., Hennrich M. L., van der Toorn H., Schwend T., Zou H., Mohammed S., Heck A. J. (2011) Enhancing the identification of phosphopeptides from putative basophilic kinase substrates using Ti (IV) based IMAC enrichment. Mol. Cell. Proteomics 10, M110.006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 17. Altelaar A. F., Frese C. K., Preisinger C., Hennrich M. L., Schram A. W., Timmers H. T., Heck A. J., Mohammed S. (2013) Benchmarking stable isotope labeling based quantitative proteomics. J. Proteomics 88, 14–26 [DOI] [PubMed] [Google Scholar]
- 18. Frese C. K., Altelaar A. F., Hennrich M. L., Nolting D., Zeller M., Griep-Raming J., Heck A. J., Mohammed S. (2011) Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 10, 2377–2388 [DOI] [PubMed] [Google Scholar]
- 19. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 20. Rigbolt K. T., Vanselow J. T., Blagoev B. (2011) GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell. Proteomics 10, O110.007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 22. Mi H., Muruganujan A., Casagrande J. T., Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jorgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oberprieler N. G., Lemeer S., Kalland M. E., Torgersen K. M., Heck A. J., Tasken K. (2010) High-resolution mapping of prostaglandin E2-dependent signaling networks identifies a constitutively active PKA signaling node in CD8+CD45RO+ T cells. Blood 116, 2253–2265 [DOI] [PubMed] [Google Scholar]
- 27. Swaney D. L., McAlister G. C., Coon J. J. (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 29. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 30. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marchion D. C., Cottrill H. M., Xiong Y., Chen N., Bicaku E., Fulp W. J., Bansal N., Chon H. S., Stickles X. B., Kamath S. G., Hakam A., Li L., Su D., Moreno C., Judson P. L., Berchuck A., Wenham R. M., Apte S. M., Gonzalez-Bosquet J., Bloom G. C., Eschrich S. A., Sebti S., Chen D. T., Lancaster J. M. (2011) BAD phosphorylation determines ovarian cancer chemosensitivity and patient survival. Clin. Cancer Res. 17, 6356–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sreeramkumar V., Fresno M., Cuesta N. (2012) Prostaglandin E2 and T cells: friends or foes? Immunol. Cell Biol. 90, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Soderblom E. J., Philipp M., Thompson J. W., Caron M. G., Moseley M. A. (2011) Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Anal. Chem. 83, 3758–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Montoya A., Beltran L., Casado P., Rodriguez-Prados J. C., Cutillas P. R. (2011) Characterization of a TiO(2) enrichment method for label-free quantitative phosphoproteomics. Methods 54, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kettenbach A. N., Gerber S. A. (2011) Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal. Chem. 83, 7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.